Abstract
The clinical signs and symptoms of acute respiratory tract infections (RTIs) are not pathogen specific. Highly sensitive and specific nucleic acid amplification tests have become the diagnostic reference standard for viruses, and translation of bacterial assays from basic research to routine clinical practice represents an exciting advance in respiratory medicine. Most recently, molecular diagnostics have played an essential role in the global health response to the novel coronavirus pandemic. How best to use newer molecular tests for RTI in combination with clinical judgment and traditional methods can be bewildering given the plethora of available assays and rapidly evolving technologies. Here, we summarize the current state of the art with respect to the diagnosis of viral and bacterial RTIs, provide a practical framework for diagnostic decision making using selected patient-centered vignettes, and make recommendations for future studies to advance the field.
Keywords: respiratory viruses, molecular diagnostics, utilization
Molecular assays have revolutionized the diagnosis of acute respiratory tract infections. However, many unanswered questions about the optimal use and cost-effectiveness of these tests remain. Additional prospective diagnostic studies are needed to measure impact on medical decision making and clinical outcomes.
BOX 1. PEDIATRIC VIGNETTE
A 4-year-old fully immunized girl with no significant past medical history presents to her pediatrician’s office in July of 2019 with cough, runny nose, and fever of 3 days’ duration. Several other preschool classmates are ill with similar symptoms. The patient has a fever but other vital signs are normal. She is breathing comfortably without signs of respiratory distress. On examination, lungs sounds are coarse with good air movement and there are no other focal findings. No respiratory testing is ordered. Instead, the patient and her family are reassured.
BOX 2. ADULT VIGNETTE
A 47-year-old male liver transplant recipient is admitted to the intensive care unit in December of 2019 with fever, respiratory distress, and new bilateral infiltrates. Empiric vancomycin, cefepime, and oseltamivir are initiated and the next day bronchoscopy is performed. Gram stain of bronchoalveolar lavage (BAL) fluid shows ≥2 gram-positive cocci with many polymorphonuclear cells. A rapid multiplex polymerase chain reaction (PCR) panel targeting viruses and bacteria detects mecA positive Staphylococcus aureus (107 genome copies/mL) and Haemophilus influenza (104 genome copies/mL). BAL cultures remain negative.
The number of Food and Drug Administration (FDA)–cleared molecular diagnostics for acute respiratory tract infection (RTI) has increased significantly over the last decade (Table 1). In addition, the FDA has granted Emergency Use Authorization for a number of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid amplification tests (NAATs) [1]. Highly sensitive and specific NAATs capable of detecting 1 or more viruses have become the diagnostic “gold standard” in clinical virology. In addition, several of the newest assays also detect and identify the most common causes of bacterial pneumonia along with selected drug-resistance determinants. Clinicians and microbiology laboratories now have multiple testing options that generate results within minutes to hours. Deciding which assay or combination of assays to choose, and when to use them, depends on a variety of factors including the clinical setting, institutional resources, workflow, and cost.
Table 1.
Landscape of Food and Drug Administration–Cleared Diagnostic Tests for Acute Respiratory Tract Infection
| Targetsa | Approved Specimen Types | Timeb | Costc |
|---|---|---|---|
| CLIA-waived assays | |||
| Influenza A/B only | NS direct, NPS direct, NP, NPS | 15–30 minutes | $$–$$$ |
| RSV only | NPS direct, NS, NPS | 15 minutes | $$$ |
| Flu A/B plus RSV | NS, NPS | 20–30 minutes | $$–$$$ |
| Multiple viruses plus atypical bacteria | NPS | 60 minutes | $$$$ |
| Moderate- to high-complexity assays | |||
| Influenza A/B only | NS, NPS | 0.5–2 hours | $$ |
| PIV only | NPS | 3.5 hours | $$ |
| Flu A/B plus RSV | NS, NPS, NPA, NW | 0.5–3.5 hours | $$–$$$$ |
| RSV plus hMPV | NS, NPS | 0.75 hours | $S |
| AdV, hMPV plus RV | NPS | 3.5 hours | $$ |
| Multiple viruses plus atypical bacteria | NPS | 0.75–5 hours | $$$$ |
| Multiple bacteria with resistance | ETA | 4–5 hours | $$$$$ |
| Multiple viruses and bacteria with resistance | S, ETA, BAL | 60 hours | $$$$$ |
The FDA’s website contains a comprehensive list of cleared molecular microbial tests: https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests. Definitions: Assays vary in the type of specimens approved by the FDA and in the number of organisms they can detect: “Direct” testing uses a swab, without transport media; “Atypical” bacteria may include Bordetella pertussis, Bordetella parapertussis Chlamydia pneumoniae and/or Mycoplasma pneumoniae; “Multiple viruses” may include AdV, coronaviruses, hMPV, influenza A/B, PIV, RSV, and RV; “Multiple bacteria” may include Acinetobacter calcoaceticus-baumannii complex, Citrobacter freundii, Enterobacter cloacae complex, Escherichia coli, Haemophilus influenzae, Klebsiella pneumoniae group, K. oxytoca group, K. variicola group, Legionella pneumophila, Moraxella catarrhalis, Morganella morganii, Proteus species, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Stenotrophomonas maltophilia, Streptococcus agalactiae, S. pneumoniae, and S. pyogenes; antimicrobial “resistance” genes may include tem, mecA/C, MREJ, CTX-M, KPC, NDM, Oxa-48-like, Oxa-23, Oxa-24, Oxa-58, IMP, and VIM.
Abbreviations: AdV, adenoviruses; BAL, bronchoalveolar lavage; CLIA, Clinical Laboratory Improvement Amendments; ETA, endotracheal aspirate; FDA, Food and Drug Administration; hMPV, human metapneumovirus; NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab; NS, nasal swab; NW, nasal wash; PIV, parainfluenza viruses; RSV, respiratory syncytial virus; RV, rhinovirus; S, induced/expectorated sputum.
a The FDA categorizes diagnostic tests by their complexity. Nonlaboratory staff can perform waived tests because they are deemed simple to use and the FDA has determined there is little chance the test will provide wrong information or cause harm if done incorrectly. Moderate- to high-complexity tests must be performed in qualified laboratories or sites that meet certain regulatory requirements and quality standards.
bAssay run time is displayed in minutes or hours. It is important to differentiate run time from total turnaround time to results, which includes the time from specimen collection to issuance of results by the laboratory.
cApproximate cost (US dollars) is derived from the quoted list price for reagents plus controls per test reaction. Instrument costs, depreciation, and labor are not included. $ = 1–25, $$ = 26–50, $$$ = 51–100, $$$$ = 101–150, $$$$$ =151–200.
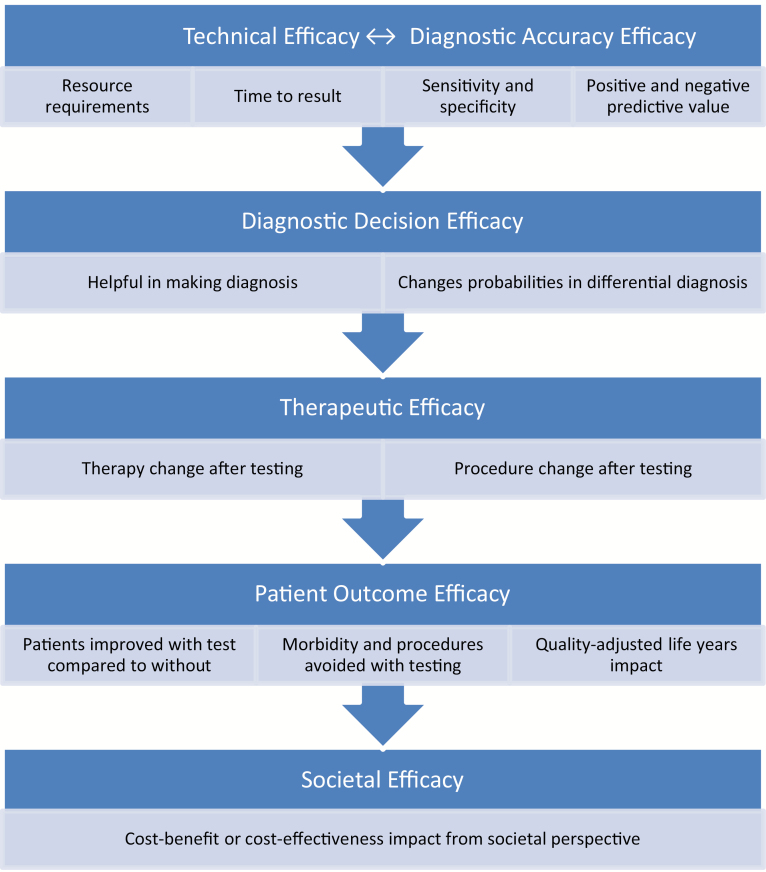
Recent studies have examined the potential impact of respiratory NAAT on clinical outcomes and resource utilization. Most publications have focused on viral testing, with the majority evaluating influenza testing only. Rapid molecular testing for influenza has the potential to reduce unnecessary antibiotic use [2–4], improve antiviral prescribing [2, 5–7], limit additional ancillary testing [3, 8], shorten hospital or emergency department (ED) lengths of stay [2–4, 8, 9], and optimize infection-control practices [7]. Molecular testing for multiple respiratory viruses simultaneously may also be more cost-effective than traditional antigen- or culture-based methods from a laboratory perspective, especially given certain thresholds of disease prevalence [10, 11]. However, not all molecular studies have reported demonstrable improvements in outcomes or cost savings [12–14]. This lack of clarity stems from the heterogeneity and variable quality of published studies. Small sample sizes and comparisons to historical controls are common weaknesses of the respiratory diagnostic literature. Furthermore, complexities in results interpretation combined with variable infrastructure to provide results in a timely manner are real-life challenges. On-the-ground effectiveness may depend as much on the logistics of testing and response to results as it does on the intrinsic accuracy of the NAATs themselves. A conceptual model of the critical components of diagnostic test efficacy is depicted in Figure 1.
Figure 1.
Conceptual hierarchical model of efficacy for molecular diagnostics. Adapted with permission from Fryback and Thornbury [15].
The Diagnostics Committee of the Infectious Diseases Society of America (IDSA) conducted a comprehensive review of the respiratory molecular diagnostic literature. The aims of the project were as follows: (1) to categorize clinical situations for which the available body of evidence supports viral and/or bacterial testing, (2) to highlight nuances in results interpretation that impact patient management and antimicrobial stewardship (AS), and (3) to identify critical knowledge gaps to guide future research. Queries of MEDLINE, Embase, and the Cochrane Library, with an emphasis on peer-reviewed manuscripts published in the last 5 years (2015–2019), identified recent outcome and cost-effectiveness studies. An update was performed in March 2020 to focus specifically on the growing SARS-CoV-2 literature. Through a standardized assessment of individual articles, we formulated key clinical questions pertaining to the rational use of current FDA-cleared molecular tests. Practical issues and unmet diagnostic needs are discussed in the context of 2 patient vignettes (Boxes 1 and 2).
QUESTION 1: TO TEST OR NOT TO TEST? THAT IS THE FIRST QUESTION
The first question to consider when deciding whether to test a patient with suspected RTI is “how will the results affect my clinical management?” The answer to this question depends on a variety of factors, including the patient’s severity of illness, duration of symptoms, comorbidities, net state of immunosuppression, availability of other ancillary test results at time of presentation, and anticipated turnaround time to results. In addition, disease prevalence (ie, the pretest probability of a given pathogen) is integral to diagnostic decision making since it affects the positive- and negative-predictive values of these assays.
Testing for Viral Pathogens
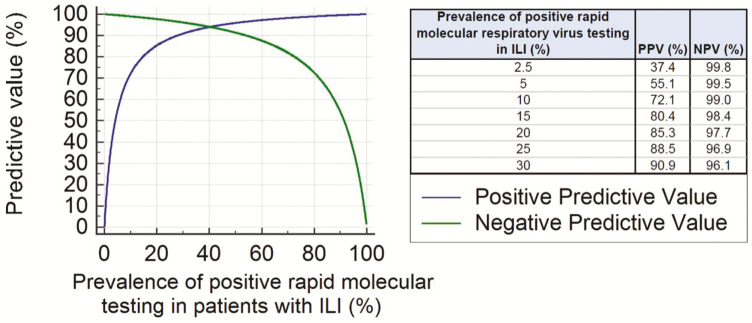
In the first vignette (Box 1), which predates the novel coronavirus pandemic, an otherwise healthy child presenting with an upper respiratory infection during a period of low influenza activity does not necessarily need influenza-specific testing. Figure 2 illustrates the impact of influenza prevalence on the predictive value of NAAT. During periods of low prevalence, positive results have a high likelihood of being falsely positive. Furthermore, since this patient has no strong indication for anti-influenza therapy [16], influenza-specific testing is unlikely to affect the decision to prescribe antiviral therapy. Alternatively, if the patient were significantly immunocompromised, had a severe influenza-like illness, or if the detection of another respiratory virus would influence the decision to prescribe an antiviral or withhold antibiotics, then syndromic testing for multiple viruses would be indicated.
Figure 2.
The importance of pretest probability. The predictive value of rapid molecular testing is displayed over the course of a typical influenza season given the published sensitivity and specificity of current influenza molecular assays. Abbreviations: ILI, influenza-like illness; NPV, negative-predictive value; PPV, positive-predictive value.
The IDSA and Centers for Disease Control and Prevention (CDC) have published influenza testing algorithms for adults and children [16–18]. Although relatively few studies have assessed the cost-effectiveness of molecular testing, modeling suggests that an approach of testing and then treating is generally preferred to empiric anti-influenza treatment during periods of moderate disease prevalence or when risk for severe disease is moderate to high [19, 20]. To be most useful in the outpatient setting, influenza results should be available during the patient visit. In the ED, the greatest impact is observed when results are issued in under 2 hours [21, 22].
Best practices for the diagnosis of coronavirus disease 2019 (COVID-19) are rapidly evolving. In the setting of ongoing community transmission, testing all symptomatic individuals is optimal for informing isolation practices, contact tracing, and evaluating the changing epidemiology. However, collection device and nucleic acid extraction reagent shortages have affected the availability of testing in some areas of the United States. In response, the IDSA [23] and others have developed expert recommendations for prioritized testing when resources are limited.
Similar guidance for other non–influenza viruses exists only in selected immunocompromised host guidelines [24–27], where initial testing for multiple viruses in addition to influenza is endorsed, and in the American Academy of Pediatrics recommendation against routine viral testing for children with bronchiolitis [28]. In a general adult population, non–influenza virus detections may not have the same influence on antibiotic prescribing and/or lengths of stay as do influenza results [2, 3, 29, 30]. Limited sample sizes, however, preclude drawing firm conclusions here and few studies have specifically assessed impact on the management of immunocompromised hosts [31]. Future studies need to be powered to measure the clinical impact of non–influenza virus detections, especially for those being evaluated in the ED or on admission to the hospital.
Testing for Bacterial Pathogens
Until recently, commercially available NAATs were limited to viral pathogens plus a few “atypical” bacteria including Mycoplasma pneumoniae, Chlamydia pneumoniae, and Bordetella species. It is clear that the presence of new or worsening infiltrates on chest X-ray is an independent predictor of antibiotic use irrespective of respiratory virus detection [29]. Thus, confidently excluding bacterial coinfection in patients with a suspected viral infection may help reduce unnecessary antibiotic use. In addition, rapid tests that accurately rule in or out additional bacterial pathogens such as Pseudomonas, methicillin-resistant Staphylococcus aureus (MRSA), Legionella, or multidrug-resistant gram-negative organisms should have value for initial management decisions as long as they can reliably discriminate between infection and respiratory tract colonization.
Current community-acquired pneumonia (CAP) [32] and hospital- or ventilator-associated pneumonia (HAP and VAP) guidelines [33] do not address molecular testing for bacterial pathogens other than a recommendation for nasal MRSA screening in patients with HAP/VAP. Since multiplex bacterial pneumonia panels are so new, their test performance and potential impact on clinical decision making are not yet established. In the absence of high-quality data, bacterial NAAT may prove most useful in situations where patients have new or worsening lung infiltrates, are moderately to severely ill, have received empiric antibiotics before obtaining cultures, and/or there is concern for multidrug-resistant bacteria or a polymicrobial infection. A recent meeting abstract highlights the importance of combined viral and bacterial testing, where clinician-directed testing would have missed potentially important viruses [34].
The second vignette (Box 2) is an example of the type of patient most likely to benefit from broad syndromic testing for viruses and bacteria at the same time. Molecular detection of high quantities of MRSA supports continuation of vancomycin in this case despite negative culture results. In addition, negative influenza results combined with detection of Haemophilus influenzae allow consideration of more-targeted gram-negative coverage along with cessation of antiviral therapy. Uncertainties associated with the interpretation of bacterial NAAT from lower respiratory tract (LRT) are discussed further under Question 3.
QUESTION 2: IF I DECIDE TO TEST, WHICH APPROACH IS BEST?
Testing for Influenza
Simple sample-to-answer molecular platforms and point-of-care devices enable high-performance influenza testing with a rapid turnaround time. In a recent meta-analysis, the pooled sensitivity and specificity of rapid viral NAAT were 90.9% and 96.1%, respectively [35]. The CDC and IDSA influenza guidelines both favor molecular detection of influenza, as opposed to antigen testing, in the outpatient and hospital setting [16–18]. Whether molecular influenza testing for all patients is the most cost-effective approach remains uncertain. For example, initial testing with a less expensive digital influenza immunoassay followed by molecular confirmation of negative results for high-risk or hospitalized patients is an alternative strategy that warrants additional study.
Testing for SARS-CoV-2
The World Health Organization declared a public health emergency of international concern on 30 January 2020. Shortly thereafter, the US Secretary of Health and Human Services announced that circumstances existed justifying authorization of the emergency use of SARS-CoV-2 diagnostics. More than 2 dozen different NAATs have received Emergency Use Authorization from the FDA [1]. Known concentrations of inactivated virus were used to determine the analytical characteristics of these tests. In contrast, clinical test performance (ie, sensitivity and specificity) has yet to established.
Available evidence suggests that SARS-CoV-2 is detectable in nasopharyngeal (NP) and oropharyngeal (OP) specimens, with peak levels typically measurable during the first week of symptoms [36–38]. The NP samples may be more sensitive than OP samples [36, 39], but detection rates appear to vary from patient to patient and change over the course of illness [37, 39]. Some patients with pneumonia, for example, have negative NP/OP samples but positive lower airway samples [40, 41]. Like other viral diseases, shedding of viral RNA in respiratory secretions may persist beyond resolution of symptoms and seroconversion [37]. Whether these patients remain infectious to others is uncertain. Much work remains to define the optimal approach to COVID-19 diagnosis, and comparisons across assays and specimen types are important unmet needs.
Testing for a Broad Range of Respiratory Pathogens Simultaneously
Upfront multiplex testing for multiple viruses may be most cost-effective in pediatric patients, where it can reduce unnecessary antibiotics as well as chest radiographs [42]. In contrast, a Veterans Affairs study evaluating a multiplex NAAT assay in adult outpatients during influenza season suggested that testing for influenza alone may be more cost-effective than a syndromic approach in this patient population [30]. Multiplex viral NAAT (potentially combined with bacterial NAAT) also makes clinical sense for immunocompromised and critically ill patients with pneumonia as well as for those with exacerbations of airway disease [43]. These are situations where treatment of non–influenza viruses such as respiratory syncytial virus (RSV) or adenovirus may be considered (eg, in a stem-cell-transplant patient) and rapid test results are most likely to influence subsequent modifications of empiric broad-spectrum antibiotics.
While the analytic sensitivity of multiplex NAAT decreases the likelihood that an important pathogen will be missed, enhanced detection also complicates interpretation. Prolonged virus shedding detectable by NAAT, but not culture, is described in immunocompromised individuals [44], and children often asymptomatically shed respiratory viruses [45]. In addition, nonviable bacteria may be detected by NAAT. These phenomena have important implications for hospital infection control and treatment decisions. Co-detection of multiple bacteria, viruses, or bacteria plus viruses is also common using NAAT, occurring in up to 30–40% of cases [46, 47]. Available studies on the medical significance of mixed infections have reported variable results. Additional studies are needed to understand whether coinfections portend poorer prognosis.
High analytic sensitivity also translates to high negative-predictive values (ie, generally >97%, depending on prevalence), but there may be important differences among individual panel targets or across manufacturers. It is incumbent on clinicians and laboratorians to understand the test characteristics of each individual panel target, especially if the results inform antibiotic de-escalation in high-acuity settings. Even the largest multiplex panels do not detect all potential pathogens, and the optimal multiplex panel design remains a matter of debate. As a result, current tests are not yet a replacement for bacterial and fungal culture with antimicrobial susceptibility testing. Culture also remains essential for epidemiologic studies, vaccine-related decisions, and local antibiograms.
Current bacterial pneumonia panels are intended for use with LRT samples, but FDA approval for specific specimen types (eg, sputum, endotracheal, and/or bronchial) varies by assay. Studies comparing diagnostic yield using different sample types collected concurrently from the same patient are currently underway. This sort of comparison will be useful for assessing the overall predictive value of test results. Factors to consider here include the higher likelihood of sputum samples to contain upper airway commensals and the theoretic benefit of site-directed bronchoalveolar lavage (BAL) over blind endotracheal suctioning in mechanically ventilated patients. In addition, negative upper respiratory tract (URT) testing for viruses and atypical bacteria is not sufficient to rule out LRT infection. In severe influenza, for example, viral shedding lasts a median of 6 days in URT as compared with a median of 11 days in LRT specimens [48] and certain strains of influenza including H1N1 and H5N1 preferentially or exclusively infect the LRT [49, 50]. A study of immunocompromised adults with RSV also observed significantly better sensitivity with LRT versus URT specimens [51]. Consequently, LRT sampling after negative URT testing is advisable when there is strong clinical suspicion for influenza or RSV and is recommended for immunocompromised patients with lung infiltrates.
QUESTION 3: HOW DO I INTERPRET THE SIGNIFICANCE OF BACTERIAL DNA DETECTIONS IN THE LOWER RESPIRATORY TRACT?
Molecular diagnostics generally detect more bacterial pathogens than culture [12, 46, 47, 52, 53]. This likely reflects the inherent sensitivity of NAAT combined with potential detection of dead, fastidious, colonizing, or metabolically impaired organisms. Assessing previous receipt of antibiotics at the time of specimen collection will be critical for interpreting NAAT-positive/culture-negative results. It is also important to remember that neither culture nor NAAT separates airway colonizers from invasive pathogens. However, use of quantitative methods may improve the clinical specificity of culture for VAP [33, 54], with higher values being more predictive of true infection. One of the FDA-cleared multiplex pneumonia panels does report relative organism abundance for 15 of its bacterial targets [55]. Bacterial detections are grouped into semiquantitative bins of 104, 105, 106 and more than 107 genomic copies/mL, which are calculated relative to calibrator material in the assay. Values below 103.5 copies/mL are reported as “not detected.” In general, there is moderate correlation between genomic units and culture quantitation, with genome copies/mL tending to be higher than the corresponding colony forming units/mL measurements.
In the second vignette (Box 2), the patient’s BAL contained much higher concentrations of MRSA nucleic acid than H. influenzae. It is possible that the low-level H. influenzae detection simply represents airway colonization and the negative culture is a result of previous antibiotics. However, in a critically ill immunocompromised patient, the consequences of not treating a potential pathogen likely outweigh the risk of toxicity from targeted antimicrobial therapy. Whether detection of high versus low concentrations of potential pathogens has prognostic value deserves additional study and this will vary by organism. A small single-center study did observe increased lengths of intensive care unit stay and more discharge diagnosis codes for pneumonia in patients with higher NAAT genomic copies/mL, which suggests that binning may have clinical value and potentially help clinicians distinguish true infections from colonization [56].
QUESTION 4: DOES PARTNERSHIP WITH ANTIBIOTIC STEWARDSHIP ENHANCE THE IMPACT OF RESPIRATORY DIAGNOSTIC TESTING?
Antimicrobial stewardship guidelines advocate rapid testing for broad panels of respiratory viruses as an important intervention to reduce the use of inappropriate antibiotics [57]. Nevertheless, there have yet to be any interventional studies assessing the safety and efficacy of antimicrobial de-escalation based on multiplex NAAT results. Active AS studies have mostly used pre- versus postintervention designs and have focused primarily on viral testing. The highest rates of antibiotic discontinuation (51%) with prospective audit and feedback were observed when a virus is detected, bacterial cultures are negative, and chest imaging is normal [58]. Otherwise, only modest antibiotic discontinuation rates (14–24% of cases) with active AS in the setting of viral RTIs were accomplished [59–61]. This is likely due to the inability to exclude bacterial coinfection with confidence in a meaningful time frame. The combination of respiratory virus NAAT with a serum biomarker (eg, procalcitonin) or host immune-response profile suggestive of a viral, but not bacterial, infection may be useful in this regard [62, 63].
The fact that molecular testing of respiratory specimens for bacteria detects more organisms than traditional culture has led to concerns that multiplex NAAT may paradoxically increase antimicrobial use. Moreover, early experience suggests that genotypic resistance aligns relatively poorly with phenotypic susceptibility results [52, 53, 64]. Molecular methods detect only the most common resistance determinants and resistance genes present in “off panel” organisms will complicate interpretation. Guidance from an AS team will likely be required to derive maximal benefit from LRT respiratory NAAT, just as it has for bloodstream infections [65]. Whether combined use of biomarkers or host immune-response plus molecular pathogen testing and AS monitoring can promote safe reductions in antibiotic use requires additional exploration. Targeting interventions to lower-risk patients with a virus detection, where prescribers may be more adherent to recommendations, may be the most pragmatic place to start.
CONCLUSIONS AND CLINICAL RECOMMENDATIONS
Molecular diagnostics have revolutionized the detection of respiratory viruses. Compared with classical culture- and antigen-based methods, these tests have high sensitivity and there is potential for a clinically meaningful turnaround time to actionable results that may reduce diagnostic uncertainty and help guide early management decisions. Newer molecular assays now target SARS-CoV-2 as well as common causes of bacterial pneumonia. Whether SARS-CoV-2 will become a seasonal virus is unknown, but we are likely to see this virus included as a part of syndromic respiratory diagnostic panels in the future.
In general, respiratory NAAT is most useful in the setting of intermediate pretest probability and intermediate disease severity. This is a situation where a negative test may permit withholding of initial empiric coverage of a potential pathogen, whereas a positive test can allow therapy to be focused against a particular pathogen, thus increasing therapeutic efficacy, decreasing avoidable drug toxicity, and potentially reducing unnecessary additional testing. Under conditions of high pretest probability and/or high risk of an adverse outcome, these tests generally lack sufficient sensitivity for a clinician not to empirically “cover” a potentially life-threatening pathogen. When there is a low pretest probability for a particular pathogen and/or low risk of adverse clinical outcomes, available tests may not add sufficient clinical value to constitute an efficient use of limited medical resources. Decisions regarding deployment of molecular diagnostics at the level of the hospital laboratory, and for a hospital system as a whole, should also consider their value in guiding protocols and policy—for example, in hospital epidemiology and antibiotic stewardship. Policies for effective use must be evaluated in an ongoing fashion as technology evolves and evidence in support of best practice emerges.
UNMET DIAGNOSTIC NEEDS AND FUTURE DIRECTIONS
Rapid molecular diagnostics are powerful tools for the evaluation and management of patients with suspected RTI. However, optimal testing algorithms and the potential impact that these tests have on patient management decisions and outcomes require further study in a variety of clinical settings. Table 2 summarizes the investigations that we believe are required to address the most important knowledge gaps and unmet diagnostic needs for RTIs. Well-designed research is especially needed in the areas of novel assay development, cost-effectiveness, and test utilization combined with AS. Outcome studies should ideally be prospective and include large enough numbers of patients to make statistically meaningful comparisons. Last, funding for interventional trials of respiratory diagnostics is a priority.
Table 2.
Committee Recommendations for Future Respiratory Diagnostic Studies
| Development of New and Innovative Diagnostics | Cost-effectiveness Studies of Available Tests | Definition of Optimal Testing Algorithms and AS Interventions |
|---|---|---|
| Novel biomarker discovery and host-response signatures that help separate viral, bacterial, fungal, and coinfections from colonization or no infection. | Prospective studies that capture both clinical outcomes and costs. | Studies combining host-response signatures or biomarkers with pathogen detection and active AS. |
| Continued refinement and analytical evaluation of unbiased next-generation sequencing platforms for use in clinical settings. Targeted tests for fungi, nontuberculous mycobacteria, and Nocardia. | Specific assessments of the impact of non–influenza virus detections, mixed infections, and bacterial pneumonia panels with antibiotic- resistance markers. | Prospective studies of AS interventions in conjunction with NAAT results and testing algorithms in the outpatient clinic, intensive care unit, and immunocompromised host settings. |
Abbreviations: AS, antimicrobial stewardship; NAAT, nucleic acid amplification test.
Notes
Disclaimer. The contents of this document are the authors’ opinions and do not necessarily reflect those of the Department of Veterans Affairs or the US government.
Potential conflicts of interest. A. M. C. is an employee of the Department of Veterans Affairs. R. B. reports grants from the National Institutes of Health. R. B. reports grants from bioMérieux, Biofire, and Roche, outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. US Food and Drug Administration. Coronavirus disease 2019 (COVID-19) emergency use authorizations in vitro diagnostics Available at: https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations#covid19ivd. Accessed 31 March 2020. [PubMed]
- 2. Brendish NJ, Malachira AK, Armstrong L, et al. . Routine molecular point-of-care testing for respiratory viruses in adults presenting to hospital with acute respiratory illness (ResPOC): a pragmatic, open-label, randomised controlled trial. Lancet Respir Med 2017; 5:401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rappo U, Schuetz AN, Jenkins SG, et al. . Impact of early detection of respiratory viruses by multiplex PCR assay on clinical outcomes in adult patients. J Clin Microbiol 2016; 54:2096–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rogers BB, Shankar P, Jerris RC, et al. . Impact of a rapid respiratory panel test on patient outcomes. Arch Pathol Lab Med 2015; 139:636–41. [DOI] [PubMed] [Google Scholar]
- 5. Andrews D, Chetty Y, Cooper BS, et al. . Multiplex PCR point of care testing versus routine, laboratory-based testing in the treatment of adults with respiratory tract infections: a quasi-randomised study assessing impact on length of stay and antimicrobial use. BMC Infect Dis 2017; 17:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu HY, Englund JA, Huang D, et al. . Impact of rapid influenza PCR testing on hospitalization and antiviral use: a retrospective cohort study. J Med Virol 2015; 87:2021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Rijn AL, Nijhuis RHT, Bekker V, et al. . Clinical implications of rapid ePlex(R) respiratory pathogen panel testing compared to laboratory-developed real-time PCR. Eur J Clin Microbiol Infect Dis 2018; 37:571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wabe N, Li L, Lindeman R, et al. . Impact of rapid molecular diagnostic testing of respiratory viruses on outcomes of adults hospitalized with respiratory illness: a multicenter quasi-experimental study. J Clin Microbiol 2019; 57:e01727-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soto M, Sampietro-Colom L, Vilella A, et al. . Economic impact of a new rapid PCR assay for detecting influenza virus in an emergency department and hospitalized patients. PLoS One 2016; 11:e0146620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahony JB, Blackhouse G, Babwah J, et al. . Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J Clin Microbiol 2009; 47:2812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dundas NE, Ziadie MS, Revell PA, et al. . A lean laboratory: operational simplicity and cost effectiveness of the Luminex xTAG respiratory viral panel. J Mol Diagn 2011; 13:175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oosterheert JJ, van Loon AM, Schuurman R, et al. . Impact of rapid detection of viral and atypical bacterial pathogens by real-time polymerase chain reaction for patients with lower respiratory tract infection. Clin Infect Dis 2005; 41:1438–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schechter-Perkins EM, Mitchell PM, Nelson KP, et al. . Point-of-care influenza testing does not significantly shorten time to disposition among patients with an influenza-like illness. Am J Emerg Med 2019; 37:873–8. [DOI] [PubMed] [Google Scholar]
- 14. Wishaupt JO, Russcher A, Smeets LC, Versteegh FG, Hartwig NG. Clinical impact of RT-PCR for pediatric acute respiratory infections: a controlled clinical trial. Pediatrics 2011; 128:e1113–20. [DOI] [PubMed] [Google Scholar]
- 15. Fryback DG, Thornbury JR. The efficacy of diagnostic imaging. Med Decis Making 1991; 11:88–94. [DOI] [PubMed] [Google Scholar]
- 16. Uyeki TM, Bernstein HH, Bradley JS, et al. . Clinical practice guidelines by the Infectious Diseases Society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenza. Clin Infect Dis 2019; 68:e1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Centers for Disease Contr ol and Prevention. Guide for considering influenza testing when influenza viruses are circulating in the community. Available at: https://www.cdc.gov/flu/professionals/diagnosis/consider-influenza-testing.htm. Accessed 31 March 2020. [Google Scholar]
- 18. Centers for Disease Control and Prevention. Algorithm to assist in the interpretation of influenza testing results and clinical decision-making during periods when influenza viruses are circulating in the community. Available at: https://www.cdc.gov/flu/professionals/diagnosis/algorithm-results-circulating.htm. Accessed 31 March 2020. [Google Scholar]
- 19. Dugas AF, Coleman S, Gaydos CA, Rothman RE, Frick KD. Cost-utility of rapid polymerase chain reaction-based influenza testing for high-risk emergency department patients. Ann Emerg Med 2013; 62:80–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee BY, McGlone SM, Bailey RR, et al. . To test or to treat? An analysis of influenza testing and antiviral treatment strategies using economic computer modeling. PLoS One 2010; 5:e11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brendish NJ, Malachira AK, Beard KR, Ewings S, Clark TW. Impact of turnaround time on outcome with point-of-care testing for respiratory viruses: a post hoc analysis from a randomised controlled trial. Eur Respir J 2018; 52:1800555. [DOI] [PubMed] [Google Scholar]
- 22. Echavarría M, Marcone DN, Querci M, et al. . Clinical impact of rapid molecular detection of respiratory pathogens in patients with acute respiratory infection. J Clin Virol 2018; 108:90–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Infectious Diseases Society of America. COVID-19 prioritization of diagnostic testing Available at: https://www.idsociety.org/news--publications-new/articles/2020/idsa-statement-on-covid-19-testing-recommendations/ Accessed 6 April 2020.
- 24. Dignan FL, Clark A, Aitken C, et al. ; Haemato-oncology Task Force of the British Committee for Standards in Haematology ; British Society for Blood and Marrow Transplantation and the UK Clinical Virology Network. BCSH/BSBMT/UK clinical virology network guideline: diagnosis and management of common respiratory viral infections in patients undergoing treatment for haematological malignancies or stem cell transplantation. Br J Haematol 2016; 173:380–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirsch HH, Martino R, Ward KN, Boeckh M, Einsele H, Ljungman P. Fourth European Conference on Infections in Leukaemia (ECIL-4): guidelines for diagnosis and treatment of human respiratory syncytial virus, parainfluenza virus, metapneumovirus, rhinovirus, and coronavirus. Clin Infect Dis 2013; 56:258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Manuel O, Estabrook M; American Society of Transplantation Infectious Diseases Community of Practice RNA respiratory viral infections in solid organ transplant recipients: guidelines from the American society of transplantation infectious diseases community of practice. Clin Transplant 2019; 33:e13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. von Lilienfeld-Toal M, Berger A, Christopeit M, et al. . Community acquired respiratory virus infections in cancer patients—guideline on diagnosis and management by the Infectious Diseases Working Party of the German Society for Haematology and Medical Oncology. Eur J Cancer 2016; 67:200–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ralston SL, Lieberthal AS, Meissner HC, et al. ; American Academy of Pediatrics Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014; 134:e1474–502. [DOI] [PubMed] [Google Scholar]
- 29. Semret M, Schiller I, Jardin BA, et al. . Multiplex respiratory virus testing for antimicrobial stewardship: a prospective assessment of antimicrobial use and clinical outcomes among hospitalized adults. J Infect Dis 2017; 216:936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Green DA, Hitoaliaj L, Kotansky B, Campbell SM, Peaper DR. Clinical utility of on-demand multiplex respiratory pathogen testing among adult outpatients. J Clin Microbiol 2016; 54:2950–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Campbell AP, Guthrie KA, Englund JA, et al. . Clinical outcomes associated with respiratory virus detection before allogeneic hematopoietic stem cell transplant. Clin Infect Dis 2015; 61:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metlay JP, Waterer GW, Long AC, et al. . Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019; 200:e45–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalil AC, Metersky ML, Klompas M, et al. . Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 2016; 63:e61–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Faron ML, Mahmutoglu D, Huang A, et al. . Clinical evaluation of a semi-quantitative multiplex molecular assay for the identification of bacteria, viruses, and fungi in lower respiratory specimens. Clinical Virology Symposium; Savannah, GA; 2017. [Google Scholar]
- 35. Vos LM, Bruning AHL, Reitsma JB, et al. . Rapid molecular tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review of diagnostic accuracy and clinical impact studies. Clin Infect Dis 2019; 69:1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zou L, Ruan F, Huang M, et al. . SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with C OVID-2019. Nature 2020. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 38. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 2020; 20:411–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang W, Xu Y, Gao R, et al. . Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Winichakoon P, Chaiwarith R, Liwsrisakun C, et al. . Negative nasopharyngeal and oropharyngeal swab does not rule out COVID-19. J Clin Microbiol 2020; 58:e00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ai T, Yang Z, Hou H, et al. . Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Subramony A, Zachariah P, Krones A, Whittier S, Saiman L. Impact of multiplex polymerase chain reaction testing for respiratory pathogens on healthcare resource utilization for pediatric inpatients. J Pediatr 2016; 173:196–201.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brendish NJ, Mills S, Ewings S, Clark TW. Impact of point-of-care testing for respiratory viruses on antibiotic use in adults with exacerbation of airways disease. J Infect 2019; 79:357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richardson L, Brite J, Del Castillo M, et al. . Comparison of respiratory virus shedding by conventional and molecular testing methods in patients with haematological malignancy. Clin Microbiol Infect 2016; 22:380, e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jansen RR, Wieringa J, Koekkoek SM, et al. . Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol 2011; 49:2631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gadsby NJ, Russell CD, McHugh MP, et al. . Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis 2016; 62:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee SH, Ruan SY, Pan SC, Lee TF, Chien JY, Hsueh PR. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect 2019; 52:920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee N, Chan PK, Wong CK, et al. . Viral clearance and inflammatory response patterns in adults hospitalized for pandemic 2009 influenza A(H1N1) virus pneumonia. Antivir Ther 2011; 16:237–47. [DOI] [PubMed] [Google Scholar]
- 49. Piralla A, Pariani E, Rovida F, et al. ; Severe Influenza A Task Force Segregation of virulent influenza A(H1N1) variants in the lower respiratory tract of critically ill patients during the 2010-2011 seasonal epidemic. PLoS One 2011; 6:e28332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yeh E, Luo RF, Dyner L, et al. . Preferential lower respiratory tract infection in swine-origin 2009 A(H1N1) influenza. Clin Infect Dis 2010; 50:391–4. [DOI] [PubMed] [Google Scholar]
- 51. Englund JA, Piedra PA, Jewell A, Patel K, Baxter BB, Whimbey E. Rapid diagnosis of respiratory syncytial virus infections in immunocompromised adults. J Clin Microbiol 1996; 34:1649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gadsby NJ, McHugh MP, Forbes C, et al. . Comparison of Unyvero P55 Pneumonia Cartridge, in-house PCR and culture for the identification of respiratory pathogens and antibiotic resistance in bronchoalveolar lavage fluids in the critical care setting. Eur J Clin Microbiol Infect Dis 2019; 38:1171–8. [DOI] [PubMed] [Google Scholar]
- 53. Ozongwu C, Personne Y, Platt G, et al. . The Unyvero P55 “sample-in, answer-out” pneumonia assay: a performance evaluation. Biomol Detect Quantif 2017; 13:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koenig SM, Truwit JD. Ventilator-associated pneumonia: diagnosis, treatment, and prevention. Clin Microbiol Rev 2006; 19:637–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. BioFire Diagnostics. Diagnostics B. FilmArraypneumonia panel instructions for use Available at: https://www.biofiredx.com/support/documents/ Accessed 19 February 2020.
- 56. Rand K, Beal S, Tremblay E, Houck H, Weber K, Sistrom C. Relationship of a multiplex molecular pneumonia panel (PP) results with hospital outcomes and clinical variables. Open Forum Infect Dis 2019; 6(Suppl 2): S299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barlam TF, Cosgrove SE, Abbo LM, et al. . Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Branche AR, Walsh EE, Jadhav N, et al. . Provider decisions to treat respiratory illnesses with antibiotics: insights from a randomized controlled trial. PLoS One 2016; 11:e0152986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abbas S, Bernard S, Lee KB, et al. . Rapid respiratory panel testing: Impact of active antimicrobial stewardship. Am J Infect Control 2019; 47:224–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mercuro NJ, Kenney RM, Samuel L, Tibbetts RJ, Alangaden GJ, Davis SL. Stewardship opportunities in viral pneumonia: why not the immunocompromised? Transpl Infect Dis 2018; 20:e12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Srinivas P, Rivard KR, Pallotta AM, et al. . Implementation of a stewardship initiative on respiratory viral PCR-based antibiotic deescalation. Pharmacotherapy 2019; 39:709–17. [DOI] [PubMed] [Google Scholar]
- 62. Branche AR, Walsh EE, Vargas R, et al. . Serum procalcitonin measurement and viral testing to guide antibiotic use for respiratory infections in hospitalized adults: a randomized controlled trial. J Infect Dis 2015; 212:1692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lydon EC, Henao R, Burke TW, et al. . Validation of a host response test to distinguish bacterial and viral respiratory infection. EBioMedicine 2019; 48:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pulido MR, Moreno-Martinez P, Gonzalez-Galan V, et al. . Application of BioFire FilmArray Blood Culture Identification panel for rapid identification of the causative agents of ventilator-associated pneumonia. Clin Microbiol Infect 2018; 24:1213.e1–. e4. [DOI] [PubMed] [Google Scholar]
- 65. Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64:15–23. [DOI] [PubMed] [Google Scholar]