ABSTRACT
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) around the world has led to a pandemic with high morbidity and mortality. However, there are no effective drugs to prevent and treat the disease. Transcriptome-based drug repositioning, identifying new indications for old drugs, is a powerful tool for drug development. Using bronchoalveolar lavage fluid transcriptome data of COVID-19 patients, we found that the endocytosis and lysosome pathways are highly involved in the disease and that the regulation of genes involved in neutrophil degranulation was disrupted, suggesting an intense battle between SARS-CoV-2 and humans. Furthermore, we implemented a coexpression drug repositioning analysis, cogena, and identified two antiviral drugs (saquinavir and ribavirin) and several other candidate drugs (such as dinoprost, dipivefrine, dexamethasone and (-)-isoprenaline). Notably, the two antiviral drugs have also previously been identified using molecular docking methods, and ribavirin is a recommended drug in the diagnosis and treatment protocol for COVID pneumonia (trial version 5–7) published by the National Health Commission of the P.R. of China. Our study demonstrates the value of the cogena-based drug repositioning method for emerging infectious diseases, improves our understanding of SARS-CoV-2-induced disease, and provides potential drugs for the prevention and treatment of COVID-19 pneumonia.
Keywords: SARS-CoV-2; COVID-19; drug repositioning; neutrophil degranulation; saquinavir, ribavirin
Transcriptome-based drug repositioning for COVID-19 recovered two antiviral drugs and identified several candidate drugs, including saquinavir, ribavirin, dinoprost and dexamethasone.
INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, previously named 2019 novel coronavirus or 2019-nCoV) has spread for several month since December 2019. The SARS-CoV-2 virus is a betacoronavirus, which includes Middle East respiratory syndrome CoV (MERS-CoV) and severe acute respiratory syndrome CoV (SARS-CoV). SARS-CoV-2 shares 95% sequence identity with a bat coronavirus, indicating that bats are likely the reservoir hosts for the virus (Zhou et al. 2020) and pangolins are possible intermediate hosts for SARS-CoV-2 (Lam et al. 2020). Both SARS-CoV-2 and SARS-CoV use angiotensin-converting enzyme II (ACE2) as the cell entry receptor (Walls et al. 2020; Wang et al. 2020; Zhou et al. 2020). The genome of SARS-CoV-2 encodes nonstructural enzymes (such as 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), helicase and RNA-dependent RNA polymerase (RdRp)) and structural and accessory proteins (such as spike glycoprotein, envelope protein, matrix protein, nucleocapsid protein) (Ashour et al. 2020; Li and De Clercq 2020).
The disease caused by SARS-CoV-2 is named coronavirus disease 2019 (COVID-19). SARS-CoV-2 can infect the upper and lower respiratory tracts of humans, and infection with SARS-CoV-2 can initiates severe and life-threatening pneumonia, probably causing the development of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). The major initial symptoms of COVID-19 include fever, cough, muscle pain or fatigue and lymphopenia (Guan et al. 2020; Liu et al. 2020). SARS-CoV-2 is highly contagious, as the basic reproduction number, R0, is approximately 2–3.5, as previously reported (Ashour et al. 2020; Zhao et al. 2020).
Although approximately 90 antiviral drugs have been approved for the treatment of nine kinds of human infectious disease (De Clercq and Li 2016), there are currently no vaccines or therapeutic drugs to prevent SARS-CoV-2 infection. Therefore, the prevention and treatment of COVID-19 is urgently needed worldwide. Drug repositioning is an efficient approach to tackle this problem. By integrating several aspects of the drugs, diseases and target, drug repositioning could produce convincing predictions, reducing the drug development cost and time (Wang et al. 2013, 2014, 2019). Several researchers have used molecular docking, high-throughput screening, network analysis or knowledge graphs to search for effective drugs (Ge et al. 2020; Matsuyama et al. 2020). Cepharanthine, selamectin and mefloquine hydrochloride were identified as potential drugs for treating 2019-nCoV infection using a pangolin coronavirus-based cell model (Fan et al. 2020). Teicoplanin, a glycopeptide antibiotic, potently prevents cell entry by SARS-CoV-2 (Zhang et al. 2020). Prulifloxacin, bictegravir, nelfinavir and tegobuvir were found to tightly bind to the viral 3CLpro in an in silico high-throughput screening assay. Nelfinavir and bictegravir are anti-HIV drugs, and tegobuvir is an anti-HCV drug (Li et al. 2020). Moreover, nelfinavir has been shown to inhibit viral 3CLpro via an integrative strategy combining homology modeling, molecular docking and binding free energy calculation.
However, there has been no transcriptome-based drug repositioning for COVID-19, which has been proven to be an efficient way to discover candidate drugs for diseases (Jahchan et al. 2013; Wang et al. 2013; Wu, Wang and Chen 2013; Arakelyan et al. 2019) and can be considered as a orthogonal and complementary method. We used bronchoalveolar lavage fluid (BALF) transcriptomics data from 8 COVID-19 patients and 20 healthy controls to explore the gene expression signature of the disease and applied a coexpression-based drug repositioning pipeline, which we previously developed, to search for candidate drugs for COVID-19. Consequently, we recovered two antiviral drugs, saquinavir and ribavirin, and identified several candidate drugs for treating COVID-19 pneumonia.
MATERIALS AND METHODS
Transcriptomic data of COVID-19 patients and healthy controls
The BALF of 8 COVID-19 patients (COVID) and healthy controls (healthy) were used to extract RNA and prepare a library for HiSeq sequencing, and data quality control, mapping and gene quantitation were performed as described in a previous work (Zhou et al. 2020). The raw data were deposited in the Genome Warehouse in the National Genomics Data Center (Members, B.I.G.D.C 2019) available at https://bigd.big.ac.cn/gsa with project number PRJCA002273.
Differential expression analysis
The raw read counts were filtered to identify low-expressed genes using the filterByExpr function in the Bioconductor edgeR package (Robinson, McCarthy and Smyth 2010), normalized using quantile normalization, and the voom method in the Bioconductor limma package (Ritchie et al. 2015). Then, differential expression analysis for the COVID and healthy groups was implemented based on the normalized gene expression level by absolute logged fold-change in normalized expression ≥ 2 and an adjusted P-value of < 0.05 to obtain differentially expressed genes (DEGs). Principal components analysis (PCA) was used to visualize the differences between two groups.
Computational drug repositioning analysis
The KEGG pathway analysis, Reactome pathway analysis and drug repositioning for the coexpressed genes were performed via the cogena package, previously developed by one of the authors (Jia et al. 2016). The KEGG and Reactome gene sets were used in the pathway analysis (Liberzon et al. 2015), and the drug-induced transcriptome in the connectivity map project (Lamb et al. 2006) was used in the cogena-based drug repositioning analysis. A core concept of cogena for drug repositioning is that identifying drugs that can revert the regulation direction of coexpressed DEGs caused by a disease and the connection between coexpressed DEGs, pathways and drugs can be used to uncover the drug mechanism of action for the treatment of the disease. In the drug repositioning analysis, a drug-induced upregulated gene set was used for the downregulated gene cluster, and a drug-induced downregulated gene set was used for the upregulated gene cluster. Particularly, the clustering method and the number of clusters were chosen using several rules, as described in the original cogena paper. Briefly, similar pathways should be enriched in a cluster, and different pathways should be enriched in different clusters. The enrichment score is the negative log2 false-discovery rate (FDR)-corrected p-value obtained via the hypergeometric test.
RESULTS AND DISCUSSION
Differentially expressed genes clearly distinguish the COVID and healthy groups
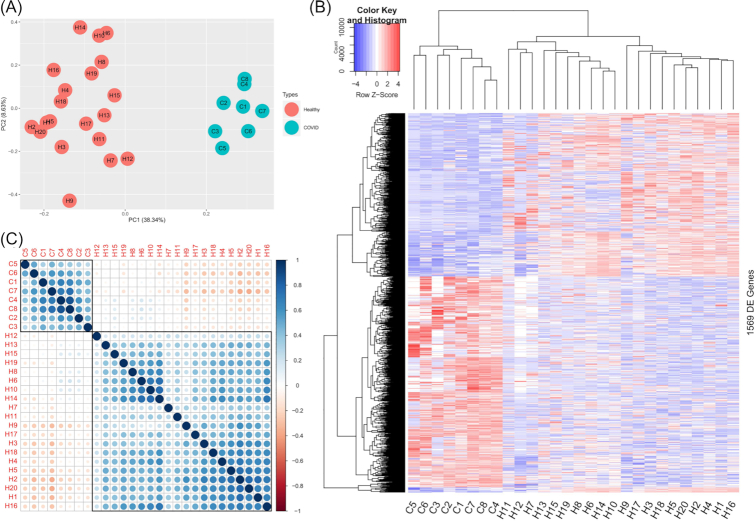
The use of lower respiratory tract specimens, such as the BALF, is important to explore the mechanisms of disease development, as the disease mainly affects the lower respiratory tract, especially the lungs. To identify the key genes affected by COVID-19, we performed a transcriptomics differential expression analysis and identified a total of 1569 differentially expressed genes (DEGs), consisting of 872 genes with upregulated expression and 697 genes with downregulated expression (Figure S1 and Table S1, Supporting Information). The two groups could be clearly separated with all the genes (Figure S2, Supporting Information) or all the DEGs (Fig. 1A) in the principal component analysis. A heatmap of these DEGs with hierarchical clustering of both the samples and genes indicated that the two groups differed from each other, as the COVID samples closely clustered and the healthy samples clustered together (Fig. 1B). The Pearson correlation between samples also presented high consistency within each group, whereas there was a marked difference between the two groups (Fig. 1C). As a result, the identified DEGs are representative of COVID.
Figure 1.
General analysis of COVID-19-induced DEGs. (A) The first two dimensions of the principal components analysis for the DEGs of COVID-19. PC1 and PC2 are principal components 1 and 2, respectively. (B) Heatmap and hierarchical clustering of DEGs. C1-C8 represent the samples of COVID-19 patients, and H1-H20 represent healthy controls. The values are shown as the normalized gene expression. (C) Correlation between all the samples. The size of the circle represents the absolute Pearson correlation coefficient, and color indicates the direction of the correlation.
Functional analysis of coexpressed genes
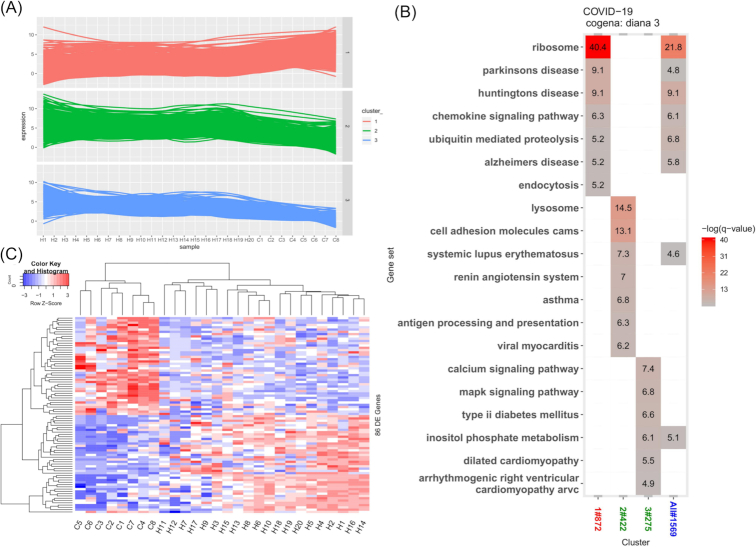
Coexpressed genes usually cooperate to implement similar functions, which is a specific way to connect key genes among DEGs with diseases and could provide an important clue to understand the pathogenesis of a disease. Three clusters of coexpressed genes were obtained via a divisive hierarchical clustering method, DIANA (Figure S3 and Table S2, Supporting Information). Compared with that of genes in the healthy group, the expression of coexpressed genes in cluster 1 in the COVID groups was increased, whereas the expression of others was decreased (Fig. 2A and Figure S3, Supporting Information). The KEGG pathway enrichment analysis for coexpressed genes is shown in Fig. 2B. Ribosome, chemokine signaling and endocytosis pathways were enriched in cluster 1. Generally, endocytosis or micropinocytosis plays a role in viral entrance into the early endosomes in cells (Zhang et al. 2020). Lysosome, renin-angiotensin system and asthma pathways were enriched in cluster 2, and calcium and MAPK signaling pathways were enriched in cluster 3. Lysosomes are involved in destroying invading viruses, and the downregulation of lysosome-associated gene expression suggests dysregulation of the innate immune system. This finding is consistent with the downregulation of the expression of lysosomal genes reported by Xiong et al. (2020).
Figure 2.
Coexpression analysis. (A) Coexpression trend in the samples. Three clusters determined by the DIANA clustering method are shown. (B) KEGG pathway analysis for coexpressed genes generated by cogena. The color indicates the degree of statistical significance, and the enrichment score is the -log (q-value). (C) Heatmap of genes in the neutrophil degranulation Reactome pathway. C1–C8 represent the samples of COVID-19 patients, and H1-H20 represent the samples of healthy controls. The values are shown as the normalized gene expression.
In the Reactome analysis (Figure S4, Supporting Information), neutrophil degranulation and the innate immune system were enriched in both clusters 1 (upregulated genes) and 2 (downregulated genes). Interestingly, neutrophils, the proportion of which was higher in the COVID group than in the healthy control group, play a role in the response to viral infection (Liu et al. 2020). Neutrophil degranulation was not fully activated or was partly blocked by the virus, as the expression of approximately half of the genes in this pathway was downregulated (Fig. 2C). The coexpression pathway analysis revealed that a large gene expression change was induced by COVID to facilitate the viral life cycle.
Computational drug repositioning for COVID-19 pneumonia
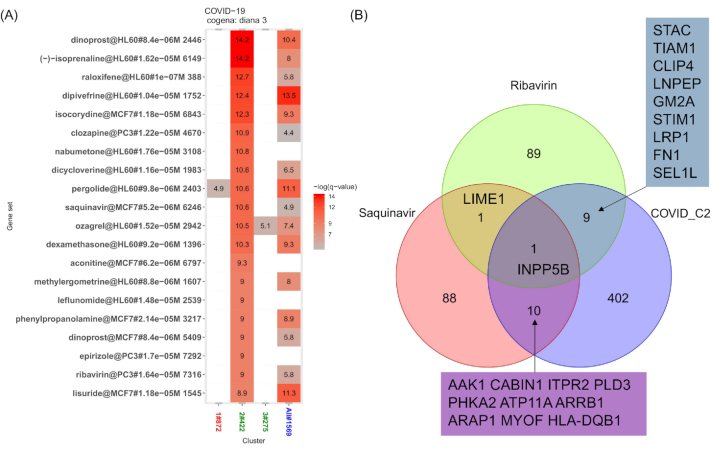
We identified two FDA-approved antiviral drugs (saquinavir and ribavirin) in the coexpression-based drug enrichment analysis (Fig. 3A). Saquinavir, a protease inhibitor ranked 10th in cluster 2, is used to help control HIV infection. Saquinavir has recently been reported as a candidate drug for treating COVID-19 by several individual researchers via molecular docking with 3CLpro, PLpro and spike proteins (Ayman et al. 2020; Ruan et al. 2020) (Table S3, Supporting Information). By tightly binding to 3CLpro and S proteins, saquinavir can inhibit the replication of this virus both extracellularly and intracellularly (Table S3, Supporting Information).
Figure 3.
Drug repositioning for COVID pneumonia. (A) Cogena-based drug repositioning using cluster 2. The top 20 enriched drugs are shown. The names of drugs followed by the cell line, concentration of drug used and instance ID in CMap data are shown on the y-axis. The color indicates the degree of statistical significance, and the enrichment score is shown as the -log (q-value). (C) Venn diagram of upregulated genes by saquinavir and ribavirin and genes in cluster 2. The overlapping numbers of genes and certain gene symbols are shown in some sets.
Ribavirin was ranked 19th in the drug enrichment analysis for cluster 2 (Fig. 3C). Ribavirin can be used in combination with other antiviral medications to treat chronic hepatitis C and for SARS-CoV and MERS-CoV infections (Chong et al. 2015; Yin and Wunderink 2018). Ribavirin was reported as a candidate drug by a team of researchers who used molecular docking or protein–protein interaction methods for PLpro and Nsp14 (Gordon et al. 2020; Li et al. 2020) (Table S3, Supporting Information). More importantly, it has been included as a possible therapeutic drug in the diagnosis and treatment protocol for COVID pneumonia (trial version 5–7) released by the National Health Commission & National Administration of Traditional Chinese Medicine of the P.R. of China on March 3, 2020 (Table S3, Supporting Information) (Commission, N.H., Medicine, N.A.o.T.C 2020), although ribavirin did not perform well at treating the virus in a prior in vitro drug evaluation experiment (Wang et al. 2020).
The recovery of the two approved antiviral drugs using our transcriptome-based drug repositioning method indicated the reliability of our method. Moreover, we have successfully applied our drug repositioning method for psoriasis and periodontal disease (Jia et al. 2016; Kang et al. 2019). Additionally, the consistency among the results produced by two totally different drug repositioning methods, our transcriptome-based and docking-based methods, further validated our results from based on data other than transcriptomics data. The former method was used to find drugs that could restore normal gene expression following viral infection, whereas the latter method aimed to find the drug that directly targets viral or human proteins and inhibits the replication or entry process of the virus. Additionally, one advantage of cogena-based drug repositioning is that it is less computationally intensive than docking methods. Consequently, the results of the cogena-based drug repositioning are robust.
Additionally, the Venn diagram between genes in cluster 2 and the upregulated genes produced by saquinavir and ribavirin is shown in Fig. 3B and Table S4 (Supporting Information). Inositol polyphosphate-5-phosphatase B (INPP5B) was shared between the 3 sets. AP2-associated protein kinase 1 (AAK1) and Arrestin Beta 1 (ARRB1), shared between saquinavir and COVID-19, are involved in endocytosis, and AAK1 is considered a target to inhibit the virus (Richardson et al. 2020). Leucyl and cystinyl aminopeptidase (LNPEP), shared between ribavirin and COVID-19, is involved in the renin-angiotensin system, and ACE2, the target of SARS-CoV-2, was also a member of this system. These genes provide insight into the mode of action of the two drugs for treating COVID-19.
Other drugs enriched in the cluster, especially those with a rank higher than that of the two antiviral drugs, could be further investigated to discover more effective drugs for COVID-19. Generally, drugs with similar indications in a cluster possess a higher confidence priority for consideration. Dinoprost, having two instances ranked 1st and 17th, is a smooth muscle activator (Fig. 3A). (-)-Isoprenaline, a bronchodilator useful in obstructive lung diseases, can dilate the bronchi and bronchioles, decreasing resistance in the respiratory airway and increasing airflow to the lungs. Nabumetone, aconitine, dexamethasone, leflunomide and epirizole are anti-inflammatory drugs. Notably, tocilizumab, a biologic binding to interleukin-6 and treatment for cytokine release syndrome, is also recommended in the diagnosis and treatment protocol for COVID pneumonia (trial version 7). Dipivefrine is an antiglaucoma drug, which was also predicted as a candidate drug for treating COVID-19 via a molecular docking method (Ayman et al. 2020). Lisuride, pergolide and clozapine are neuropsychiatric agents. We also identified enriched drugs for the other clusters (Figure S5, Supporting Information). Interestingly, the drugs discussed above have not been associated with antiviral activity thus far. This could be due to the principle of our transcriptome-based drug screening that identified drugs that are capable of restoring virus-induced gene expression dysregulation, rather than directly targeting viral or human proteins, as is the case for classical molecule docking methods. Thus, these drugs are of high priority for experimental validation.
CONCLUSION
Based on the COVID group BALF transcriptome data, we showed that several key pathways, such as endocytosis, the lysosome and neutrophil degranulation, in the disease, represent the intense battle between SARS-CoV-2 and humans. We provided several candidate drugs for treating COVID pneumonia using the cogena-based drug repositioning method. A total of two antiviral drugs, saquinavir and ribavirin, were recovered in our results. Coincidentally, these two drugs were also identified by several research groups using molecular docking methods, a totally different but classical strategy for drug development. Furthermore, several candidate drugs for preventing and treating COVID-19 were also identified. Our study shows that cogena is a powerful and efficient drug repositioning tool for emerging infectious diseases, enhances the understanding of SARS-CoV-2-induced disease and importantly provides drug candidates for the prevention and treatment of COVID-19.
AUTHOR CONTRIBUTORS
Conceptualization: K.H. and W.W.; data analysis: Z.J., J.S., X.S. Writing & Editing: Z.J. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank professor Mingkun Li and his team from Beijing Institute of Genomics, Chinese Academy of Sciences for providing the raw read count data of the transcriptomic data and helpful discussion.
Contributor Information
Zhilong Jia, Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China; Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China.
Xinyu Song, Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China; Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China.
Jinlong Shi, Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China; Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China.
Weidong Wang, Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China; Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China.
Kunlun He, Key Laboratory of Biomedical Engineering and Translational Medicine, Ministry of Industry and Information Technology, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China; Beijing Key Laboratory for Precision Medicine of Chronic Heart Failure, Medical Innovation Research Division of Chinese PLA General Hospital, Beijing, 100853, China.
DATA AVAILABILITY STATEMENT
The datasets analyzed for this study can be found in the Genome Warehouse in the National Genomics Data Center available at https://bigd.big.ac.cn/gsa with project number PRJCA002273.
FUNDING
This work was supported by the National Natural Science Foundation of China [grant number 31701155]; National Key Research and Development Program of China [grant number 2017YFC0908403].
Conflicts of Interests
None declared.
REFERENCES
- Arakelyan A, Nersisyan L, Nikoghosyan M et al. Transcriptome-guided drug repositioning. Pharmaceutics. 2019;11:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour HM, Elkhatib WF, Rahman MM et al. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks, Pathogens (Basel, Switzerland). 2020;9:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayman F, Ping W, Mahmoud A et al. Identification of FDA Approved Drugs Targeting COVID-19 Virus by Structure-Based Drug Repositioning. 2020, DOI: 10.26434/chemrxiv.12049647.ChemRxiv. Preprint. 10.26434/chemrxiv.12003930.v2. [DOI] [Google Scholar]
- Chong YP, Song JY, Seo YB et al. Antiviral treatment guidelines for middle east respiratory syndrome. Infect Chemother. 2015;47:212–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, Li G. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev. 2016;29:695–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H-H, Wang L-Q, Liu W-L et al. Repurposing of clinically approved drugs for treatment of coronavirus disease 2019 in a 2019-novel coronavirus (2019-nCoV) related coronavirus model. Chin Med J. 2020, DOI: 10.1097/CM9.0000000000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Tian T, Huang S et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting COVID-19, bioRxiv. 2020, DOI: 10.1101/2020.03.11.986836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M et al. A SARS-CoV-2-Human protein-protein interaction map reveals drug targets and potential Drug-Repurposing, bioRxiv. 2020, DOI: 10.1101/2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W-j, Ni Z-y, Hu Y et al. Clinical characteristics of 2019 novel coronavirus infection in China, medRxiv. 2020, DOI: 10.1056/NEJMoa2002032. [Google Scholar]
- Jahchan NS, Dudley JT, Mazur PK et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Liu Y, Guan N et al. Cogena, a novel tool for co-expressed gene-set enrichment analysis, applied to drug repositioning and drug mode of action discovery. BMC Genomics. 2016;17:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Jia Z, Tang D et al. Time-course transcriptome analysis for drug repositioning in Fusobacterium nucleatum-Infected human gingival fibroblasts. Front Cell Dev Biol. 2019;7:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science (New York, NY). 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- Lam TT-Y, Shum MH-H, Zhu H-C et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China, bioRxiv. 2020, DOI: 10.1101/2020.02.13.945485. [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020;19:149–50. [DOI] [PubMed] [Google Scholar]
- Li H, Wang YM, Xu JY et al. Potential antiviral therapeutics for 2019 Novel Coronavirus. Chin JTubercul Respirat Dis. 2020;43:170–2. [DOI] [PubMed] [Google Scholar]
- Liu J, Li S, Liu J et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients, medRxiv. 2020, DOI: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Fang Y-Y, Deng Y et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J. 2020, DOI: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang J, Wang N et al. Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening, bioRxiv. 2020, DOI: 10.1101/2020.01.28.922922. [Google Scholar]
- Matsuyama S, Kawase M, Nao N et al. The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15, bioRxiv. 2020, DOI: 10.1101/2020.03.11.987016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Members, B.I.G.D.C Database resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019;47:D8–D14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Health Commission and State Administration of Traditional Chinese Medicine Diagnosis and treatment protocol for novel coronavirus pneumonia (Trial Version 7). Chin Med J. 2020;133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P, Griffin I, Tucker C et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England). 2020;395:e30–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47, DOI: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z, Liu C, Guo Y et al. Potential inhibitors targeting RNA-dependent RNA polymerase activity (NSP12) of SARS-CoV-2, Preprints.org. 2020, DOI: 10.20944/preprints202003.0024.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici MA et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020, DOI: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhao S, Liu M et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism, medRxiv. 2020, DOI: 10.1101/2020.02.05.20020545. [Google Scholar]
- Wang L, Wang Y, Hu Q et al. Systematic analysis of new drug indications by drug-gene-disease coherent subnetworks. CPT Pharmacometrics Syst Pharmacol. 2014;3:e146, DOI: 10.1038/psp.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cao R, Zhang L et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen S, Chen L et al. Associating lncRNAs with small molecules via bilevel optimization reveals cancer-related lncRNAs. PLoS Comput Biol. 2019;15:e1007540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen S, Deng N et al. Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS One. 2013;8:e78518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Wang Y, Chen L. Network-based drug repositioning. Mol Biosyst. 2013;9:1268–81. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Liu Y, Cao L et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. SSRN. 2020, DOI: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma X, Yu F et al. Teicoplanin potently blocks the cell entry of 2019-nCoV, bioRxiv. 2020, DOI: 10.1101/2020.02.05.935387. [Google Scholar]
- Zhao S, Lin Q, Ran J et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Ren L, Zhang L et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–90.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed for this study can be found in the Genome Warehouse in the National Genomics Data Center available at https://bigd.big.ac.cn/gsa with project number PRJCA002273.