Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic demonstrates the need for accurate and convenient approaches to diagnose and therapeutically monitor respiratory viral infections. We demonstrated that self-sampling with mid-nasal foam swabs is well-tolerated and provides quantitative viral output concordant with flocked swabs. Using longitudinal home-based self-sampling, we demonstrate that nasal cytokine levels correlate and cluster according to immune cell of origin. Periods of stable viral loads are followed by rapid elimination, which could be coupled with cytokine expansion and contraction. Nasal foam swab self-sampling at home provides a precise, mechanistic readout of respiratory virus shedding and local immune responses.
Keywords: respiratory virus, viral diagnostics, cytokines, immune response
Self-sampling with foam swabs is well-tolerated and provides quantitative viral output concordant with flocked swabs. Using longitudinal home-based self-sampling, we demonstrate that nasal foam swab sampling provides a precise, mechanistic readout of respiratory virus shedding and local immune responses.
The coronavirus disease 2019 (COVID-19) pandemic is an unprecedented event in modern history. As of August 4, there were approximately 18.5 million documented COVID-19 cases and 700 000 deaths worldwide with rapidly expanding outbreaks ongoing in dozens of countries [1]. In all likelihood, this highly contagious and lethal respiratory virus will circulate widely for years to come [2].
A critical research priority is to develop rapid molecular tests that provide accurate diagnosis, determine infectiousness and transmissibility, and allow for monitoring of viral load during therapy [3]. For numerous viral infections, including influenza, viral load correlates with disease severity and secondary household attack rate [4–6]. Early studies suggest that peak viral load differentiates mild from severe COVID-19 [7]. Furthermore, viral load monitoring during antiviral therapy is a mainstay for various human infections including human immunodeficiency virus, hepatitis B, cytomegalovirus, and hepatitis C infections [8–14]. For viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for which severe clinical outcomes are rare, viral load may serve as a useful surrogate marker to design smaller, but still sufficiently powered treatment studies [7, 15].
Another major unmet medical need is the ability to frequently measure the local mucosal immune response during infection. It is increasingly recognized that tissue resident T cells and antigen-presenting cells are phenotypically and functionally distinct from circulating immune cells, especially in respiratory viral infections [16–18]. Therefore, measuring immune responses in blood can fundamentally misclassify the agents responsible for local viral elimination. Important shifts in the immune response against respiratory viruses likely occur rapidly and in stages during the early and late phases of viral shedding [19], and serial measurement of local cytokines may provide a window into the local cellular response [20].
Self-testing for respiratory viruses has been successfully performed both in research and primary care, but regulatory agencies have been slow to accept patient collected samples as valid, especially in the home setting. Currently licensed flocked swabs may not be optimal for patients with vulnerable mucosal membranes and low platelet counts (eg, following cytotoxic chemotherapy) because they are associated with discomfort and possible bleeding. Moreover, discomfort may deter participants from collecting longitudinal samples. Importantly, a reliable and comfortable home-based self-testing methodology is needed to prevent potentially infected individuals from entering healthcare facilities to be tested and transmitting virus to healthcare workers and other patients. Initial data on foam swabs are promising, suggesting a broader role for home-based self-swabbing for respiratory viral pandemics [21, 22].
Here we report data on a novel respiratory virus detection method using self-collected nasal foam swabs. This methodology expands our testing armamentarium with easily collected and comfortable swabs that can be applied to viral load and cytokine kinetic studies. Most importantly, they can be easily scaled and used at home in this time of severe testing shortages and dangerous transmission risk.
MATERIALS AND METHODS
Protocol
The study was approved by the Institutional Review Board at Fred Hutchinson Cancer Research Center.
Flocked vs Foam Swab Study
Participants with respiratory symptoms (Supplementary Table 1) for <3 days were enrolled in the study. Each participant completed 2 sample collections, each separated by 1 hour. At each time point, the participant collected either (i) 2 self-collected Copan flocked swabs (number 23-600-966), 1 from each nostril in the mid-turbinate space; or (ii) 2 self-collected Puritan foam swabs (Puritan Medical Red number 25-1805-SC 2), 1 from each nostril in the mid-nasal space. Swab collection was randomized by order of swab type. Details of the swab collection and processing are provided in the Supplementary Methods. Following sample collection, participants were asked to complete a survey to assess tolerability and acceptability. McNemar test with exact P values was used to compare survey responses.
Longitudinal Sampling Study
Separately, new participants with respiratory symptoms (Supplementary Table 1) for <3 days were enrolled in the study. Each participant collected 2 Puritan foam nasal swabs, 1 from each nostril, per day for 14 days after enrollment or until symptoms resolved, whichever was longer. Details of the swab collection and processing are provided in the Supplementary Methods. Participants completed a daily electronic symptom survey (Supplementary Table 1) and an end-of-study survey.
Laboratory Methods
Sample Processing
Each conical vial containing a swab was vortexed and 500 μL of buffer was removed and stored at –80°C for polymerase chain reaction (PCR) analysis. Swabs were processed and stored at –80°C for cytokine testing as described in the Supplementary Methods.
Viral Testing
Nasal swab specimens were tested using a multiplex PCR assay for 11 respiratory viruses (adenovirus A–F, human rhinovirus [HRV], influenza A and B, parainfluenza viruses 1–4, human coronavirus [CoV], bocavirus [BoV], respiratory syncytial virus [RSV], and human metapneumovirus [HMPV]) as previously described [23].
Cytokine Testing
Cytokine levels were quantified in nasal specimens using the electrochemiluminescence-based Mesoscale Discovery platform. Details of the panels used are described in the Supplementary Methods.
Statistical Analysis
PCR results that were positive but below the limit of detection (LOD) were imputed as 500 copies/mL, using the LOD divided by 2. Negative results were assigned a value of 0. The concordance correlation coefficient was used to measure agreement of quantitative results between paired samples [24]. Cytokine results below the fitted curve range were assigned the value of the lower LOD divided by 2, and results above the fitted curve range were assigned the value of the upper LOD. Symptoms are represented as the total number of symptoms present for each day, out of a total of 26 (Supplementary Table 1). SAS version 9.4 (SAS Institute, Cary, North Carolina) and Stata version 16.1 (StataCorp, College Station, Texas) software packages were used for analysis.
Cytokine Clustering
We performed a cluster analysis where each sample is an array of 20 measured cytokine concentrations. First, we checked for cluster tendency of the samples using the Hopkins statistic [25, 26], where values close to 1 indicate that the samples are highly clustered and values close to 0.5 indicate random samples. When the calculated Hopkins statistic (computed using the “get_clust_tendency” function in R statistical software) was >0.5, we did a linkage hierarchical clustering with Euclidean distances of the samples [27].
Mathematical Modeling
We used an acute viral infection model that distinguishes between early and late responses to RSV and HMPV. Full description of the definitions, model assumptions, and surrogate cytokine selection is provided in the Supplementary Methods.
RESULTS
Concordance Between Foam and Flocked Nasal Swabs for Viral Detection
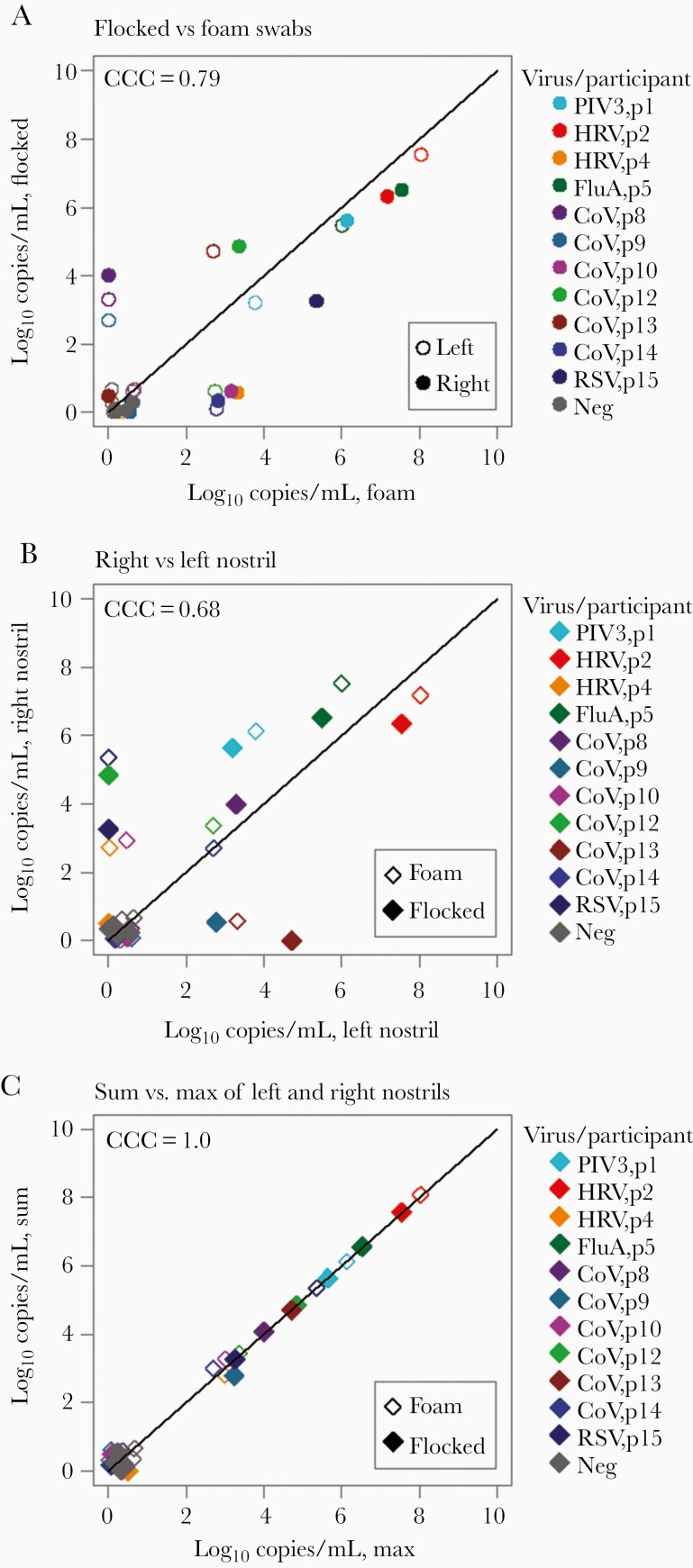
Fifteen participants were enrolled in the foam vs flocked nasal swab study. Four participants were negative for any respiratory virus from all swabs (Table 1). Combining results from both nostrils, foam and flocked swabs were concordant for viral detection in 22 of 30 samples (73.3%) (Supplementary Table 2). Discrepant results occurred exclusively in samples with low viral load (<4 log10 viral copies/mL) (Table 1). Agreement between samples collected by foam and flocked swabs from the same nostril was generally high, particularly with high viral load samples, with no evidence of higher yield with 1 method vs the other (Figure 1A).
Table 1.
Viral Loads in Matched Foam Versus Flocked Swabs in Participants With New-Onset Respiratory Symptoms
| Participant Number | Virus | Foam Right | Foam Left | Flocked Right | Flocked Left |
|---|---|---|---|---|---|
| p1 | PIV3 | 6.12 | 3.79 | 5.64 | 3.19 |
| p2 | HRV | 7.18 | 8.02 | 6.35 | 7.54 |
| p3 | Neg | 0 | 0 | 0 | 0 |
| p4 | HRV | 2.70 | 0 | 0 | 0 |
| p5 | FluA | 7.53 | 6.00 | 6.52 | 5.48 |
| p6 | Neg | 0 | 0 | 0 | 0 |
| p7 | Neg | 0 | 0 | 0 | 0 |
| p8 | CoV | 0 | 0 | 3.99 | 3.29 |
| p9 | CoV | 0 | 0 | 0 | 2.70 |
| p10 | CoV | 2.70 | 0 | 0 | 0 |
| p11 | Neg | 0 | 0 | 0 | 0 |
| p12 | CoV | 3.40 | 2.70 | 4.84 | 0 |
| p13 | CoV | 0 | 2.70 | 0 | 4.71 |
| p14 | CoV | 2.70 | 2.70 | 0 | 0 |
| p15 | RSV | 5.34 | 0 | 3.26 | 0 |
Data are presented as log10 copies/mL.
Abbreviations: CoV, coronavirus; FluA, influenza virus A; HRV, human rhinovirus; Neg, negative; PIV3, parainfluenza virus type 3; RSV, respiratory syncytial virus.
Figure 1.
Comparison between self-collected mid-nasal foam and mid-turbinate flocked swabs. A, Viral loads from the same nostril using flocked and foam swabs are concordant, particularly at higher viral loads. B, Differential viral loads with the same swab type, observed between nostrils, show moderate concordance. C, Viral load from the highest nostril strongly agrees with the sum of the 2 nostrils, suggesting that a majority of sampled virus comes from 1 side. Overlapping data points have been jittered to allow viewing of all data points. Abbreviations: CCC, concordance correlation coefficient; CoV, coronavirus; FluA, influenza A; HRV, human rhinovirus; Neg, negative; PIV3, parainfluenza virus 3; RSV, respiratory syncytial virus.
Focality of Respiratory Virus Shedding in Nasal Passages
In the same dataset, we compared swab samples obtained with the same swab type from separate nostrils with a total of 15 paired samples. Viral loads were notably higher in 1 nostril than the other and were less in agreement (Figure 1B). Moreover, the value from the highest nostril strongly agreed with the sum of the 2 nostrils, suggesting that a majority of sampled virus comes from 1 side (Figure 1C) and that sampling the other side underestimates viral load. Therefore, bilateral sampling is likely required for optimal yield and accurate quantitation.
Comfort and Ease of Self-Collected Foam Swabs
Survey responses suggested that participants found foam swabs more comfortable (9/15 participants agreed/strongly agreed that foam swabs were comfortable, vs 4/15 for flocked swabs; P = .13) and easier to collect (14/15 participants agreed/strongly agreed for foam swabs vs 11/15 for flocked swabs; P = .25). Almost all participants (14/15) would consider participating in future research using foam swabs, but only 10 of 15 if flocked swabs are used (P = .13).
Ease, Comfort, and High Compliance Associated With Longitudinal Nasal Sampling
We next enrolled 9 otherwise healthy, adult study participants who self-sampled their nasal passage serially for 14 days. One participant contributed serial samples twice. Overall compliance was high: the median number of sample days was 14 (range, 11–19 days). After study completion, the majority of participants agreed or strongly agreed that foam swabs were comfortable (70%) and easy (90%) and that they would participate in future research with foam swabs (80%). Serial home-based testing appears to be a well-accepted methodology.
Steady-State Nasal Passage Viral Load Kinetics and Correlation With Symptoms
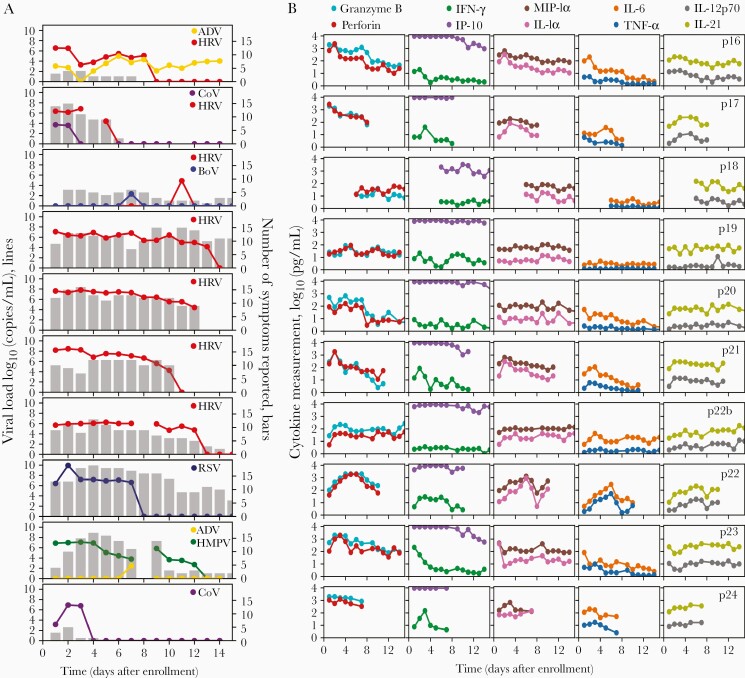
In longitudinal sampling, we were able to detect 14 viruses including 7 HRV, 2 CoV, 1 BoV, 2 adenovirus, 1 HMPV, and 1 RSV. There were 4 instances of viral coinfection, though in each case a dominant virus was evident based on greater duration of shedding and higher viral load (Figure 2A).
Figure 2.
Viral load, symptoms, and cytokine levels in serial sampling in both nostrils. Each row represents a participant. A, Viral load (lines) and quantity of symptoms (bars) are shown on left and often tracked with each other longitudinally. Serial sampling in both nostrils with foam swabs reveals a steady state for human rhinovirus, respiratory syncytial virus, and human metapneumovirus viral loads prior to rapid elimination. B, Levels for each cytokine are shown on the right. Paired cytokines show concordant expansion and clearance phases. Abbreviations: ADV, adenovirus; BoV, bocavirus; CoV, coronavirus; HMPV, human metapneumovirus; HRV, human rhinovirus; IFN-γ, interferon gamma; IL, interleukin; IP-10, Interferon gamma-induced protein 10; MIP-1α, macrophage inflammatory protein 1 alpha; RSV, respiratory syncytial virus; TNF-α, tumor necrosis factor alpha.
During most extended periods of HRV, RSV, and HMPV shedding, viral loads were remarkably stable (Figure 2A). For HRV, a pattern of viral load steady state or slight gradual decline followed by rapid elimination was noted. The case of RSV had a similar profile but with an initial high viral load peak and shorter duration of shedding. The case of HMPV had a more protracted decline with a single reexpansion phase. These transiently observed periods of steady-state viral loads are highly unlikely to occur by chance if true viral loads fluctuated or exhibited stochastic noise. Thus, the sampling method appears highly reliable. These data also suggest a brief period of equilibrium between the virus and local immune system before viral elimination.
In general, the level of symptoms appeared to track with detectable virus, particularly for CoV, HRV, and HMPV. For the single case of RSV, a high number of symptoms persisted beyond viral elimination (Figure 2A).
Stable and Surging Nasal Cytokine Levels During Respiratory Virus Infection
For several cytokines, particularly those in the Th2, Th17, and nondefined pathways (interleukin [IL] 2, IL-4, IL-5, IL-10, IL-13, IL-17A, and eotaxin), there was notable stability within and between study participants, independent of viral shedding (Figure 2B, Supplementary Figure 2B). This result demonstrates consistency in swabbing technique and again validates the precision of our approach.
Other molecules, particularly those associated with cytotoxic T-cell responses (granzyme B, perforin, tumor necrosis factor alpha [TNF-α], and interferon gamma [IFN-γ]) and macrophage responses (macrophage inflammatory protein 1 alpha [MIP-1α], IL-1α, IL-6, IL-18), showed monotonic expansion or clearance in response to most infections, with particularly dynamic shifts during RSV, HMPV, and 1 instance of HRV (p21) with the highest initial viral load (Figure 2B, Supplementary Figure 2B).
Cytokines Correlations According to Cellular Origin
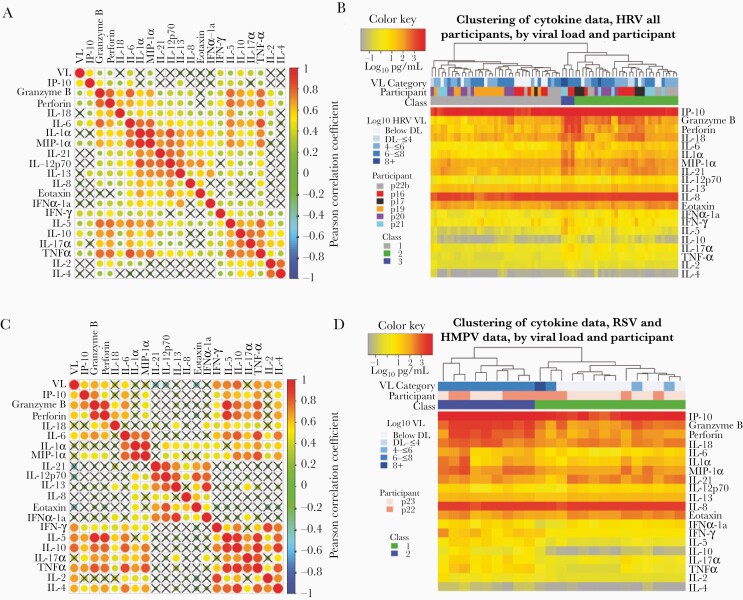
In 6 participants with HRV, we correlated cytokine patterns to infer cellular origin. High positive correlations were noted among analytes associated with a cytotoxic T-cell response (granzyme B, perforin, TNF-α, IL-6), among macrophage or epithelial cell–derived cytokines (MIP-1α, IL-1α, IL-6, IL-12p70, IL-21), and among Th2-associated responses (IL-5, IL-10, IL-17). The Th2-associated cytokines also correlated with many of the cytolytic T-cell and macrophage-associated cytokines (Figure 3A). HRV viral load was only moderately correlated with granzyme B, perforin, and interferon gamma-induced protein 10. This suggests that HRV may not induce an intense local immune response in a dose-dependent fashion. Similar results occurred with inclusion of all samples from all participants in the cohort (Supplementary Figure 3A).
Figure 3.
Cytokines correlate according to cellular origin during respiratory virus infection, whereas samples cluster according to level of inflammation. A and B, Data from participants p16, p17, p18, p19, p20, p21, and p22b who have human rhinovirus (HRV) infection. C and D, Data from participants p22 and p23 who have respiratory syncytial virus (RSV) and human metapneumovirus (HMPV), respectively. A and C, Correlation plots with strong correlation according to cell type origin. X indicates a nonsignificant correlation. Color intensity and the size of the dot are proportional to the Pearson correlation coefficient. For both datasets, strong positive correlations are noted within cytokines linked with cytolytic T-cell responses, macrophage responses, and Th2 responses. B and D, Linkage clustering analysis of samples (columns) demonstrates classes of samples based on the concentration of inflammatory cytokines. B, For HRV infections, a minority of samples (class 3) from 2 participants and with the highest levels granzyme B, perforin, interleukin 6, interleukin 1α, macrophage inflammatory protein 1α, and interferon-γ all had high viral loads. All 6 participants had samples in the least inflammatory class (class 1) and 5 participants had samples in the moderate inflammatory class (class 2). D, For RSV and HMPV, inflammatory (class 2) and noninflammatory (class 1) sample clusters are evident. The inflammatory class of samples is highly associated with the highest viral loads. Abbreviations: DL, detection limit; HMPV, human metapneumovirus; HRV, human rhinovirus; IFN-α, interferon alpha; IFN-γ, interferon gamma; IL, interleukin; IP-10, interferon gamma-induced protein 10; MIP-1α, macrophage inflammatory protein 1 alpha; RSV, respiratory syncytial virus; TNF-α, tumor necrosis factor alpha; VL, viral load.
In 2 participants infected with more inflammatory viruses, RSV and HMPV, we noted similar correlative trends as with HRV. Correlations among related pairs were higher for RSV/HMPV than for HRV (Figure 3B) and temporal kinetics were often strikingly similar, suggesting an equivalent cellular source (Figure 2B, Supplementary Figure 2B). Overall, there was a lack of correlation between cytokines associated with T-cell responses and with epithelial cells and macrophages. Viral load correlated with many cytokines of T-cell origin (Figure 3B), suggesting that RSV and HMPV may induce inflammation in a dose-dependent fashion.
Sample Clustering According to Degree of Inflammation
We sorted all HRV samples using linkage clustering analysis and demonstrated 3 classes of samples that were distinguished by levels of T-cell and macrophage-associated cytokines (Figure 3B). The minority of samples with the highest levels of granzyme B, perforin, IL-6, IL-1α, MIP-1α, and IFN-γ all had high viral loads, all from 2 participants. All 6 participants had some samples in the least inflammatory class and 5 participants had samples in the moderate inflammatory class. These data indicate that the inflammatory milieu in the HRV-infected nasal passage is dynamic over time, but tilts toward higher inflammation with higher viral loads. Similar results were observed when all samples were analyzed, though only 2 classes were distinguished (Supplementary Figure 3B).
We next sorted the RSV and HMPV samples and could not identify the optimal number of clusters. We selected 2 clusters, which were differentiated according to concentrations of most cytokines, again including granzyme B, perforin, IL-6, IL-1α, MIP-1α, and IFN-γ. Here the more inflammatory cytokine cluster clearly associated with high viral loads for both RSV and HMPV (Figure 3D).
Mathematical Modeling
We developed the ordinary differential equation model in equation (1) to link RSV and HMPV viral load and early and late immune responses, and evaluated which cytokines may track those responses (Supplementary Figures 4–6). The models suggest that IFN-γ and IL-21 may play a major role in RSV and HMPV control in vivo but do not rule out the effects of other cytokines and molecules in limiting infection.
DISCUSSION
Here we demonstrate that home self-sampling with mid-nasal foam swabs is well tolerated and provides reliable results for monitoring viral load and molecular immune responses to respiratory virus infections. These results have enormous practical implications. Self-collection at home is safe, noninvasive, and easily learned, allowing a reliable method for diagnosis and therapeutic monitoring. Because our kits could easily be used at home or in a drive-through testing environment, they provide an avenue to eliminate contact between an infected and contagious person and healthcare providers. They could also be used in the hospital or clinic setting, thereby saving personnel time and personal protective equipment. The use of comfortable, safe, and affordable foam swabs also highlights the possibility of scaling this approach to pediatric, adult, elderly, and immunocompromised populations. For the current SARS-CoV-2 pandemic, and future deadly respiratory virus epidemics, home self-swabbing will be a vital tool. The simplicity of the sampling approach also facilitates large-scale research studies of viral pathogenesis and transmission dynamics in which participants self-sample for months.
We have previously demonstrated increased sensitivity of self-collected foam nasal swabs compared to nasal washes in immunocompetent adults with respiratory viral infections [21], and in longitudinal studies in solid organ transplant recipients [22], with good compliance and participants reporting no issues with swab discomfort. The specific swab used in these prior studies and our present study were custom designed to limit discomfort while maintaining adequate sensitivity; we have demonstrated stability with these swabs with and without transport media after storage at room temperature for 7 days [21], making them ideal for home self-testing followed by shipment directly to a testing laboratory. Our data also demonstrate that bilateral swabbing increases yield and allows for more accurate quantification than swabbing a single nostril.
We also demonstrate an ability to accurately sample local cytokines with our swab, present at picogram levels. The combination of precise virologic and immunologic readouts of local infection is highly relevant for developing clinical severity scores and biomarkers. While studies are beginning to show that viral load may be predictive of COVID-19 severity [7], it is equally plausible that the intensity and phenotype of the early local cellular immune response plays a causal role in limiting the extent of infection [28]. By following the molecular immune response closely with daily sampling intervals, we also provide adequate data for mathematical models that can link specific arms of the cellular immune response to pathogen control in real time [20], a goal that has been difficult to attain for a majority of viral infections in humans.
Our study demonstrates several novel features of respiratory virus kinetics. RSV infection achieves a brief, extremely high, viral load, followed by a steady state and a final rapid phase of elimination. HRV also has a remarkably stable viral load in most participants before being rapidly eliminated. During a majority of our observed episodes, viral shedding is strongly correlated with symptoms. As viral load decreases, symptoms tend to dissipate.
Certain molecular immune responses are constitutively expressed, and vary little between and within participants, particularly those associated with Th2 mechanisms that are unlikely to play a role in elimination of virally infected cells. On the other hand, cytokines associated specifically with tissue-resident T-cell responses such as granzyme B, perforin, and IFN-γ, and macrophages such as IL-6 and IL-1 expand and contract during the course of viral shedding, particularly with more severe infections such as RSV and HMPV. Our technique therefore overcomes a fundamental limitation of human immunological studies, which is the inability to sample over temporally granular time intervals at the mucosal site of viral replication.
Further application of our technique is demonstrated with mathematical modeling that links expression of certain cytokines with early and late elimination of virus, although we were only able to model data from single participants with RSV and HMPV. Larger-scale studies may be able to link surges in different cytokines with specific respiratory viruses, including SARS-CoV-2, and to differentiate severity using these techniques.
There are important limitations to our study. Correlations between foam and flocked swabs were weaker at low viral loads. However, stochastic variation in low viral load samples is inherent to quantitation of viruses that replicate in mucosa. Additional variables such as storage temperature may have contributed to viral quantification variability. Our sample size for longitudinal episodes is relatively small, particularly on a per virus basis. A greater number of participants will be required to definitively differentiate kinetics patterns of different respiratory viruses and cytokine profiles associated with their containment. Selection of cytokines was incomplete and may have missed critical responders to viral infection. Finally, our mathematical models dramatically oversimplify the coordinated immune responses but do generate testable hypotheses that IFN-γ and IL-21 are vital for early and late containment of infection.
In summary, we establish a foam swab-based sampling method that is optimal for patient self-testing, both at home and in the clinical setting, permits serial therapeutic monitoring, and is suitable for tracking the natural virologic and immunologic course of respiratory virus infections. We recommend that this method be adapted to future clinical and research applications, including for the study of SARS-CoV-2.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We would like to thank our study participants.
Author contributions. A. W., J. T. S., and M. B. designed the experiments. E. M. K. and S. B. performed statistical analysis. E. V. performed data analysis. T. L. and E. L. C. enrolled participants and performed experiments. U. P. performed the cytokine analysis. K. R. J. designed the respiratory virus polymerase chain reaction assay, and A. L. G. and J. K. performed the assay. J. T. S. designed the mathematical modeling, and S. B., D. B. R., and E. F. C. O. performed the mathematical modeling. A. W., M. B., J. T. S., and E. R. D. wrote the manuscript.
Financial support. This work was supported by the National Institutes of Health (grant numbers K24 HL093294-06 to M. B. and K23 AI114844-02 to A. W.); and the Fred Hutchinson Cancer Center Vaccine and Infectious Diseases Faculty Initiative Fund (to M. B. and J. S.).
Potential conflicts of interest. A. W. reports personal fees from Kyorin and research support from Ansun, VB Tech, Amazon, and Allovir, all outside the submitted work. A. G. reports personal fees from Abbott Molecular, outside the submitted work. M. B. reports personal fees from Kyorin, Gilead, ReViral, Janssen, Ansun, Moderna, Vir Bio, GSK, Pulmocide, Pulmotect, Bavarian Nordic, ADMA, Allovir, and EvrysBio; and research support from Gilead, Janssen, Ansun, Vir Bio,VB Tech, and Amazon, all outside the submitted work. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Alpana Waghmare, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Pediatrics, University of Washington, Seattle, Washington, USA; Center for Clinical and Translational Research, Seattle Children’s Research Institute, Seattle, Washington, USA.
Elizabeth M Krantz, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Subhasish Baral, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Emma Vasquez, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Tillie Loeffelholz, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
E Lisa Chung, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Urvashi Pandey, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Obstetrics and Gynecology, University of Washington, Seattle, Washington, USA.
Jane Kuypers, Department of Laboratory Medicine, University of Washington, Seattle, Washington, USA.
Elizabeth R Duke, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Medicine, University of Washington, Seattle, Washington, USA.
Keith R Jerome, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Laboratory Medicine, University of Washington, Seattle, Washington, USA.
Alexander L Greninger, Department of Laboratory Medicine, University of Washington, Seattle, Washington, USA.
Daniel B Reeves, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Florian Hladik, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Obstetrics and Gynecology, University of Washington, Seattle, Washington, USA; Department of Medicine, University of Washington, Seattle, Washington, USA.
E Fabian Cardozo-Ojeda, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Michael Boeckh, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Medicine, University of Washington, Seattle, Washington, USA; Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
Joshua T Schiffer, Vaccine and Infectious Diseases Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA; Department of Medicine, University of Washington, Seattle, Washington, USA; Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, USA.
References
- 1. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 2020; 20:533–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gutierrez P. Coronavirus mapped: which countries have the most cases and deaths?https://www.theguardian.com/world/2020/mar/27/coronavirus-mapped-map-which-countries-have-the-most-cases-and-deaths. Accessed 30 March 2020.
- 3. Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of Covid-19—studies needed. N Engl J Med 2020; 382:1194–6. [DOI] [PubMed] [Google Scholar]
- 4. Clark TW, Ewings S, Medina MJ, et al. Viral load is strongly associated with length of stay in adults hospitalised with viral acute respiratory illness. J Infect 2016; 73:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hijano DR, Brazelton de Cardenas J, Maron G, et al. Clinical correlation of influenza and respiratory syncytial virus load measured by digital PCR. PLoS One 2019; 14:e0220908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fry AM, Goswami D, Nahar K, et al. Effects of oseltamivir treatment of index patients with influenza on secondary household illness in an urban setting in Bangladesh: secondary analysis of a randomised, placebo-controlled trial. Lancet Infect Dis 2015; 15:654–62. [DOI] [PubMed] [Google Scholar]
- 7. Yang Y, Yang M, Shen C, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv [Preprint]. Posted online 17 February 2020. 10.1101/2020.02.11.20021493. Accessed 22 April 2020. [DOI] [Google Scholar]
- 8. Agyemang E, Magaret AS, Selke S, Johnston C, Corey L, Wald A. Herpes simplex virus shedding rate: surrogate outcome for genital herpes recurrence frequency and lesion rates, and phase 2 clinical trials end point for evaluating efficacy of antivirals. J Infect Dis 2018; 218:1691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duke ER, Gilbert PB, Stevens-Ayers TL, et al. Viral kinetic correlates of cytomegalovirus disease and death after hematopoietic cell transplant. Biol Blood Marrow Transplant 2018; 24:S20. [Google Scholar]
- 10. Green ML, Leisenring W, Xie H, et al. Cytomegalovirus viral load and mortality after haemopoietic stem cell transplantation in the era of pre-emptive therapy: a retrospective cohort study. Lancet Haematol 2016; 3:e119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Natori Y, Alghamdi A, Tazari M, et al. CMV Consensus Forum . Use of viral load as a surrogate marker in clinical studies of cytomegalovirus in solid organ transplantation: a systematic review and meta-analysis. Clin Infect Dis 2018; 66:617–31. [DOI] [PubMed] [Google Scholar]
- 12. Murray JS, Elashoff MR, Iacono-Connors LC, Cvetkovich TA, Struble KA. The use of plasma HIV RNA as a study endpoint in efficacy trials of antiretroviral drugs. AIDS 1999; 13:797–804. [DOI] [PubMed] [Google Scholar]
- 13. Feld JJ, Jacobson IM, Hézode C, et al. ASTRAL-1 Investigators . Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med 2015; 373:2599–607. [DOI] [PubMed] [Google Scholar]
- 14. Feld JJ, Wong DK, Heathcote EJ. Endpoints of therapy in chronic hepatitis B. Hepatology 2009; 49:S96–102. [DOI] [PubMed] [Google Scholar]
- 15. Walker P, Whittaker C, Watson O, et al. The global impact of COVID-19 and strategies for mitigation and suppression. 10.25561/77735. Accessed 22 April 2020. [DOI]
- 16. Topham DJ, Reilly EC. Tissue-resident memory CD8+ T cells: from phenotype to function. Front Immunol 2018; 9:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng M, Hu S. Lung-resident γδ T cells and their roles in lung diseases. Immunology 2017; 151:375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kandasamy M, Furlong K, Perez JT, Manicassamy S, Manicassamy B. Suppression of cytotoxic T cell functions and decreased levels of tissue resident memory T cell during H5N1 infection. J Virol 2020; 94:e00057–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith AP, Moquin DJ, Bernhauerova V, Smith AM. Influenza virus infection model with density dependence supports biphasic viral decay. Front Microbiol 2018; 9:1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roychoudhury P, Swan DA, Duke E, et al. Tissue-resident T cell-derived cytokines eliminate herpes simplex virus-2-infected cells. J Clin Invest 2020; 130:2903–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Campbell AP, Kuypers J, Englund JA, Guthrie KA, Corey L, Boeckh M. Self-collection of foam nasal swabs for respiratory virus detection by PCR among immunocompetent subjects and hematopoietic cell transplant recipients. J Clin Microbiol 2013; 51:324–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Preiksaitis CM, Kuypers JM, Fisher CE, et al. A patient self-collection method for longitudinal monitoring of respiratory virus infection in solid organ transplant recipients. J Clin Virol 2015; 62:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuypers J, Wright N, Ferrenberg J, et al. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol 2006; 44:2382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989; 45:255– 68. [PubMed] [Google Scholar]
- 25. Hopkins B, Skellam JG. A new method for determining the type of distribution of plant individuals. Ann Bot 1954:213–27. [Google Scholar]
- 26. Banerjee A, Dave RN. Validating clusters using the Hopkins statistic. In: 2004 IEEE International Conference on Fuzzy Systems (IEEE catalog number 04CH37542), Budapest, Hungary, 2004:149–53. [Google Scholar]
- 27. R Core Team . R: a language and environment for statistical computing.http://www.R-project.org/. Accessed 22 April 2020.
- 28. Schiffer JT, Abu-Raddad L, Mark KE, et al. Mucosal host immune response predicts the severity and duration of herpes simplex virus-2 genital tract shedding episodes. Proc Natl Acad Sci U S A 2010; 107:18973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.