Abstract
Background
Although the mechanisms of adaptive immunity to pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are still unknown, the immune response to the widespread endemic coronaviruses HKU1, 229E, NL63, and OC43 provide a useful reference for understanding repeat infection risk.
Methods
Here we used data from proactive sampling carried out in New York City from fall 2016 to spring 2018. We combined weekly nasal swab collection with self-reports of respiratory symptoms from 191 participants to investigate the profile of recurring infections with endemic coronaviruses.
Results
During the study, 12 individuals tested positive multiple times for the same coronavirus. We found no significant difference between the probability of testing positive at least once and the probability of a recurrence for the betacoronaviruses HKU1 and OC43 at 34 weeks after enrollment/first infection. We also found no significant association between repeat infections and symptom severity, but found strong association between symptom severity and belonging to the same family.
Conclusions
This study provides evidence that reinfections with the same endemic coronavirus are not atypical in a time window shorter than 1 year and that the genetic basis of innate immune response may be a greater determinant of infection severity than immune memory acquired after a previous infection.
Keywords: endemic coronaviruses, waning immunity to endemic coronavirus, repeated endemic coronavirus infection
Through direct measurement of natural endemic coronavirus infections in a cohort of children and adults, this study confirms previous evidence that immunity developed upon infection with endemic coronaviruses is short-lived and reinfection is common within 1 year.
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appears to have emerged in humans in the Hubei province of China during November 2019 [1]. Human-to-human transmission was confirmed in early January, and since then the virus has rapidly spread to all continents except Antarctica. The outbreak was declared a pandemic by the World Health Organization on 11 March 2020. As of 4 July 2020, it has spread to >180 countries, with 10 922 324 confirmed cases and 523 011 deaths reported [2].
Symptoms associated with SARS-CoV-2 vary from none to extremely severe, with elder adults and people with underlying medical conditions more at risk for developing severe and potentially fatal disease [3]. At present, there is no vaccine or approved antiviral treatment for SARS-CoV-2, and therapies rely principally on symptom management. Many institutions across the world are working to develop a SARS-CoV-2 vaccine, and clinical trials with some vaccine candidates have already begun [4].
As the pandemic progresses, infecting millions of people across the world, a key question is whether individuals upon recovery are prone to repeat infection. There have been reports of individuals again testing positive by polymerase chain reaction (PCR) weeks after recovering from a SARS-CoV-2 infection. However, in Korea, as reported by the Korean Centers for Disease Control and Prevention, viable SARS-CoV-2 was not isolated in cell culture of respiratory samples from potentially reinfected individuals [5]; thus, these subsequent positive results may have been due to inactive genetic material detected by molecular testing. A recent animal challenge study showed evidence of (at least) short-term protection against reinfections in rhesus macaques experimentally reinfected 4 weeks after first infection [6].
The immune response following reexposure to a virus depends highly on the pathogen and on host–pathogen interactions. Some viruses such as measles elicit lifelong immunity; others, like influenza, do not. Moreover, when a reinfection is prone to occur, there can be differences in symptom severity. Reinfection with the same virus may be less symptomatic, as shown for subsequent influenza infections among children [7]. However, reinfection can also result in a more severe disease via antibody-dependent enhancement, a phenomenon in which antibodies raised against a virus bind with but are unable to neutralize a different strain of the same virus [8]
Two main processes appear to be responsible for the short-lived immunity engendered against some pathogens: (1) waning of antibodies and memory cells in the host system; and (2) antigenic drift of the pathogen that enables escape from the immunity built against previous strains.
Reinfections with the respiratory viruses have been reported in previous studies, in which individuals were infected in 2 sequential challenges with the same H1N1 virus [7, 9]. Studies focusing on respiratory syncytial virus have provided evidence of subsequent reinfection with very similar strains or with the same strain in <1 year [10, 11]. Serological studies have documented subsequent infections with endemic coronaviruses [12]. Sequential rhinovirus infections have also been reported in a number of studies; however, this finding could be due to the multitude (>150) of antigenically distinct types of rhinovirus in circulation [13].
To contextualize the issue of protective immunity to SARS-CoV-2, we here present findings from a recent proactive sampling project carried out in New York City that documented rates of infection and reinfection among individuals shedding seasonal CoV (types: HKU1, 229E, NL63, and OC43). The results are discussed and analyzed in the broader context of coronavirus infections.
METHODS
Data are derived from sampling performed between October 2016 and April 2018 as part of the Virome project, a proactive sampling of respiratory virus infection rates, associated symptom self-reports, and rates of seeking clinical care. We enrolled 214 healthy individuals from multiple locations in the Manhattan borough of New York City. Cohort composition is described in [14] and includes children attending 2 daycares, along with their siblings and parents; teenagers and teachers from a high school; adults working at 2 emergency departments (a pediatric and an adult hospital); and adults working at a university medical center. The cohort was obtained using convenience sampling, and all participants were <65 years of age. While the study period spanned 19 months from October 2016 to April 2018, some individuals enrolled for a single cold and flu season (October–April) and others for the entire study period. Participants (or their guardians, if minors) provided informed consent after reading a detailed description of the study (Columbia University Medical Center institutional review board AAAQ4358).
Nasopharyngeal samples were collected by study coordinators once a week irrespective of participant symptoms. Samples were screened using the GenMark eSensor respiratory viral panel (RVP) system for 18 different respiratory viruses, including coronavirus 229E, NL63, OC43, and HKU1. Sample collection and extraction followed the same protocol as shown in [15].
In addition, participants completed daily self-reports rating 9 respiratory illness–related symptoms (fever, chills, muscle pain, watery eyes, runny nose, sneezing, sore throat, cough, chest pain), each of which was recorded on a Likert scale (0 = none, 1 = mild, 2 = moderate, 3 = severe); see Supplementary Text 1, Supplementary Tables 1 and 2, and Galanti et al [14] for further survey details.
For this analysis, only the 191 participants who contributed at least 6 separate pairs of nasopharyngeal samples in the same season were included. We defined an infection (or viral) episode as a group of consecutive weekly specimens from a given individual that were positive for the same virus (allowing for a 1-week gap to account for false negatives and temporary low shedding). We classified all infection episodes as symptomatic or asymptomatic according to individual symptom scores in the days surrounding the date of the first positive swab of an episode. We considered multiple criteria for discriminating between symptomatic and asymptomatic episodes, as a standard definition for symptomatic infection does not exist in the literature. Table 1 reports the 5 symptomatic thresholds used; all of the symptom scores are described in reference to a –3 to +7-day window around the date of the initial positive swab for each infection episode. The daily symptom score is defined as the sum of the 9 individual symptoms (range, 0–27) on a given day. Total symptom score is the daily symptom score summed over the –3 to +7-day window. See Supplementary Text 1 for details and examples on how symptom scores were calculated (Supplementary Tables 1 and 2) per the definitions in Table 1.
Table 1.
Definitions of Symptomatic Infections
| Definition | |
|---|---|
| Definition 1 | At least 1 day with a daily score >3 |
| Definition 2 | Minimum 2 individual symptoms >0 and at least 1 symptom >1 |
| Definition 3 | Total symptom score >9 |
| Definition 4a | Total symptom score greater than twice the weekly average for the infected individual |
| Definition 5 | Total symptom score >0 (ie, any reported symptom) |
All symptom definitions are described in reference to a –3/+7-day window around the date of the initial positive swab for an infection episode.
aDefinition 4 is relative to an individual’s long-term average total symptom score.
We applied standard methods of survival analysis to our longitudinal records of infections to estimate (1) the probability of infection with each endemic coronavirus type; and (2) the probability of being reinfected with the same coronavirus type following a previous documented infection. More specifically, the probability of infection and reinfection by time t, ,was estimated as:
where S(t) is the standard Kaplan–Meier estimator and time t is measured in either weeks from enrollment in the cohort, for the first analysis, or weeks from the previous documented infection (with a specific coronavirus type), for the second analysis. Here are the participants testing positive exactly i weeks after enrollment (after first infection) and are the participants who are still enrolled i weeks after enrollment (after first infection). The denominator corrects for participants withdrawing from the study at different times by right censoring.
The Kaplan–Meier estimators are compared statistically using the log-rank test. We used Fisher exact test to analyze the difference between symptoms developed during subsequent infections and analysis of variance (ANOVA) comparison to test differences in symptom scores reported by different family clusters. We restricted the last analysis to the family clusters within the cohort that presented at least 3 coronavirus infections during the study.
RESULTS
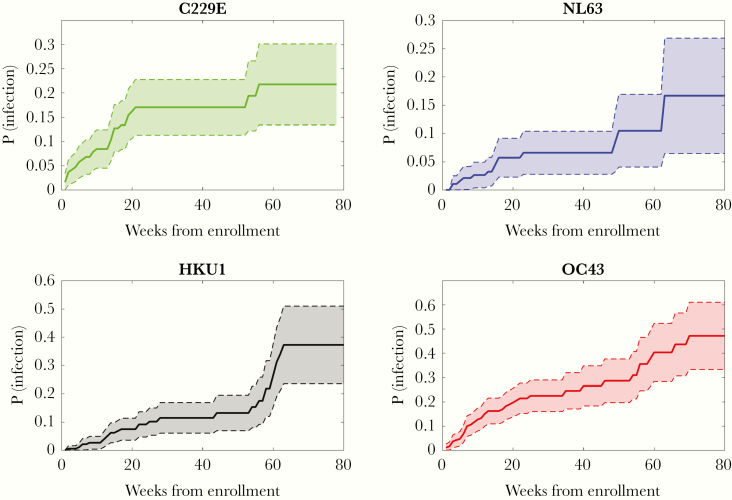
Among all participants enrolled, 86 individuals tested positive at least once during the study for any coronavirus infection. Forty-eight individuals tested positive at least once for OC43, 31 tested positive for 229E, 15 tested positive for NL63, and 28 tested positive for HKU1. Figure 1 shows a Kaplan–Meier plot estimating the probability of becoming infected with each coronavirus within x weeks following enrollment (see Supplementary Table 3 for the number of individuals infected and censored at each time point). OC43 was the most widely diffused virus; the probability of testing positive following 80 weeks in the study was 0.47. In contrast, NL63 was the least frequently isolated coronavirus type; the probability of testing positive after 80 weeks was 0.17. Among the study participants, 12 individuals tested positive multiple times during the study for the same coronavirus: 9 tested positive multiple times for OC43, 2 tested positive twice for HKU1, 1 tested positive twice for 229E, and none tested positive multiple times for NL63. Among the 9 participants with multiple OC43 infections, 3 individuals experienced 3 separate infection episodes, and the other 6 experienced 2 separate episodes. The median time between reinfection events was 37 weeks. The shortest time for a reoccurrence of infection was 4 weeks (OC43), and the longest was 48 weeks (OC43). Among the 12 individuals testing positive multiple times for the same coronavirus, 9 were children aged between 1 and 9 years at enrollment, and 3 were adults aged between 25 and 34 years (see Supplementary Table 4 and Supplementary Figure 1 for characteristics and timelines of the repeated infections).
Figure 1.
Kaplan–Meier plots showing the probability of testing positive within x weeks after enrollment for each of the 4 types of seasonal coronavirus. The shaded area is the 95% confidence interval. In the case of individuals testing positive multiple times for the same coronavirus type, we only considered the time to the first occurrence in this plot. Abbreviation: P(infection), probability of testing positive.
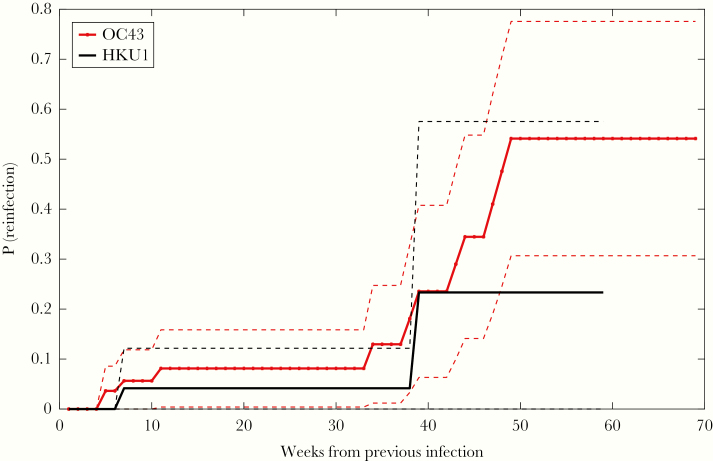
Figure 2 shows a Kaplan–Meier plot estimating the probability of becoming reinfected with the same betacoronavirus (OC43 and HKU1) within x weeks after a previously documented infection (see Supplementary Table 5 for the number of individuals infected and censored at each time point). A comparison between the data shown in Figure 2 and Figure 1 found no significant differences between the probability of testing positive at least once and the probability of a recurrence for both HKU1 and OC43 at 34 weeks after enrollment/first infection.
Figure 2.
Probability of becoming reinfected with the same betacoronavirus type (OC43 in red crossed line and HKU1 in black straight line) within x weeks after a first documented infection. Dashed lines show the 95% confidence interval. Here, only individuals testing positive multiple times for the same coronavirus type are considered. For each occurrence, we calculated the time distance from the previous infection. Abbreviation: P(reinfection), probability of testing positive again after a previous documented infection with the same coronavirus type.
To control for false-positive PCR results, we tested the sensitivity of the findings to different choices of the positivity threshold used in RVP testing (see Supplementary Text 2 and Supplementary Figures 2–5). The probability of reinfection with betacoronaviruses at >38 weeks after prior infection was robust across different thresholds, whereas short-term reinfection signals could be an artifact due to PCR amplification. This shifted threshold also yields a statistically significant difference between the probability of testing positive at least once and the probability of a recurrence after first infection until week 43 (P = .04).
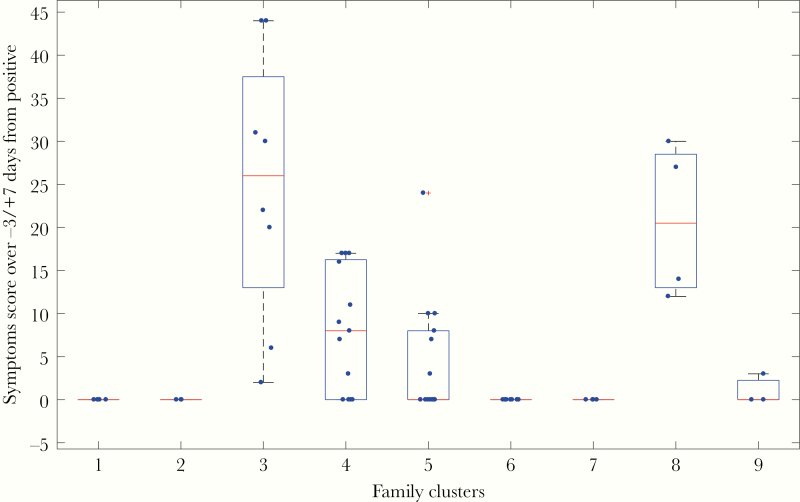
There was no significant difference in the likelihood of experiencing symptomatic infection between the first and subsequent infection episodes by any of the 5 definitions provided in Table 1; that is, severity of symptoms neither increased nor decreased significantly upon second infection. In particular, all of the individuals who were completely asymptomatic during the first recorded occurrence did not report any symptoms during subsequent infection(s) with the same coronavirus type. However, there was a significant association between severity of symptoms associated with any coronavirus infection and belonging to the same family cluster (P < .0001, 1-way ANOVA). Figure 3 shows the total symptom score associated with any coronavirus infection for infections grouped by family cluster.
Figure 3.
Total symptom score associated with infections by any coronavirus type. The score is calculated as the sum of daily symptom scores for the –3/+7 day window around the test date, as indicated for definition 3 in Table 1. Each point represents an infection event, and each cluster represents a family group. Each family group 1 to 9 is composed of a parent and 1–4 children. For each box, the red line indicates the median, and the bottom and top edges of the blue box are the 25th and 75th percentiles. The dashed lines extend to the most extreme data points that are not outliers, whereas the outliers are indicated by the red “+” symbol.
DISCUSSION
As the SARS-CoV-2 pandemic spreads to millions of individuals worldwide, it is extremely important to understand the mechanisms of protective immunity elicited by infection. Until direct observations of adaptive immune response to SARS-CoV-2 become available, analyses of protective immunity elicited by other coronaviruses may offer useful insights.
Several studies in the last 4 decades have shown that infections with the 4 endemic coronaviruses 229E, OC43, NL63, and HKU are common in the general population [12, 16]. Infection with these viruses generally produces mild and even asymptomatic infection [17]. Serological studies have shown that >90% of the population presents a baseline level of antibodies against these endemic coronaviruses, with first seroconversion occurring at a young age [16, 18]. Shortly after infection, baseline antibody titers increase sharply; this response has been demonstrated for both natural and experimentally induced infections [12, 19, 20].
Antibody titers start increasing roughly 1 week following infection, reach a peak after about 2 weeks [20], and by 4 months to 1 year have returned to baseline levels [12, 20]. A challenge study [20] showed that the likelihood of developing an infection after inoculation correlated with participants’ concentration of antibodies at enrollment. Moreover, a positive correlation has been shown between antibody rise after infection, severity of clinical manifestation, and viral shedding [19], with milder cases linked to less substantial postinfection antibody rises.
Instances of natural reinfections with the same virus type have been documented previously [12] in which repeated infections with OC43 and 229E were recorded by serological testing. Subsequent infections were separated by at least 8 months, though study participants were tested every 4 months. Participants in a separate challenge study were inoculated with coronavirus 229E and then rechallenged with the same virus after 1 year [20]. In most cases, reinfection occurred, though it presented with decreased symptom severity and shortened duration of shedding.
The adaptive immune response to coronavirus is mainly directed toward the most variable part of the virus, a region that is not conserved across types; consequently, cross-reactive protection between different types does not appear to be an important factor [21, 22]. In addition, the effects of antigenic drift on reinfection have not been elucidated [23], and more studies are warranted to understand whether repeat infections are ascribable to rapid virus evolution rather than a decline in antibody titers.
The mild pathogenicity of seasonal coronavirus infection (with immune response often localized to the upper respiratory trait) is also often regarded as the reason for short-lived immunity. Coronavirus infections, and the adaptive immunity acquired toward them, have also been studied in animals. In a study on porcine respiratory coronavirus, which causes subclinical infections in pigs, antibody titers waned approximately 1 year after experimental infection [24]. In contrast, an experimental study on murine coronavirus, which produces severe, systemic infections in mice, has shown an interplay between virus-specific antibodies and T cells, that upon survival in the host lead to lifelong protection against reinfection [25]. Similarly, a longer immunity profile has been hypothesized for severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) due to their increased severity and to the systemic response that infection induces [21]. Specific antibodies were detectable for at least 2 years in SARS and MERS survivors [26, 27]. Although longitudinal studies on SARS survivors have not detected specific SARS immunoglobulin G antibody persistence 5 years after infection, they have found that specific memory T cells persist in the peripheral blood of recovered SARS patients, and at higher levels in patients who experienced severe disease [28]. Whether the presence of these memory T cells would be enough to induce a fast, protective response upon reinfection with SARS has not been assessed.
Our study confirms that seasonal coronaviruses are widespread in the general population, with infections directly documented for a large fraction of the participants in our study. The methods for our analysis are based on the hypothesis that infection probabilities are comparable among participants enrolled at different times in the study. However, the seasonality of endemic coronaviruses, which are mostly absent during the summer months, and the relative magnitude across years of seasonal coronavirus epidemics are limitations. In the United States, the prevalence of OC43 during the 2016–2017 season was much higher than during the 2017–2018 season, whereas the opposite trend was observed for HKU1 [29]. Moreover, our estimates of infection and reinfection probabilities must be considered as a lower bound, due to the occurrence of weekly swabs missed by the participants and due to the design of the study itself, which may have missed infections of short duration in between consecutive weekly tests. Nevertheless, this study confirms that reinfections with the same coronavirus type occur in a time window shorter than 1 year, and finds no significant association between repeat infections and symptom severity. Instead, it suggests the effect of possible genetic determinants of innate immune response, as individuals asymptomatic during first infection did not experience symptoms during subsequent infections, and members of the same families reported similar symptom severity. Genetic variations associated with immune responses have been associated with increased severity and exacerbation of symptoms due to respiratory infections [30, 31].
We recognize that the self-reporting of symptoms is an important limitation in this analysis and that parents reported symptoms for their dependents, which possibly introduced bias. Moreover, the majority of the repeated coronavirus infections were found in children, a cohort more vulnerable to infection because of their immature immune system [32], and 26% of the episodes in the repeated infections were coinfections with other respiratory viruses (see Supplementary Table 2). Another potential limitation of our study is the high sensitivity of PCR tests, which can amplify very small amounts of genetic material, possibly not ascribable to active infections. However, the occurrence of repeated infections separated by at least 38 weeks was corroborated by repeating the analysis with different positivity thresholds for the RVP. Still, without virus sequencing, we cannot exclude the possibility that subsequent positives are the resurgence of the same infection rather than new infections, especially for infections reoccurring within a short time window. Additional analyses involving viral sequencing and serological testing would be necessary to confirm repeated infections and to help disentangle the effects of antigenic drift and antibody waning.
More studies analyzing the genetic basis of individual response to coronavirus infections are also warranted. Even though endemic coronaviruses are very rarely associated with severe disease, their widespread diffusion, together with the fact that OC43 and HKU1 belong to the same Betacoronavirus genus as SARS-CoV-2, offers important opportunities for investigation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The funders had no role in study design, data collection and analysis, preparation of the manuscript, or decision to submit the manuscript for publication.
Financial support. This work was supported by the Defense Advanced Research Projects Agency (contract number W911NF-16-2-0035).
Potential conflicts of interest. J. S. and Columbia University disclose partial ownership of SK Analytics. J. S. also discloses consulting for BNI. M. G. reports no potential conflicts of interest.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med 2020; 26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Situation report–166.2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200704-covid-19-sitrep-166.pdf?sfvrsn=6247972_2. Accessed 4 July 2020.
- 3. Centers for Disease Control and Prevention. Coronavirus disease 2019: symptoms 2020. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html. Accessed 15 June 2020.
- 4. National Institutes of Health. NIH clinical trials of investigational vaccine for COVID-19 begins. 2020. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-investigational-vaccine-covid-19-begins. Accessed 15 June 2020.
- 5. Korea Centers for Disease Control and Prevention. Press releases 2020. https://www.cdc.go.kr/board/board.es?mid=a30402000000&bid=0030. Accessed 15 June 2020.
- 6. Bao L, Deng W, Gao H, et al. . Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv [Preprint]. Posted 14 March 2020. https://www.biorxiv.org/content/10.1101/2020.03.13.990226v1. Accessed 15 June 2020. [Google Scholar]
- 7. Davies JR, Grilli EA, Smith AJ. Influenza A: infection and reinfection. J Hyg 1984; 92:125–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tirado SM, Yoon KJ. Antibody-dependent enhancement of virus infection and disease. Viral Immunol 2003; 16:69–86. [DOI] [PubMed] [Google Scholar]
- 9. Memoli MJ, Han A, Walters K. Influenza A reinfection in sequential human challenge: implications for protective immunity and “universal” vaccine development. Clin Infect Dis 2020; 70:748–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall CB, Walsh EE, Long CE, Schnabel KC. Immunity to and frequency of reinfection with respiratory syncytial virus. J Infect Dis 1991; 163:693–8. [DOI] [PubMed] [Google Scholar]
- 11. Scott PD, Ochola R, Ngama M, et al. . Molecular analysis of respiratory syncytial virus reinfections in infants from coastal Kenya. J Infect Dis. 2006; 193:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt OW, Allan ID, Cooney MK, Foy HM, Fox JP. Rises in titers of antibody to human coronaviruses OC43 and 229E in Seattle families during 1975–1979. Am J Epidemiol 1986; 123:862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zlateva KT, De Vries JJC, Coenjaerts FEJ, et al. . Prolonged shedding of rhinovirus and re-infection in adults with respiratory tract illness. Euro Respir J 2014; 44:169–77. [DOI] [PubMed] [Google Scholar]
- 14. Galanti M, Birger R, Ud-Dean M, et al. . Longitudinal active sampling for respiratory viral infections across age groups. Influenza Other Respir Viruses 2019; 13:226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shaman J, Morita H, Birger R, et al. . Asymptomatic summertime shedding of respiratory viruses. J Infect Dis 2018; 217:1074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou W, Wang W, Wang H, Lu R, Tan W. First infection by all four non-severe acute respiratory syndrome human coronaviruses takes place during childhood. BMC Infect Dis 2013; 13:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galanti M, Birger R, Ud-Dean M, et al. . Rates of asymptomatic respiratory virus infection across age groups. Epidemiol Infect 2019; 147:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Severance EG, Bossis I, Dickerson FB, et al. . Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin Vaccine Immunol 2008; 15:1805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kraaijeveld CA, Reed SE, Macnaughton MR. Enzyme-linked immunosorbent assay for detection of antibody in volunteers experimentally infected with human coronavirus strain 229 E. J Clin Microbiol 1980; 12:493–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990; 105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perlman S, Vijay R. Middle East respiratory syndrome vaccines. Int J Infect Dis 2016; 47:23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Macnaughton MR, Hasony HJ, Madge MH, Reed SE. Antibody to virus components in volunteers experimentally infected with human coronavirus 229E group viruses. Infect Immun 1981; 31:845–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monto AS, Cowling BJ, Peiris JSM. Coronaviruses. In: Kaslow R, Stanberry L, Le Duc J, eds. Viral infections of humans. Boston, MA: Springer, 2014. [Google Scholar]
- 24. Wesley R. Neutralizing antibody decay and lack of contact transmission after inoculation of 3- and 4-day-old piglets with porcine respiratory coronavirus. J Vet Diagn Invest 2002; 14:525–7. [DOI] [PubMed] [Google Scholar]
- 25. Williamson JS, Stohlman SA. Effective clearance of mouse hepatitis virus from the central nervous system requires both CD4+ and CD8+ T cells. J Virol 1990; 64:4589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mo H, Zeng G, Ren X, et al. . Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology 2006; 11:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Payne DC, Ibrahim I, Rha B, et al. . Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis 2016; 22:1824–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang F, Quan Y, Xin ZT, et al. . Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol 2011; 186:7264–8. [DOI] [PubMed] [Google Scholar]
- 29. Centers for Disease Control and Prevention. National Respiratory and Enteric Virus Surveillance System 2019. https://www.cdc.gov/surveillance/nrevss/coronavirus/index.html. Accessed 15 June 2020.
- 30. Janssen R, Bont L, Siezen CL, et al. . Genetic susceptibility to respiratory syncytial virus bronchiolitis is predominantly associated with innate immune genes. J Infect Dis 2007; 196:826–34. [DOI] [PubMed] [Google Scholar]
- 31. Loisel DA, Du G, Ahluwalia TS, et al. . Genetic associations with viral respiratory illnesses and asthma control in children. Clin Exp Allergy 2016; 46:112–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci 2015; 282:20143085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.