Abstract
For patients with COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the damages to multiple organs have been clinically observed. Since most of current investigations for virus–host interaction are based on cell level, there is an urgent demand to probe tissue-specific features associated with SARS-CoV-2 infection. Based on collected proteomic datasets from human lung, colon, kidney, liver, and heart, we constructed a virus-receptor network, a virus-interaction network, and a virus-perturbation network. In the tissue-specific networks associated with virus–host crosstalk, both common and different key hubs are revealed in diverse tissues. Ubiquitous hubs in multiple tissues such as BRD4 and RIPK1 would be promising drug targets to rescue multi-organ injury and deal with inflammation. Certain tissue-unique hubs such as REEP5 might mediate specific olfactory dysfunction. The present analysis implies that SARS-CoV-2 could affect multi-targets in diverse host tissues, and the treatment of COVID-19 would be a complex task.
Keywords: SARS-CoV-2, proteome-wide, tissue-specific
Introduction
The outbreak of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has gradually expanded to pandemic (Wang et al., 2020a; World Health Organization, 2020b), due to the strong infectivity of the virus. It is an urgent demand to explore the mechanism of infection and treatment of the disease. According to the clinical observation, ∼14% experienced severe disease and 5% were critically ill (World Health Organization, 2020a). In addition to the typical symptoms of pulmonary infection, severe patients also show damage to multiple organs, including kidney, liver, heart, etc. (Wang et al., 2020b). In order to better understand the mechanism of viral infection and to treat patients more specifically, it is a matter of concern whether there are differences in the susceptibility and response of different organs to the virus. Since ACE2 is the major receptor mediating virus entry to host cell, previous studies mostly focused on transcription level on ACE2 (Zou et al., 2020). However, the well-known discordance of transcriptome and proteome levels proposes that study at the protein level could reveal the direct circumstances of host responses to virus infection. Moreover, the protein–protein interaction (PPI) triggered by virus invasion and replication in host cells would be complex and tissue-specific. There are hundreds of proteins that could be interacted with or perturbated by virus, while the drugs should work on major players. Thus, the network analysis is more helpful to understand virus perturbation and select drugs.
Due to the similarity of spike proteins in coronavirus, SARS-CoV-2 could invade in host cells possibly like SARS-CoV, through three potential receptors namely ACE2, DC-SIGN (genotype CD209), and L-SIGN (genotype CLEC4M) (Cai et al., 2020). In addition, TMPRSS2 has been reported to prime the spike protein of SARS-CoV-2 (Hoffmann et al., 2020). When virus entered host cells, a number of host proteins could be hijacked and regulated to facilitate virus activity, which could be detected by proteomic approach. Most recent study revealed interactome of SARS-CoV-2 in 293T cells using AP−MS technology (Gordon et al., 2020). Another work identified proteins in host cells regulated by SARS-CoV-2 infection (Bojkova et al., 2020). However, these studies based on cells could not provide specific features of diverse tissues. In order to extend the cell-based data to tissue-specific mode, we collected population-based proteomic datasets, from human lung (Mertins et al., 2018), colon (Vasaikar et al., 2019), kidney (Clark et al., 2019), liver (Gao et al., 2019), and heart (Doll et al., 2017). The first four datasets are from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC), which include various normal tissues. Data from hearts include two parts from cavities and vessels, respectively. Based on the PPI skeleton or protein–protein expression correlation, we construct the first tissue-specific proteome-wide network associated with SARS-CoV-2 infection.
Results
Protein–RNA discordance of virus receptors in diverse tissues
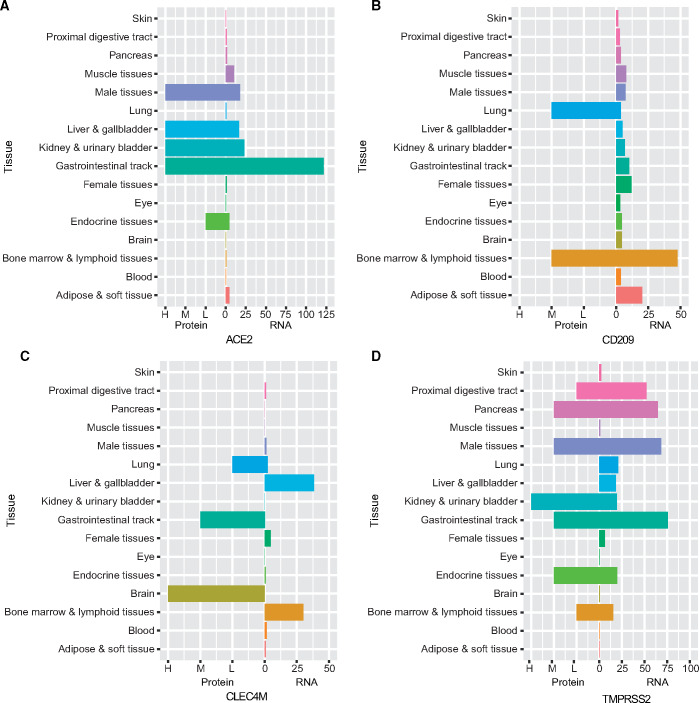
First, we checked RNA and antibody-based protein data for three receptors and TMPRSS2, from a variety of tissues in the Human Protein Atlas (HPA) database (https://www.proteinatlas.org/). We can clearly see that ACE2 has high transcription levels in gastrointestinal tract, kidney and bladder, liver and gallbladder, male tissue (including ductus deferens, testis, epididymis, seminal vesicle, and prostate), and muscle tissue (Figure 1A). CD209 has higher RNA levels in adipose, soft tissue, bone marrow, and lymphoid tissue (Figure 1B). CLEC4M has more RNA transcription in bone marrow, lymphoid tissue, liver, and gallbladder tissue (Figure 1C). However, their transcription level in lung is not high. The protein expression level is not consistent with the RNA transcription level in most tissues. For example, in lung tissue, CD209 and CLEC4M protein expression is found (Figure 1B and C), while ACE2 is undetectable by antibody (Figure 1A). In addition, we found CD209 protein expression in bone marrow and lymphoid tissue (Figure 1B), CLEC4M in brain and gastrointestinal tract (Figure 1C), and ACE2 in gastrointestinal tract, liver and gallbladder, kidney and bladder, male tissue as well as endocrine tissue (Figure 1A). Compared to the three receptors, TMPRSS2 has higher RNA transcription and protein expression levels in more tissues. In lung, a certain degree of RNA transcription but no protein expression is observed. Higher RNA transcription and protein expression levels are found in gastrointestinal tract, male tissues, and pancreas. Although RNA transcription levels are not very high in kidney, urinary bladder, and endocrine tissues, high protein expression levels are observed (Figure 1D). The obvious discordance between RNA and protein levels demonstrates that it is essential to measure protein expression itself in different tissues, in order to evaluate the influence by SARS-CoV-2 in human body.
Figure 1.
Protein expression and RNA transcription of SARS-CoV-2 receptors (ACE2, CD209, and CLEC4M) and the serine protease TMPRSS2. For protein data, each bar represents the highest expression score found in a group of tissues. RNA expression summary shows the consensus RNA data based on normalized expression data. Details are in Materials and methods.
Virus-receptor network
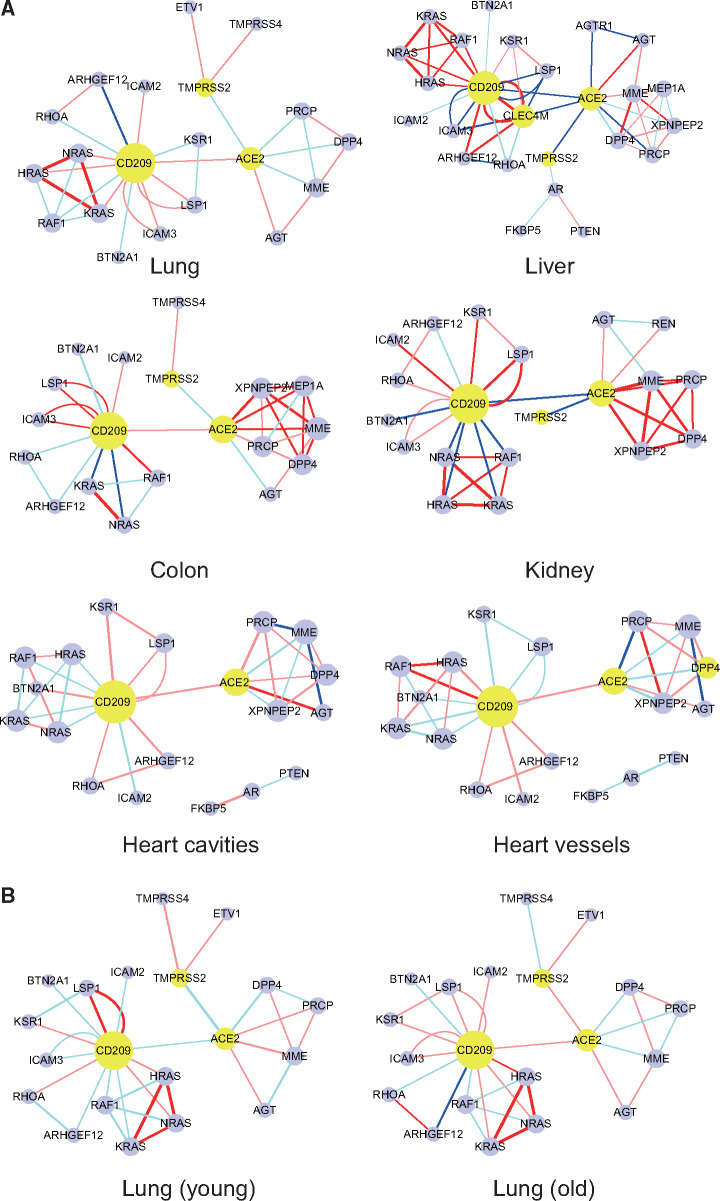
Then, we collected mass spectrometry-based proteomic data of normal tissues of four organs from CPTAC database and a set of data of normal heart (Doll et al., 2017), including two parts, respectively, from cavities and vessels. We constructed PPI network with the three potential receptors and TMPRSS2 as the center. The correlation between each two proteins across individuals in each dataset was used as the weight of the edge. Each kind of tissue has its specific network structure. First, we notice that ACE2 and CD209 could be detected in all tissues, while the expression of CLEC4M is only observed in liver (Figure 2A). The number of protein–protein pairs in each tissue network ranges from 33 to 55, with the least in lung network and the most in liver network (Figure 2A). Specifically, we focus on the interactions among three potential receptors. ACE2 and CD209 are positively correlated in lung, colon, and two parts of heart (cavities and vessels). They are negatively correlated in liver and kidney, suggesting that there may be some competition or substitution between the two receptors in these two organs. The correlation between CLEC4M and CD209 is positive in liver, while both of them show negative correlation with ACE2 (Figure 2A). Assumingly, the virus entry to liver cell might also be through CLEC4M and CD209 coordination. On the whole, there are more positive correlations between the receptor and other proteins in most tissues, indicating a possibility that after the virus binds to the receptor, it is easy to coordinate the network partners, such as the correlation between ACE2 and AGT. Some correlations change in different tissues, such as CD209−KRAS correlation that is positive in lung and liver but negative in colon, kidney, and heart (Figure 2A). There are a lot of differences between heart cavities and vessels, indicating different reactions of the two parts of heart. According to the threshold that we set, TMPRSS2 is not associated with the three receptors in the String database and TMPRSS2 is not found in two tissues of the heart. However, according to the previous report (Hoffmann et al., 2020), TMPRSS2 is an important protein for initiating the S protein of SARS-CoV-2. We assume that there are some connections between the two proteins, so the connection between ACE2 and TMPRSS2 is added on the network. Unexpectedly, the ACE2 and TMPRSS2 showed negative correlation in all tissues. In lung, the correlation between TMPRSS2 and ACE2 was found to be weakly negative (Pearson correlation coefficient −0.191, P-value 0.237). Considering the mixed population, the relationship between TMPRSS2 and ACE2 might be affected by various factors such as age. We divided the population into young group and old group at the age of 55 years old, and the numbers are 16 and 86, respectively. TMRPSS2 and ACE2 have a strong negative correlation (Pearson correlation coefficient −0.596, P-value 0.069) in the young group, and a weak-positive correlation (Pearson correlation coefficient 0.121, P-value 0.525) in the old group (Figure 2B). These observations may interpret the different responses to SARS-CoV-2 in young and elderly population. In lung and colon, we found a positive correlation between TMPRSS2 and TMPRSS4 (Figure 2A), which may indicate a synergistic effect of these two serine proteases (TMPRSSs) in these tissues.
Figure 2.
Receptor-centered PPI networks in six normal tissues of five organs. The yellow ones are the receptors. The red edge in the network indicates a significant or strong-positive correlation between the two proteins, while pink indicates a non-significant or weak-positive correlation. Dark blue indicates a significant or strong-negative correlation, while light blue indicates a non-significant or weak-negative correlation. The thickness of the edge is determined by the correlation coefficient.
Virus-interaction network
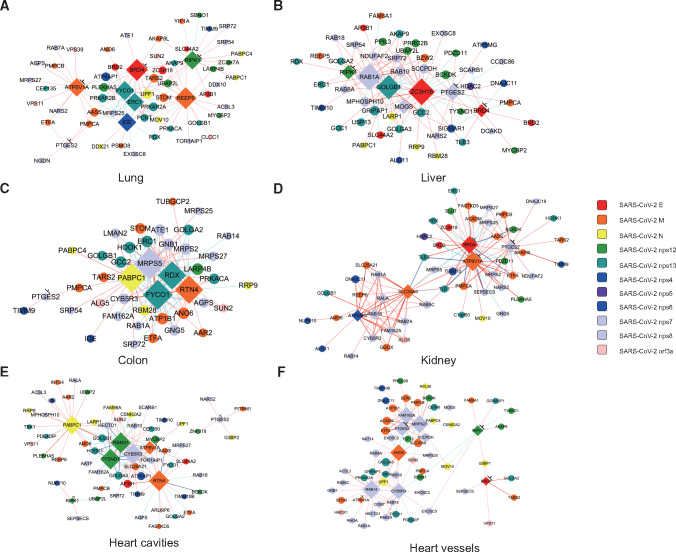
A most recent study revealed the interactome of SARS-CoV-2 proteins in 293T cells using AP-MS technology, including 332 proteins with 66 targets by 69 available drugs (Gordon et al., 2020). In the 332 proteins interacted with SARS-CoV-2, 188 of them may interact with the major virus components. We searched for the existence of the 188 proteins in each kind of tissue, respectively, and then calculated the correlation between each two proteins. We took the relationship with a P-value <0.01 as a relatively reliable relationship. The top hubs were defined by ranking according to the numbers of connections with other proteins. The top 5 hubs and the top 10 drug target proteins were selected to construct the tissue-specific network (Figure 3). Hubs often interact with E, N, M, nsp7, nsp8, nsp12, and nsp13 proteins of SARS-CoV-2. Some of these hubs are potential drug targets, such as bromodomain-containing protein 4 (BRD4), ATP6V1A, PTGES2, receptor-interacting protein kinase 1 (RIPK1), etc. (Supplementary Table S1). Each network contains at least one hub that can be targeted by available drugs, which provides a reference for accurate medication. Each tissue shows specific structure of network, indicating the complex interaction of virus in different tissues. In lung, drug-targeting hubs, such as BRD4, ATP6V1A, and RIPK1, have extensive interactions with other proteins, and these interactions are mainly positively related. Therefore, inhibiting these hubs might be crucial to regulate virus–host interaction (Figure 3). BRD4 could interact with viral structural E protein, and RIPK interacts with nsp12. Viral nsp12 and E proteins are vital components for SARS-CoV-2 replication and package. Another hub receptor accessory protein 5 (REEP5), which interacts with viral structural M protein and is only found as the hub in lung, which seems playing unique function in lung-related tissues. In addition, FYCO1, PABPC1, CYB5R3, reticulon 4 (RTN4), and other proteins repeatedly appear as hubs in a variety of tissues. Although there are no suitable drugs to target these proteins, they may play an important role in the infection and pathogenesis of the virus.
Figure 3.
Correlation networks consisting of hub proteins interacted with SARS-CoV-2 in six tissues. Each color represents a specific SARS-CoV-2 protein that interacts with the nodes (shown on the right). Major hubs are marked by diamond. Hubs that can be drug targets are marked with ‘⊥’, including ATP6V1A, BRD4, PTGES2, and RIPK1 in lung, BRD4, PTGES2, and RIPK1 in liver, PTGES2 in colon, ATP6V1A, BRD4, and PTGES2 in kidney, ATP6V1A, PTGES2, and RIPK1 in heart cavities, as well as ATP6V1A, BRD4, PTGES2, and RIPK1 in heart vessels. The coloring principle of edges is the same as described in Figure 2.
Virus-perturbation network
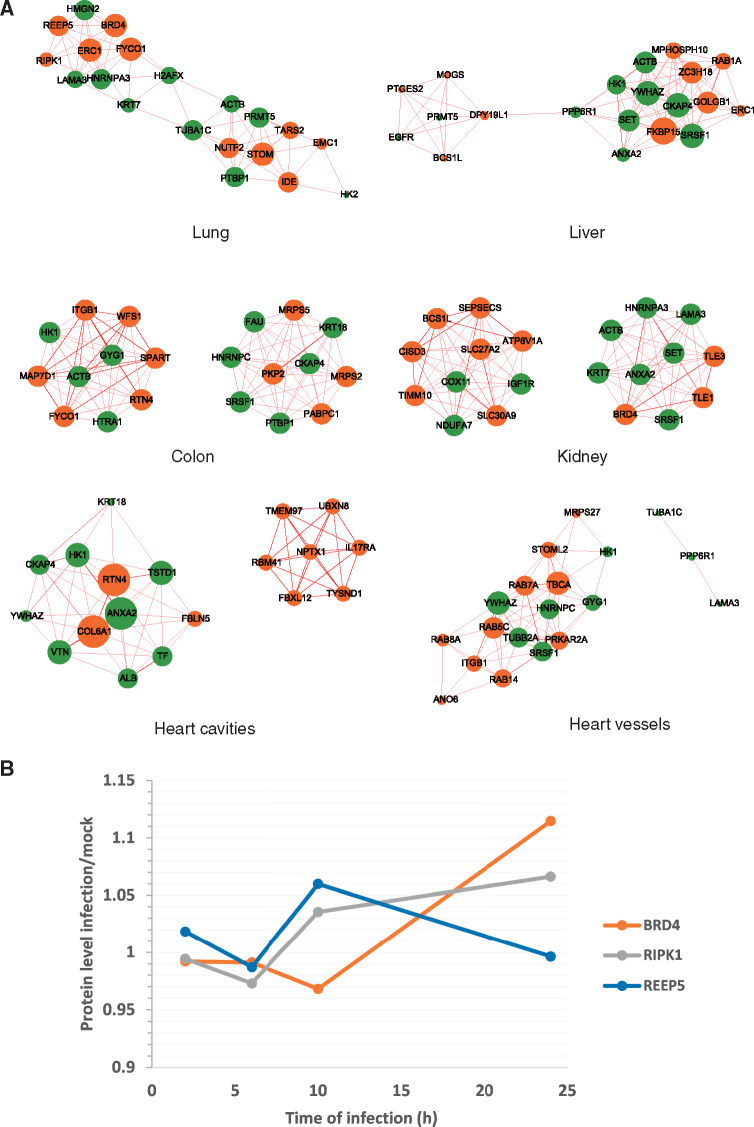
Next, we extracted 45 upregulated proteins in host cells affected by 24-h SARS-CoV-2 infection from a recently reported dataset (Bojkova et al., 2020). By analyzing the proteomic data of different tissues, 30–42 differential proteins were matched in various tissues. After calculating the correlation between the proteins, the edge of correlation networks with a P-value <0.01 was taken as a reliable edge. Ranging from 54 to 433, the number of edges of networks varies greatly. Both of the two networks of heart are small, in which the smallest is cavity network with only 54 edges. In comparison, the largest is the liver network that contains 433 edges (Supplementary Figure S1). We extracted the five nodes with the highest degree from each network as hubs and only took the edges related to these hubs to construct the core sub-network. Consistent with the overall networks, the heart core sub-networks have fewer edges, while the kidney and liver core sub-networks have more edges. It is worth noting that multiple hubs appear multiple times in different tissues, such as ANXA2, GYG1, YWHAZ, KRT7, ACTB, etc. These proteins may play an important role in the process of viral infection and pathogenicity (Supplementary Figure S2).
We combined the top 10 proteins with the highest degree in the virus-interaction network (Supplementary Table S2) and the top 10 proteins with the highest degree in the virus-perturbation network as the core proteins and only kept the edges that connected each two core proteins. Since the dominant edge in some networks is the positively correlated edge, we filtered out the negatively correlated edge. All the edges can be seen in Supplementary Figure S3. There are also interactions between the nodes obtained from the two datasets. In addition to the nodes mentioned before, proteins such as RTN4 and PTBP1 also appear in the networks of different tissues more than once and are worthy of further study (Figure 4A). In addition, by analyzing the same set of data from Bojkova et al. (2020), we identified changes of hubs essential for lung at different time points after SARS-CoV-2 infection. After 24 h of infection, the overall abundances of BRD4 and RIPK1 slightly increased. The expression level of REEP5 slightly increased at 10 h after infection, and then began to decrease after 10 h (Figure 4B).
Figure 4.
The hubs–hubs network and the changes of key hubs over time upon SARS-CoV-2 infection. (A) Core positive correlation networks consisting of hub proteins affected by SARS-CoV-2 (green) and hub proteins interacted with SARS-CoV-2 (orange) in six tissues. The coloring principle of edges is the same as described in Figure 2. (B) The change curves of selected hub proteins in lung, according to the proteome data after SARS-CoV-2 infection in human Caco-2 cells. The X-axis indicates different time points, and the Y-axis indicates the fold change of hub proteins at each time point (infection/control).
Key hubs regulated by virus
By constructing different networks, we have found several key proteins. Among them, both BRD4 and RIPK1 can interact with SARS-CoV-2 proteins. They are widely found in various organs and tract our attention as drug targets. BRD4 plays an important role in gene expression. It can bind to acetylated lysine residues of histones, recruit positive transcription elongation factor b (P-TEFb) to activate RNA polymerase II (RNA Pol II), and start gene transcription (Figure 5A). RIPK1 is a double-faced molecule, and its characteristics have been well characterized in the tumor necrosis factor (TNF) signaling cascade. On one hand, it can promote the activation of the nuclear factor kappa B (NF-κB) pathway and regulate the expression of pro-inflammatory and pro-survival genes. On the other hand, it can paradoxically participate in Fas-associated protein with death domain (FADD)–caspase 8-mediated apoptosis and RIPK3–mixed lineage kinase domain-like (MLKL)-mediated necroptosis (Figure 5B). RIPK1 is widely expressed in lung and other tissues. It is worthwhile to probe the role of RIPK1 in lung inflammation and injury caused by SARS-CoV-2. In addition, REEP5 as a member of REEPs has also attracted our attention, because REEPs are related to olfactory signaling pathway (Figure 5C).
Figure 5.
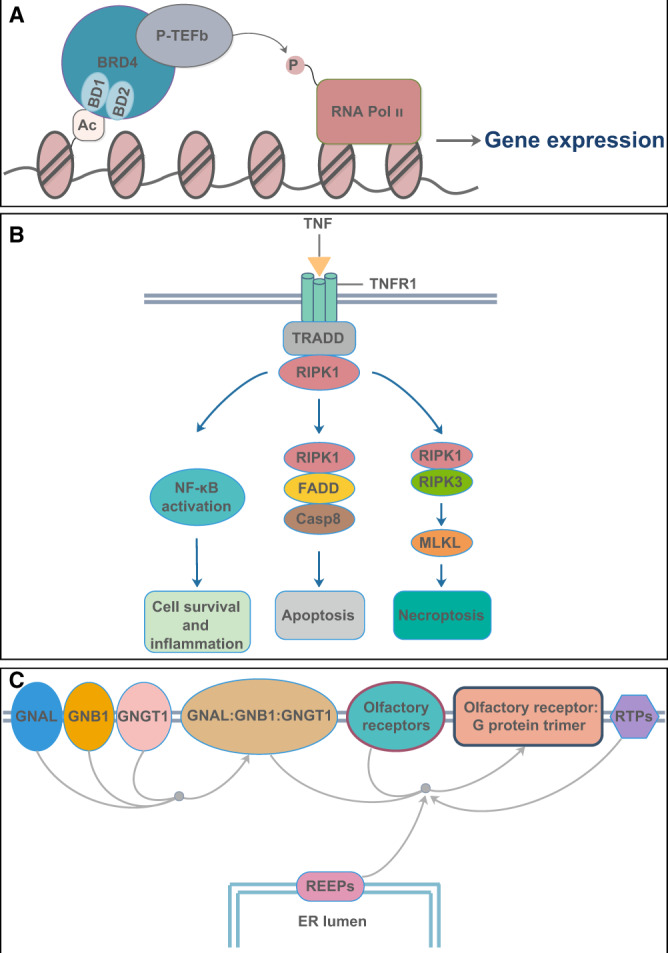
Brief functional diagram of three key proteins. (A) The functional diagram of BRD4. (B) RIPK1 participates in NF-κB activation, FADD–caspase 8-mediated apoptosis, and RIPK3–MLKL-mediated necroptosis pathway. Casp8, caspase 8; TNFR1, tumor necrosis factor receptor 1; TRADD, tumor necrosis factor receptor-associated protein with death domain. (C) The olfactory signal pathway involved in REEPs. ER, endoplasmic reticulum; GNAL, guanine nucleotide-binding protein Golf subunit alpha; GNB1, guanine nucleotide-binding protein GI/GS/GT subunit beta-1; GNGT1, guanine nucleotide-binding protein GT subunit gamma-T1; RTPs, receptor transporting proteins.
Discussion
In the proteome data of Caco-2 infected by SARS-CoV-2 (Bojkova et al., 2020), we screened the top 10 hubs based on the interaction. Among them, HNRNPC, SRSF1, and HNRNPA3 are related to mRNA processing (Huang et al., 1994; Kohtz et al., 1994; Sébillon et al., 1995; Ma et al., 2002; Kim et al., 2003; Shetty, 2005). In addition, HK2 plays a role in the initial step of glycolysis (Nawaz et al., 2018) and plays a key role in maintaining the integrity of the outer mitochondrial membrane (Chiara et al., 2008). The expression changes of these proteins under the influence of SARS-CoV-2 further indicate the extensive effects on cell translation, splicing, and metabolism after the virus invades the host cell. However, we found that the perturbation of SARS-CoV-2 to host cell is much smaller than SARS-CoV as we reported (Jiang et al., 2005). The mild disruption to host cell but lasting replication with SARS-CoV-2 itself would make the treatment more complicated and uncertain.
In the SARS-CoV-2 PPI map data (Gordon et al., 2020), we screened the hubs in different tissues based on the interactions. Some of these proteins can be regulated by drugs, including TMEM97, ATP6V1A, FKBP15, PTGES2, RIPK1, and BRD4 (Supplementary Table S2). Among them, TMEM97, FKBP15, PTGES2, and RIPK1 are all related to protein binding. TMEM97 is a conserved integral membrane protein that plays a role in controlling cellular cholesterol levels. In addition, TMEM97 corresponds to the sigma-2 receptor, which is thought to play an important role in regulating cell survival, morphology, and differentiation (Bartz et al., 2009; Huang et al., 2014; Guo and Zhen, 2015). Chloroquine could target this process; however, TMEM97 is the hub only in heart cavities, and sigma receptors are not hubs in any tissue. Thus, the efficiency and safety of chloroquine need more clinical investigations. FKBP15 is involved in the transport of early endosomes at the level of transition between microfilament-based and microtubule-based movements (Viklund et al., 2009). PTGES2 is the hub in liver, which is a membrane-associated protein that catalyzes the conversion of PGH2 primarily to PGE2 (Watanabe et al., 2003, 2008). ATP6V1A is the hub in kidney, which is catalytic subunit of the peripheral V1 complex of vacuolar ATPase (V-ATPase) responsible for acidifying a variety of intracellular compartments in eukaryotic cells. In aerobic conditions, V-ATPase is involved in intracellular iron homeostasis, thus triggering the activity of Fe2+ prolyl hydroxylase enzymes and leading to HIF1α hydroxylation and subsequent proteasomal degradation (Miles et al., 2017).
BRD4 is the hub of kidney and lung. BRD4 is an important functional protein in the bromodomain and extra-terminal domain (BET) protein family that includes BRD2, BRD3, BRD4, and BRDT. BRD4 contains two conserved N-terminal bromodomains, an extra-terminal domain, and a carboxyterminal domain (Shi and Vakoc, 2014; Wang and Filippakopoulos, 2015). Both BRD4 and other members of the BET family can bind to histones and non-histone acetylated lysine residues to regulate genomic activity (Hajmirza et al., 2018). In terms of transcriptional regulation, two bromine domain modules of BRD4 are involved in the recognition of acetylated lysine. The binding of BRD4 to acetylated histones recruits the P-TEFb complex to chromatin. BRD4 also mediates the formation of an active form of P-TEFb, which in turn phosphorylates and activates RNA Pol II to initiate gene transcription (Sahai et al., 2016). The combination of SARS-CoV-2 transmembrane protein E with bromine-containing proteins BRD2 and BRD4 prompted people to speculate that protein E can mimic histone to disrupt its interaction with BRD2 and induce changes in host protein expression, which has a beneficial effect on virus proliferation (Gordon et al., 2020). In our data, compared to BRD2, BRD4 has more general expression and protein interactions in all tissues. As a hub, BRD4 is affected by SARS-CoV-2, which may lead to more cascade reactions in host cells to adversely affect the host.
RIPK1, as a member of the RIP family, is the hub in lung. RIPK1 participates in many important signaling pathways (Holler et al., 2000; Weinlich and Green, 2014; Kondylis, 2017; Wegner et al., 2017) and is also involved with bacterial and viral infection (Cho et al., 2009; Wang et al., 2014; Dondelinger et al., 2019). Infectious viruses that spread through the respiratory tract can trigger apoptosis or necrosis, causing the death of epithelial cells in the respiratory tract and lung, causing tissue damage, and causing serious damage to the host (Rodrigue-Gervais et al., 2014). RIPK1 is a double-faced molecule, with function to inhibit cell death and promote inflammation whereas also related to enhancing necroptosis and apoptosis (Holler et al., 2000; Weinlich and Green, 2014; Kondylis, 2017; Wegner et al., 2017). The roles of RIPK1 would be dependent on the circumstances and stages in viral infections. RIPK1 consists of an N-terminal serine/threonine kinase domain, an intermediate domain, and a C-terminal death domain. The kinase activity encoded by the N-terminal kinase domain is essential for necroptosis and RIPK1-dependent apoptosis induced by TNFα (Degterev et al., 2008; Dondelinger et al., 2015; Geng et al., 2017). As an interacted protein with virus and a hub in host−cell network, RIPK1 might have an important correlation with inflammation or tissues damage caused by SARS-CoV-2 infection.
REEP5 is the hub only in lung. REEPs are members of the DP1 family (Yao et al., 2018). Six REEPs have been found in humans and other mammals, including REEP1–REEP6 (Park et al., 2010). Among them, REEP1 is thought to promote cell surface expression of odorant receptors in 293T cells (Saito et al., 2004). Olfactory receptors belong to the G protein-coupled receptor (GPCR) family and are expressed in various human tissues including skin (Tsai et al., 2017). Accessory proteins play an important role in the functional expression of GPCRs on the cell surface (Cooray et al., 2009; Roux and Cottrell, 2014). Among SARS-CoV-2-positive hospitalized patients, 33.9% of patients developed taste or/and olfactory disorders (Giacomelli et al., 2020). REEP5 was found in human olfactory cleft mucus as a secreted protein (Yoshikawa et al., 2018). REEP5 has also been shown to regulate cardiac function, as well as the growth and metastasis of lung cancer cells (Park et al., 2016; Yao et al., 2018). Whether SARS-CoV-2 infection-caused adverse effect on patients’ olfactory function is related to REEP5 or REEP family will be an interesting research direction.
In addition to the previously demonstrated proteins that can be used as drug targets, some proteins that interact with SARS-CoV-2 also deserve attention. PABPC1 is a hub in colon, which can bind to mRNA and regulate mRNA metabolic pathways. It is also related to translation and maintenance of mRNA stability (Grosset et al., 2000; Patel and Bag, 2006; Schäffler et al., 2010; Lim et al., 2014). RTN4 is a hub in colon and heart cavities. RTN family proteins influence nuclear envelope expansion, nuclear pore complex formation, and proper localization of inner nuclear membrane proteins (Christodoulou et al., 2016). In addition, knockdown of RTN4-A in cardiomyocytes markedly attenuated hypoxia/reoxygenation-induced apoptosis (Sarkey et al., 2011). RAB1A is a hub in liver. It mainly regulates the transport of vesicle proteins from the endoplasmic reticulum to the Golgi compartment and to the cell surface and plays a role in the secretion of IL-8 and growth hormone. It is also related to the assembly of autophagosomes and the cellular defense reactions against pathogenic bacteria (Wang et al., 2010; Zhuang et al., 2010; Mukhopadhyay et al., 2011; Dong et al., 2012). SLC30A9 is a hub found in kidney and acts as a zinc transporter involved in intracellular zinc homeostasis (Perez et al., 2017). The roles of these host proteins interacting with SARS-CoV-2 in the process of transcription, secretion, and apoptosis suggest that these proteins may be related to the virus propagation process.
By building the networks associated with SARS-CoV-2, we have found some key proteins in the network that frequently appear in different tissues and interact extensively with other proteins. Some of them are common hubs in multiple tissues (such as BRD4 and RIPK1), which could be promising drug targets to rescue multi-organ injury and deal with inflammation. Some of them are tissue-unique hubs such as REEP5, possibly mediating specific phenotype. All these observations will help us to understand the infection and perturbation mechanism of SARS-CoV-2 in different tissues.
Since the amount of data in this study is still limited, there is uncertainty in this network. However, the selected data sets with large sample size could largely ensure the reliability of the analysis through horizontal comparison among organs. The proteome-wide analysis could provide tissue-specific features associated with SARS-CoV-2 infection. The perturbation of SARS-CoV-2 to host tissues is multi-targeting and complicated, and thus the treatment of COVID-19 would be a complex task.
Materials and methods
Data preprocessing
We collected proteomic data of five organs, including four tissues from the CPTAC database and two parts of the heart. Data can be downloaded from the following websites:
ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2017/11/PXD006675.
The data downloaded from CPTAC database provide relative protein abundance by sample. The ‘XXXX Log Ratio’ columns contain the relative abundance of sample XXXX and were used in the analysis, with respect to the pooled reference sample, as log ratios (base 2). For the heart data, we used z-scored protein abundances (LFQ intensities). We filtered out proteins with too much NA (≥2/3 of the samples). The sample size and protein numbers of processed data are in Supplementary Table S3. Part data used in this publication were generated by the CPTAC.
Comparison of three receptors of SARS-CoV-2
We downloaded the protein and RNA data of a variety of tissues from the HPA database (https://www.proteinatlas.org). For protein data, each bar represents the highest expression score found in a group of tissues. Protein expression scores are based on a best estimate of the ‘true’ protein expression from a knowledge-based annotation. For genes where >1 antibody has been used, a collective score is set displaying the estimated true protein expression. RNA expression summary shows the consensus RNA data based on normalized expression data from three different sources, internally generated HPA RNA-seq data, RNA-seq data from the Genotype−Tissue Expression (GTEx) project, and CAGE data from the FANTOM5 project (https://www.proteinatlas.org/about/assays+annotation#ihk).
Construction of receptor-centered PPI network
For PPI networks, we took the three receptors and TMPRSS2 to find the relationships in String database (https://string-db.org/). The correlation of score value >0.8 was conserved to build a PPI network, and then the Pearson correlation coefficient of each two proteins was calculated. P-value <0.05 was considered as significant correlation.
Construction of correlation networks of proteins interacted with SARS-CoV-2 and the core sub-networks
We took 332 proteins to calculate the Pearson correlation coefficient of each two proteins, and the correlation of P-value <0.01 was conserved. Correlation coefficient >0.8 was considered as strong correlation. Then we ranked the proteins by degree and chose the top 10 to check whether they were drug targets.
In the 332 proteins interacted with SARS-CoV-2, 188 of them could interact with the major virus components (e.g. SARS-CoV2 E, SARS-CoV2 M, SARS-CoV2 N, SARS-CoV2 nsp12, SARS-CoV2 nsp13, SARS-CoV2 nsp4, SARS-CoV2 nsp6, SARS-CoV2 nsp3, SARS-CoV2 nsp5, SARS-CoV2 nsp7, SARS-CoV2 nsp8, and SARS-CoV2 orf3a). We then calculated the Pearson correlation coefficient of each two proteins among the 188 candidates, and the correlation of P-value <0.01 was conserved. Correlation coefficient >0.8 was considered as strong correlation. Then we ranked the proteins by degree and chose the top 5.
The potential drug targets in top 10 of the 332 proteins and top 5 of the 188 proteins were used as hubs to construct the core sub-networks. Only the hubs and the nodes connected with these hubs are shown in the sub-networks. Due to the large differences in the size of the sub-networks, only 120 edges with the highest correlation coefficient are shown.
Construction of correlation networks of proteins changed in cells infected by SARS-CoV-2 and the core sub-networks
We took 45 upregulated proteins to calculate the Pearson correlation coefficient of each two proteins, and the correlation of P-value <0.01 was conserved. Correlation coefficient >0.8 was considered as strong correlation. In Supplementary Figure S1, all the edges are shown, while in Supplementary Figure S2, only the edges related to the top 5 nodes with the largest degree are shown.
Construction of virus-perturbation networks
We used the top 10 proteins interacted with the virus ranked by the degree in all 332 proteins and the top 10 proteins changed in the cells infected by the virus as two groups of hubs and constructed the correlation networks only composed by these 20 nodes. The correlation of P-value <0.01 was conserved. Correlation coefficient >0.8 was considered as strong correlation.
Network plots
Cytoscape (Shannon et al., 2003) was used to draw the network plots.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by grants from the Ministry of Science and Technology (2017YFA0505500) and the Strategic CAS Project (XDA12010000 and XDB38000000).
Conflict of interest: none declared.
Supplementary Material
References
- Bartz F., Kern L., Erz D., et al. (2009). Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 10, 63–75. [DOI] [PubMed] [Google Scholar]
- Bojkova D. Klann K. KochB., et al ( . 2020). Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583, 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Cui X., Zhu X., et al. (2020). A hint on the COVID-19 risk: population disparities in gene expression of three receptors of SARS-CoV. Preprints, doi: 10.20944/preprints202002.0408.v1.
- Chiara F., Castellaro D., Marin O., et al. (2008). Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels. PLoS One 3, e1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.S., Challa S., Moquin D., et al. (2009). Phosphorylation-driven assembly of the RIP1–RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou A., Santarella-Mellwig R., Santama N., et al. (2016). Transmembrane protein TMEM170A is a newly discovered regulator of ER and nuclear envelope morphogenesis in human cells. J. Cell Sci. 129, 1552–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.J., Dhanasekaran S.M., Petralia F., et al. (2019). Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell 179, 964–983.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooray S.N., Chan L., Webb T.R., et al. (2009). Accessory proteins are vital for the functional expression of certain G protein-coupled receptors. Mol. Cell. Endocrinol. 300, 17–24. [DOI] [PubMed] [Google Scholar]
- Degterev A., Hitomi J., Germscheid M., et al. (2008). Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S., Dressen M., Geyer P.E., et al. (2017). Region and cell-type resolved quantitative proteomic map of the human heart. Nat. Commun. 8, 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y., Delanghe T., Priem D., et al. (2019). Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 10, 1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondelinger Y., Jouan-Lanhouet S., Divert T., et al. (2015). NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell 60, 63–76. [DOI] [PubMed] [Google Scholar]
- Dong N., Zhu Y., Lu Q., et al. (2012). Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Gao Q., Zhu H., Dong L., et al. (2019). Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell 179, 561–577.e22. [DOI] [PubMed] [Google Scholar]
- Geng J., Ito Y., Shi L., et al. (2017). Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli A., Pezzati L., Conti F., et al. (2020). Self-reported olfactory and taste disorders in SARS-CoV-2 patients: a cross-sectional study. Clin. Infect. Dis. 2020, ciaa330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosset C., Chen C.-Y.A., Xu N., et al. (2000). A mechanism for translationally coupled mRNA turnover: interaction between the Poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103, 29–40. [DOI] [PubMed] [Google Scholar]
- Guo L., Zhen X.C. (2015). Sigma-2 receptor ligands: neurobiological effects. Curr. Med. Chem. 22, 989–1003. [DOI] [PubMed] [Google Scholar]
- Hajmirza A., Emadali A., Gauthier A., et al. (2018). BET family protein BRD4: an emerging actor in NFκB signaling in inflammation and cancer. Biomedicines 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N., Zaru R., Micheau O., et al. (2000). Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 1, 489–495. [DOI] [PubMed] [Google Scholar]
- Huang M., Rech J.E., Northington S.J., et al. (1994). The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 14, 518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-S., Lu H.-L., Zhang L.-J., et al. (2014). Sigma-2 receptor ligands and their perspectives in cancer diagnosis and therapy. Med. Res. Rev. 34, 532–566. [DOI] [PubMed] [Google Scholar]
- Jiang X.S., Tang L.Y., Dai J., et al. (2005). Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics 4, 902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Paek K.Y., Choi K., et al. (2003). Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23, 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J.D., Jamison S.F., Will C.L., et al. (1994). Protein−protein interactions and 5'-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Kondylis V. (2017). RIPK1 and allies in the battle against hepatocyte apoptosis and liver cancer. Transl. Cancer Res. 6, S569–S577. [Google Scholar]
- Lim J., Ha M., Chang H., . et al. (2014). Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A.S., Moran-Jones K., Shan J., et al. (2002). Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 277, 18010–18020. [DOI] [PubMed] [Google Scholar]
- Mertins P., Tang L.C., Krug K., et al. (2018). Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 13, 1632–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles A.L., Burr S.P., Grice G.L., et al. (2017). The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1α prolyl hydroxylation by regulating cellular iron levels. eLife 6, e22693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Nieves E., Che F.-Y., et al. (2011). Proteomic analysis of endocytic vesicles: Rab1a regulates motility of early endocytic vesicles. J. Cell Sci. 124, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M.H., Ferreira J.C., Nedyalkova L., et al. (2018). The catalytic inactivation of the N-half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation. Biosci. Rep. 38, BSR20171666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.R., You D.-J., Park S., et al. (2016). The accessory proteins REEP5 and REEP6 refine CXCR1-mediated cellular responses and lung cancer progression. Sci. Rep. 6, 39041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Zhu P.-P., Parker R.L., et al. (2010). Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 120, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G.P., Bag J. (2006). IMP1 interacts with poly(A)-binding protein (PABP) and the autoregulatory translational control element of PABP-mRNA through the KH III−IV domain. FEBS J. 273, 5678–5690. [DOI] [PubMed] [Google Scholar]
- Perez Y., Shorer Z., Liani-Leibson K., et al. (2017). SLC30A9 mutation affecting intracellular zinc homeostasis causes a novel cerebro-renal syndrome. Brain 140, 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue-Gervais I.G., Labbé K., Dagenais M., et al. (2014). Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe 15, 23–35. [DOI] [PubMed] [Google Scholar]
- Roux B.T., Cottrell G.S. (2014). G protein-coupled receptors: what a difference a ‘partner’ makes. Int. J. Mol. Sci. 15, 1112–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, V., Redig, A.J., Collier, K.A., al (2016). Targeting BET bromodomain proteins in solid tumors. Oncotarget 7, 53997–54009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Kubota M., Roberts R.W., et al. (2004). RTP family members induce functional expression of mammalian odorant receptors. Cell 119, 679–691. [DOI] [PubMed] [Google Scholar]
- Sarkey J.P., Chu M., McShane M., et al. (2011). Nogo-A knockdown inhibits hypoxia/reoxygenation-induced activation of mitochondrial-dependent apoptosis in cardiomyocytes. J. Mol. Cell. Cardiol. 50, 1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffler K., Schulz K., Hirmer A., et al. (2010). A stimulatory role for the La-related protein 4B in translation. RNA 16, 1488–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sébillon P., Beldjord C., Kaplan J.C., et al. ., (1995). A T to G mutation in the polypyrimidine tract of the second intron of the human β-globin gene reduces in vitro splicing efficiency: evidence for an increased hnRNP C interaction. Nucleic Acids Res. 23, 3419–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty S. (2005). Regulation of urokinase receptor mRNA stability by hnRNP C in lung epithelial cells. Mol. Cell. Biochem. 272, 107–118. [DOI] [PubMed] [Google Scholar]
- Shi J., Vakoc C.R. (2014). The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai T., Veitinger S., Peek I., et al. (2017). Two olfactory receptors—OR2A4/7 and OR51B5—differentially affect epidermal proliferation and differentiation. Exp. Dermatol. 26, 58–65. [DOI] [PubMed] [Google Scholar]
- Vasaikar S., Huang C., Wang X., et al. (2019). Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 177, 1035–1049.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund I.-M., Aspenström P., Meas-Yedid V., et al. (2009). WAFL, a new protein involved in regulation of early endocytic transport at the intersection of actin and microtubule dynamics. Exp. Cell Res. 315, 1040–1052. [DOI] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., et al. (2020. a). A novel coronavirus outbreak of global health concern. Lancet 395, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yoo Y., Fan H., et al. (2010). Regulation of Integrin β1 recycling to lipid rafts by Rab1a to promote cell migration. J. Biol. Chem. 285, 29398–29405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-Y., Filippakopoulos P. (2015). Beating the odds: BETs in disease. Trends Biochem. Sci. 40, 468–479. [DOI] [PubMed] [Google Scholar]
- Wang T., Du Z., Zhu F., et al. (2020. b). Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet 395, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Jiang W., Yan Y., et al. (2014). RNA viruses promote activation of the NLRP3 inflammasome through a RIP1–RIP3–DRP1 signaling pathway. Nat. Immunol. 15, 1126–1133. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ito S., Yamamoto S. (2008). Studies on membrane-associated prostaglandin E synthase-2 with reference to production of 12L-hydroxy-5,8,10-heptadecatrienoic acid (HHT). Biochem. Biophys. Res. Commun. 367, 782–786. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ohkubo H., Niwa H., et al. (2003). Essential 110Cys in active site of membrane-associated prostaglandin E synthase-2. Biochem. Biophys. Res. Commun. 306, 577–581. [DOI] [PubMed] [Google Scholar]
- Wegner K.W., Saleh D., Degterev A. (2017). Complex pathologic roles of RIPK1 and RIPK3: moving beyond necroptosis. Trends Pharmacol. Sci. 38, 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich R., Green D.R. (2014). The two faces of receptor interacting protein kinase-1. Mol. Cell 56, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2020. a). Coronavirus disease 2019 (COVID-19) situation report–41. World Health Organization. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200301-sitrep-41-covid-19.pdf?sfvrsn=6768306d_2
- World Health Organization. (2020. b). Coronavirus disease 2019 (COVID-19) situation report–51. World Health Organization. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10
- Yao L., Xie D., Geng L., et al. (2018). REEP5 (receptor accessory protein 5) acts as a sarcoplasmic reticulum membrane sculptor to modulate cardiac function. J. Am. Heart Assoc. 7, e007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K., Wang H., Jaen C., et al. (2018). The human olfactory cleft mucus proteome and its age-related changes. Sci. Rep. 8, 17170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Adipietro K.A., Datta S., et al. (2010). Rab1 small GTP-binding protein regulates cell surface trafficking of the human calcium-sensing receptor. Endocrinology 151, 5114–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., et al. (2020). Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 14, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.