Abstract
The effective treatment of carbapenemase-producing Klebsiella pneumoniae infection has been limited and required novel potential agents. Due to the novel drug development crisis, using old antimicrobial agents and combination therapy have been highlighted. This study focused on fosfomycin which inhibits cell wall synthesis and has potential activity on Enterobacteriaceae. We evaluated fosfomycin activity against carbapenemase-producing K. pneumoniae and characterized fosfomycin resistance mechanisms. Fosfomycin revealed effective activity against only 31.8% of carbapenemase-producing K. pneumoniae isolates. The major resistance mechanism was FosA3 production. The co-occurrence of FosA3 overexpression with the mutation of glpT (or loss of glpT) and/or uhpT was mediated high-level resistance (MIC>256 mg/L) to fosfomycin. Moreover, fosA3 silenced in sixteen fosfomycin-susceptible isolates and the plasmid carrying fosA3 of these isolates increased 32- to 64-fold of fosfomycin MICs in Escherichia coli DH5α transformants. The in vitro activity of fosfomycin combination with amikacin by checkerboard assay showed synergism and no interaction in six (16.2%) and sixteen isolates (43.3%), respectively. No antagonism of fosfomycin and amikacin was observed. Notably, the silence of aac (6)’-Ib and aphA6 was observed in amikacin-susceptible isolates. Our study suggests that the combination of fosfomycin and amikacin may be insufficient for the treatment of carbapenemase-producing K. pneumoniae isolates.
Introduction
Klebsiella pneumoniae, a Gram-negative bacilli pathogen causes hospital-acquired infections including lower respiratory tract, urinary tract, and bloodstream infections. Since the emergence of carbapenem-resistant K. pneumoniae in clinical settings globally, effective treatment options have been limited [1, 2]. Colistin (polymyxin E) is a last potential agent against carbapenem-resistant K. pneumoniae, but it should be well-inform due to the rise of resistance rate and toxicity [3]. Therefore, infections caused by carbapenem-resistant K. pneumoniae urgently require novel potential agents for treatment. Nevertheless, the number of novel antibiotics introduced for treatment strongly decline when compared to the emergence of antibiotic resistance. During this crisis, various old antibiotics including fosfomycin which were previously effective, have been reassessed and used [4].
Fosfomycin inhibits the initial step of bacterial cell wall synthesis by covalent binding to UDP-N-acetylglucosamine-3-enolpyruvyltransferase (MurA). Fosfomycin is considerably effective against not only Escherichia coli, but also other Enterobacteriaceae including K. pneumoniae [4]. However, the range of fosfomycin susceptibility against carbapenem-resistant K. pneumoniae (39.2% to 66.2%) has been broader than that against carbapenem-resistant E. coli (83.3% to 100%) [4, 5]. K. pneumoniae is resistant to fosfomycin by the production of various fosfomycin-modifying enzymes (such as FosA, FosA3, and FosA5) to catalyze fosfomycin. Several fosA genes co-carry with carbapenemase genes (blaNDM, blaOXA-48, blaIMP, and blaKPC) in plasmids resulting in resistance to both fosfomycin and carbapenems [4]. The target alteration by the mutation of MurA particularly at the active site (Cys115 to Asp) strongly confers fosfomycin resistance in E. coli [6]. Moreover, fosfomycin resistance is also mediated by reduced fosfomycin uptake by alteration of glycerol-3-phosphate (G3P) or glucose-6-phosphate (G6P) transporters (GlpT and UhpT transporters, respectively) [4, 7].
Although fosfomycin activity is variable against carbapenem-resistant K. pneumoniae, synergy has been observed when combined with carbapenems, colistin, or aminoglycosides [4]. However, the synergism of fosfomycin and aminoglycosides is controversial among different carbapenemase-producing K. pneumoniae. Fosfomycin and amikacin combination exhibits less synergistic activity against KPC producers than OXA-48 and/or NDM producers [8, 9]. These studies demonstrated that the activity of fosfomycin either alone or combination with aminoglycosides is unpredictable against carbapenem-resistant K. pneumoniae. In this study, we aimed to determine the activity of fosfomycin alone and in combination with amikacin together with characterization of fosfomycin and amikacin resistance mechanisms among carbapenem-resistant K. pneumoniae clinical isolates.
Materials and methods
Bacterial isolates
A total of 66 carbapenemase-producing K. pneumoniae which isolated from the patients was obtained from the routine laboratory’s stocks at the King Chulalongkorn Memorial Hospital, Bangkok, Thailand during 2017–2018. This study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University (IRB 221/62). Neither human nor animal was involved in this study. The need for consent was waived by the ethics committee.
Antimicrobial susceptibility testing
Susceptibility testing to imipenem (Apollo Scientific, Manchester, UK) meropenem (Sigma-Aldrich, Steinheim, Germany) and amikacin (Sigma-Aldrich, Steinheim, Germany) was determined by agar dilution method. Fosfomycin (Wako Pure Chemical Industries, Osaka, Japan) susceptibility testing was performed on Mueller-Hinton agar (MHA) (Becton Dickinson BBL, MD, USA) supplemented with 25 mg/L glucose-6-phosphate (G6P) (Sigma-Aldrich, Steinheim, Germany). E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as reference control strains. Susceptibility of imipenem, meropenem, and fosfomycin was interpreted according to the breakpoints and guidance of the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2020).
Detection of antimicrobial resistance genes
The metallo-carbapenemase genes (blaIMP-like and blaVIM-like) were detected by multiplex PCR as described previously [10]. Other carbapenemase genes including blaNDM-like, blaOXA-48-like, and blaKPC-like were detected by another multiplex PCR as described previously [11]. The fosfomycin-modifying enzyme genes (fosA, fosA3, fosA5, fosB, fosC2, and fosX) were detected by PCR using primers listed in S1 Table. The 16S rRNA methylase genes (armA, rmtB, and rmtC) and the aminoglycoside-modifying enzyme (AME) genes (aac (6)’-Ib and aphA6) were screened by PCR using primers listed in S1 Table.
Expression of fosfomycin-modifying enzyme genes
Total RNA of K. pneumoniae was extracted by using Monarch total RNA miniprep kit (NEB, USA) and converted to cDNA by using SuperScript® III reverse transcriptase (Thermo Fisher Scientific, USA). The numbers of fosfomycin-modifying enzyme transcripts were determined by qRT-PCR using Luna® Universal qPCR master mix (NEB, USA) and QuantStudio5 (Thermo Fisher Scientific, USA). Relative expression levels of fosfomycin-modifying enzyme genes were calculated and normalized with 16S rRNA. The qRT-PCR experiments were performed in at least three independent experiments.
Transformation of the fosA3-carrying plasmid into E. coli DH5α
K. pneumoniae plasmids were extracted by using HiYield Plasmid Mini Kit (RBC, Taipei, Taiwan) and transformed into E. coli DH5α by using the heat shock method. The E. coli DH5α transformants were selected on MHA supplemented with fosfomycin and G6P. The presence of fosA3 in E. coli transformants was confirmed by PCR. Fosfomycin susceptibility of fosA3-carrying E. coli transformants was determined by agar dilution.
Sequence analysis of murA, glpT, and uhpT gene
The entire sequences of murA, glpT, and uhpT were amplified by PCR as described previously and sequenced by using the BigDye Terminator V3.1 cycle sequencing kit from the 1st Base DNA sequencing service, Malaysia. The amino acid substitutions of MurA, GlpT, and UhpT in K. pneumoniae isolates were compared with those of wild type of K. pneumoniae isolate K68 from a previous study by Lu et al [12] and deposited in GenBank accession number KT334183, KT334186, and KT334184, respectively.
The ability of K. pneumoniae to grow on different carbohydrates
To investigate the ability of K. pneumoniae to grow in the presence of different carbohydrates, G6P, or glycerol-3-phosphate (G3P) (Sigma-Aldrich, Steinheim, Germany). Briefly, M9 minimal medium liquid supplemented with 0.2% (w/v) G6P or G3P was inoculated with K. pneumoniae suspension and incubated at 35 °C with shaking for 48 hr. The growth of K. pneumoniae was determined by measurement of the OD600nm of the cell suspension compared with M9 supplemented with G6P or G3P without bacterial inoculation.
Checkerboard assay
In vitro activity of fosfomycin in combination with amikacin against carbapenemase-producing K. pneumoniae was performed by checkerboard assay. Briefly, each well of the row in 96-well microtiter plates was contained cation-adjust Mueller-Hinton broth (CAMHB) (Becton Dickinson BBL, MD, USA) with the two-fold dilution of fosfomycin (supplemented with 25 mg/L of G6P). Besides, each well of the column in the plates was contained CAMHB with the two-fold dilution of amikacin. The checkerboard plate was inoculated with K. pneumoniae and incubated at 35 °C for 18–24 hr. The fractional inhibitory concentration index (FICI) was calculated and interpreted as synergism (FICI≤0.5), no interaction (FICI>0.5–4), and antagonism (FICI>4).
Results
Fosfomycin susceptibility of carbapenemase-producing K. pneumoniae
All K. pneumoniae exhibited resistance to imipenem and meropenem except isolate KP35 carrying blaIMP-like (Table 1). The majority of isolates co-harbored blaNDM-like and blaOXA-48-like (n = 38, 57.5%) followed by carrying blaNDM-like (n = 17, 25.8%), blaOXA-48-like (n = 9, 13.7%) and blaIMP-like (n = 2, 3.0%), respectively. Neither blaKPC-like nor blaVIM-like was found in this study. Resistance to fosfomycin was 68.2% (n = 45) of isolates. Among these, 27 isolates (60.0%) co-carried blaNDM-like and blaOXA-48-like, whereas, 9 (20.0%), 8 (17.8%), and one isolate (2.2%) carried blaNDM-like, blaOXA-48-like and blaIMP-like, respectively.
Table 1. Antimicrobial susceptibility of 66 carbapenemase-producing K. pneumoniae isolates.
| Strain | MIC (mg/L) | Carbapenemase gene | Fos gene | Strain | MIC (mg/L) | Carbapenemase gene | Fos gene | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | FOF | AK | IPM | MEM | FOF | AK | ||||||
| KP35 | 2 | 2 | 32 | 1 | blaIMP-like | fosA5 | KP12 | 32 | 128 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP71 | 32 | 16 | 128 | 1 | blaIMP-like | fosA5 | KP21 | 32 | 64 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP79 | 32 | 32 | 8 | 8 | blaNDM-like | fosA5 | KP30 | 64 | 128 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP23 | 16 | 128 | 16 | 16 | blaNDM-like | fosA5, fosA3 | KP39 | 64 | 128 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP97 | 16 | 16 | 16 | 2 | blaNDM-like | fosA5, fosA3 | KP81 | 64 | 128 | 32 | 1 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP53 | 16 | 32 | 32 | 8 | blaNDM-like | fosA5 | KP90 | 32 | 64 | 32 | 2 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP72 | 32 | 16 | 32 | 16 | blaNDM-like | fosA5 | KP3 | 32 | 64 | 64 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP78 | 128 | 32 | 32 | 16 | blaNDM-like | fosA5 | KP32 | 32 | 64 | 64 | 8 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP14 | 8 | 32 | 32 | 16 | blaNDM-like | fosA5, fosA3 | KP45 | 64 | 128 | 64 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP68 | 64 | 128 | 32 | 16 | blaNDM-like | fosA5, fosA3 | KP46 | 64 | 128 | 64 | 2 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP34 | 128 | 128 | 64 | 256 | blaNDM-like | fosA5 | KP49 | 64 | 128 | 64 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP48 | 32 | 64 | 64 | 16 | blaNDM-like | fosA5, fosA3 | KP52 | 64 | 256 | 64 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP54 | 32 | 64 | 64 | 1 | blaNDM-like | fosA5, fosA3 | KP60 | >256 | 256 | 64 | 4 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP61 | 64 | 128 | 64 | 16 | blaNDM-like | fosA5, fosA3 | KP73 | 32 | 128 | 64 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP64 | 64 | 128 | 64 | 16 | blaNDM-like | fosA5, fosA3 | KP95 | 64 | 128 | 64 | >256 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP87 | 128 | 128 | 64 | 16 | blaNDM-like | fosA5, fosA3 | KP36 | 128 | 256 | 128 | 128 | blaNDM-like, blaOXA-48-like | fosA5 |
| KP70 | 16 | 32 | 128 | 8 | blaNDM-like | fosA5, fosA3 | KP94 | >256 | 256 | 128 | 2 | blaNDM-like, blaOXA-48-like | fosA5 |
| KP82 | 16 | 16 | 128 | 16 | blaNDM-like | fosA5, fosA3 | KP57 | 64 | 128 | 128 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP58 | 256 | 256 | >256 | 16 | blaNDM-like | fosA5, fosA3 | KP84 | 64 | 128 | 256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP44 | 4 | 16 | 32 | 8 | blaOXA-48-like | fosA5, fosA3 | KP4 | 32 | 64 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP27 | 128 | 128 | 64 | 256 | blaOXA-48-like | fosA5 | KP6 | 32 | 64 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP29 | 4 | 4 | 128 | 8 | blaOXA-48-like | fosA5, fosA3 | KP15 | 32 | 64 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP62 | 16 | 32 | >256 | >256 | blaOXA-48-like | fosA5 | KP18 | 256 | 256 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP11 | 8 | 32 | >256 | >256 | blaOXA-48-like | fosA5 | KP19 | >256 | 256 | >256 | 32 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP26 | 8 | 32 | >256 | >256 | blaOXA-48-like | fosA5 | KP43 | 32 | 64 | >256 | 8 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP63 | 8 | 32 | >256 | >256 | blaOXA-48-like | fosA5, fosA3 | KP47 | 32 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP88 | 64 | 256 | >256 | 32 | blaOXA-48-like | fosA5, fosA3 | KP50 | 64 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP51 | 8 | 32 | >256 | >256 | blaOXA-48-like | fosA5, fosA3 | KP55 | 64 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP31 | 16 | 8 | 16 | 8 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 | KP56 | 128 | 128 | >256 | 1 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP1 | 32 | 64 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 | KP59 | 128 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP2 | 32 | 64 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 | KP89 | >256 | 256 | >256 | 32 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP5 | 32 | 64 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 | KP92 | 32 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
| KP8 | 32 | 64 | 32 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 | KP7 | 64 | 128 | >256 | 16 | blaNDM-like, blaOXA-48-like | fosA5, fosA3 |
IPM: Imipenem; MEM: Meropenem; FOF: Fosfomycin; AK: Amikacin
Prevalence and mRNA expression level of fosfomycin-modifying enzyme genes
Intrinsic fosfomycin-modifying enzyme gene, fosA5 was found in all K. pneumoniae isolates (Table 1). Among forty-five fosfomycin-resistant isolates, 37 isolates (82.2%) carried fosA3 with fosfomycin MIC range of 64–>256 mg/L. Interestingly, the silence of fosA3 was also present in 16 fosfomycin-susceptible isolates with the MIC range of 16–32 mg/L.
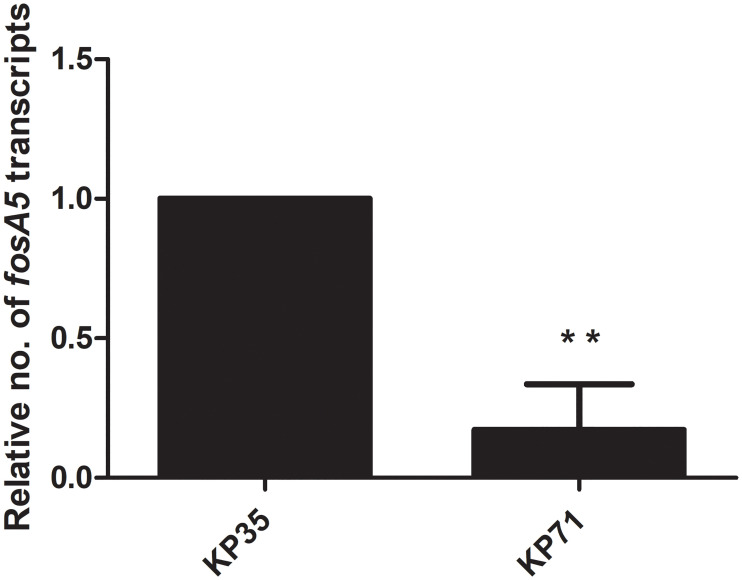
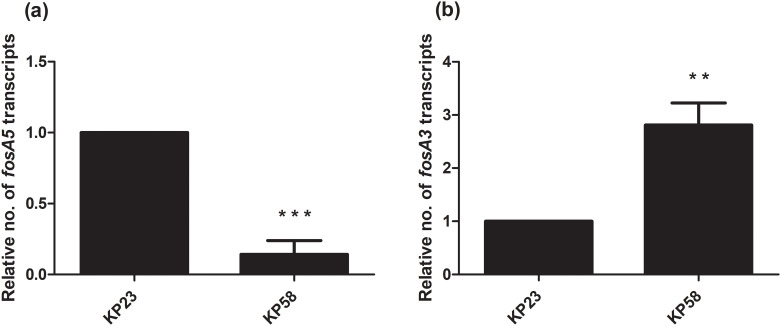
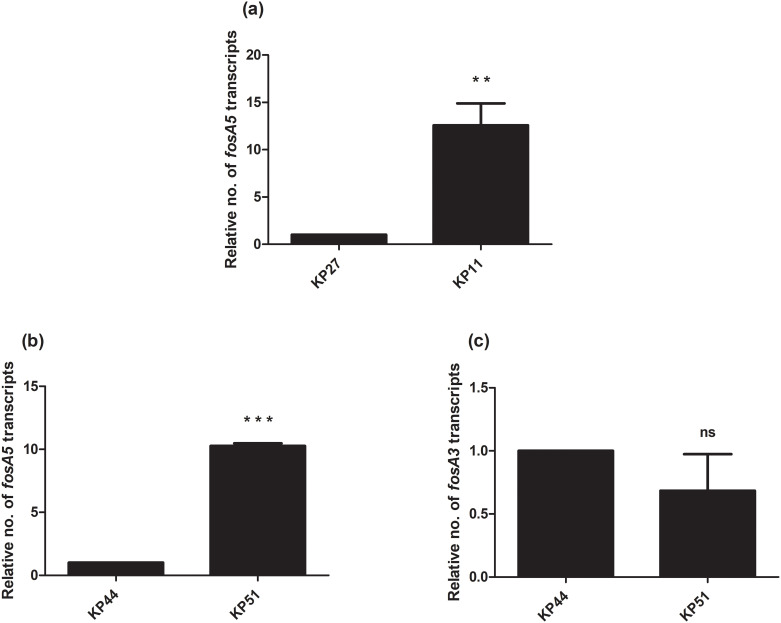
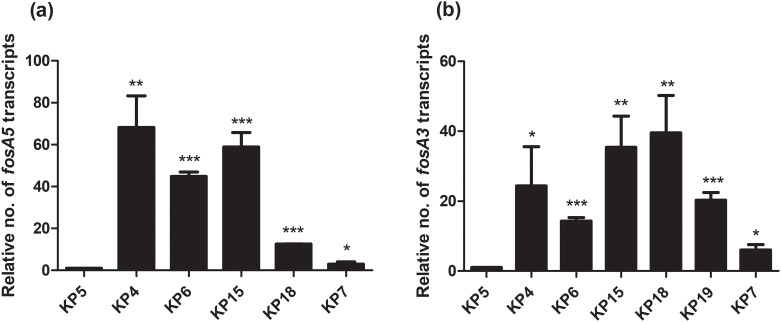
Due to the presence of fosA5 and fosA3 in K. pneumoniae with different fosfomycin MICs, we determined the relationship of expression levels of fosA5 and fosA3 and fosfomycin susceptibility among each group of carbapenemase production. In IMP-producing strains, interestingly, fosA5 expression of fosfomycin-resistant strain (KP71, MIC = 128 mg/L) was lower than that of KP35 which was susceptible to fosfomycin (Fig 1). In NDM-producing isolates, fosA3 expression of fosfomycin-resistant isolate (KP58, MIC = >256 mg/mL) was significantly higher than that of susceptible isolate (Fig 2). In the case of NDM producers, fosfomycin-resistant isolates (KP11 and KP51) showed higher fosA3 expression levels but a similar level of fosA5 expression to fosfomycin-susceptible isolates (Fig 3). In contrast, fosfomycin-resistant strains with co-producing NDM and OXA-48 revealed a strongly higher expression level of fosA5 as well as fosA3 (Fig 4).
Fig 1. Relative fosA5 expression levels of IMP-producing K. pneumoniae.
qRT-PCR assay of fosA5 expression was performed in K. pneumoniae isolate KP35 and KP71. The relative number of transcripts of fosA5 were normalized to 16S rRNA expression and calculated using the 2-ΔΔct method compared to the expression of fosfomycin-susceptible K. pneumoniae isolate KP35. p-values were calculated using unpaired t-test (*, p-value <0.05; **, p-value <0.01; ***, p-value <0.001 and ns, non-significant).
Fig 2. Relative fosA5 and fosA3 expression levels of NDM-producing K. pneumoniae.
qRT-PCR assay of fosA5 (a) and fosA3 (b) expression was performed in K. pneumoniae isolate KP23 and KP58. The relative number of transcripts of fosA5 and fosA3 were normalized to 16S rRNA expression and calculated using the 2-ΔΔct method compared to the expression of fosfomycin-susceptible K. pneumoniae isolate KP23. p-values were calculated using unpaired t-test (*, p-value <0.05; **, p-value <0.01; ***, p-value <0.001 and ns, non-significant).
Fig 3. Relative fosA5 and fosA3 expression levels of OXA-48-producing K. pneumoniae.
qRT-PCR assay of fosA5 (a) and fosA3 (b, c) expression was performed in K. pneumoniae isolate KP27, KP11, KP44, and KP51. The relative number of transcripts of fosA5 and fosA3 were normalized to 16S rRNA expression and calculated using the 2-ΔΔct method compared to the expression of fosfomycin-susceptible K. pneumoniae isolate KP27 or KP44. p-values were calculated using unpaired t-test (*, p-value <0.05; **, p-value <0.01; ***, p-value <0.001 and ns, non-significant).
Fig 4. Relative fosA5 and fosA3 expression levels of NDM and OXA-48-coproducing K. pneumoniae.
qRT-PCR assay of fosA5 (a) and fosA3 (b) expression was performed in K. pneumoniae isolate KP5, KP4, KP6, KP15, KP18, KP19, and KP7. The relative number of transcripts of fosA5 and fosA3 were normalized to 16S rRNA expression and calculated using the 2-ΔΔct method compared to the expression of fosfomycin-susceptible K. pneumoniae isolate KP5. p-values were calculated using unpaired t-test (*, p-value <0.05; **, p-value <0.01; ***, p-value <0.001 and ns, non-significant).
Impact of fosA3 on fosfomycin susceptibility
The fosfomycin MICs of E. coli DH5α transformants carrying fosA3 plasmids were evaluated. The transformants carrying fosA3 plasmid from fosfomycin-susceptible isolates (KP23, KP44, and KP5) revealed 32- to 64-fold increased fosfomycin MICs (Table 2). Moreover, fosfomycin MICs of the transformants receiving fosA3 plasmid from resistant strains exhibited a 64-fold to >512-fold increase in MICs from the baseline.
Table 2. Antimicrobial susceptibility of K. pneumoniae clinical isolates and E. coli DH5α carrying fosA3.
| Strain | MIC (mg/L) | ||
|---|---|---|---|
| Fosfomycin | Imipenem | Meropenem | |
| KP23 | 16 | 16 | 128 |
| KP58 | >256 | 256 | 256 |
| KP44 | 32 | 4 | 16 |
| KP51 | >256 | 8 | 32 |
| KP5 | 32 | 32 | 64 |
| KP4 | >256 | 32 | 64 |
| KP6 | >256 | 32 | 64 |
| KP15 | >256 | 32 | 64 |
| KP18 | >256 | 256 | 256 |
| KP19 | >256 | >256 | 256 |
| KP7 | >256 | 64 | 128 |
| DH5α | 0.5 | 0.25 | 0.015 |
| DH5α/fosA3_KP23 | 32 | 0.25 | 0.015 |
| DH5α/fosA3_KP58 | 128 | 0.25 | 0.015 |
| DH5α/fosA3_KP44 | 16 | 0.25 | 0.015 |
| DH5α/fosA3_KP51 | >256 | 0.5 | 0.03 |
| DH5α/fosA3_KP5 | 32 | 0.25 | 0.015 |
| DH5α/fosA3_KP4 | 32 | 0.25 | 0.015 |
| DH5α/fosA3_KP6 | 128 | 0.5 | 0.015 |
| DH5α/fosA3_KP15 | 128 | 0.25 | 0.03 |
| DH5α/fosA3_KP18 | 64 | 0.25 | 0.03 |
| DH5α/fosA3_KP19 | 128 | 0.25 | 0.015 |
| DH5α/fosA3_KP7 | 256 | 0.5 | 0.03 |
Amino acid substitutions in MurA, GlpT, and UhpT
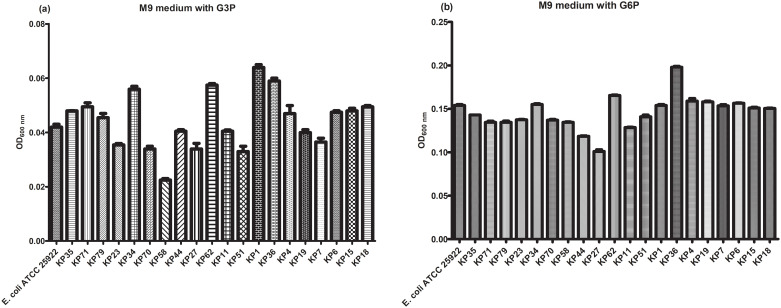
Although fosA3 plasmids of K. pneumoniae had an impact on fosfomycin MIC in E. coli transformants, the prevalence of fosA3 among fosfomycin-resistant K. pneumoniae was only 82.2% indicating that other mechanisms may involve in fosfomycin resistance. Amino acid sequences of MurA, GlpT, and UhpT were characterized in 37 isolates. The Thr287Asn substitution in MurA and Arg171Gly in UhpT were detected in both susceptible and resistant isolates (Table 3). Absence of glpT gene was detected in two fosfomycin-resistant isolates (KP58 and KP19) and these isolates showed reduced growth in M9 medium supplemented with G3P (compare to KP70 and KP4, respectively) suggesting an impair function of GlpT transporter (Fig 5). Various amino acid substitutions in GlpT (His147Gln, Pro212Leu, Gly386Ser, Gly386Ile, Phe112Ser, and Pro97Arg) and absence of glpT gene were detected in fosfomycin-resistant isolates. However, isolates carrying Gly386Ser, Gly386Ile, Phe112Ser, or Pro97Arg substitutions or absence of GlpT showed decreased growth in the presence of G3P as a carbon source (compare to KP70, KP27, and KP1, respectively). There were other substitutions in UhpT detected in highly resistant isolates including Ala176Pro in KP58 and Leu132Val in KP62 but their growth in the presence of G6P was similar to that of fosfomycin-susceptible isolate KP70 and KP27, respectively. Our results suggest that the overexpression level of fosA3 with the mutation or lack of glpT may be responsible for high-level resistance to fosfomycin.
Table 3. Fosfomycin resistance mechanisms of carbapenemase-producing K. pneumoniae.
| Strain | Fosfomycin MIC (mg/L) | Fos gene | Amino acid substitution | ||
|---|---|---|---|---|---|
| MurA | GlpT | UhpT | |||
| IMP-producing strain | |||||
| KP35 | 32 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP71 | 128 | fosA5 | Thr287Asn | none | Arg171Gly |
| NDM-producing strain | |||||
| KP79 | 8 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP23 | 16 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP97 | 16 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP53 | 32 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP72 | 32 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP78 | 32 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP14 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP68 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP34 | 64 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP70 | 128 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP58 | >256 | fosA5, fosA3 | Thr287Asn | Not detected | Arg171Gly, Ala176Pro |
| OXA-48-producing strain | |||||
| KP44 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP27 | 64 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP62 | >256 | fosA5 | Thr287Asn | none | Leu132Val, Arg171Gly |
| KP11 | >256 | fosA5 | Thr287Asn | His147Gln | Arg171Gly |
| KP26 | >256 | fosA5 | Thr287Asn | His147Gln | Arg171Gly |
| KP63 | >256 | fosA5, fosA3 | Thr287Asn | Pro212Leu | Arg171Gly |
| KP51 | >256 | fosA5, fosA3 | Thr287Asn | Pro212Leu | Arg171Gly |
| NDM and OXA-48-coproducing strain | |||||
| KP1 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP2 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP5 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP8 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP12 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP21 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP30 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP39 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP81 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP90 | 32 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP36 | 128 | fosA5 | Thr287Asn | none | Arg171Gly |
| KP4 | >256 | fosA5, fosA3 | Thr287Asn | Gly386Ser | Arg171Gly |
| KP6 | >256 | fosA5, fosA3 | Thr287Asn | none | Arg171Gly |
| KP15 | >256 | fosA5, fosA3 | Thr287Asn | Gly386Ile | Arg171Gly |
| KP18 | >256 | fosA5, fosA3 | Thr287Asn | Phe112Ser | Arg171Gly |
| KP19 | >256 | fosA5, fosA3 | Thr287Asn | Not detected | Arg171Gly |
| KP7 | >256 | fosA5, fosA3 | Thr287Asn | Pro97Arg | Arg171Gly |
Fig 5. Growth of K. pneumoniae isolates in M9 minimal medium supplemented with G3P or G6P as a carbon source.
The ability of K. pneumoniae growth was determined by measurement of the OD600nm of the cell suspension, normalized to the OD600nm of M9 supplemented with G6P or G3P without bacterial inoculation and compared to growth ability of E. coli ATCC 25922.
The activity of fosfomycin and amikacin combination
Amikacin alone was less effective against carbapenemase-producing K. pneumoniae than fosfomycin alone (Table 1). Among 66 isolates, only 18 (27.3%) were susceptible to amikacin. Resistance to amikacin was unlikely related to fosfomycin resistance. High-level resistance to amikacin (MIC>256 mg/L) was observed in OXA-48-producing isolates and NDM with OXA-48-co-producing isolates. However, high-level resistance to both amikacin and fosfomycin was found in OXA-48-producing isolates. The most common aminoglycoside-modifying enzyme gene, aac (6)’-Ib was detected in K. pneumoniae isolates with amikacin MIC range of 2–>256 mg/L (Table 3). Highly amikacin-resistant strains (KP34, KP27, and KP36) also carried armA.
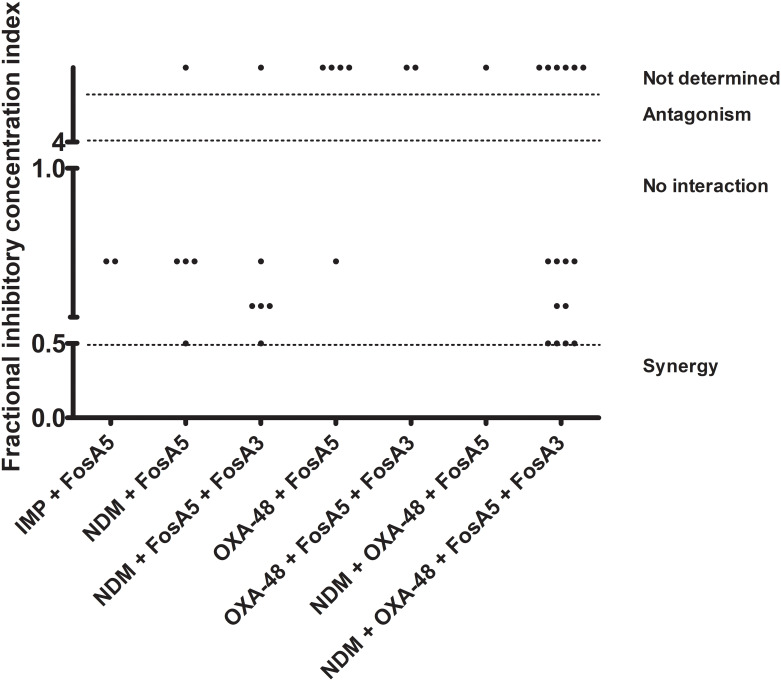
In vitro activity of fosfomycin combination with amikacin was performed in 37 carbapenemase-producing isolates by checkerboard assay. The FICIs of this combination are shown in Table 4 and Fig 6. The synergistic effect was observed in only 6 isolates (16.2%) including two isolates producing NDM and four isolates producing NDM with OXA-48. Fifteen isolates with high-level resistance to either fosfomycin or amikacin (MIC>256 mg/L) were excluded from the synergistic study by checkerboard assay. However, fosfomycin and amikacin MICs of these isolates were only two-fold reduced when compared to those of each antibiotic alone. Moreover, the synergism of fosfomycin and amikacin did not correlate with the presence of aminoglycoside-modifying enzymes. Our results suggest that amikacin unlikely enhances fosfomycin activity against carbapenemase-producing K. pneumoniae. Fortunately, antagonism was not found in our study.
Table 4. The activity of amikacin in combination with fosfomycin against carbapenemase-producing K. pneumoniae.
| Strain | Fosfomycin MIC (mg/L) | Amikacin MIC (mg/L) | Aminoglycoside-modifying enzyme gene | Fosfomycin MIC in combination (mg/L) | Amikacin MIC in combination (mg/L) | FICI |
|---|---|---|---|---|---|---|
| IMP-producing strain | ||||||
| KP35 | 32 | 1 | NF | 16 | 0.25 | 0.75 |
| KP71 | 128 | 1 | NF | 64 | 0.25 | 0.75 |
| NDM-producing strain | ||||||
| KP79 | 8 | 8 | aac (6)’-Ib | 4 | 2 | 0.75 |
| KP23 | 16 | 16 | aac (6)’-Ib | 8 | 4 | 0.75 |
| KP97 | 16 | 2 | aac (6)’-Ib, aphA6 | 2 | 1 | 0.63 |
| KP53 | 32 | 8 | aac (6)’-Ib, aphA6 | 8 | 2 | 0.75 |
| KP72 | 32 | 16 | aac (6)’-Ib, aphA6 | 8 | 4 | 0.5 |
| KP78 | 32 | 16 | aac (6)’-Ib, aphA6 | 8 | 8 | 0.75 |
| KP14 | 32 | 16 | aac (6)’-Ib | 4 | 8 | 0.63 |
| KP68 | 32 | 16 | aac (6)’-Ib | 4 | 8 | 0.63 |
| KP34 | 64 | >256 | aac (6)’-Ib, aphA6, armA | 16 | 256 | ND |
| KP70 | 128 | 8 | aac (6)’-Ib | 32 | 2 | 0.5 |
| KP58 | >256 | 16 | aac (6)’-Ib | >256 | 16 | ND |
| OXA-48-producing strain | ||||||
| KP44 | 32 | 8 | aac (6)’-Ib | 8 | 4 | 0.75 |
| KP27 | 64 | >256 | aac (6)’-Ib, aphA6, armA | 16 | 256 | ND |
| KP62 | >256 | >256 | aac (6)’-Ib | >256 | >256 | ND |
| KP11 | >256 | >256 | aac (6)’-Ib | >256 | >256 | ND |
| KP26 | >256 | >256 | aac (6)’-Ib | 128 | >256 | ND |
| KP63 | >256 | >256 | aac (6)’-Ib | 128 | >256 | ND |
| KP51 | >256 | >256 | aac (6)’-Ib | 128 | >256 | ND |
| NDM and OXA-48-coproducing strain | ||||||
| KP1 | 32 | 16 | aac (6)’-Ib | 16 | 4 | 0.75 |
| KP2 | 32 | 16 | aac (6)’-Ib | 4 | 8 | 0.63 |
| KP5 | 32 | 16 | aac (6)’-Ib | 8 | 4 | 0.5 |
| KP8 | 32 | 16 | aac (6)’-Ib | 16 | 4 | 0.75 |
| KP12 | 32 | 16 | aac (6)’-Ib | 4 | 8 | 0.63 |
| KP21 | 32 | 16 | aac (6)’-Ib | 8 | 4 | 0.5 |
| KP30 | 32 | 16 | aac (6)’-Ib | 8 | 4 | 0.5 |
| KP39 | 32 | 16 | aac (6)’-Ib | 16 | 4 | 0.75 |
| KP81 | 32 | 16 | aac (6)’-Ib | 4 | 16 | 0.75 |
| KP90 | 32 | 1 | NF | 8 | 0.25 | 0.5 |
| KP36 | 128 | >256 | aac (6)’-Ib, aphA6, armA | 32 | 128 | ND |
| KP4 | >256 | 16 | aac (6)’-Ib | >256 | 8 | ND |
| KP6 | >256 | 16 | aac (6)’-Ib | 128 | 8 | ND |
| KP15 | >256 | 16 | aac (6)’-Ib | >256 | 8 | ND |
| KP18 | >256 | 16 | aac (6)’-Ib | >256 | 8 | ND |
| KP19 | >256 | 32 | aac (6)’-Ib | >256 | 32 | ND |
| KP7 | >256 | 16 | aac (6)’-Ib | 32 | 8 | ND |
FICI: Fraction Inhibitory Concentration Index; NF: Not Found; ND: Not Determined
Fig 6. Distribution of Fraction Inhibitory Concentration Index (FICI) of K. pneumoniae isolates.
The FICIs of K. pneumoniae were plotted with different carbapenemase and fosA genes and categorized by synergism (FICI≤0.5), no interaction (0.5<FICI≤4), antagonism (FICI>4) and not determined for isolates with fosfomycin and/or amikacin MIC >256 mg/L.
Discussion
The occurrence of carbapenemase-producing K. pneumoniae has been globally mediated by carbapenemase productions (including KPC, OXA-48, NDM, IMP, and VIM). NDM and OXA-48-co-producing strains have been endemic especially in Asia including Thailand [1, 13, 14]. The emergence of carbapenem-resistant K. pneumoniae leads to limit treatment options and requires novel active agents or combination therapy. Fosfomycin has been proposed as an effective agent against multidrug-resistant K. pneumoniae as well as carbapenem-resistant K. pneumoniae [4, 15]. Our results demonstrated that 68.2% of carbapenemase-producing K. pneumoniae isolates were resistant to fosfomycin, while the resistance rate from worldwide is approximately 39.2%-66.2% [1]. These data suggest that fosfomycin monotherapy is unsuitable for being empirical therapy and requires susceptibility testing.
The major mechanism of fosfomycin resistance in Enterobacteriaceae is the production of fosfomycin modifying enzymes, particularly FosA, which catalyze fosfomycin [4]. The gene encoding FosA, called fosAKP or fosA5, is present in 99.7% of K. pneumoniae genomes [16]. However, the intrinsic fosA5 gene was found in both susceptible and resistant isolates suggesting that other mechanisms involve in fosfomycin resistance in K. pneumoniae. Plasmid-mediated fosA3 which originates from the chromosome of Kluyvera georgiana mobilized to plasmid and spread in Enterobacteriaceae including K. pneumoniae resulting in fosfomycin resistance [17, 18]. Notably, 16 fosfomycin-susceptible isolates in our study exhibited silence of fosA3. Plasmid-mediated fosA3 of these isolates elevated fosfomycin MICs of E. coli DH5α transformants supporting that fosfomycin monotherapy might be inappropriate for treatment of carbapenemase-producing K. pneumoniae infection. High prevalence of fosfomycin resistance in KPC-producing isolates from China is associated with plasmid co-harboring fosA3 with blaKPC [19]. Moreover, co-carriage of fosA3 and blaNDM has been found in Salmonella enterica serovar Corvallis in Germany [20] and plasmid co-carrying fosA3 and an ESBL gene, blaCTX-M has been reported in E. coli [3] indicating co-transfer of multiple resistance genes. Although E. coli DH5α transformants were selected on MHA supplemented with only fosfomycin, we also found blaOXA-48 in the transformants carrying fosA3 indicating co-transfer of carbapenemase gene and fosA3 (S2 Table). However, co-carriage of these resistance genes was not determined.
Apart from FosA production, fosfomycin resistance is also mediated by amino acid substitutions in fosfomycin target (MurA) and transporter proteins (GlpT and UhpT). The mutation at the active site (Cys115) of MurA has never been reported in K. pneumoniae including in our isolates indicating high fitness cost of the mutation [12, 21]. Thr287Asn substitution was found in both fosfomycin susceptible and resistant isolates suggesting that this substitution is unable to confer fosfomycin resistance. However, Thr287Asn in MurA was able to elevate fosfomycin MIC of E. coli DH5α transformant [12]. GlpT and UhpT, the transporter of G3P, and G6P, respectively in K. pneumoniae are also mediated fosfomycin influx and defective in these transporters involves in fosfomycin resistance [4, 7]. Therefore, impair growth of K. pneumoniae in M9 minimal medium supplemented with G3P or G6P indicates the defective transporter which related to fosfomycin resistance. In our study, many amino acid substitutions (Gly386Ser, Gly386Ile, Phe112Ser, and Pro97Arg) or loss of GlpT reduced growth of high-level fosfomycin-resistant isolates in the presence of G3P as a carbon source. The Arg171Vla substitution in UhpT was also detected in both fosfomycin-susceptible and fosfomycin-resistant isolates. The addition of Ala176Pro and Leu132Val substitutions were observed in high-level resistant isolates. Therefore, our results strongly suggest that FosA3 overexpression together with mutations or loss of GlpT or/and UhpT transporters are associated with high-level fosfomycin resistance in carbapenemase-producing K. pneumoniae.
Due to inadequate monotherapy, fosfomycin has been frequently combined with other active agents including carbapenems, colistin, and amikacin which exhibit synergism against carbapenem-resistant K. pneumoniae [8, 9, 22]. In our study, we focused on non-carbapenem and non-colistin, amikacin which has been reported to be an effective antibiotic against carbapenem-resistant K. pneumoniae [4]. Unfortunately, both amikacin-susceptible and amikacin-resistant isolates carried aac (6)’-Ib and aphA6 which are common AME genes conferring amikacin resistance in K. pneumoniae [23, 24]. High-level amikacin-resistant carrying armA isolates have been co-carried with blaKPC, [25] and blaCTX-M [26]. Although fosfomycin and amikacin combination has been demonstrated to be a potential combination against KPC-producing K. pneumoniae [22]. Unfortunately, synergism was rarely found in our study especially against isolates with high resistance to fosfomycin or/and amikacin. This discrepancy might be due to 1) different carbapenemases and K. pneumoniae strains, 2) co-carriage of multiple resistance genes and 3.) silence of resistance genes. However, these presumptions require further investigation.
In summary, the majority of carbapenem-resistant K. pneumoniae isolates are co-producing NDM and OXA-48 isolates. Overexpression of fosA3 with the mutations of glpT (even loss of glpT) and/or uhpT are associated with high-level resistance to fosfomycin. The silence of fosA3 frequently occurs in fosfomycin-susceptible K. pneumoniae. Amikacin cannot restore the activity of fosfomycin to achieve synergism against carbapenemase-producing K. pneumoniae, particularly isolates which are highly resistant to either fosfomycin or amikacin. Our results suggest that fosfomycin monotherapy may be inadequate due to the silence of fosA3 and high resistance rate. Moreover, fosfomycin with amikacin may also be insufficient combination therapy against carbapenemase-producing K. pneumoniae isolates.
Supporting information
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Ratchadapisek Somphot Fund for Postdoctoral Fellowship, Chulalongkorn University to Uthaibhorn Singkham-in, the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphiseksompot Endowment Fund) from Graduate School, Chulalongkorn University to Netchanok Muhummudaree, and the Ratchadapiseksompotch Fund from Faculty of Medicine, Chulalongkorn University to Tanittha Chatsuwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pitout JD, Nordmann P, Poirel L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob Agents Chemother. 2015;59(10):5873–84. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart A, Harris P, Henderson A, Paterson D. Treatment of Infections by OXA-48-Producing Enterobacteriaceae. Antimicrob Agents Chemother. 2018;62(11). 10.1128/AAC.01195-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol. 2016;7:895 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, Vouloumanou EK, Samonis G, Vardakas KZ. Fosfomycin. Clin Microbiol Rev. 2016;29(2):321–47. 10.1128/CMR.00068-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maraki S, Samonis G, Rafailidis PI, Vouloumanou EK, Mavromanolakis E, Falagas ME. Susceptibility of urinary tract bacteria to fosfomycin. Antimicrob Agents Chemother. 2009;53(10):4508–10. 10.1128/AAC.00721-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DH, Lees WJ, Kempsell KE, Lane WS, Duncan K, Walsh CT. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35(15):4923–8. 10.1021/bi952937w [DOI] [PubMed] [Google Scholar]
- 7.Silver LL. Fosfomycin: Mechanism and Resistance. Cold Spring Harb Perspect Med. 2017;7(2). 10.1101/cshperspect.a025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu W, Shen P, Bao Z, Zhou K, Zheng B, Ji J, et al. In vitro antibacterial activity of fosfomycin combined with other antimicrobials against KPC-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2017;50(2):237–41. 10.1016/j.ijantimicag.2017.03.011 [DOI] [PubMed] [Google Scholar]
- 9.Erturk Sengel B, Altinkanat Gelmez G, Soyletir G, Korten V. In vitro synergistic activity of fosfomycin in combination with meropenem, amikacin and colistin against OXA-48 and/or NDM-producing Klebsiella pneumoniae. Journal of Chemotherapy. 2020:1–7. 10.1080/1120009X.2020.1745501 [DOI] [PubMed] [Google Scholar]
- 10.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321–2. 10.1093/jac/dkl481 Epub 2006 Dec 21. [DOI] [PubMed] [Google Scholar]
- 11.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–23. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 12.Lu PL, Hsieh YJ, Lin JE, Huang JW, Yang TY, Lin L, et al. Characterisation of fosfomycin resistance mechanisms and molecular epidemiology in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates. Int J Antimicrob Agents. 2016;48(5):564–8. 10.1016/j.ijantimicag.2016.08.013 [DOI] [PubMed] [Google Scholar]
- 13.Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013;19(9):E421–3. 10.1111/1469-0691.12247 [DOI] [PubMed] [Google Scholar]
- 14.Laolerd W, Akeda Y, Preeyanon L, Ratthawongjirakul P, Santanirand P. Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae from Bangkok, Thailand, and Their Detection by the Carba NP and Modified Carbapenem Inactivation Method Tests. Microb Drug Resist. 2018;24(7):1006–11. 10.1089/mdr.2018.0080 [DOI] [PubMed] [Google Scholar]
- 15.Kaase M, Szabados F, Anders A, Gatermann SG. Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J Clin Microbiol. 2014;52(6):1893–7. 10.1128/JCM.03484-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, et al. Widespread Fosfomycin Resistance in Gram-Negative Bacteria Attributable to the Chromosomal fosA Gene. mBio. 2017;8(4). 10.1128/mBio.00749-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito R, Pacey MP, Mettus RT, Sluis-Cremer N, Doi Y. Origin of the plasmid-mediated fosfomycin resistance gene fosA3. J Antimicrob Chemother. 2018;73(2):373–6. 10.1093/jac/dkx389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, et al. Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother. 2012;67(12):2843–7. 10.1093/jac/dks319 [DOI] [PubMed] [Google Scholar]
- 19.Jiang Y, Shen P, Wei Z, Liu L, He F, Shi K, et al. Dissemination of a clone carrying a fosA3-harbouring plasmid mediates high fosfomycin resistance rate of KPC-producing Klebsiella pneumoniae in China. Int J Antimicrob Agents. 2015;45(1):66–70. 10.1016/j.ijantimicag.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 20.Villa L, Guerra B, Schmoger S, Fischer J, Helmuth R, Zong Z, et al. IncA/C Plasmid Carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica Serovar Corvallis Strain Isolated from a Migratory Wild Bird in Germany. Antimicrob Agents Chemother. 2015;59(10):6597–600. 10.1128/AAC.00944-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, Chen S, Wu ZY, Qi M, Li XY, Liu CX. Mechanisms of fosfomycin resistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae. J Glob Antimicrob Resist. 2020. 10.1016/j.jgar.2019.12.019 [DOI] [PubMed] [Google Scholar]
- 22.Yu W, Zhou K, Guo L, Ji J, Niu T, Xiao T, et al. In vitro Pharmacokinetics/Pharmacodynamics Evaluation of Fosfomycin Combined with Amikacin or Colistin against KPC2-Producing Klebsiella pneumoniae. Front Cell Infect Microbiol. 2017;7:246 10.3389/fcimb.2017.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YT, Jang JH, Kim HC, Kim H, Lee KR, Park KS, et al. Identification of strain harboring both aac(6')-Ib and aac(6')-Ib-cr variant simultaneously in Escherichia coli and Klebsiella pneumoniae. BMB Rep. 2011;44(4):262–6. 10.5483/BMBRep.2011.44.4.262 [DOI] [PubMed] [Google Scholar]
- 24.Galani I, Nafplioti K, Adamou P, Karaiskos I, Giamarellou H, Souli M, et al. Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect Dis. 2019;19(1):167 10.1186/s12879-019-3801-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zacharczuk K, Piekarska K, Szych J, Zawidzka E, Sulikowska A, Wardak S, et al. Emergence of Klebsiella pneumoniae coproducing KPC-2 and 16S rRNA methylase ArmA in Poland. Antimicrob Agents Chemother. 2011;55(1):443–6. 10.1128/AAC.00386-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, Lai YK, et al. Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2009;53(1):104–11. 10.1128/AAC.00852-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.