Abstract
This study reveals a new microfluidic biosensor consisting of a multi-constriction microfluidic device with embedded electrodes for measuring the biophysical attributes of single cells. The biosensing platform called the iterative mechano-electrical properties (iMEP) analyzer captures electronic records of biomechanical and bioelectrical properties of cells. The iMEP assay is used in conjunction with standard migration assays, such as chemotaxis-based Boyden chamber and scratch wound healing assays, to evaluate the migratory behavior and biophysical properties of prostate cancer cells. The three cell lines evaluated in the study each represent a stage in the standard progression of prostate cancer, while the fourth cell line serves as a normal/healthy counterpart. Neither the scratch assay nor the chemotaxis assay could fully differentiate the four cell lines. Furthermore, there was not a direct correlation between wound healing rate or the migratory rate with the cells’ metastatic potential. However, the iMEP assay, through its multiparametric dataset, could distinguish between all four cell line populations with p-value < 0.05. Further studies are needed to determine if iMEP signatures can be used for a wider range of human cells to assess the tumorigenicity of a cell population or the metastatic potential of cancer cells.
Keywords: Cell deformability, microfluidics, impedance, cancer, migration assays
1. Introduction
Chemotherapeutic treatment of cancer has focused on the inhibition of invasion and, consequently, metastasis. The scratch wound healing assay and Boyden chamber are common and well-established methods of evaluating cell migration in vitro [1–5]. The wound healing assay involves creating a gap within a cell monolayer and capturing time-lapse imagery of cell migration. The rate at which the cells travel to “heal” the wound is indicative of cell-matrix and cell-cell interactions during cell migration [2–5]. The Boyden chamber assay, also known as the transwell assay, consists of two compartments separated by a membrane with micropores, where pore sizes can range from 3–12 μm to suit cells of interest [1, 6]. The cells are seeded in the top region and the bottom region contains chemo-attractants. Using this method, migratory behavior is quantified using a plate reader or by staining cells and taking images of the cells [1, 4, 6]. Drawbacks of the wound healing method are lengthy experimentation time of the assay, the influence of cell proliferation, and lack of chemo-attractants. In contrast, the Boyden chamber method typically consumes less time, is independent of proliferation, and allows for the usage of chemotactic agents. Additionally, microfabricated migration assays have also been developed using microfluidic platforms, where researchers have evaluated cell migratory behavior and kinetics of endothelial and cancer cells [7–10].
Deformability of cells in the presence of a compressive force is a mechanical property that can be used to phenotype cells with different sizes ranging from red blood cells to tumor cells [11–14]. The deformability of a cell is influenced by its intracellular structures and is particularly dependent upon the cytoskeleton, the kinetic framework of the cell. The compliancy of a cell can be used to classify cells according to specific phenotypes, most notably, diseased versus healthy tissues. In this regard, phenotyping can provide information regarding the immune response, stem cell therapies, cancer diagnostics, etc. Deformability assays have become increasingly common in cancer research as it has been observed that in most cases more aggressive cancer cells exhibit decreased structural rigidity most likely benefitting cellular locomotion and metastasis [11, 13, 15–17].
In order to accommodate the urgency placed upon health-related diagnostic practices, a high-throughput method to analyze the compliance and migratory behavior of single cells is required. Methods of determining cell deformability already in practice are atomic force microscopy (AFM) [18, 19], micropipette aspiration [20], hydrodynamic stretchers [21], magnetic tweezers [22], and optical stretchers [13]; however, these techniques require specialized equipment and require time-consuming pre-processing, experimentation, and post-processing times. In addition, these procedures involve tools that can cause damage to the cells through contact or fixing and staining. Standard migration assays also share similar drawbacks, including time-consuming preparation and experimentation. To alleviate these issues, constriction-based microfluidic flow cytometry, which operates at a much greater throughput, has proven to be a viable method of deformability and migratory analysis with the added benefits of cost-effectiveness and low-complexity [11, 12, 15, 16].
Flow cytometry is a versatile tool commonly employed to study cell surface markers and biophysical properties under laminar flow using optical scattering or fluorescent tags [23–26]. Deformability can be observed when a constriction slightly smaller than cell diameter is incorporated into the channel, which is the principle of constriction-based microfluidics. Data collected from these systems describe the passage time of single cells. Passage time is composed of entry time, the time required for a cell to deform and enter the constriction, and transit time, the time elapsed between the cell entering the constriction and exiting it. Transit time is influenced by friction between the cell and the channel wall, as well as fluid flow pressure [11, 15, 16]. Constriction-based technologies have been improved by employing a cyclic deformation assay, which involves sequential deformation regions separated by relaxation regions. Previously, our group has shown that cyclic deformations can improve the detection of breast cancer cells from their normal counterparts [15–17]. The traditional method of observing cell velocities as they pass through these constriction points is with a high-speed camera; however, this practice is time-consuming in data analysis and the equipment is expensive. The post-processing time is further amplified by incorporating multiple constriction microchannels as multiple time points need to be obtained for each constriction region. These disadvantages are improved through incorporation of electrode sensors embedded within the device. Electrodes can measure impedance within the constriction channel and impedance peak profiles can be utilized to measure transit times [27–33].
Bioelectrical properties of cells are emerging as label-free markers which allow for significant differentiation between similar cell types [34–38]. This is due to variations in electrical properties of cells, such as membrane capacitance and cytoplasm resistance, attributed to differing physiologies between cells. Recently, research has emerged combining electrical analysis of cells with deformability microfluidics to produce microfluidic impedance devices [27–31]. In this study, it was hypothesized that measuring the impedance of single cells in combination with deformability, represented by transit times, allows for greater differentiation between similar prostate cancer cell types: LNCaP, LNCaP-C4–2, and LNCaP-C4–2B. These cell lines have clinical relevance as they represent the progression that human prostate cancer typically goes through, where the prostate cancer gains androgen-independence then metastasizes to bone. Thus, they provide a good model to help understand the mechanisms of androgen-independence and bone metastasis. Additionally, a normal/healthy prostate cell line, PWR-1E, has been included in the study to compare its biophysical attributes to those of various cancer cells.
The biosensor reported in this paper consists of five sequential constriction channels separated by relaxation regions where biomechanical and bioelectrical attributes of cells are obtained simultaneously. Yang et al. developed a multi-constriction microfluidic device with embedded 3D electrodes to obtain biomechanical and bioelectrical data of single cells [33]. Our work differs in terms of the device configuration, application and post-processing of data. The 3D electrode configuration is advantageous with respect to elimination of alignment and sensitivity, however they do not have the flexibility for an inexpensive off-chip device via planar electrodes [39]. Device configuration also differs as they do not have a built-in anti-clogging mechanism. Their work compared chemically treated MCF-7 cells, while our platform is used to compare various prostate cell lines with differing cancer progression. Lastly, their data analysis requires complex neural network post-processing compared to our quick and simple post-processing. We report the first instance of studying biophysical attributes of the prostate progression model of LNCaP cell lines, and how it compares to a normal prostate cell line counterpart. Additionally, migratory behavior has been assessed through scratch wound healing and chemotaxis Boyden chamber assays, and results are compared to biophysical attributes obtained from our microfluidic sensor. The coupling of these assays provides a comprehensive analysis of cells at different stages of prostate progression model and how it compares to a healthy prostate cell line.
2. Materials and Methods
2.1. Cell lines
LNCaP is a human prostate cancer cell line; derived from a metastatic site- the left supraclavicular lymph node of a Caucasian 50-year-old male. The C4 cell line constitutes the in vitro cultured subline grown from the murine host’s tumor. When the C4 sub-line was subsequently co-inoculated with MS osteosarcoma fibroblasts in a castrated athymic male nude mouse host for another 12 weeks, prostatic epithelial cells cultured from the resultant tumor in this host constituted the C4–2 subline which is androgen-independent. The LNCaP-C4–2B cell was obtained from bone metastatic C4–2 cells grown in a castrated mouse and is also androgen-independent [40]. LNCaP and LNCaP-C4–2B were gifts from Dr. Leland W.K. Chung via Dr. Simon W. Hayward and Dr. Magda M. Grabowska (Case Western Reserve University). LNCaP-C4–2 cells were a gift from Dr. Warren D. Heston (Cleveland Clinic). The PWR-1E cell line is prostate epithelial line derived from a Caucasian male at 67 years of age and was purchased and verified by ATCC. All lines have been confirmed negative for mycoplasma.
2.2. Sample Preparation
Human prostate cancer cell lines LNCaP, LNCaP-C4–2, and LNCaP-C4–2B were grown in RPMI-1640 with 10% fetal bovine serum and 1% Pen-Strep ( 100 U/mL penicillin and 100 ug/mL streptomycin)(GIBCO). The human immortalized prostate epithelial cell line, PWR-1E, was cultured in keratinocyte serum-free medium (K-SFM) supplemented with 0.05 mg/mL of bovine pituitary extract and 5 ng/mL of human recombinant epidermal growth factor provided in the K-SFM kit (GIBCO). Cell monolayers were grown in T-25 cm2 culture flasks at 37 °C in a combination of 95% air and 5% CO2 until cells reached proper confluency. To process cell monolayers for experimentation, the cells were detached from the inner surface of the flask with a trypsin-ethylenediaminetetraacetic acid solution for ~5 minutes at 37 °C and gently aspirated to create a single cell suspension. Cell suspensions are spun down, rinsed, and resuspended in 1× PBS. Cell counts were ~ 10 × 104 cells/mL for each respective cell line. Cell diameters for LNCaP, LNCaP-C4–2, LNCaP-C4–2B, and PWR-1E ranged from 10–16 μm, 10–16 μm, 10–15 μm, and 10–15 μm, respectively [41].
2.3. Device Fabrication
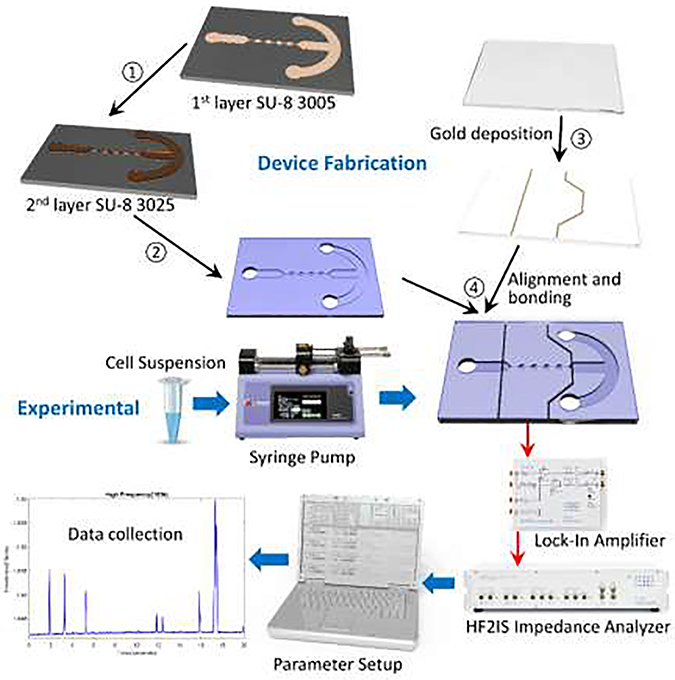
Fabrication of the electrodes (Figure 1) starts with the patterning of a glass wafer with photoresist (S1813, MicroChem, Newton, MA) by means of photolithography. Using electron-beam (e-beam) evaporation, layers of chromium (~40 μm) and gold (~80 μm) are evaporated onto the patterned wafer and electrodes are created using standard metal lift-off techniques. To create the microfluidic channels (Figure 1), we patterned a silicon wafer with two layers of SU-8 (SU-8 3005 and SU-8 3025, MicroChem, Newton, MA) through photolithography to create a master mold. The SU-8 3005 was used to build the constriction channels with a height of 8 μm; and the SU-8 3025 was used to build the remaining microfluidic channels with a total height of ~30 μm. The mold is then coated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-tricholosilane (TFOCS, Fisher Scientific) for easy release of polydimethylsiloxane (PDMS). Using soft-lithography techniques, we use the master mold to create the PDMS microchannel. Through plasma activated bonding we align and bond the PDMS microchannels onto the glass with electrodes. Each electrode has its own bond pad so that wires can be soldered in order to be connected to the impedance spectroscope.
Figure 1.
Overview of iMEP device fabrication and microfluidic experimental setup.
2.4. Device Design
The sensor consists of two separate channels, delivery and constriction, to prevent cell accumulation at the constriction and clogging (Figure 1). The sample of single cells suspended in 1× PBS passes through the channel from an inlet to an outlet. Once cells have been introduced to the delivery channel, a vacuum pump applies a negative pressure at the outlet. Consequently, cells flow through the delivery channel due to pressure differences. The entrance of the constriction channel is located at the center of the delivery channel. A separate negative pressure applied at the end of the constriction channel, via a Harvard Apparatus syringe pump, in order to initiate flow through the constriction channel. Flow through the constriction channel is initiated by applying a constant negative pressure of ΔP= ~−100 Pa. Once a cell has entered the constriction channel, the flow coming from the delivery channel stops until the cell trapped in the constriction channel has passed through completely. The constriction channel consists of five constriction regions and four relaxation regions, where each region is 50 μm in length, 8 μm in height and 45 μm in width, respectively. The device also consists of a pair of two electrodes 20 μm-wide and 120 nm-tall with a spacing of 625 μm. The electrodes are aligned ~85–90 μm from the entrance and exit of the constriction channel.
2.5. Experimental Setup
2.5.1. Scratch Wound Assay
Scratch wound migration assays were performed using the 96 well image lock plates (Sartorius) and the wound maker from IncuCyte ZOOM Live-Cell Analysis System (Sartorius). Cells were plated into each well at 4.5 × 104 - 7.5 × 104 in 100 μL of medium and allowed to adhere overnight. Cells were wounded using the wound maker, washed, and 200 μL of new media added. Plates were placed in the IncuCyte and scanned every two hours until wounds closed, media was changed every 2–4 days.
2.5.2. Chemotaxis Migration Assay
LNCaP, LNCaP-C4–2, LNCaP-C4–2B, and PWR-1E cells were plated, in ClearView 96-well chemotaxis plates (Sartorius) with 8 μm pore size, in biologically equivalent sextuplicates at 60–120 cells per well using complete RPMI-1640 or complete K-SFM and incubated overnight to allow for adherence. To establish a chemical gradient, media was changed inside the upper reservoir from complete RPMI-1640 to serum-reduced RPMI-1640 supplemented with 1% FBS and complete K-SFM to growth factor-reduced K-SFM supplemented with 0.01 mg/mL BPE and 1 ng/mL. Cell migration towards complete media was monitored using the IncuCyte ZOOM Live-Cell Analysis System (Sartorius) by continuous imaging every 2 hours for 72 hours. Migrated cell count was quantified using IncuCyte software analysis (Sartorius) and normalized to the initial top value.
2.5.3. Microfluidic iMEP Assay
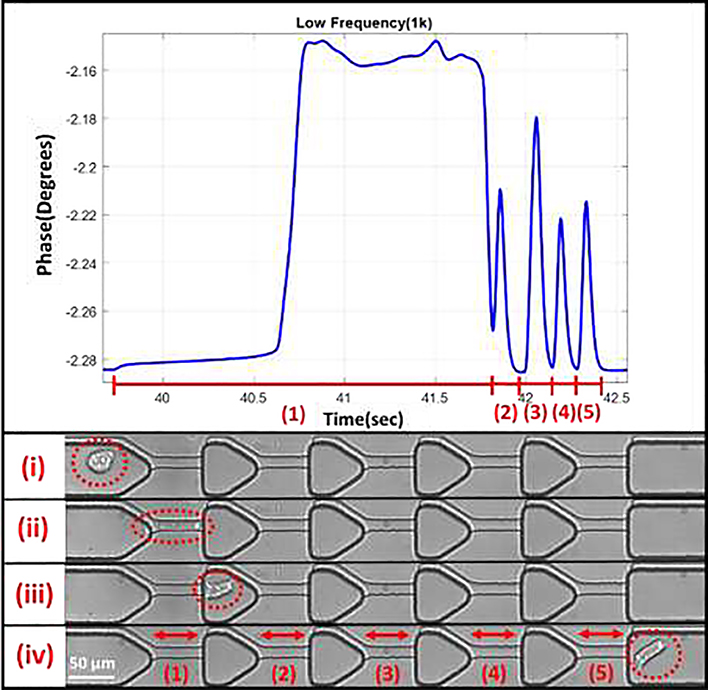
The iterative mechanoelectrical properties (iMEP) device is mounted on an inverted microscope (Zeiss Axio Observer LSM-510, Thornwood, NY) with a lens magnification of 20×. During experimentation, the electrical impedance across the channel was obtained by applying 1V AC signal at 8 different frequencies of 0.5, 1, 5, 10, 50, 100, 500, and 1000 kHz using the HF2IS Impedance Spectroscope (Zurich Instruments, Zurich, Switzerland). Two devices on separate days were used to obtain the biomechanical and bioelectrical results, where a minimum of three runs for each cell type was used on each individual device. Figure 2 shows a representation of the impedance peak collection for a single cell passing through the five constrictions. The electrical properties are represented by shifts in magnitude and phase, where the max peak is subtracted from the baseline. High-speed videos were obtained at 100 frames/sec via the Motion Xtra NX4-S3 high-speed camera (IDT, Tallahassee, FL) for cross-verification of cell transit through the constriction channels. Data was obtained from the impedance analyzer using Python 3.6.
Figure 2.
Representative impedance plot of a single cell passing through the sequential constriction channels. Each constriction results in its own peak, where timing and impedance information can be obtained. (i) Cell before deformation. (ii) Cell within the first deformation. (iii) Cell after first deformation. (iv) Cell after transit through all constriction channels.
2.5.4. Statistical Analysis
Statistical significance was determined by two-way or one-way analysis of variance (ANOVA) with Tukey post-test using the GraphPad Prism 7 software. For experiments over time, significance was confirmed using linear regression models to test for differences and test interactions between cell type and slope of the relationship with time with the SAS software. Error bars represent the SEM of experiments. * p<0.05, ** p<0.005, and *** p<0.001, and **** p<0.0001.
3. Results
3.1. Scratch Wound Assay
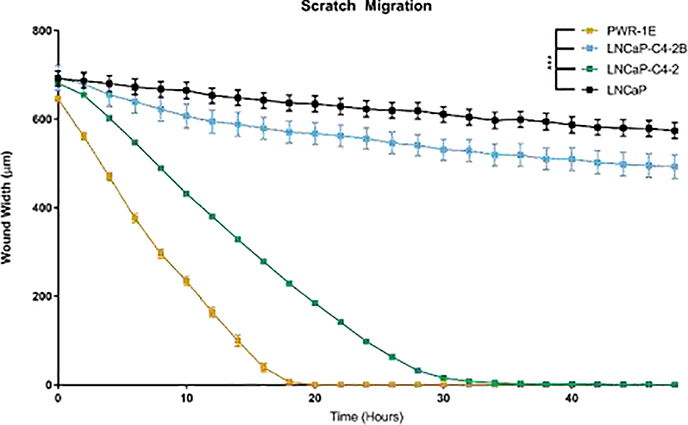
To evaluate sheet migration behavior of normal prostate epithelial cells and androgen-dependent or independent prostate cancer cells, we performed a scratch wound healing assay. Results depicted in Figure 3 below indicate that the normal prostate cell line PWR1-E healed the wound the quickest out of all cell lines: 5-fold faster than LNCaP, 1.6-fold faster than LNCaP-C4–2, and 4.4-fold faster than LNCaP-C4–2B cells. However, comparing the prostate cancer cell lines show that the LNCaP-C4–2 heal the wound the quickest, followed by LNCaP-C4–2B (2.7-fold slower than LNCaP-C4–2) and LNCaP (3-fold slower than LNCaP-C4–2). Thus, the two-dimensional migration demonstrates altered migratory behavior dependent on the cell line tested with the normal epithelial cells healing the wound the fastest.
Figure 3.
Normal prostate epithelial cells migrate quickly in two dimensions. Width of remaining wound after scratch represented as mean ± SEM. *** represents p<0.001 by two-way ANOVA with Tukey post-test.
3.2. Chemotaxis Migration Assay
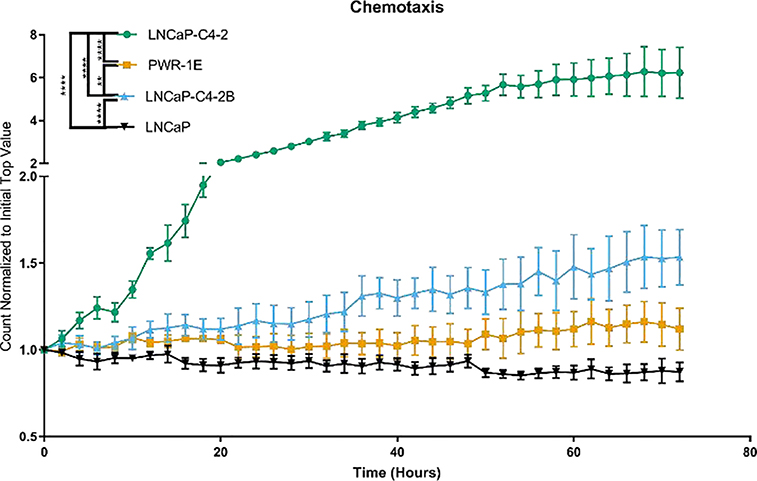
To evaluate the chemotactic potential of normal prostate epithelial cells and androgen-dependent or independent prostate cancer cells, we performed a chemotactic migration assay using PWR-1E, LNCaP, LNCaP-C4–2, and LNCaP-C4–2B (results are shown in Figure 4). LNCaP-C4–2 cells demonstrated the fastest migration through the pores: 3.5-fold faster than PWR-1E, 3-fold faster than LNCaP-C4–2B, and 4-fold faster than LNCaP cells. LNCaP-C4–2B and PWR-1E cells migrated more quickly than LNCaP cells, 1.4-fold, and 1.2-fold, respectively. Thus, in contrast to the two-dimensional migration, the normal epithelial cells and androgen-dependent prostate cancer cells migrated more slowly than the androgen-independent prostate cancer cell lines through the pores.
Figure 4.
Androgen independent prostate cancer cell lines demonstrate higher chemotaxis compared with the normal epithelial cells and androgen-dependent prostate cancer cell lines. Number of cells in the bottom well normalized to the initial top well represented as mean ± SEM. * represents p<0.05, ** represents p<0.005, *** represents p<0.001, **** represents p<0.0001 by two-way ANOVA with Tukey post-test.
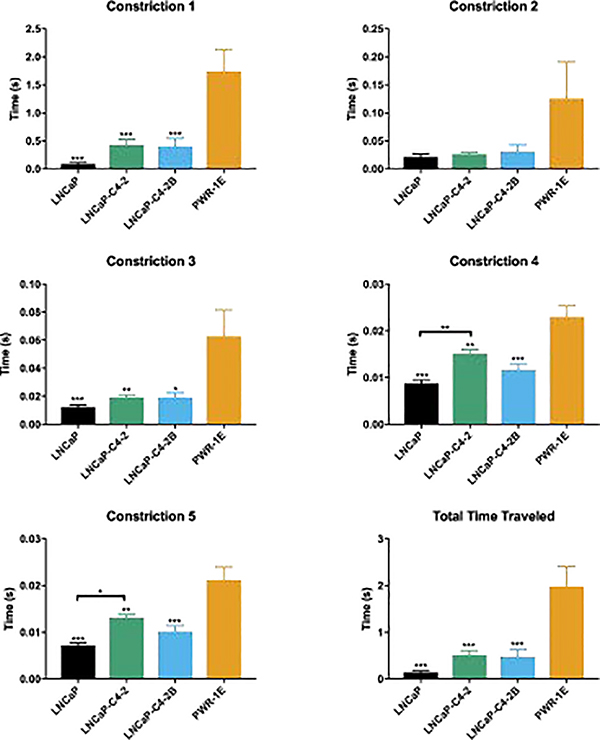
3.3. Microfluidic iMEP Assay
Cell timing information in the iMEP device was obtained in all 5 constriction regions in order to compare the four cells lines based on their response to the constriction channel. Average values +/− standard error of the mean (SEM) are shown in Figure 5. Comparing the timing in constriction #1, which represents both entry and transit time, showed that the LNCaP and PWR1E cells are the most and least deformable, respectively. Additionally, the LNCaP-C4–2 and LNCaP-C4–2B have comparable constriction 1 values. Lower and higher timing in the first constriction are indicative of cells with more and less deformability, respectively. However, in the remaining four constriction channels, the cell’s ability to regain its shape in the relaxation region and deformability both play a role. After experiencing the first constriction, LNCaP cells timing information approached those of the other LNCaP derivatives. The PWR-1E cells tended to have higher transit times regardless of the constriction region.
Figure 5.
Passage time differentiates normal and cancerous cells. Average time traveled for single cells represented as mean ± SEM. * represents p<0.05, ** represents p<0.005, *** represents p<0.001 by one-way ANOVA with Tukey post-test. Cell count for each cell line is N = 60, 66, 41, and 48, respectively.
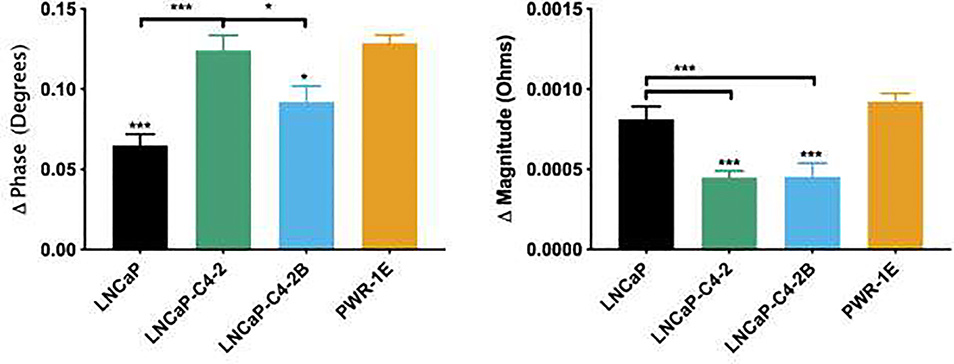
In addition to timing information, electrical properties were obtained for each cell line where the average +/− SEM of phase and magnitude, measured at respective frequencies of 50 kHz and 100 kHz, is shown in Figure 6. Although phase and magnitude were obtained for frequencies ranging from 0.5–1,000 kHz, we have included a single example of each due to redundancies in data and for the purpose of brevity. Looking at both mean values of phase and magnitude, it is clear that each cell line has a distinct difference in bioelectrical properties.
Figure 6.
Bioelectrical measurements differentiate between all four cell lines with cell count N = 60, 45, 41, and 48, respectively. Phase shift and magnitude for single cells represented as mean ± SEM. * represents p<0.05, ** represents p<0.005, *** represents p<0.001 by one-way ANOVA with Tukey post-test.
4. Discussion
In vitro migration assays are proven techniques for evaluating cell motility, chemotherapeutic response, and metastatic potential of tumor cells [1–6]. Here, scratch wound, chemotaxis, and constriction-based microfluidic assays have been used to study prostate cancer cells of varying aggressiveness in addition to a healthy prostate cell line. Each assay provides both differing and overlapping information regarding the cells of interest. Scratch wound healing assays operate by creating an opening in a cell monolayer and monitoring how long it takes for the cells to heal the wound. Here, cells maintain their cell-to-cell junctions and probe the collective cell migration called sheet migration [2, 5]. Our data demonstrate that normal prostate epithelial cells migrate faster than cancer cell lines in sheet migration. This is likely due to the role of epithelial cells in healing wounded tissues. In contrast, the chemotaxis and our constriction-based microfluidic assay probed the single cells by their movement through pores and microchannels where the cell is required to deform. This deformation is more relevant to the type of movement required for cancer progression. The chemotactic Boyden chamber assay, which relies on chemical stimuli to drive cell movement, measures the combination of the cells’ response to the chemicals and its ability to deform and transit through the micropores [1, 6]. Our data show that the androgen-independent prostate cancer cell lines migrate more quickly through the pores and toward serum as a chemoattractant compared with androgen-dependent prostate cancer and normal prostate epithelial cells. Our microfluidic biosensor measures the ability of the cells to deform and transit through sequential constriction channels separated by relaxation regions that allow cells to have the opportunity to regain its original spherical-like shape from the rod-like shaped caused by the constriction channel. We demonstrate that normal prostate epithelial cells move slowly through the constriction channels, while the androgen-dependent prostate cancer cell line can deform and recover the most. Additionally, the electrical impedance of the cells is obtained, which relate to cell biophysical properties such as cell size, membrane capacitance, and resistance. These data were most effective in differentiating the different cell lines and when combined with constriction deformability in our iMEP device could differentiate among the metastatic cells. Figure S1 in the supplementary information illustrates an example of how a multi-parametric analysis can be utilized for single cell differentiation. Additionally, this figure shows how combining biomechanical and bioelectrical parameters of cells can further distinguish cells. None of the standard techniques used in this study could provide this type of single cell differentiation.
The scratch wound healing and chemotaxis assay show that metastatic potential cannot be assessed using these methods. In brief, these methods are robust in terms of determining sheet migration behavior, response to chemo-attractants, and deformability; however, they cannot solely be used to study the metastatic potential of cancer cell lines. For the scratch wound assays, the results on the sheet migratory behavior do not correspond with aggressiveness in all cases. When comparing the LNCaP-C4–2 and LNCaP-C4–2B to the parental LNCaP cell line, the assay correctly indicates that the former cell lines are more aggressive than the parental cell line. However, it is not a good indicator of metastatic aggressiveness when comparing LNCaP-C4–2 and LNCaP-C4–2B; however, as these two cell lines are closely related their grouping together may be more indicative of their similarities as androgen-independent prostate cancer cell lines. Additionally, when comparing the normal PWR-1E and LNCaP cell lines, it shows that the normal cell line migrates much faster than all three cell lines. This is likely due to the role of epithelial cells in healing wounded tissues and their ability to migrate as a sheet. For the chemotaxis migration assay, we were not able to distinguish between the LNCaP and PWR-1E cell lines with statistical significance. The chemotaxis results also show that this assay is not always a good indicator of cancer aggressiveness, because the more bone-trophic LNCaP-C4–2B cell migrates at a much lower rate than the less aggressive LNCaP-C4–2 cells. We hypothesize that this could be due to changes in the cell generated by exposure to their metastatic niche: the bone microenvironment; however, further studies will be needed to tease out this association.
Cell stiffness and deformability have been used by our lab and other research groups to distinguish cancer cells from their non-tumorigenic counterparts, and untreated versus drug-treated cells [13, 16, 27, 32]. Cell deformability can be probed while attached to a surface such as in atomic force microscopy, or while suspended in liquid solution, such as microfluidic-based techniques. Microfluidic technologies are advantageous compared to alternative methods in terms of cost, throughput, and experimentation time. Researchers have commonly claimed that cell deformability is higher for cancer cells in comparison to normal cells and directly related to the aggressiveness of the cancer [42–48]. However, contradicting results have been presented, which counteract the narrative that cell stiffness is directly correlated to cell aggressiveness [49, 50]. For instance, Bastatas et al. compared stiffnesses of lowly and highly metastatic human prostate cancer cells via AFM, where the more metastatic cells were reported to be stiffer than the lowly metastatic counterpart [49]. Our iMEP results of prostate cells in some cases agree, and others disagree with this contradicting narrative, which proves that deformability or stiffness alone is not a reliable method of evaluating cell metastatic potential. More specifically, using the biomechanical properties manifested in cell transit times, we were not able to accurately predict the migratory behavior and metastatic potential of the cancer cells. However, we could clearly distinguish between the normal and cancerous populations. Furthermore, we could identify all four cell lines using the bioelectrical properties of deformed cells by combining the results of phase and magnitude shifts. The timing information was able to separate the androgen-dependent LNCaP and androgen-independent LNCaP-C4–2 cell lines providing prognostic information on cancer severity. The later constriction points demonstrated increasing separation between the androgen-dependent and independent cell lines indicating that additional constriction points may lead to an improved ability to separate these two cell lines.
The uniqueness of our proposed assay, when compared to the aforementioned standard techniques, is its multi-parametric output. Through a combination of cell mechanical modulatory behavior extracted by the deformation-relaxation transit times and the bioimpedance data of deformed cells, our assay is capable of not only distinguishing between normal and cancer cells but also between cancer cells having different metastatic potential. Further experimentation and analysis can be done to map these biophysical properties to cell attributes for other prostate cancer cells and other cancer types to determine if iMEP can be used as a standalone tool to analyze cancer cell invasiveness.
5. Conclusion
Each assay presented in this paper provides differing information about cell biological and biophysical properties. For instance, the chemotaxis assay provides information about how cell populations migrate through constrictive pores in response to a chemo-attractant gradient, while the scratch wound assay probes cell populations’ sheet migration behavior. Although these two standard migration assays provide information about migratory behavior, they do not provide the whole story of the metastatic potential. The iMEP device provides information about the cell deformability and dielectric properties, but also cannot reliably predict metastatic potential. However, the iMEP device is advantageous over the other standard migration assays because we can obtain information about the single cells. Experimentation time is another advantage of our iMEP assay, as it can analyze a population of cells in a matter of minutes. To conclude, our iMEP device has the capability of identifying tumor cells from their normal counterparts, as well as distinguishing between closely related cancer cells using the average bioelectrical properties of the deformed cells. In contrast, the scratch wound assays showed significant differences between the normal and cancer cell lines, but the results with the three cancer cell lines did not correspond to the cells’ metastatic potential. Additionally, the chemotaxis assay results were not able to distinguish between the normal and cancer cell lines, and the results did not correspond to the metastatic potential for all cancer cell lines. Thus, the iMEP device presented here represents one method to differentiate between normal and cancerous cells and with additional development may be able to provide prognostic information on the metastatic potential of cancer cells.
Supplementary Material
Research Highlights.
Prostate cancer cell lines of differing cancer progression and its normal/healthy counterpart have varying biomechanical/bioelectrical characteristics, wound healing times, and chemotaxis response.
The iMEP assay rapidly obtains a multiparametric dataset on single cell biophysical characteristics to distinguish between four different prostate cell lines with p-value < 0.05.
Individual migration assays are not sufficient for assessing the tumorigenicity of a cell population or the metastatic potential of cancer cells.
Acknowledgments
We thank Drs. Leland W.K. Chung, Simon W. Hayward, Warren D. Heston, and Magda M. Grabowska for the kind gift of the prostate cancer cell lines. The authors also acknowledge the Micro & Nano Fabrication Laboratory at Virginia Tech for the equipment support.
Funding
The research performed in the VT MEMS Lab was funded by the National Institute of Health (NIH) National Cancer Institute, grant number R21CA210216. Research performed in the Kerr Lab was supported by an NIH/NCI grant R00 CA175291 and by the Wake Forest Baptist Comprehensive Cancer Center Shared Resources grant (NIH/NCI CCSG P30 CA012197).
Author Bios
Parham Ghassemi is currently pursuing his Ph.D. in Electrical Engineering at Virginia Tech. He received his B.S. in Electrical Engineering from George Mason University and M.S. in Electrical Engineering from Virginia Tech in 2015 and 2017, respectively. His research focuses on microfluidic sensors for biological applications. Specifically, he utilizes bioelectrical and biomechanical characteristics of cells to study and evaluate cancer cells and circulating tumor cells in blood.
Koran Harris received a Bachelor of Science in biology at North Carolina Agricultural and Technical State University in 2017. He is currently completing a Ph.D. in Molecular and Cellular Biosciences, with a concentration in cancer biology, at Wake Forest School of Medicine under the supervision of Dr. Bethany Kerr. His current studies focus on developing in vitro models to understand the driving forces contributing to prostate cancer-bone metastasis.
Xiang Ren received the B.S. degree in electrical engineering from the University of Electronic Science and Technology of China, Chengdu, China, in 2009, the M.S. degree in electrical and computer engineering from Johns Hopkins University, Baltimore, MD, USA, in 2010, and the Ph.D. degree in mechanical engineering and mechanics from Drexel University, Philadelphia, PA, USA, in 2016. He was a Research Assistant with the Advanced Manufacturing Laboratory and BioMEMS and Lab-on-a-Chip Laboratory, Drexel University. He continued Post-Doctoral Research in electrical and computer engineering with the VT-MEMS Laboratory, Virginia Tech, Blacksburg, VA, USA and a second postdoctoral research in aerospace and mechanical engineering at the University of Notre Dame. His research interests are BioMEMS and microfluidics, and additive manufacturing. He received the Outstanding Promise Award upon graduation of Ph.D. from Drexel University.
Brittni M. Foster received her M.S. in Chemistry from Western Carolina in 2016. Currently she is a PhD candidate at Wake Forest School of Medicine in the Molecular Medicine and Translational Science program. Her research focuses on prostate cancer metastasis to bone and platelet-tumor interactions.
Carl D. Langefeld, Ph.D. is a Professor in the Department of Biostatistics and Data Science, Division of Public Health Sciences, Wake Forest School of Medicine. He is the former Director of the Center for Public Health Genomics, and former Section Head of Statistical Genetics and Bioinformatics. Dr. Langefeld has three broad areas of research: 1) the identification of genomic (genetic, epigenetic, transcriptomic), metabolomic, and microbiome variation that predisposes to traits related to autoimmune disease, cancer, stroke, renal disease, diabetes, and obesity, 2) the development of analytic and bioinformatic methods designed to assist in the identification and interpretation of this variation, and 3) precision medicine. He has a strong interest and history of translational research in minority populations.
Dr. Bethany Kerr works to understand how cancers metastasize or spread to the bone. She utilizes cancer cell biology, live imaging, animal models, and tissue-engineered models to uncover the mechanisms controlling metastasis to develop new treatments to prevent the spread of cancer. She is an Assistant Professor at the Wake Forest School of Medicine in the Departments of Cancer Biology, Orthopaedic Surgery, Biomedical Engineering, and Urology and is a member of the Wake Forest Baptist Comprehensive Cancer Center. She has received several awards for her academic work and has been funded by an NIH F32 Individual NRSA, an NIH K99/R00 Pathway to Independence Award, and a Pardee Foundation Award. Dr. Kerr has a B.E. double majoring in Biomedical Engineering and Chemical Engineering from Vanderbilt University and a Ph.D. in Regenerative Medicine from Thomas Jefferson University.
Masoud Agah is a Virginia Microelectronics Consortium Professor in the Bradley Department of Electrical and Computer Engineering with a courtesy appointment in the Department of Mechanical Engineering at Virginia Tech. In 2005, he established the VT MEMS Laboratory and has focused his research on environmental and biomedical applications of MEMS. Dr. Agah received the National Science Foundation CAREER Award in 2008 for his research on micro gas chromatography and the Virginia Tech’s College of Engineering Outstanding New Assistant Professor Award in 2009. He is a senior member of the Institute of Electrical and Electronic Engineers (IEEE), as well as its Electron Devices and its Solid-State Circuits Societies. He received his B.S. and M.S. degrees in electrical engineering from Sharif University of Technology (SUT), Iran, in 1996 and 1998, respectively, and his Ph.D. degree from the University of Michigan, Ann Arbor, in 2005.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen H-C, Boyden chamber assay, in Cell migration. 2005, Springer; p. 15–22. [DOI] [PubMed] [Google Scholar]
- 2.Jonkman JE, et al. , An introduction to the wound healing assay using live-cell microscopy. Cell adhesion & migration, 2014. 8(5): p. 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang C-C, Park AY, and Guan J-L, In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nature protocols, 2007. 2(2): p. 329. [DOI] [PubMed] [Google Scholar]
- 4.Ramesh A, Pattabhi A, and Ravi M, Assays Used in vitro to Study Cancer Cell Lines. Life Science Research, 2016. 1(01): p. 19–24. [Google Scholar]
- 5.Rodriguez LG, Wu X, and Guan J-L, Wound-healing assay, in Cell Migration. 2005, Springer; p. 23–29. [DOI] [PubMed] [Google Scholar]
- 6.Kramer N, et al. , In vitro cell migration and invasion assays. Mutation Research/Reviews in Mutation Research, 2013. 752(1): p. 10–24. [DOI] [PubMed] [Google Scholar]
- 7.Chaw K, et al. , Multi-step microfluidic device for studying cancer metastasis. Lab on a Chip, 2007. 7(8): p. 1041–1047. [DOI] [PubMed] [Google Scholar]
- 8.Chung S, et al. , Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab on a Chip, 2009. 9(2): p. 269–275. [DOI] [PubMed] [Google Scholar]
- 9.Hosseini Y, et al. , A Silicon-Based Porous Thin Membrane as a Cancer Cell Transmigration Assay. Journal of Microelectromechanical Systems, 2017. 26(2): p. 308–316. [Google Scholar]
- 10.Zervantonakis IK, et al. , Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proceedings of the National Academy of Sciences, 2012. 109(34): p. 13515–13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adamo A, et al. , Microfluidics-based assessment of cell deformability. Analytical chemistry, 2012. 84(15): p. 6438–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bow H, et al. , A microfabricated deformability-based flow cytometer with application to malaria. Lab on a Chip, 2011. 11(6): p. 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guck J, et al. , Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophysical journal, 2005. 88(5): p. 3689–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Vliet K, Bao G, and Suresh S, The biomechanics toolbox: experimental approaches for living cells and biomolecules. Acta materialia, 2003. 51(19): p. 5881–5905. [Google Scholar]
- 15.Babahosseini H, Strobl JS, and Agah M, Microfluidic iterative mechanical characteristics (iMECH) analyzer for single-cell metastatic identification. Analytical Methods, 2017. 9(5): p. 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren X, et al. , Single-cell mechanical characteristics analyzed by multiconstriction microfluidic channels. ACS sensors, 2017. 2(2): p. 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren X, et al. , Kernel-based microfluidic constriction assay for tumor sample identification. ACS sensors, 2018. 3(8): p. 1510–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirmizis D and Logothetidis S, Atomic force microscopy probing in the measurement of cell mechanics. International journal of nanomedicine, 2010. 5: p. 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, et al. , Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PloS one, 2012. 7(10): p. e46609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Q, Park S, and Ma H, Microfluidic micropipette aspiration for measuring the deformability of single cells. Lab on a Chip, 2012. 12(15): p. 2687–2695. [DOI] [PubMed] [Google Scholar]
- 21.Gossett DR, et al. , Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proceedings of the National Academy of Sciences, 2012. 109(20): p. 7630–7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan V, et al. , Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer research, 2011. 71(15): p. 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basiji DA, et al. , Cellular image analysis and imaging by flow cytometry. Clinics in laboratory medicine, 2007. 27(3): p. 653–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh D, et al. , Microfluidics for flow cytometric analysis of cells and particles. Physiological measurement, 2005. 26(3): p. R73. [DOI] [PubMed] [Google Scholar]
- 25.Khan SS, Solomon MA, and McCoy JP Jr, Detection of circulating endothelial cells and endothelial progenitor cells by flow cytometry. Cytometry Part B: Clinical Cytometry: The Journal of the International Society for Analytical Cytology, 2005. 64(1): p. 1–8. [DOI] [PubMed] [Google Scholar]
- 26.Spitzer MH and Nolan GP, Mass cytometry: single cells, many features. Cell, 2016. 165(4): p. 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babahosseini H, et al. , The impact of sphingosine kinase inhibitor-loaded nanoparticles on bioelectrical and biomechanical properties of cancer cells. Lab on a Chip, 2016. 16(1): p. 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, et al. , Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab on a Chip, 2011. 11(18): p. 3174–3181. [DOI] [PubMed] [Google Scholar]
- 29.Cheung KC, et al. , Microfluidic impedance- based flow cytometry. Cytometry Part A, 2010. 77(7): p. 648–666. [DOI] [PubMed] [Google Scholar]
- 30.Song H, et al. , A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab on a Chip, 2013. 13(12): p. 2300–2310. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, et al. , A microfluidic system enabling continuous characterization of specific membrane capacitance and cytoplasm conductivity of single cells in suspension. Biosensors and Bioelectronics, 2013. 43: p. 304–307. [DOI] [PubMed] [Google Scholar]
- 32.Ren X, et al. , Biophysical phenotyping of cells via impedance spectroscopy in parallel cyclic deformability channels. Biomicrofluidics, 2019. 13(4): p. 044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D, et al. , Biophysical phenotyping of single cells using a differential multiconstriction microfluidic device with self-aligned 3D electrodes. Biosensors and Bioelectronics, 2019. 133: p. 16–23. [DOI] [PubMed] [Google Scholar]
- 34.Campbell CE, et al. , Monitoring viral-induced cell death using electric cell–substrate impedance sensing. Biosensors and Bioelectronics, 2007. 23(4): p. 536–542. [DOI] [PubMed] [Google Scholar]
- 35.Giaever I and Keese CR, Monitoring fibroblast behavior in tissue culture with an applied electric field. Proceedings of the National Academy of Sciences, 1984. 81(12): p. 3761–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szulcek R, Bogaard HJ, and van Nieuw Amerongen GP, Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. JoVE (Journal of Visualized Experiments), 2014(85): p. e51300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran TB, et al. , Electrical dual-sensing method for real-time quantitative monitoring of cell-secreted MMP-9 and cellular morphology during migration process. Biosensors and Bioelectronics, 2016. 77: p. 631–637. [DOI] [PubMed] [Google Scholar]
- 38.Zudaire E, et al. , The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. The Journal of clinical investigation, 2008. 118(2): p. 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghassemi P, et al. , Post-enrichment circulating tumor cell detection and enumeration via deformability impedance cytometry. Biosensors and Bioelectronics, 2020. 150: p. 111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thalmann GN, et al. , LNCaP progression model of human prostate cancer: Androgen-independence and osseous metastasis. The Prostate, 2000. 44(2): p. 91–103. [DOI] [PubMed] [Google Scholar]
- 41.Park S, et al. , Morphological differences between circulating tumor cells from prostate cancer patients and cultured prostate cancer cells. PloS one, 2014. 9(1): p. e85264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alibert C, Goud B, and Manneville JB, Are cancer cells really softer than normal cells? Biology of the Cell, 2017. 109(5): p. 167–189. [DOI] [PubMed] [Google Scholar]
- 43.Faria EC, et al. , Measurement of elastic properties of prostate cancer cells using AFM. Analyst, 2008. 133(11): p. 1498–1500. [DOI] [PubMed] [Google Scholar]
- 44.Ketene AN, et al. , The effects of cancer progression on the viscoelasticity of ovarian cell cytoskeleton structures. Nanomedicine: Nanotechnology, Biology and Medicine, 2012. 8(1): p. 93–102. [DOI] [PubMed] [Google Scholar]
- 45.Lekka M, Discrimination between normal and cancerous cells using AFM. Bionanoscience, 2016. 6(1): p. 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lekka M and Pabijan J, Measuring elastic properties of single cancer cells by AFM, in Atomic Force Microscopy. 2019, Springer; p. 315–324. [DOI] [PubMed] [Google Scholar]
- 47.Lekka M, et al. , Cancer cell recognition–mechanical phenotype. Micron, 2012. 43(12): p. 1259–1266. [DOI] [PubMed] [Google Scholar]
- 48.Park S and Lee YJ, AFM-based dual nano-mechanical phenotypes for cancer metastasis. Journal of biological physics, 2014. 40(4): p. 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastatas L, et al. , AFM nano-mechanics and calcium dynamics of prostate cancer cells with distinct metastatic potential. Biochimica et Biophysica Acta (BBA)-General Subjects, 2012. 1820(7): p. 1111–1120. [DOI] [PubMed] [Google Scholar]
- 50.Liu C-Y, et al. , Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget, 2015. 6(18): p. 15966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.