Abstract
BACKGROUND.
Treatment of prostate cancer (PCa) may be improved by identifying biological mechanisms of tumor growth that directly impact clinical disease progression. We investigated whether genes associated with a highly tumorigenic, drug resistant, progenitor phenotype impact PCa biology and recurrence.
METHODS.
Radical prostatectomy (RP) specimens (±disease recurrence, N = 276) were analyzed by qRT-PCR to quantify expression of genes associated with self-renewal, drug resistance, and tumorigenicity in prior studies. Associations between gene expression and PCa recurrence were confirmed by bootstrap internal validation and by external validation in independent cohorts (total N = 675) and in silico. siRNA knockdown and lentiviral overexpression were used to determine the effect of gene expression on PCa invasion, proliferation, and tumor growth.
RESULTS.
Four candidate genes were differentially expressed in PCa recurrence. Of these, low AXIN2 expression was internally validated in the discovery cohort. Validation in external cohorts and in silico demonstrated that low AXIN2 was independently associated with more aggressive PCa, biochemical recurrence, and metastasis-free survival after RP. Functionally, siRNA-mediated depletion of AXIN2 significantly increased invasiveness, proliferation, and tumor growth. Conversely, ectopic overexpression of AXIN2 significantly reduced invasiveness, proliferation, and tumor growth.
CONCLUSIONS.
Low AXIN2 expression was associated with PCa recurrence after RP in our test population as well as in external validation cohorts, and its expression levels in PCa cells significantly impacted invasiveness, proliferation, and tumor growth. Given these novel roles, further study of AXIN2 in PCa may yield promising new predictive and therapeutic strategies.
Keywords: prostate cancer, AXIN2, biomarker, cancer stem-like cells, radical prostatectomy
INTRODUCTION
Prostate cancer is one of the most commonly diagnosed malignancies in men, with over 230,000 men diagnosed in the United States in 2014 [1]. More than 80% of these cases present as localized disease, and a central therapeutic dilemma in this disease state is whom to treat and how aggressively [2]. Currently, radical prostatectomy (RP) is the most common treatment for localized PCa, but up to a third of men experience disease recurrence after surgery [3]. Predictors of recurrence include preoperative prostate-specific antigen (PSA), Gleason score, extracapsular extension, positive surgical margins, seminal vesicle invasion, lymph node involvement, and treatment year; [4] however, there remains significant variability in patients with similar clinical and pathologic characteristics [5]. There is, therefore, a continuing need to identify accurate markers of PCa recurrence that can better direct the use of adjuvant therapies and lead to improved survival and reductions in morbidity.
Molecular markers to help stratify PCa risk have been proposed. Targets from blood (KLK2-KLK3 SNP rs2735839, 17p12 SNP rs4054823) and primary tissue (immunohistochemistry: p53, Ki67, PTEN loss; fluorescent in situ hybridization: MYC amplification, TMPRSS2-ERG chromosome fusion, PTEN deletion; RNA: cell cycle progression score, miRNA predictor, high grade signatures) have been examined [6]. A few of these have been utilized after RP to predict recurrence and have demonstrated promise [7,8]. In general, however, integration of molecular characterization of PCa into clinical decision-making has been challenging and may reflect the need for genes that play a more integral role in the biology of PCa tumor renewal and progression. One recent example of this approach in the advanced disease setting was the report that expression levels of a truncated variant of androgen receptor (ARV7) was associated with resistance to AR inhibition in metastatic PCa [9].
The past decade has witnessed a renewed interest in tumor heterogeneity, specifically the presence of cancer cell populations marked by a self-renewing “stem-like” gene expression, drug resistance, and high serial tumorigenicity in mice. We and others have isolated cell subpopulations with these cancer stem-like properties from prostate tumors using low attachment growth conditions and cell surface markers [10,11]. Notably, the same markers used to identify such cells can play a direct functional role in their aggressive phenotype. For example, CD44 is a cell surface marker that mediates the adhesion of PCa cells to bone marrow derived endothelial cells [12]. CD44high cancer cells with a CSC phenotype possess features indicative of epithelial to mesenchymal transition (EMT), a process by which cells gain migratory and invasive properties [13]. Furthermore, some studies in other malignancies suggest an association between the presence of highly tumorigenic drug resistant subpopulations and tumor grade, degree of invasiveness, and survival outcomes in cancer patients [14].
The precise identity and derivation of highly tumorigenic drug resistant cells remains the subject of intense study. We and others have shown that cancer cells can exhibit phenotypic plasticity, converting in and out of a tumorigenic, drug-resistant state [15–17]. Based on these insights—the existence of highly tumorigenic drug resistant cell subpopulations in prostate tumors, their association with poor outcomes in other malignancies, and the potential plasticity of this phenotype—we hypothesized that PCa recurrence may be associated with tumor mRNA levels of genes putatively linked to the onset of, or reversion to, a highly tumorigenic, drug resistant, progenitor state. In this study we sought to identify such genes, to validate their clinical significance in external cohorts, and to further analyze their biologic role using in vitro and in vivo models of PCa.
MATERIALS AND METHODS
This study met REMARK criteria for analysis of a prognostic biomarker [18].
Discovery Cohort
We identified 1,468 men who underwent RP with bilateral pelvic lymph node dissection for localized PCa at the University of Southern California (USC) in a prospectively maintained Institutional Review Board-approved, clinically annotated PCa tissue repository. All RP specimens in the repository were collected longitudinally at the time of surgery, processed expeditiously within the Department of Pathology, formalin-fixed, paraffin embedded (FFPE), and stored in the bio-specimen bank. The sample selection process for this nested case-control analysis is shown in the CONSORT diagram in Supplementary Figure S1. Patients were excluded if they had neoadjuvant androgen deprivation therapy (ADT), follow-up <5 years, or non-adenocarcinoma pathology. Cases were defined as patients who experienced biochemical or clinical recurrence, and controls consisted of patients without recurrence. Biochemical recurrence was defined as two consecutive PSA rises (interval 3–4 months) above the contemporary undetectable level (<0.3ng/ml from July 1988 to July 1994; <0.05ng/ml from July 1994 to March 2005; and <0.03ng/ml from March 2005 to present) in men with a postoperative undetectable PSA. Clinical recurrence was defined as local disease confirmed by biopsy or distant recurrence confirmed by biopsy or imaging. Controls were frequency-matched to cases by D’Amico risk group, which includes PSA, Gleason score and clinical T stage [19]. The cohort included 276 men (152 cases and 124 controls) with RP specimens that underwent gene expression analysis.
All the FFPE tumor block samples that were selected for this study were reviewed for quality and tumor content by dedicated genitourinary pathologists. Study pathologists ensured that the selected tumor for analysis matched the prior pathology report Gleason grade. Ten micrometer thick sections were obtained from areas with the highest tumor concentration, and micro-dissection was performed with a scalpel under direct visualization to isolate representative tumor tissue estimated to contain ≥80% cancer cells by the pathologist.
Candidate Gene Selection
The selection of genes putatively linked to a highly tumorigenic, drug resistant cancer stem-like phenotype was based on the results of a comprehensive literature search focused on original research articles reporting on cancer stem-like phenotypes and associated gene transcripts in malignancy. The search strategy included the terms PCa, RP, CSCs, and biomarkers. Because the cancer stem-like terminology is used broadly in the literature to connote a functional definition of self-renewal, tumorigenicity, drug resistance, and invasiveness, we aimed to select representative genes in pathways that were repeatedly found to impart these properties in prostate and other cancers. Based on this strategy, we selected 12 frequently cited genes (ALDH1A1, AXIN2, BMI1, CD133, CD44, CTNNB1/β-catenin, ITGA2/integrin α2, NANOG, NKX3-1, NOTCH1, POU5F1, TACSTD2/TROP2).
Discovery Cohort mRNA Expression Levels
Slide sections containing tumor per genitourinary pathologist review were processed in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory at Response Genetics (Los Angeles, CA) for mRNA gene expression analysis by qRT-PCR as previously described [20]. Primer sequences are shown in Supplementary Table SI. Gene expression analysis was performed only on specimens that met pre-designated quality control measures based on threshold expression of housekeeping genes. Study investigators were blinded to the identity of cases and controls. qRT-PCR analyses yielded values that were expressed as ratios between two absolute measurements: the gene of interest and an internal reference gene, β-actin. The threshold expression of β-actin was defined as a threshold cycle (Ct)<30.
Statistical Analysis-Discovery Cohort
Prostate cancer recurrence status was the primary endpoint. Given that gene expression levels were continuous, but the distribution was not normal, the Wilcoxon rank sum two-sample test, recursive partitioning, logistic regression analysis, and bootstrap internal validation were used to measure and confirm associations of gene expression levels with cancer recurrence. In a descriptive exploratory analysis, a classification and regression tree method based on recursive partitioning was used to identify homogenous subgroups for recurrence.
To adjust for multiple testing and control for the false positive rate, bootstrap internal validation was performed for both the univariable two-sample Wilcoxon rank-sum tests and the multivariable recursive partitioning analyses. One hundred bootstrap samples of 249 observations each were drawn from the original cohort using simple random sampling with replacement. The full set of analyses (univariable Wilcoxon tests and the recursive partitioning) was rerun for each of the 100 bootstrap simulated samples. The number of simulations out of 100 with a significant P value (P < 0.05) in the univariable analysis or selection in the recursive partitioning tree analysis was reported for each gene. For both the univariable and multivariable analyses, the consistency of the initial findings was supported by validation bootstrap analysis when the initially selected transcripts were also selected in greater than 50% of the bootstrap samples.
After AXIN2 was identified as significantly associated with PCa recurrence, this gene expression was evaluated for an association with lymphovascular invasion in the primary prostatectomy specimen. The comparison was made with the Kruskal–Wallis test.
All reported P values were two-sided (P ≤ 0.05). All analyses were performed using the SAS statistical package version 9.2 (SAS Institute Inc., Cary, NC) and the rPART function in Splus 7.0.
External Validation
Clinically annotated RP cohorts with gene expression information from the Affymetrix Human Exon 1.0 ST Microarray were used to test for univariable and multivariable associations between AXIN2 expression and biochemical recurrence (BCR) or metastasis-free survival (MFS) from two previously published databases [21,22]. The Single Channel Array Normalization (SCAN) algorithm was used to normalize and summarize the expression data at the core transcript cluster level [23]. The expression of the transcript cluster (3767480) mapping to AXIN2 was used in this analysis. The Thomas Jefferson University PCa Database (TJU) included 130 patients who underwent RP with high-risk pathologic features (pT3 or positive surgical margins) that received adjuvant radiotherapy [21]. There were 51 (39%) patients who experienced BCR. In patients with undetectable PSA prior to radiotherapy, BCR was defined as a PSA ≥0.4ng/dl. For those with detectable PSA prior to radiation therapy, recurrence was defined as three consecutive increases in PSA over at least 6 weeks. The Mayo Clinic (MC) Radical Prostatectomy Registry included 545 high-risk PCa patients after RP [22]. There were 388 (71%) who experienced BCR and 212 (39%) who developed distant metastasis. Biochemical recurrence was defined as two successive increases in PSA above 0.02ng/ml with the subsequent measurement 0.05 ng/ml above the first measurement.
Kaplan Meier curves were used to estimate BCR free survival and MFS, where the patient populations for comparison are defined by AXIN2 expression being above or below the median AXIN2 expression. Wilcoxon rank sum test and multivariable logistic regression were used to compare expression differences between the patients with and without BCR and with and without distant metastatic recurrence. For both of the databases, multivariable analysis included Gleason score (<8 or ≥8), pathologic extracapsular extension (pT3a), pathologic seminal vesicle invasion (pT3b), surgical margin status, PSA (≤20 or >20), and administration of ADT. The MC database analysis also included pathologic lymph node involvement and administration of radiotherapy.
The Oncomine microarray data compendium, an online data-mining platform [24], was queried using a variety of search filters, and differential expression of AXIN2 in relevant studies was compiled and annotated for significance using either the statistical measures provided by the Oncomine platform or, in one highly matched case, by re-analyzing the primary data using the t test and Wilcoxon test.
In Vitro Analyses
Cell culture.
DU145, LNCaP, and PC3 PCa cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Omega), penicillin (100 units/ml), and streptomycin (100 μg/ml). Cells were maintained at 37°C, 5% CO2.
Tet-inducible AXIN2 lentiviral system.
A tetracycline-inducible AXIN2 overexpression system was created. AXIN2 was inserted into the BamHI/NotI site of the pLVX-TRE3G-IRES lentivirus [25,26]. PCa cells (105) were co-infected with the Tet3G and AXIN2 lentivirus supplemented with polybrene (8 μg/ml). As a control, cells were co-infected with Tet3G lentivirus and lentivirus containing no insert. The Tet3G lentivirus included a G418 resistance gene while the AXIN2 and empty vector viruses included a puromycin resistance gene. Medium was changed after one day of infection and antibiotic selection with puromycin (0.5 μg/ml) and G418(500 μg/ml) was performed after three days. Single cell clone isolation was performed with DU145 cells, whereas pooled samples were used for LNCaP and PC3 cells. With 400 ng/dl doxycycline treatment, there was 166-fold induction of AXIN2 expression in DU145 cells, 300-fold induction in LNCaP cells, and 150-fold induction in PC3 cells
AXIN2 knockdown.
To establish a robust baseline AXIN2 expression phenotype, cells were treated with 1 μM of 6-bromoindirubin-3’-oxime (BIO), a competitive inhibitor of GSK-3α/β that activates β-catenin signaling, resulting in maximal (dose-dependent) AXIN2 expression. Supplementary Figure S2a demonstrates the dose-responsiveness in DU145 cells. Knockdown of AXIN2 mRNA was accomplished with 50 nM pooled On-Target Plus siRNA (Thermo Scientific, Waltham, MA). The siRNA was placed in the DharmaFECT™ transfection formulation #1 and combined with PCa cells. Transfection was performed for 24 hr with parallel transfection with non-targeting siRNA as a control (siControl). Knockdown efficiency was tested by cell lysis with RNA Bee, RT and qRT-PCR for AXIN2 and is shown in Supplementary Figure S2b–d. AXIN2 mRNA depletion was sustained for at least 5 days.
Cell invasiveness.
Cell invasiveness was measured using BD Biocoat™ Matrigel™ Invasion Chambers (BD Biosciences). DU145 cells were serum-starved for 24 hr, and 5000 cells were placed into the invasion chamber with serum-free media. For AXIN2 knockdown experiments, siRNA transfection was performed 24 hr prior to invasion assays. For AXIN2 overexpression experiments, doxycycline (400 ng/dl) was placed within the invasion chamber. The invasion chambers were placed into wells with RPMI 1640 media supplemented with 10% fetal bovine serum (Omega) and the chemoattractant stromal cell-derived factor 1 (100ng/ml, R&D systems). Cells were allowed to migrate for 48 hr at 37°C and 5% CO2. Cells that did not migrate were removed by scraping the superior portion of the Matrigel with a cotton swab. The migrated cells on the bottom surface of the Matrigel were fixed with 100% methanol for 15 min and stained with Giemsa (Sigma-Aldrich) for 1 hr. The Matrigel was washed with sterile water and dried at room temperature. Migrated cells were counted by microscopy and compared using the paired t test.
Cell proliferation.
Cell numbers were measured using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (Promega) reagent on DU145 cells plated at 1000 cells/well in triplicate in 96 well plates. For AXIN2 knockdown experiments, DU145 cells were re-plated 24 hr after siRNA transfection. For AXIN2 overexpression experiments, doxycycline (400ng/ml) was added at the time of plating and medium was changed every 2 days. At each time point, 20 μl of MTS was added to each well containing 100μl of media. The plate was incubated for 90 min at 37°C and 5% CO2, and absorbance was read at 490 nm using Hidex Multilable Detection Program and MikroWin 2000 was used to analyze the data. Absorbance between groups was compared with the t test.
mRNA expression by qRT-PCR.
Cells were lysed with RNA Bee, and RNA extraction was performed with Zymo Direct-zol™ RNA MiniPrep kit. cDNA was generated by reverse transcription (Takara Prime-Script RT Master Mix). The cDNA was used for qRT-PCR with B-R SYBR Green qRT-PCR supermix (Quanta Biosciences) using Bio-Rad MyiQ single color Real-Time PCR Detection System (Bio-Rad) and Bio-Rad iQ2 (Bio-Rad). The conditions for PCR have been previously described [15]. Expression levels were referenced against β-actin and compared with the paired t test.
Tumor growth.
For all in vivo experiments, NOD-SCID-gamma (NSG) male mice were utilized under a protocol approved by the USC Institutional Animal Care and Use Committee (IACUC). The mice were subcutaneously injected with 5 × 105 DU145 cells suspended in 30 ml of matrigel. AXIN2 overexpression was tested using either DU145 cells infected with the tet-inducible AXIN2 construct (n = 4) or with empty vector (n = 4). All eight mice received doxycycline for the duration of the experiment. AXIN2 knockdown was tested using either DU145 cells that were transfected with anti-AXIN2 siRNA (n = 3) or siControl (n = 3). Mice were inoculated with the cells one day after transfection. Tumors were palpated and measured three times a week for 5 weeks until tumor size met the humane endpoint for sacrifice. Tumor growth (diameter) patterns by arms were compared using a mixed regression model after square root transformation. Tumors were excised, measured, weighed, and compared between arms using the exact Wilcoxon two-sample test.
RESULTS
Identification and Internal Validation of Candidate Biomarkers
Primary tissue mRNA was evaluable in 241 of 276 men (87%), and the cases and controls were similar with regard to median age, D’Amico risk group, and year of surgery (all comparisons P > 0.05, Supplementary Table SII). Univariable analysis (Table I) identified four genes with differential mRNA expression when comparing patients with or without disease recurrence. In patients with recurrence, expression of AXIN2, CD44, and TACSTD2 (TROP2) was lower, whereas expression of POU5F1 (OCT 4) was higher. Recursive partitioning was performed to identify any gene sets that may be associated with recurrence. All 12 genes were tested, and AXIN2, NANOG, and CTNNB1 jointly resulted in a classification and regression tree diagram with cut-points yielding odds ratios as high as 8.46 (Supplementary Fig. S3). Subsequent internal bootstrapping validation (Table I) confirmed four genes from the univariable analysis and one gene, AXIN2, in the multivariable recursive partitioning analysis.
TABLE I.
Discovery Cohort Gene Expression and Validation
| mRNA expression |
Internal bootstrap analysis N=number of bootstrap samples validation |
||||||
|---|---|---|---|---|---|---|---|
| No recurrence |
Cancer recurrence |
Univariate |
Multivariable |
||||
| Gene | Median | Range | Median | Range | P-value | N | N |
| ALDH1A1 | 0.01 | 0.001–45.908 | 0.416 | 0.001–58.435 | 0.82 | 6 | 11 |
| AXIN2 | 2.476 | 0.001–8.812 | 1.558 | 0.001–25.316 | <0.001 | 95 | 71 |
| Bmil | 2.541 | 0.001–13.726 | 2.802 | 0.001–17.036 | 0.34 | 14 | 24 |
| CD133 | 0.001 | 0.001–6.568 | 0.001 | 0.001–3.031 | 0.53 | 10 | 1 |
| CD44 | 1.848 | 0.001–10.102 | 1.381 | 0.001–30.536 | 0.036 | 53 | 19 |
| CTNNB1 | 0.705 | 0.001–2.957 | 0.596 | 0.001–2.611 | 0.28 | 18 | 8 |
| ITGA2 | 0.001 | 0.001–1.789 | 0.001 | 0.001–2.169 | 0.31 | 19 | 0 |
| NANOG | 5.965 | 0.001–31.189 | 3.766 | 0.001–81.742 | 0.22 | 23 | 24 |
| Nkx3 1 | 24.642 | 0.001–109.928 | 27.315 | 3.223–184.399 | 0.36 | 23 | 25 |
| Notch1 | 0.001 | 0.001–0.641 | 0.001 | 0.001–0.572 | 0.3 | 18 | 8 |
| POU5F1 | 0.001 | 0.001–8.145 | 1.005 | 0.001–14.404 | 0.017 | 64 | 48 |
| TACSTD2 | 86.601 | 19.276–251.149 | 65.594 | 8.006–293.531 | 0.008 | 70 | 47 |
For each of the 12 genes tested, median normalized gene expression scores and range are shown for patients with versus without disease recurrence. Genes that were differentially expressed with statistical significance between patients with recurrence versus without are in BOLD (AXIN2, CD44, POU5F1, TACSTD2). Of these, all four remained significant (N > 50) in the univariate bootstrap analysis, and AXIN2 also remained significant in the multivariable analysis.
External Validation of AXIN2
Of the genes tested in the discovery cohort, low AXIN2 expression was found to predict PCa recurrence in univariable, multivariable, and bootstrap analyses. Therefore, we selected this gene for further analysis in external clinically annotated RP databases. Examination of the TJU database demonstrated that low expression of AXIN2 was associated with biochemical recurrence (BCR) free survival in a univariable analysis (P = 0.026) (Fig. 1a). The MC database was used to measure associations between AXIN2 and the outcomes BCR free survival and metastasis free survival (MFS) [22]. Low AXIN2 expression was significantly associated with BCR free survival and MFS on univariable analysis (P = 0.041 and 0.044, respectively) (Fig. 1b and c).
Fig. 1.
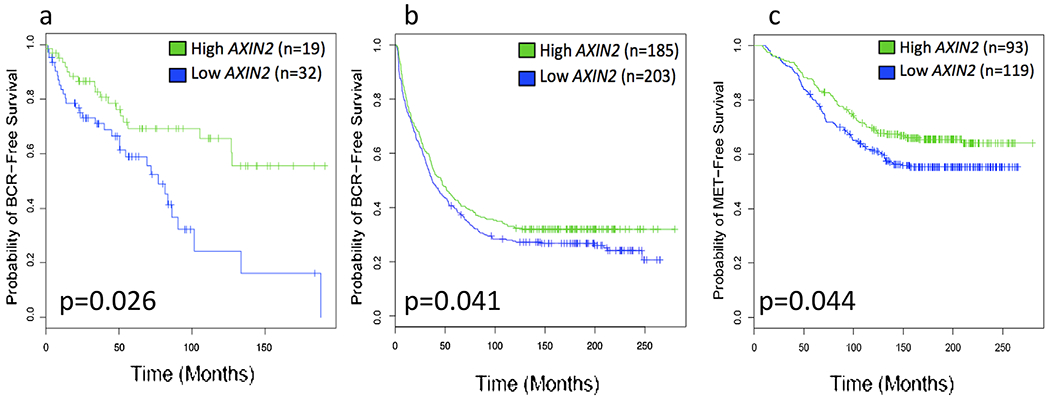
Association between AXIN2 expression and clinical outcomes in external validation prostatectomy cohorts. Kaplan Meier curves for (a) biochemical recurrence-free survival in Thomas Jefferson University cohort, (b) biochemical recurrence-free survival in Mayo Clinic cohort, and (c) distant metastasis-free survival in Mayo Clinic cohort.
The results of the multivariable analysis of the external RP cohorts are shown in Table II. In the TJU cohort, low AXIN2 was associated with BCR (P = 0.020), even after adjusting for clinical variables known to impact recurrence risk (e.g., Gleason score, pathologic grade, PSA, and treatment). Similarly, in the MC database, low AXIN2 was independently associated with distant metastasis (P = 0.015) after adjusting for the same covariates. In the MC database, low AXIN2 was not found to be independently associated with risk of BCR (P = 0.12, data not shown).
TABLE II.
Multivariable Analyses of AXIN2 in Validation Cohorts
| Thomas Jefferson University —Biochemical recurrence |
Mayo Clinic Registry—Distant metastasis |
||||
|---|---|---|---|---|---|
| OR (97.5%CI) | P-value | OR (97.5%CI) | P-value | ||
| AXIN2 expression | 0.13 (0.02–0.67) | 0.020 | AXIN2 expression | 0.33 (0.12–0.83) | 0.015 |
| Gleason 7 | 3.29 (0.75–23.11) | 0.15 | Gleason 7 | 1.90 (0.71–5.87) | 0.23 |
| Gleason ≥8 | 5.71 (1.20–42.32) | 0.046 | Gleason ≥8 | 4.90 (1.77–15.55) | 0.0037 |
| pT3a | 2.06 (0.63–7.66) | 0.025 | pT3a | 1.07 (0.66–1.73) | 0.78 |
| pT3b | 1.44 (0.59–3.53) | 0.42 | pT3b | 1.45 (0.86–2.45) | 0.16 |
| Lymph node positive | 1.23 (0.63–2.39) | 0.55 | |||
| Positive surgical margins | 1.00 (0.37–2.76) | 0.99 | Positive surgical margins | 0.56 (0.34–0.90) | 0.019 |
| PSA 10–20 | 1.44 (0.49–4.25) | 0.51 | PSA 10–20 | 0.59 (0.32–1.05) | 0.073 |
| PSA > 20 | 2.36 (0.66–9.25) | 0.20 | PSA > 20 | 0.57 (0.32–1.01) | 0.056 |
| Administration of ADT | 2.71 (1.02–7.47) | 0.048 | Administration of ADT | 16.78 (9.31–31.90) | <0.001 |
| Administration of radiotherapy | 4.05 (2.36–7.12) | <0.001 | |||
Association of low AXIN2 expression with clinical outcomes (left, TJU: biochemical recurrence; right, MC: distant metastasis), after adjustment for other clinical variables known to impact recurrence.
We interrogated the Oncomine microarray compendium with progressively broad search filters. We found one highly similar study with gene expression data on patients with cancer recurrence from prostatectomy specimens [27]. We re-analyzed the raw data from this study, and univariable analysis revealed that low AXIN2 was associated with PCa recurrence (Table III, panel 1) [27]. Five studies were identified with an association between AXIN2 expression and pathologic features in localized and locally advanced PCa (Table III, panel 2). [27–31] These studies allowed for seven comparisons between expression in high and low Gleason scores as well as between positive and negative lymph nodes. In all seven comparisons, AXIN2 expression was lower in more advanced disease, and this was statistically significant in four of these. In light of this observation, we conducted a post-hoc analysis within the original USC discovery cohort and found that low AXIN2 was significantly associated with greater risk of lymphovascular invasion in the prostatectomy specimen (P = 0.023, data not shown). Lastly, Oncomine was queried for AXIN2 expression in metastatic tissue compared with the primary tumor (Table III, panel 3). [27,29,31–35] AXIN2 levels were lower in metastatic tissue relative to primary tumor in six of the seven studies and the expression was significantly lower in four of these.
TABLE III.
AXIN2 In Silico Analysis
| Panel 1. Re-analysis of prostate samples from Taylor et al [27]. Clinical criteria for sample selection: radical prostatectomy, no neoadjuvant or adjuvant hormones. Search filter: AXIN2, prostate cancer, recurrence versus no recurrence, mRNA, gene rank top 10%ile | ||||
|---|---|---|---|---|
| [27] | Recurrence (N = 32) | Expression mean (range): −0.057 (−0.165, 0.051) | P-value 0.003 (t-test) | |
| No recurrence (N = 102) | Expression mean (range): 0.102 (0.053, 0.150) | |||
| Recurrence (N = 32) | Expression median (range): −0.129 (−0.706, 0.572) | P-value 0.008 (Wilcoxon) | ||
| No recurrence (N = 102) | Expression median (range): 0.101 (−0.583, 0.651) | |||
| Panel 2. Search filter: AXIN2, prostate cancer, pathology subtype, mRNA, gene rank top 10%ile | ||||
| Ref | Description (N) | Fold Change | Gene Rank | P-value |
| [27] | Gleason Score 9 (7) versus 8 (8)versus 7 (74) versus 6 (41) | −0.235 | 590 | 0.007 |
| Lymph node+ (6) versus Lymph node— (102) | −1.223 | 1160 | 0.01 | |
| [28] | Stage 4 (3) versus Stage 3 (23) versus Stage 2 (20) | −0.266 | 1838 | 0.037 |
| [29] | Gleason Score 9 (5) versus 8 (10) versus 7 (18) versus 6 (23) | −0.0101 | 3160 | 0.231 |
| Lymph node+ (5) versus Lymph node— (48) | −1.048 | 5127 | 0.395 | |
| [30] | Gleason Score 7 (7) versus 6 (3) | −1.947 | 2211 | 0.086 |
| [31] | Gleason Score 9 (15) versus 6 (12) | −1.095 | 4237 | 0.034 |
| Panel 3. Search filter: AXIN2, prostate cancer, metastasis versus primary, mRNA, gene rank top 10%ile | ||||
| Ref | Description (N) | Fold Change | Gene Rank | P-value |
| [32] | Metastasis (35) versus Primary (59) | −3.288 | 1465 | 2.6 × 10−9 |
| [29] | Metastasis (7) versus Primary (57) | −1.758 | 961 | 0.005 |
| [27] | Metastasis (19) versus Primary (131) | −1.212 | 1776 | 0.007 |
| [33] | Metastasis (6) versus Primary (7) | −3.448 | 68 | 2.3 × 10−7 |
| [34] | Metastasis (17) versus Primary (10) | 1.622 | 1060 | 2.8 × 10−8 |
| [31] | Metastasis (5) versus Primary (27) | −1.091 | 2364 | 0.07 |
| [35] | Metastasis (12) versus Primary (23) | −1.217 | 3601 | 0.133 |
Association of low AXIN2 with clinical outcome defined as recurrence versus no recurrence (Panel 1, re-analysis of raw data), pathologic features (Panel 2, Gleason score and lymph node involvement), and disease dissemination (Panel 3, metastasis versus primary tumor).
AXIN2 Effects on Prostate Cancer Cells In Vitro and In Vivo
We evaluated the impact of AXIN2 on cell invasiveness using an in vitro matrigel invasion chamber assay. In DU145 and PC3 cells, AXIN2 mRNA expression was found to be significantly lower in invasive than in non-invasive cells (Fig. 2a). Consistent with this, siRNA depletion of AXIN2 increased cell invasiveness (Fig. 2b), whereas lentiviral overexpression of AXIN2 reduced invasiveness (Fig. 2c). A similar reduction in invasiveness was observed with lentiviral AXIN2 overexpression in LNCaP cells (from 140 cells down to 40 cells, P < 0.05, data not shown). Ectopic overexpression of AXIN2 in DU145 cells resulted in reduced cell proliferation whereas siRNA depletion of AXIN2 resulted in greater cell proliferation (Supplementary Fig. S4a, b). In vivo, AXIN2- depleted DU145 cells formed tumors that grew at significantly faster rates than controls (Fig. 3a, P = 0.016) and trended towards greater final wet weights (Fig. 3c, P = 0.20). Conversely, AXIN2-over-expressing cells formed tumors that grew at significantly slower rates than controls (Fig. 3b, P = 0.008) and formed tumors that had significantly lower wet weights (Fig. 3d, P = 0.029).
Fig. 2.
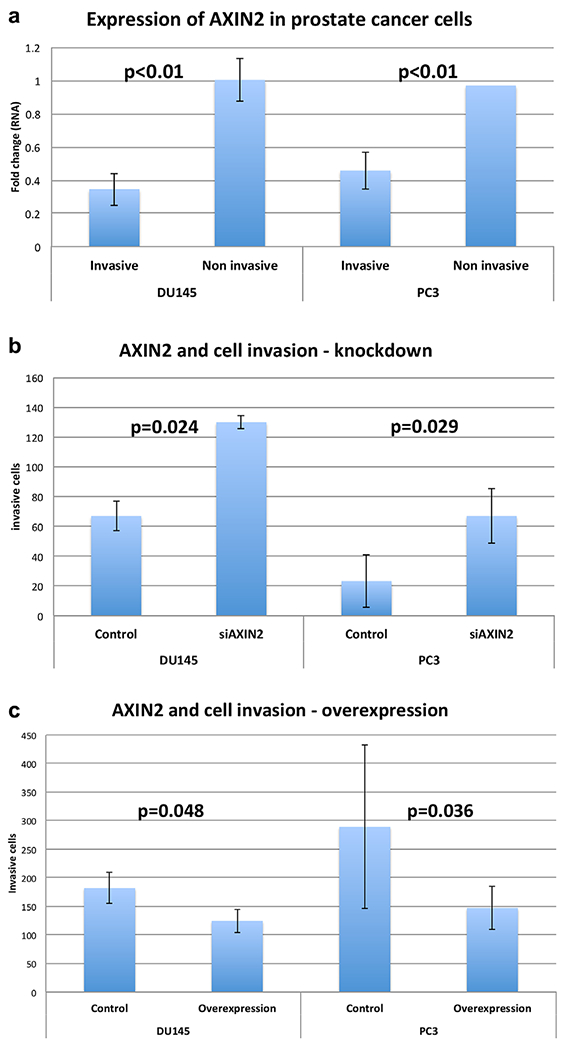
AXIN2 expression and cell invasiveness in PCa cell lines. a: AXIN2 expression in invasive versus non-invasive PCa cells expressed as fold change from unselected parental cell line; b: Impact of AXIN2 knockdown on PCa cell invasion; c: Impact of AXIN2 overexpression on PCa cell invasion.
Fig. 3.
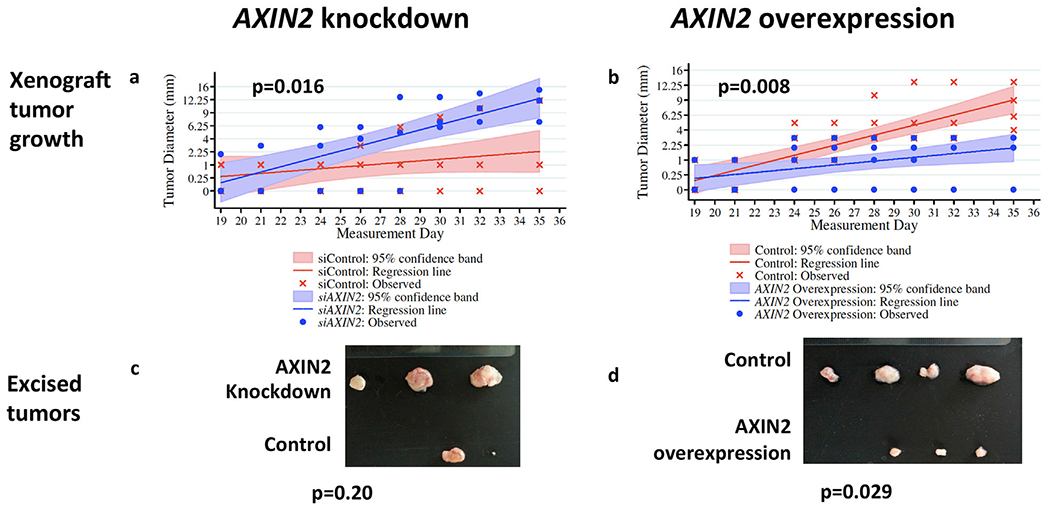
Effect of AXIN2 expression on DUI45 PCa xenograft tumor growth. a-b: Growth (square root transformed) of tumors formed by AXIN2-depleted cells (a) or AXIN2-overexpressing cells (b); c-d: Final wet weights of tumor formed from AXIN2-depleted cells (c) or AXIN2-overexpressing cells (d).
DISCUSSION
The high prevalence of PCa compared with the relatively low risk of PCa death presents a central dilemma of whom to treat and with what therapies. Currently, externally validated nomograms based on clinical and pathologic factors facilitate risk stratification, though they are far from definitive [4]. Recently, gene expression studies have yielded panels of genes associated with disease progression and whose clinical utility is being assessed in prospective validation cohorts [22,38]. In this study, we have taken a different but complementary strategy: rather than an agnostic search, we pursued a hypothesis-driven approach that leverages existing mechanistic insights to identify genes that may serve not only as biomarkers but also as potential therapeutic targets with known function in PCa progression. Specifically, we and others have shown that prostate tumors—like other malignancies —contain subpopulations of cells marked by highly tumorigenic, drug resistant, cancer stem-like properties [10,11]. Recently, some genes associated with this phenotype were studied in PCa models, but with limited translation of in vitro findings to clinically meaningful endpoints [36,39]. Our study began by examining a group of these genes in large cohorts to assess their clinical value, followed by experiments to identify a direct functional role in PCa biology.
Using mRNA extracted from 276 RP specimens in the USC discovery cohort, we identified low AXIN2 expression as predicting PCa recurrence after RP in univariable, multivariable, and bootstrap modeling analyses. We assessed the predictive value of AXIN2 in several external cohorts analyzed by gene expression microarrays and found low AXIN2 expression was independently prognostic of BCR even when adjusting for standard clinical and pathologic variables, thus emerging as a robust predictor in this population. Notably, all patients in the TJU RP cohort also received adjuvant radiotherapy, suggesting a possible role for AXIN2 in modulating radioresistance; indeed, such a role has been ascribed previously to prostate cancer cells marked by a cancer stem-like phenotype [40]. The association of AXIN2 expression with recurrence was further bolstered by external validation in a second cohort, the raw Taylor et al. [27] data extracted from the Oncomine compendium, wherein low AXIN2 expression was associated with significantly higher rates of PCa recurrence. Several additional cohorts extracted from Oncomine similarly demonstrated a significant association between low AXIN2 and high Gleason score and lymph node involvement portending local progression.
Our findings suggest that AXIN2 expression may have a significant association not only with local recurrence, but also with disease progression across the spectrum of PCa. The strongest evidence was observed in the MC cohort, where AXIN2 was independently predictive of distant metastasis. This result corresponds with our Oncomine analysis wherein AXIN2 levels were lower in metastatic tissue when compared with primary tumors. The relevance of AXIN2 across disease states suggests that, rather than signifying the presence of a distinct cancer cell subpopulation ("cancer stem cells" per se), its expression level may potentiate a more aggressive cancer stem-like phenotype along multiple states of cancer progression ranging from localized to metastatic disease. Several studies have demonstrated an association between "stem-like" or "progenitor-like" gene expression patterns and survival in solid malignancies [37] though only a few in PCa [41,42]. In a microarray analysis of 281 PCa specimens from a Swedish watchful-waiting cohort, patients were classified on the basis of their mRNA microarray signature profile, and those with tumors manifesting stem-like signatures together with p53 and PTEN inactivation had the worst survival [43]. Another study examined RP specimens from patients treated with or without neoadjuvant docetaxel and found that differential expression of EMT markers reflecting a cancer stem-like phenotype was associated with poor outcomes [41]. In a third report, a 3-gene embryonal stem cell signature from PCa biopsy specimens helped predict overall survival [42]. In aggregate, these data support the hypothesis that expression patterns of genes associated with a stem-like phenotype can be used for molecular profiling and identification of high-risk patients.
We determined that low expression of AXIN2, singly, was predictive of disease recurrence using the REMARK criteria for evaluation of biomarkers. Functionally, we demonstrated that depletion of AXIN2 levels in PCa cells significantly promoted invasiveness, proliferation, and tumor growth, whereas these phenotypes were attenuated by AXIN2 overexpression. While it is well-recognized as a key component in Wnt/β-catenin signaling and hence in carcinogenesis, stem cell renewal and EMT [44], more than one function has been ascribed to AXIN2 in various malignancies. AXIN2’s canonical role is as a negative regulator of Wnt signaling and thus a tumor suppressor: It is a transcriptional target of b-catenin and encodes the AXIN2 protein, which acts as a scaffold that promotes the assembly of a multiprotein complex composed of adenomatous polyposis coli (APC), the serine/threonine glycogen synthase kinase (GSK)-3β, CK1, Axin1, and β-catenin. Once associated with this complex, AXIN2 efficiently supports the (GSK)-3β-dependent phosphorylation of b-catenin, which subsequently marks β-catenin for β-TrCP-dependent ubiquitination and proteosomal degradation [45]. Consistent with this tumor suppressive role, mutations in AXIN2 in colorectal cancer led to increased β-catenin activity and defective mismatch repair [46], and therapeutics to increase AXIN2 levels by targeting tankyrase-1/2 have been proposed [45,47]. In contrast to this canonical tumor-suppressive role, AXIN2 was recently found to have a potential tumor-promoting role through direct stabilization of Snail1 in colorectal [48] and breast cancer [49]; hence, a comprehensive picture of AXIN2’s role in these malignancies is still emerging.
In PCa, fewer AXIN2 mechanistic studied have been performed, but most suggest a role consistent with its more established, canonical tumor suppressive function: one study demonstrated that the AXIN2 protein co-localizes with prostate progenitor cells in murine prostate models [50], and at least two reports have identified single nucleotide polymorphisms in AXIN2 coding regions associated with an increased incidence of PCa, possibly by disrupting AXIN2-β-catenin complex binding [51,52]. While by no means conclusive, these PCa studies complement our own findings to support a hypothesis wherein dysregulation of AXIN2, whether from low expression or a mutation, can disrupt β-catenin degradation and increase nuclear translocation and transcriptional activity, thereby potentiating an aggressive phenotype marked by proliferation, invasiveness, and tumor formation.
CONCLUSION
Using a discovery cohort and several validation cohorts, we found that expression of AXIN2, a transcriptional target and protein partner of β-catenin, is significantly associated with prostate cancer outcomes ranging from clinical and biochemical recurrence to metastasis-free survival and progression. Moreover, we showed that depletion of AXIN2 mRNA in prostate cancer cells leads to increased proliferation, invasion, and tumor formation, whereas its ectopic overexpression has opposite effects. Therefore, the clinical and biologic roles of AXIN2 in prostate cancer merit further study, which may lead to valuable new predictive and therapeutic strategies in this disease.
Supplementary Material
Acknowledgments
Grant sponsor: NCI; Grant number: 5 P30CA014089-39.
Footnotes
Conflict of interest statement: The authors disclose no potential conflicts of interest.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- 1.SEER Cancer Statistics Factsheets: Prostate Cancer. National Cancer Institute; Bethesda, MD: www.seer.cancer.gov/statfacts/html/prost.html [Google Scholar]
- 2.Li J, Djenaba JA, Soman A, Rim SH, Master VA. Recent trends in prostate cancer incidence by age, cancer stage, and grade, the United States, 2001–2007. Prostate Cancer 2012;2012:691380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol 2003;169(2):517–523. [DOI] [PubMed] [Google Scholar]
- 4.Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ Jr., Dotan ZA, DiBlasio CJ, Reuther A, Klein EA, Kattan MW. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol 2005;23(28):7005–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephenson AJ, Kattan MW, Eastham JA, Bianco FJ Jr., Yossepowitch O, Vickers AJ, Klein EA, Wood DP, Scardino PT. Prostate cancer-specific mortality after radical prostatectomy for patients treated in the prostate-specific antigen era. J Clin Oncol 2009;27(26):4300–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choudhury AD, Eeles R, Freedland SJ, Isaacs WB, Pomerantz MM, Schalken JA, Tammela TL, Visakorpi T. The role of genetic markers in the management of prostate cancer. Eur Urol 2012;62(4):577–587. [DOI] [PubMed] [Google Scholar]
- 7.Haldrup C, Mundbjerg K, Vestergaard EM, Lamy P, Wild P, Schulz WA, Arsov C, Visakorpi T, Borre M, Hoyer S, Orntoft TF, Sorensen KD. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. J Clin Oncol 2013;31 (26):3250–3258. [DOI] [PubMed] [Google Scholar]
- 8.Cooperberg MR, Simko JP, Cowan JE, Reid JE, Djalilvand A, Bhatnagar S, Gutin A, Lanchbury JS, Swanson GP, Stone S, Carroll PR. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol 2013;31(11):1428–1434. [DOI] [PubMed] [Google Scholar]
- 9.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, Lotan TL, Zheng Q, De Marzo AM, Isaacs JT, Isaacs WB, Nadal R, Paller CJ, Denmeade SR, Carducci MA, Eisenberger MA, Luo J. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Eng J Med 2014;371(11):1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu T, He K, Wang L, Goldkorn A. Prostate tumor cells with cancer progenitor properties have high telomerase activity and are rapidly killed by telomerase interference. Prostate 2011;71 (13):1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65(23):10946–10951. [DOI] [PubMed] [Google Scholar]
- 12.Desai B, Rogers MJ, Chellaiah MA. Mechanisms of osteopontin and CD44 as metastatic principles in prostate cancer cells. Mol Cancer 2007;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133(4):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008;40(5):499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He K, Xu T, Goldkorn A. Cancer cells cyclically lose and regain drug-resistant highly tumorigenic features characteristic of a cancer stem-like phenotype. Mol Cancer Ther 2011;10(6):938–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He K, Xu T, Xu Y, Ring A, Kahn M, Goldkorn A. Cancer cells acquire a drug resistant, highly tumorigenic, cancer stem-like phenotype through modulation of the PI3K/Akt/beta-catenin/ CBP pathway. Int J Cancer 2014;134(1):43–54. [DOI] [PubMed] [Google Scholar]
- 17.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA 2011;108(19):7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97(16):1180–1184. [DOI] [PubMed] [Google Scholar]
- 19.D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Schnall M, Tomaszewski JE, Wein A. Combined modality staging of prostate carcinoma and its utility in predicting pathologic stage and postoperative prostate specific antigen failure. Urology 1997;49(3A Suppl):23–30. [DOI] [PubMed] [Google Scholar]
- 20.Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, Sanchez JJ, Danenberg KD, Danenberg PV, Rosell R. Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 2002;8(7):2286–2291. [PubMed] [Google Scholar]
- 21.Den RB, Feng FY, Showalter TN, Mishra MV, Trabulsi EJ, Lallas CD, Gomella LG, Kelly WK, Birbe RC, McCue PA, Ghadessi M, Yousefi K, Davicioni E, Knudsen KE, Dicker AP. Genomic prostate cancer classifier predicts biochemical failure and metastases in patients after postoperative radiation therapy. Int J Radiat Oncol Biol Phys 2014;89(5):1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z, Zimmermann B, Sierocinski T, Ballman KV, Triche TJ, Black PC, Karnes RJ, Klee G, Davicioni E, Jenkins RB. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics 2012;100(6):337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9(2):166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldkorn A, Blackburn EH. Assembly of mutant-template telomerase RNA into catalytically active telomerase ribonucleo-protein that can act on telomeres is required for apoptosis and cell cycle arrest in human cancer cells. Cancer Res 2006;66(11):5763–5771. [DOI] [PubMed] [Google Scholar]
- 26.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 1992;89(12):5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bittner M International Genomics Consortium. www.intgen.org/expo; 2005.
- 29.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004;101(3):811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog 2002;33(1):25–35. [DOI] [PubMed] [Google Scholar]
- 31.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res 2003;63(14):3877–3882. [PubMed] [Google Scholar]
- 32.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487(7406):239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005;8(5):393–406. [DOI] [PubMed] [Google Scholar]
- 34.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007;7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Furihata M, Tsunoda T, Ashida S, Takata R, Obara W, Yoshioka H, Daigo Y, Nasu Y, Kumon H, Konaka H, Namiki M, Tozawa K, Kohri K, Tanji N, Yokoyama M, Shimazui T, Akaza H, Mizutani Y, Miki T, Fujioka T, Shuin T, Nakamura Y, Nakagawa H. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res 2007;67(11):5117–5125. [DOI] [PubMed] [Google Scholar]
- 36.Shi X, Gipp J, Dries M, Bushman W. Prostate progenitor cells proliferate in response to castration. Stem Cell Res 2014;13(1):154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matchett KB, Lappin TR. Concise reviews: cancer stem cells: from concept to cure. Stem Cells 2014;32(10):2563–2570. [DOI] [PubMed] [Google Scholar]
- 38.Knezevic D, Goddard AD, Natraj N, Cherbavaz DB, Clark-Langone KM, Snable J, Watson D, Falzarano SM, Magi-Galluzzi C, Klein EA, Quale C. Analytical validation of the Oncotype DX prostate cancer assay—a clinical RT-PCR assay optimized for prostate needle biopsies. BMC Genomics 2013;14:690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu C, Li Z, Bi L, Li K, Zhou B, Xu C, Huang J, Xu K. NOTCH1 signaling promotes chemoresistance via regulating ABCC1 expression in prostate cancer stem cells. Mol Cell Biochem 2014;393(1–2):265–270. [DOI] [PubMed] [Google Scholar]
- 40.Ni J, Cozzi PJ, Hao JL, Beretov J, Chang L, Duan W, Shigdar S, Delprado WJ, Graham PH, Bucci J, Kearsley JH, Li Y. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 2014;74(6):602–617. [DOI] [PubMed] [Google Scholar]
- 41.Marin-Aguilera M, Codony-Servat J, Reig O, Lozano JJ, Fernandez PL, Pereira MV, Jimenez N, Donovan M, Puig P, Mengual L, Bermudo R, Font A, Gallardo E, Ribal MJ, Alcaraz A, Gascon P, Mellado B. Epithelial-to-mesenchymal transition mediates docetaxel resistance and high risk of relapse in prostate cancer. Mol Cancer Ther 2014;13(5):1270–1284. [DOI] [PubMed] [Google Scholar]
- 42.Peng Z, Skoog L, Hellborg H, Jonstam G, Wingmo IL, Hjalm-Eriksson M, Harmenberg U, Cedermark GC, Andersson K, Ahrlund-Richter L, Pramana S, Pawitan Y, Nister M, Nilsson S, Li C. An expression signature at diagnosis to estimate prostate cancer patients’ overall survival. Prostate Cancer Prostatic Dis 2014;17(1):81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markert EK, Mizuno H, Vazquez A, Levine AJ. Molecular classification of prostate cancer using curated expression signatures. Proc Natl Acad Sci USA 2011;108(52):21276–21281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008;8(5):387–398. [DOI] [PubMed] [Google Scholar]
- 45.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009;461(7264):614–620. [DOI] [PubMed] [Google Scholar]
- 46.Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C, Christensen E, Schmidt SS, Roche PC, Smith DI, Thibodeau SN. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet 2000;26(2):146–147. [DOI] [PubMed] [Google Scholar]
- 47.Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS ONE 2012;7(11):48670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu ZQ, Brabletz T, Fearon E, Willis AL, Hu CY, Li XY, Weiss SJ. Canonical Wnt suppressor, Axin2, promotes colon carcinoma oncogenic activity. Proc Natl Acad Sci USA 2012;109(28):11312–11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 2006;8(12):1398–1406. [DOI] [PubMed] [Google Scholar]
- 50.Ontiveros CS, Salm SN, Wilson EL. Axin2 expression identifies progenitor cells in the murine prostate. Prostate 2008;68(12):1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinarbasi E, Gunes EG, Pinarbasi H, Donmez G, Silig Y. AXIN2 polymorphism and its association with prostate cancer in a Turkish population. Med Oncol 2011;28(4):1373–1378. [DOI] [PubMed] [Google Scholar]
- 52.Ma C, Liu C, Huang P, Kaku H, Chen J, Guo K, Ueki H, Sakai A, Nasu Y, Kumon H, Shimizu K, Watanabe M. Significant association between the Axin2 rs2240308 single nucleotide polymorphism and the incidence of prostate cancer. Oncol Lett 2014;8(2):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


