Abstract
DCs are a critical component of immune responses in cancer primarily due to their ability to cross-present tumor-associated antigens. Cross-presentation by DCs in cancer is impaired, which may represent one of the obstacles for the success of cancer immunotherapies. Here, we report that polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) blocked cross-presentation by DCs without affecting direct presentation of antigens by these cells. This effect did not require direct cell-cell contact and was associated with transfer of lipids. Neutrophils (PMN) and PMN-MDSC transferred lipid to DCs equally well; however, PMN did not affect DC cross-presentation. PMN-MDSC generate oxidatively truncated lipids previously shown to be involved in impaired cross-presentation by DCs. Accumulation of oxidized lipids in PMN-MDSC was dependent on myeloperoxidase (MPO). MPO-deficient PMN-MDSC did not affect cross-presentation by DCs. Cross-presentation of tumor-associated antigens in vivo by DCs was improved in MDSC-depleted or tumor-bearing MPO-KO mice. Pharmacological inhibition of MPO in combination with checkpoint blockade reduced tumor progression in different tumor models. These data suggest MPO-driven lipid peroxidation in PMN-MDSC as a possible non–cell autonomous mechanism of inhibition of antigen cross-presentation by DCs and propose MPO as potential therapeutic target to enhance the efficacy of current immunotherapies for patients with cancer.
Keywords: Immunology, Oncology
Keywords: Antigen presenting cells, Cancer immunotherapy
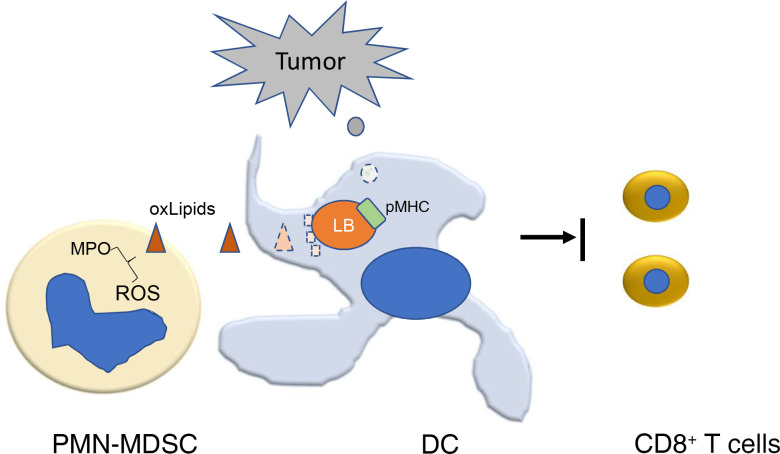
Accumulation of oxidized lipids in polymorphonuclear myeloid-derived suppressor cells may inhibit cross-presenting activity of DCs in cancer.
Introduction
DCs are highly specialized antigen-presenting cells (APC) (1, 2). They can be broadly divided into 3 subsets: classical DCs (cDC), plasmacytoid DCs, and inflammatory DCs. cDCs consist of 2 large groups: cDC type 1 (cDC1) and type 2 (cDC2). cDC1 are key players in the regulation of cancer immune responses due to their ability to cross-present antigens to CD8+ T cells and to generate cytotoxic effector T cell responses (3–6). Their presence is critically important for the success of immunotherapies, such as adoptive T cell transfer and checkpoint blockade (7–9). Defective function of cDC1, which was described in several types of cancer, contributed to ineffective immune responses (3, 5). We and others have found that accumulation of lipids dramatically affected the antigen cross-presentation by cDC1, resulting in an impaired induction of specific antitumor CD8+ T cell responses (10–13). Recent studies demonstrated a specific role of lipid bodies (LB) containing oxidatively truncated species of lipids in negative regulation of cross-presentation by cDC1 (10). However, the source of oxidized lipids in DCs remained unclear, because the machinery for lipid oxidation is largely missing in cDC1.
Myeloid-derived suppressor cells (MDSC) are pathologically activated neutrophils (PMN) and monocytes (Mon) with potent immune-suppressive activity. These cells are vital components of the immunosuppressive network that supports tumor progression and limits the efficacy of cancer therapies (14, 15). MDSC consist of 2 main subsets of cells: monocytic (M-MDSC) and polymorphonuclear (PMN-MDSC) cells. PMN-MDSC are most abundant population of MDSC. We and others have shown that accumulation of lipids in PMN-MDSC is a critical driver of their suppressive functions (16–19). PMN-MDSC are characterized by high production of reactive oxygen species as well as elevated levels of myeloperoxidase (MPO), which may contribute to the formation of various species of oxidized lipids. In this study, we tested the hypothesis that PMN-MDSC could regulate cross-presentation by cDC1.
Results
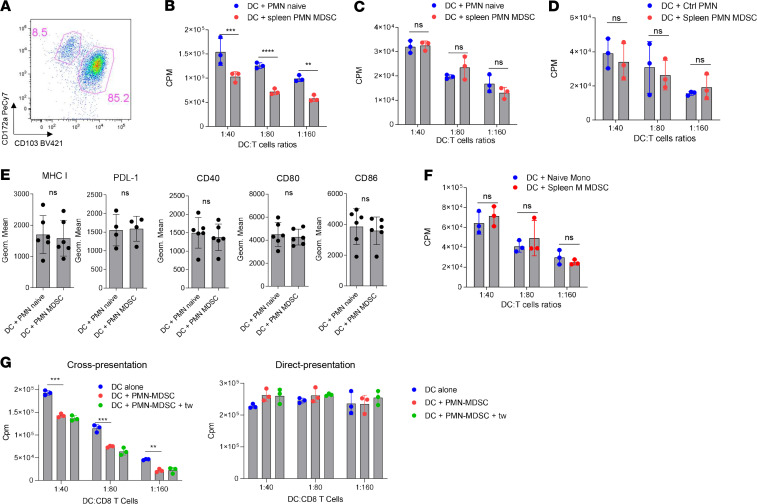
We generated CD172a–CD103+ cDC1 in vitro from BM hematopoietic progenitor cells (HPC) of naive mice with GM-CSF and FLT3L (Figure 1A). cDC1 were cultured with PMN from control mice and PMN-MDSC or M-MDSC from spleens of Lewis lung carcinoma (LLC) tumor-bearing (TB) mice. Immune-suppressive activity of PMN-MDSC was confirmed in an antigen-specific suppression assay (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138581DS1). After 24 hours, cDC1 were isolated from the coculture with MDSC and loaded with the OVA-derived long-peptide (Pam)2-KMFVESIINFEKL, which requires processing and cross-presentation or the OVA-derived short-peptide (SIINFEKL) that directly binds to MHC class I H2Kb (pMHC). Coculture of cDC1 with PMN did not affect cross-presentation or direct presentation of antigens by DCs as compared with DCs cultured alone (Supplemental Figure 2A). In contrast, PMN-MDSC dramatically reduced the ability of cDC1 to present OVA-derived antigens to peptide-specific OT-1 CD8+ T cells (Figure 1B) without impairing the direct presentation of short peptide (Figure 1C). We asked whether coculture with DCs affected the viability of PMN and PMN-MDSC differently. We assessed the number of granulocytes at start of the culture and after 24 hours culture with DCs. No differences in the total number of granulocytes or fold changes from baseline were found (Supplemental Figure 2B). Similarly, no differences from background (DCs cultured alone) were found in the number of DCs or cDC1 after coculture with PMN-MDSC or PMN (Supplemental Figure 2C).
Figure 1. PMN-MDSC inhibit antigen cross-presentation by CD103+ DCs.
(A) Example of gating strategy for the identification of cDC1 (CD172–CD103+ DC) and cDC2 (CD172a+CD103– DC) in cultures of DCs differentiated in presence of GM-CSF and FLT3L. (B) Proliferation of OT1 CD8+ T cells in the presence of CD103+ DCs loaded with OVA-derived long peptides after coculture with PMN or PMN-MDSC (n = 5, each experiment has been performed in triplicate). (C) Proliferation of OT1 CD8+ T cells after stimulation with CD103+ DC loaded with OVA-derived short peptide after coculture with PMN or PMN-MDSC (n = 5, each experiment has been performed in triplicate). (D) Proliferation of allogenic CD8+ T cells after stimulation with CD103+ DCs after coculture with PMN or PMN-MDSC (n = 3, each experiment has been performed in triplicate). (E) Analysis of surface markers by flow cytometry in CD103+ DCs after coculture with PMN (n = 6) or PMN-MDSC (n = 6). (F) Proliferation of OT1 CD8+ T cells after stimulation with CD103+ DCs loaded with OVA-derived long peptides after coculture with monocytes or M-MDSC (n = 3, each experiment has been performed in triplicate). (G) Proliferation of OT1 CD8+ T cells after stimulation with CD103+ DCs loaded with OVA-derived long peptides (left) or short derived peptides (right) after coculture with PMN-MDSC separated in Transwell system. n = 3. (B–D and F and G) Representative experiments are shown. Proliferation was measured by 3[H]-thymidine uptake in triplicates. In all experiments, mean ± SEM is shown. **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired 2-tailed Student’s t test or ANOVA with corrections for multiple comparisons when more than 2 groups were compared.
cDC1 stimulated allogeneic CD8+ T cells equally well after coculture with PMN or PMN-MDSC (Figure 1D). No differences were seen in the expression of molecules involved in T cell stimulation: MHC I, CD40, CD80, CD86, and PD-L1 on the surface of cDC1 cultured with PMN and PMN-MDSC (Figure 1E). M-MDSC did not affect cross-presentation of DCs as compared with control Mon (Figure 1F). Thus, only PMN-MDSC, but not PMN or M-MDSC, selectively inhibited DC cross-presentation. We asked, whether direct cell-cell contact was required for PMN-MDSC–mediated inhibition of DC cross-presentation. Separation of PMN-MDSC and CD103+ cDC1 using semipermeable Transwell (0.4 μm) membrane did not abrogate the negative effect of PMN-MDSC on cross-presentation by cDC1. Direct presentation of peptide was not changed (Figure 1G).
Transwell data suggested that PMN-MDSC affected DC cross-presentation via soluble factors. PMN-MDSC release different cytokines and factors (arginase, S100A8/A9, IL-6, PGE2, etc.) that can inhibit DC function. However, they are known to affect broad functional activity of cells (20–23), rather than selectively affect cross-presentation, as observed in our experiments. We did not find changes in the ability of DCs to activate T cell proliferation after direct presentation of short peptide or stimulation of allogeneic T cell proliferation (a hallmark of DC activity) as well as in the expression of MHC class I and major costimulatory molecules or PD-L1 (Figure 1, C–E).
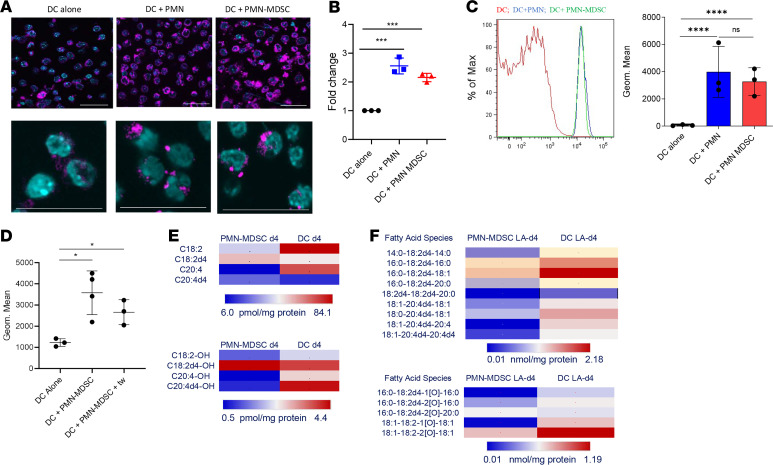
Since, the accumulation of lipids was previously shown to be responsible for the defective antigen cross-presentation by DCs (10) and for the regulation of PMN-MDSC–suppressive functions (18), we hypothesized that PMN-MDSC might inhibit antigen cross-presenting ability of DCs by acting as a source of transferable lipids. To test this hypothesis, we assessed the lipids in cDC1 after coculture with PMN or PMN-MDSC and BODIPY 493/503 staining. By using confocal microscopy (Figure 2A) and flow cytometry (Figure 2B) we found that coculture with PMN or PMN-MDSC caused very substantial but similar accumulation of lipids in cDC1. To confirm these data, we stained PMN or PMN-MDSC with BODIPY 493/503 before the coculture with DC, washed the excess of BODIPY and cultured for 24 hours with DCs. Both PMN and PMN-MDSC were able to transfer lipids to cDC1 (Figure 2C). Transfer of lipids did not require direct cell-cell contact. PMN-MDSC were stained with BODIPY 493/503. Excess of BODIPY was washed away, and cells then were cocultured with cDC1 separated by Transwells (0.4 μm). Transwells did not prevent the transfer of lipids from PMN-MDSC to DCs (Figure 2D).
Figure 2. Lipid transfer from PMN-MDSC to DCs.
(A) Confocal image of lipid bodies (LB) using BODIPY in cDC1 after coculture with PMN or PMN-MDSC. Scale bar: 50 μm. (B) Flow cytometric analysis of lipid content in cDC1 stained with BODIPY after coculture with PMN and PMN-MDSC, expressed as fold change to the geometric mean of Bodipy expression in DCs alone (n = 3). (C) Flow cytometric analysis of lipid transfer from PMN-MDSC to cDC1 after coculture with BODIPY-labeled PMN and PMN-MDSC. Left: representative example of staining. Right: cumulative results of the experiments (n = 3). (D) Flow cytometric analysis of lipid content in cDC1 stained with BODIPY after coculture with PMN-MDSC with or without Transwell system (n = 3). (E) Heatmap showing the content of nonoxidized fatty acids LA, LA-d4, AA, and AA-d4 elongated from LA-d4 (top) and their monooxygenated species (bottom) in PMN-MDSC and DCs after coculture of DCs with PMN-MDSC LA-d4. (F) Heatmap showing the content of nonoxidized TAG molecular species containing LA-d4 and elongated AAd4 (top) and their oxygenated species (bottom) in PMN-MDSC and DCs after coculture of DCs with PMN-MDSC LA-d4. (B–D) In all experiments, mean ± SD is shown. *P < 0.05, ***P < 0.001, ****P < 0.0001, ANOVA test with corrections for multiple comparisons.
To quantitatively characterize the transfer of lipids from PMN-MDSC to DCs, we loaded PMN-MDSC with deuterium labeled linoleic acid (LA-d4) and assessed the incorporation of LA-d4 into different types of lipids in DCs. PMN-MDSC–derived free LA-d4 and its elongation product, free arachidonic acid (AA-d4), were found in DCs as nonoxidized fatty acids as well as their mono-oxygenated species (Figure 2E and Supplemental Figure 3). Moreover, LA-d4 and AA-d4 esterified into nonoxidized triacylglycerols (TAGs), and oxidized TAGs were detected in both PMN-MDSC and, importantly, in DCs (Figure 2F and Supplemental Figure 3). Thus, taken together, these data indicate that although both PMN and PMN-MDSC can transfer lipids to CD103+ cDC1, only PMN-MDSC affected cross-presentation by DCs. This suggested that the nature of lipids released from PMN-MDSC could be a factor that affects cross-presentation.
Previous studies implicated accumulation of LB in defective DC cross-presentation (10, 11). We evaluated whether PMN and PMN-MDSC were able to facilitate accumulation of LB in DCs. Coculture of cDC1 with either PMN or PMN-MDSC resulted in accumulation of LB in cDC1 (Figure 2A), indicating that appearance of large LB was not enough for defective cross-presentation. Previous studies demonstrated that only LB containing oxidatively truncated lipids, particularly TAGs, impaired DC cross-presentation (10). Thus, it appears that the chemical nature, rather than the amounts of accumulated lipids, may have an effect on DC cross-presentation. Therefore, we used quantitative LC-MS analysis to assess the amounts of oxidatively truncated TAGs in PMN and PMN-MDSC. We found that PMN-MDSC from TB mice had markedly higher amounts of oxidatively truncated lipids than PMN from tumor-free mice (Figure 3A). Similar species of lipids accumulated in DCs in the presence of tumor explant supernatants (Supplemental Figure 4), consistent with a previous observation (10).
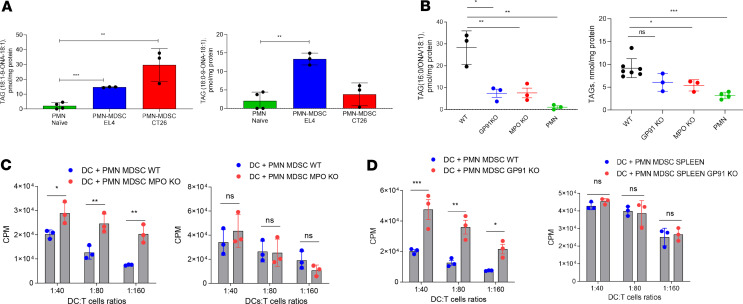
Figure 3. MPO and NADPH oxidase in PMN-MDSC affect cross-presentation by CD103+ DCs.
(A) Content of oxidatively truncated TAG molecular species in PMN-MDSC from indicated tumor-bearing and PMN from tumor-free mice (n = 3). ONA, 9-oxononanoic acid. (B) Content of oxidatively truncated TAG molecular species (left) and total TAGs (right) in PMN-MDSC isolated from spleens of EL4 tumor-bearing WT, GP91-KO, and MPO-KO mice and PMN from tumor-free mice. Please note 1000-fold difference in the scales between left and right graphs in B. (n = 3). (C) Proliferation of OT1 CD8+ T cells after stimulation with CD103+ DCs after coculture with WT PMN-MDSC or MPO-KO PMN-MDSC and loading with OVA-derived long peptides (left) or OVA-derived short peptide (right) (n = 3, each experiment has been performed in triplicate). (D) Proliferation of OT1 CD8+ T cells after stimulation with CD103+ DCs after coculture with WT PMN-MDSC or GP91-KO PMN-MDSC and loading with OVA-derived long peptides (left) or OVA-derived short peptide (right) (n = 3, each experiment has been performed in triplicate). (C and D) Representative experiments are shown. Proliferation was measured by 3[H]-thymidine uptake in triplicates. In all experiments, mean ± SD is shown. (A and B) *P < 0.05, **P < 0.01, ***P < 0.001, 2-sided Student’s t test and ANOVA with correction for multiple comparisons. (C and D) *P < 0.05, **P < 0.01, ***P < 0.001, 2-sided Student’s t test.
PMN-MDSC have efficient oxidative machinery, with MPO and NADPH oxidase as its key components. To determine whether MPO or NADPH oxidase are responsible for the generation of oxidized lipids in PMN-MDSC, we analyzed the lipid profile in PMN-MDSC isolated from spleens of EL4 TB WT, MPO-, or GP91-KO (component of NADPH oxidase complex) mice. We found that the amount of oxidatively truncated TAG was dramatically reduced to essentially the same low levels in both types of KO mice. In contrast, only a modest decrease in the amount of total TAG was detected (Figure 3B).
Next we sought to determine the effect of MPO and GP91 on the ability of PMN-MDSC to block cross-presentation by DCs. We cocultured cDC1 with PMN-MDSC obtained from spleens of LLC and EL4 TB MPO-KO or GP91-KO mice. cDC1 were then loaded with OVA long or short peptides and used to stimulate OT-1 CD8+ T cells. The amount of lipids transferred to cDC1 by WT and KO PMN-MDSC was the same (Supplemental Figure 5A). Coculture with WT PMN-MDSC altered cross-presenting ability of cDC1. However, coculture with PMN-MDSC from MPO-KO or GP91-KO mice did not have an effect cross-presentation of DCs (Figure 3, C and D). The direct presentation of short peptide by cDC1 (Figure 3, C and D) and the expression of MHC I, CD40, CD80, CD86, and PDL-1 molecules on the DC surface were not affected (Supplemental Figure 5B).
Next, we sought to assess the effect of MDSC on DC cross-presentation in vivo by depleting MDSC using anti-DR5 antibody described previously (24, 25). Lung cancer cells with expression of OVA (LLC-OVA) were injected s.c. Two weeks later when tumors reached 0.5 cm, mice were treated with control immunoglobulin or DR5 antibody twice with a 3 day interval. Reduction of PMN-MDSC and M-MDSC in spleens was verified by flow cytometry (Supplemental Figure 6A). Short treatment with DR5 antibody did not significantly affect tumor growth (Supplemental Figure 6B). cDC1 (CD11c+MHCII+CD103+CD11b–CD172a–) and cDC2 (CD11c+MHCII+CD103-CD11b+CD172a+) were isolated from draining lymph nodes (dLNs) and used to stimulate OT-1 CD8+ T cells. cDC1 isolated from mice with MDSC depletion showed a higher ability to stimulate the proliferation of specific CD8+ T cells, compared with cDC1 isolated from untreated mice (Supplemental Figure 6C).
DR5 antibody reduced both populations of MDSC. Although those results were suggestive of potential role of MDSC in DC cross-presentation, we tested more specific mechanisms associated with PMN-MDSC function and formation of oxidized lipids by using MPO-KO mice. We did not test this hypothesis in GP91-KO mice because superoxide is important for the DC cross-presentation and GP91-KO DCs showed defective cross-presentation (26). Thus, it would have made difficult the interpretation of data.
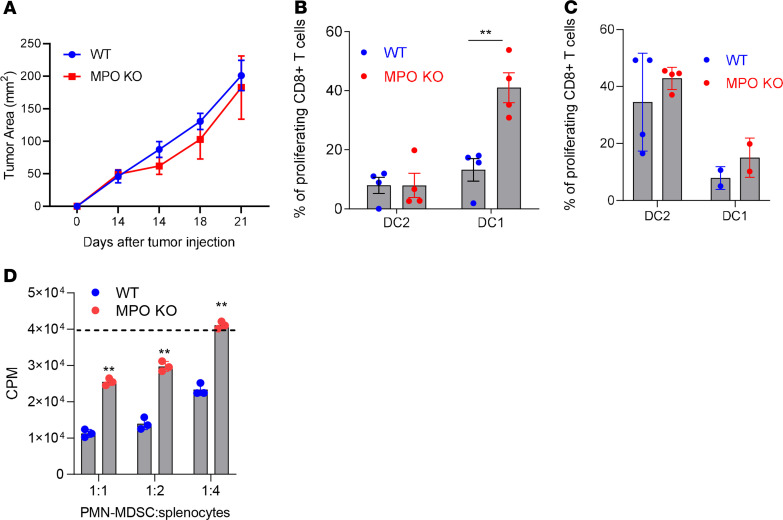
LLC-OVA tumors were established s.c. Two weeks later when tumors reached 1 cm in diameter (tumor sizes in WT and MPO-KO mice were similar) (Figure 4A), cDC1 and cDC2 were isolated from dLNs, and their ability to stimulate OVA-specific OT-1+ T cells was assessed. cDC1 isolated from MPO-KO LLC-OVA TB mice had a markedly higher ability to cross-present OVA-derived peptide than cDC1 from WT mice (Figure 4B). As expected, cDC2 had low cross-presenting activity, and it was similar in WT and MPO-KO mice (Figure 4B). Both DC populations from WT and KO mice showed similar stimulation of allogeneic T cells (Figure 4C). PMN-MDSC from KO mice were not able to suppress antigen-specific CD8+ T cell responses in vitro compared with WT PMN-MDSC (Figure 4D).
Figure 4. Effect of MPO deletion on cross-presentation by DCs and pharmacological inhibition of MPO on tumor growth.
(A) LLC-OVA tumor growth in MPO-KO mice (n = 3). (B) Proliferation of OT1 CD8+ T cells stimulated with DC1 (CD11c+MHCII+CD103+CD11b–CD172a–) and DC2 (CD11c+MHCII+CD103–CD11b+CD172a+) isolated from draining lymph nodes of WT and MPO-KO mice bearing LLC-OVA tumor (n = 4). (C) Proliferation of allogeneic CD8+ T cells stimulated with CD103+ DC (cDC1) and CD172a+ DC (cDC2) isolated from draining lymph nodes of WT and MPO-KO mice bearing LLC-OVA tumors. Proliferation of CD8+ T cells measured at 72 hours by CFSE dilution in triplicates (n = 4). (D) Suppressive activity of PMN-MDSC isolated from spleens of WT or MPO-KO tumor-bearing mice. The dashed line represents the proliferation of CD8+ T cells in absence of MDSC. Proliferation was measured by 3[H]-thymidine uptake in triplicates. A typical example of 3 performed experiments is shown. In all experiments, mean ± SD is shown. **P < 0.01, unpaired 2-tailed Student’s t test between compared groups.
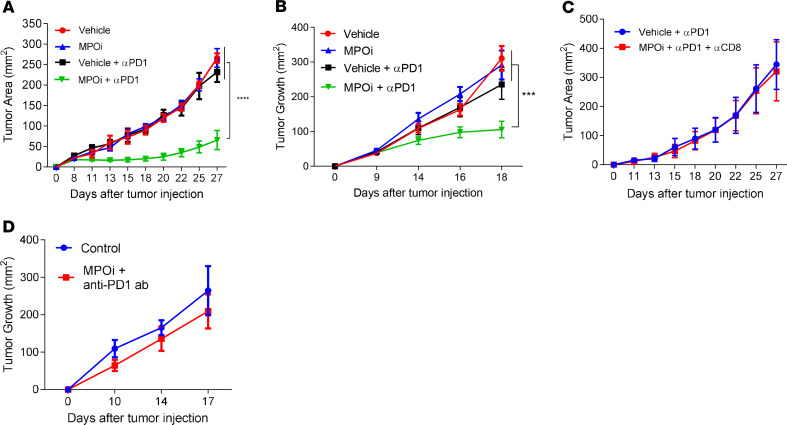
These results suggested that deletion of MPO in PMN-MDSC may have strong effect on cross-presentation by DCs. Therefore, we investigated whether the pharmacological inhibition of MPO affected the efficacy of immunotherapy in TB mice. Mice were injected s.c. with LLC (Figure 5A) or B16F10 (Figure 5B) tumor cells and treated with MPO inhibitor 4-ABAH (4-aminobenzoic hydrazide 95%) twice per day starting from day 5 and PD1 antibody twice per week, starting from day 10. Neither PD1 antibody nor MPO inhibition had antitumor effects in these models. However, the combination markedly decreased tumor growth in both models (Figure 5, A and B). The antitumor effect of combination of MPO inhibitor and PD1 antibody was abrogated in mice depleted of CD8+ T cells (Figure 5C), indicating that the observed antitumor effect was dependent on the generation of antitumor CD8+ T cells. To ascertain that observed effect was dependent on cross-presentation, we used BATF3-KO mice, which are depleted of cDC1 (27). In the absence of cDC1, i n BATF3-KO mice the effect of combined treatment was abrogated (Figure 5D).
Figure 5. Effect of pharmacological inhibition of MPO on tumor growth.
(A) Tumor growth in LLC TB mice, treated with anti-PD1 and MPO inhibitor (each group n = 5). (B) Tumor growth in B16F10 TB mice, treated with anti-PD1 and MPO inhibitor (each group n = 8). (C) Tumor growth in LLC-bearing mice depleted of CD8+ T cells and treated with anti-PD1 and MPO inhibitor (each group n = 4). (D) Tumor growth in LLC-bearing BATF3-KO mice, treated with anti-PD1 and MPO inhibitor (each group n = 4). Mean ± SEM is shown. ***P < 0.001, ****P < 0.0001, 2-way ANOVA with corrections for multiple comparison.
Discussion
Our study suggests a mechanism of negative regulation of cross-presentation by DCs in cancer, involving possible transfer of oxidized lipids from PMN-MDSC to cDC1. PMN-MDSC are dramatically expanded in many tumor models in mice and different types of human cancers and infiltrate tumors and lymphoid organs (15, 28). These cells are characterized by immune-suppressive activity and distinct biochemical and transcriptomics profiles compared with normal PMNs (15). Classical PMN respond to various pathogens and trauma by respiratory burst and degranulation with the release of antibacterial and antiviral enzymes. They rapidly die in the process. As a result, even activated classical PMN usually lack immune-suppressive activity. However, their antitumor activity is also difficult to detect. In contrast, PMN-MDSC produce a number of factors (arginase 1, prostaglandin E2, sustained level of superoxide and peroxynitrite, etc.) that inhibit the function of T cells and other cells of immune system (15). As a result, these cells may blunt antitumor response and promote tumor progression and limit the effect of immune therapy of cancer. In mice, tumor PMN-MDSC are more suppressive than in spleens due to upregulation of Nos2 expression and several other suppressive mechanisms regulated by hypoxia as well as other factors present in tumors (28). It is known that spleen PMN-MDSC have relatively weak suppression of T cell function as compared with M-MDSC and tumor-associated macrophages (14, 28). Therefore, the well-established association between the increased presence of PMN-MDSC in blood and negative outcomes in patients with cancer was rather puzzling. Our study showed that PMN-MDSC can blunt the ability of DCs to cross-present antigens and thus demonstrated potential role of PMN-MDSC in blocking the priming phase of immune responses in cancer by affecting cross-presenting cDC1 and thus contributing to the resistance to checkpoint blockade therapy.
The conclusion that lipid transfer from PMN-MDSC to DCs can inhibit cross-presentation is based on several previous and current findings. (a) LB containing oxidatively truncated lipids blocked cross-presentation of DCs (10, 11). LB containing electrophilic oxidatively truncated lipids covalently bound to chaperone heat shock protein 70. This interaction prevented the translocation of peptide–MHC class I complexes to cell surface by causing their accumulation inside late endosomes/lysosomes. As a result, DCs were no longer able to stimulate adequate CD8+ T cells responses (10). Loading of DCs with oxidized but not with nonoxidized fatty acids inhibited cross-presentation by DCs (10). (b) PMN-MDSC and PMN transfer lipids to DCs, but only PMN-MDSC blocked cross-presentation. (c) PMN-MDSC, in contrast to PMN, are enriched for oxidized lipids. This effect depends on MPO and NADPH oxidase activity, which is also known to be substantially increased in PMN-MDSC as compared with PMN (14, 29, 30). (d) In the absence of MPO or NADPH oxidase, PMN-MDSC lose the ability to block cross-presentation and inhibition of MPO in PMN-MDSC in TB mice dramatically improved cross-presentation by DCs and antitumor effect of immunotherapy.
Although MPO is detected in Mon, its amount and activity are much higher in PMNs, which may explain why M-MDSC lack the ability to block cross-presentation by DCs. MPO expression is generally lost during differentiation of Mon to macrophages (31, 32). MPO is not expressed in DCs, which further supports the role of PMN-MDSC in blocking DC cross-presentation. It is important to point out that MPO deficiency also affects PMN-MDSC–suppressive activity. However, at the time of evaluation, MPO KO and WT mice had similar tumor size, which excludes the effect of differences in tumor burden on DC cross-presentation. Moreover, a previous study demonstrated that MPO can directly inhibit expression of activation markers in DC and the ability of DCs to stimulate allogeneic responses (33). The fact that PMN-MDSC had a selective effect only on cross-presenting ability of DCs strongly suggested that this effect was not mediated via release of MPO. MPO and NADPH oxidase may affect not only lipids but also various proteins, so it is possible that regulation of cross-presentation by PMN-MDSC is more complex process than lipid transfer. However, unlike oxidized lipids, oxidized proteins are not released from live PMN-MDSC. Our data indicate that most of PMN-MDSC remained alive during coculture with DCs, which make this mechanism less likely. More studies will be needed to clarify this mechanism. However, regardless of the specific mechanism, our findings suggest that targeting of MPO enhanced the effect of check-point inhibitor, which may suggest novel therapeutic opportunity.
Methods
Mouse models.
Animal experiments were approved by The Wistar Institute Animal Care and Use Committee. BALB/c or C57BL/6 mice (female, 4–6 week old) were obtained from Charles River; OT-I TCR-transgenic mice [C57BL/6-Tg(TCRaTCRb)1100mjb] (female, 4–6 week old), PMEL TCR-transgenic mice [B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J] (female, 4–6 week old), MPO KO (B6.129X1-Mpotm1Lus/J) (female, 4–6 week old), GP91-KO mice (B6.129S-Cybbtm1Din/J) (female, 4–6 week old), CD204-KO mice (B6.Cg-Msr1tm1Csk/J) (female, 4–6 week old), and Batf3-KO mice (129S-Batf3tm1Kmm/J) (female, 4–6 week old) were purchased from The Jackson Laboratory.
Reagents and cell lines.
Tumor cell lines, including EL4 lymphoma, CT26 (colon carcinoma) and LLC were maintained in DMEM medium supplemented with 10% FBS (Atlanta) at 37°C, 5% CO2. LLC-OVA were maintained in complete DMEM supplemented with 0.5 mg/mL G418 (Gibco). Tumors were injected s.c. on the flanks of the animal at 5 × 105 cells per mouse. Tumor cell lines were tested for mycoplasma contamination by using a Universal Mycoplasma detection kit (ATCC). SIINFEKL peptide and control peptide RAHYNIVTF were obtained from the American Peptide Company. (Pam)2−KMFVESIINFEKL peptide (derived from OVA) and (Pam)2-KMFVKVPRNQDWL (derived from gp100) were obtained from DBA Synthetic biomolecules. Anti-mouse CD103 biotin–conjugated beads were purchased from BioLegend and used for CD103+ DC purification. APC-conjugated anti-mouse CD11c, Percp 5.5–conjugated anti-mouse MHC I, Apc-cy7–conjugated anti-mouse CD11b, Pe-conjugated anti-mouse Ly6G, Apc-conjugated anti-mouse Ly6C, Pe-cy7–conjugated anti-mouse CD3 and AF700, and FITC anti-mouse CD45 antibodies were purchased from BD Biosciences. FITC-conjugated anti-mouse F4/80 and Percp 5.5–conjugated anti-mouse CD11c were purchased from Invitrogen. Pe-cy7 anti-mouse MHC II–, CD172-, CD40-, and AF700-conjugated anti-mouse CD86; BV421-conjugated anti-mouse CD103 or XCR; and Apc-conjugated anti-mouse PDL-1 were purchased from BioLegend. Aqua live was obtained from Invitrogen. BODIPY lipid dye 493/503 was obtained from Invitrogen. A list of antibodies is provided in Supplemental Table 1.
Cell phenotype and lipid contents by flow cytometry.
DCs were incubated with FC-block (BD Biosciences) for 5 minutes, and surface staining was performed at 4°C for 15 minutes. Cells were run on Celesta flow cytometer (BD Biosciences), and data were analyzed by FlowJo (Tristar). For lipid staining, cells were resuspended in 500 μl Bodipy 493/503 at 0.25 μg/mL in PBS. Cells were stained for 15 minutes at room temperature in the dark, then washed twice, resuspended in PBS, and run immediately on Celesta. For lipid transfer staining, PMN-MDSC (or other cells) were resuspended in 500 μl Bodipy 493/503 at 0.25 μg/mL in PBS, then stained for 15 minutes at room temperature in the dark, washed twice, resuspended in complete media, and cocultured with unstained DCs. After 24 hours cells were stained for surface staining. At least 10,000 cells were collected for subsequent analysis.
Generation of DCs.
Mouse DCs were generated from enriched BM HPCs with 10 ng/mL GM-CSF (PeproTech) and 100 ng/mL FLT3-L (PeproTech). Briefly, HPCs were isolated from mouse BM by using a Lineage depletion kit (Miltenyi), according to the manufacturer’s instructions. Cells were seeded at 3 × 104 cell/mL in 24-well plates; GM-CSF (10 ng/mL) and FLT3-L (100 ng/mL) were added to the culture at day 0 and day 3. At day 5, cells were split and cultured with fresh media with cytokines at 7.5 × 105 cell/mL in 6-well plates. Cytokines were then added every 3 days. Cells were finally harvested and used between day 11 and day 14. DC phenotype was analyzed before performing each experiment to check the percentage of CD103+ DC (80%–90%).
Cocultures.
Harvested DCs were cocultured with FACS-sorted (BD FACSAria II) PMN-MDSC (CD45+CD11b+Ly6G+) from spleens or tumors of TB mice, PMN naive (CD45+CD11b+Ly6G+) from BM or spleens of naive mice, m-MDSC (CD45+CD11b+Ly6C+) from spleens of TB mice, naive Mon (CD45+CD11b+Ly6C+) from BM or spleens of naive mice, in the ratio 1:4 (DC/other cells). Sorted cells were obtained from both LLC and EL-4 TB mice sacrificed between day 21 and day 28 after tumor challenge. When comparing WT TB mice with KO TB mice, animals with similar sized tumors were used. Spleens were mechanically dissociated. BM was flushed with precision needle and 1 mL syringes. Cells were plated at the density of 1 × 106 cells/mL of complete media containing GM-CSF (10 ng/mL) and FLT3-L (100 ng/mL). For cocultures with Transwell system, 0.4 μm pore size Transwell inserts for 6-well plates (Corning) were used. DCs were plated in the lower compartment, while other cells were plated in the upper compartment. Cells were plated at the density of 1 × 106 cells/mL of complete media containing GM-CSF (10 ng/mL) and FLT3-L (100 ng/mL). After 24 hours of coculture, cells were harvested and CD103+ DCs were isolated with anti-mouse CD103 biotin–conjugated beads (BioLegend) and streptavidin Microbeads (Miltenyi). The purity has been checked for each cell type coculture (≥95%).
Cross-presentation of OVA-derived long peptides.
Isolated CD103+ DCs were loaded for 16–18 hours with 5 μg/mL long peptides. Splenocytes were isolated from spleens of responder mice by using mechanical digestion and then plated at 2.5 × 104 to 10 × 104 splenocytes per well (depending on the experiment). DCs and splenocytes were mixed at different ratios. DCs were loaded with 0.5 μg/mL SIINFEKL or Gp100 control peptide for 1 hour at 37°C. After antigen loading, DCs have been washed twice in complete media to remove the excess of peptide. In some experiments, DCs were used to stimulate allogeneic splenocytes isolated from spleens of BALB/c mice. Before mixing DCs with splenocytes, some DCs for each condition have been analyzed for surface activation markers. After 30 hours (for direct presentation) or 48 hours (for cross-presentation) or 96 hours (for allogenic MLR), 3[H]-thymidine was added at 1 μCi per well for an additional 18 hours, followed by cell harvesting and a radioactivity count on liquid scintillation counter.
Cross-presentation in vivo.
LLC OVA was injected s.c. on both the flanks of the mice at 5 × 105 cells per mouse. After 20 days, mice were euthanized and dLNs (for each flank: 2 axillar, 1 inguinal) were collected. LN were then digested for 30 minutes at 37°C with collagenase A (0.5 mg/mL; MilliporeSigma) and Dnase I (0.2 mg/mL, Roche) and diluted in HBSS with Ca2+/Mg2+; 20 mM EDTA (Invitrogen) was then added for 5 minutes at room temperature to stop the reaction. Single-cell suspensions were prepared and then DCs were stained and sorted on a BD FACS Aria II (BD Biosciences). CFSE-labeled OT1 CD8+ T cells, isolated by using EasySep Mouse CD8+ T Cell Enrichment Kit (STEMCELL), were used as responders at a 1:5 ratio (5,000 DCs/25,000 T cells). Proliferation was checked 3 days after by FACS Celesta. Allogenic stimulation of CFSE-labeled CD8+ T cells isolated from spleens of BALB/c mice was used as control.
Confocal microscopy.
DCs were washed twice with PBS and resuspended in complete RPMI, and 50,000 cells were seeded on poly-L-lysine cellware 12 MM round coverslips (Corning) for 45 minutes at 37°C. After that time, cells were washed with PBS and were stained for surface markers. Briefly, cells were incubated with Fc-block (BD Biosciences) for 10 minutes, stained with unconjugated antibodies for 15 minutes at 4°C, washed twice with PBS before incubation with fluorochrome-associated secondary antibodies. Afterward, cells were fixed and permeabilized with Fixation & Permeabilization Buffers (BD Biosciences) for 15 minutes at room temperature, washed twice with wash buffer (BD Biosciences), and then blocked with PBS containing 5% FBS for 45 minutes. Cells were incubated with Fc-block for 5 minutes at room temperature and stained with different primary antibodies at 4°C for 16–18 hours. Cells were washed 3 times and incubated with fluorochrome-associated antibodies for 45 minutes at room temperature. After that time cells were washed 3 times and then stained with BODIPY to detect LB for 15 minutes at room temperature. Cells were washed and incubated with DAPI and mounted on slides using Prolong Gold antifade reagent (Life Technology). The cells were imaged with a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems).
Suppression assay.
Single-cells suspensions from spleens and tumors were prepared as described above. Then cells were stained and sorted on a BD FACS Aria BD (Biosciences). PMN-MDSC (CD45+CD11b+Ly6G+Ly6Clo) and M-MDSC (CD45+CD11b+Ly6G–Ly6Chi) were plated in U-bottom 96-well plates (3 replicates) in RPMI with 10% FBS and cocultured at different ratios with splenocytes from Pmel- or OT-1–transgenic mice in the presence of cognate peptides: OT-1 (SIINFEKL; 0.1 ng/mL) and Pmel (EGSRNQDWL; 0.1 μg/mL). After 48 hours, cells were incubated with [3H]-thymidine (PerkinElmer)
α-DR5 treatment, MPO inhibitors, and checkpoint blockade.
Mice were challenged (day 0) with LLC OVA on both the flanks at 5 × 105 cells per mouse. Fourteen days after the challenge (when the tumor reached 0.5 cm of diameter), the animals were randomly allocated in 2 different groups. One of those received α-DR5 (anti-mouse DR5 [CD262], MD5-1, Bio Cell) i.p., every 3 days starting from day 5 after the challenge, at a concentration of 100 μg/mouse in 100 μL PBS (Corning). The other group was treated with the control immunoglobulin (in 100 μL PBS). The percentage of Mon and PMN of live cells was assessed in blood.
Mice were challenged (day 0) with LLC on the flanks at 5 × 105 cells per mouse. Four days after the challenge, mice were randomly allocated in 4 different groups. MPO inhibitor 4-ABAH (MilliporeSigma) was given i.p. at a dose of 40 μg/g in 400 μL HBSS 1× (Gibco), twice a day starting from day 4 after tumor challenge. α-PD1 (anti-mouse PD1 [CD279], RMP1-14, Bio Cell) was given i.p. at a dose of 200 μg per mouse in 100 μL PBS, every 3 days starting from day 8 after tumor challenge. One group received the vehicle, one group received α-PD1 and vehicle, one group received 4-ABAH, and the last group received a combination of 4-ABAH and α-PD1. The same experiment was repeated with the addition of α-CD8 treatment. α-CD8 (anti-mouse CD8a, 53-6.7, Bio Cell) was given i.p. at a dose of 100 μg per mouse, twice a week starting from day 1 after tumor challenge.
Liquid chromatography and mass-spectral analysis of deuterated and nondeuterated free fatty acids and neutral lipids.
Lipids were extracted by the Folch procedure with slight modifications, under nitrogen atmosphere, at all steps. Free fatty acids were analyzed using a Dionex Ultimate 3000 HPLC system coupled online to Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher) using a C18 column (Acclaim PepMap RSLC, 300 μm 15 cm, Thermo Fisher). Gradient solvents (A, methanol [20%]/water [80%] [v/v], and B, methanol [90%]/water [10%] [v/v]; both containing 5 mM ammonium acetate) were used. The column was eluted at a flow rate of 12 μL/min using a linear gradient from 30% solvent B to 95% solvent B over 70 minutes, held at 95% B from 70–80 minutes followed by a return to initial conditions by 83 minutes and reequilibration for an additional 7 minutes. Spectra were acquired in negative ion mode. The scan range for MS analysis was 150–600 m/z, with a maximum injection time of 100 ms using 1 micro scan and a resolution of 140,000. An isolation window of 1.0 Da was set for the MS and MS2 scans. Capillary spray voltage was set at 2.6 kV, and capillary temperature was 250°C. The S-lens Rf level was set to 60. Analytical data were acquired and analyzed using Xcalibur software.
TAGs were analyzed as molecular ammonium adducts (TAG+NH4) using a Dionex Ultimate 3000 HPLC system coupled online to Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher) using a Luna 3 μm C18 (2) 100 A, 150 × 1.0 mm column (Phenomenex) at a flow rate of 0.065 ml/min. The column was maintained at 35°C. The analysis was performed using gradient solvents (A and B) containing 0.1% NH4OH. Solvent A was methanol, and solvent B was propanol. The column was eluted for 2 minutes from 0% B to 2% B (linear), from 3–6 minutes with a linear gradient from 2% solvent B to 3% solvent B, then isocratically from 3–18 minutes using 3% solvent B, 18–35 minutes with a linear gradient from 3% solvent B to 40% solvent B, 35–60 minutes using a linear gradient from 40%–55% solvent B, then isocratically from 60–65 minutes at 55% solvent B, then from 65–80 minutes from 55% to 0% B (linear), followed by equilibration from 80–90 minutes at 0% B. MS analysis was performed in positive ion mode at a resolution of 140,000 for the full MS scan and 17,500 for the MS2 scan in a data-dependent mode with an inclusion list for TAG. The scan range for MS analysis was 300–1200 m/z, with a maximum injection time of 128 ms using 1 micro scan. A maximum injection time of 500 ms was used for MS2 (high-energy collisional dissociation) analysis with collision energy set to 24. An isolation window of 1.0 Da was set for the MS and MS2 scans. Capillary spray voltage was set at 4.5 kV, and capillary temperature was 320°C. Sheath gas was set to 8 arbitrary units and the S-lens Rf level was set to 60.
Statistics.
Statistical analysis was performed using unpaired 2-tailed Student’s t test and 1-way ANOVA with corrections for multiple comparison when more than 2 groups were compared. Significance was determined at P < 0.05. Estimation of variation within each group of data was performed, and variance was similar between groups that were compared. Animal experiments were not blinded.
Study approval.
Animal experiments were approved by The Wistar Institute Animal Care and Use Committee.
Author contributions
AU, VAT, ENT, and LD performed experiments. YYT performed data analysis. VEK designed experiments, analyzed data, and wrote manuscript. DIG designed overall concept and experiments, analyzed data, and wrote manuscript. FV performed and designed experiments, analyzed data, and wrote manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA R01CA165065, PO1 CA114046, AI156924, AI145406, and HL114453 and by animal and flow cytometry core facilities of The Wistar Institute.
Version 1. 06/25/2020
In-Press Preview
Version 2. 08/06/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(15):e138581.https://doi.org/10.1172/jci.insight.138581.
Contributor Information
Alessio Ugolini, Email: alessiougolini217@gmail.com.
Laxminarasimha Donthireddy, Email: Donthireddy@Wistar.org.
Dmitry I. Gabrilovich, Email: dgabrilovich@wistar.org.
Filippo Veglia, Email: Filippo.Veglia@moffitt.org.
References
- 1.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 3.Böttcher JP, Reis e Sousa C. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer. 2018;4(11):784–792. doi: 10.1016/j.trecan.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Audsley KM, McDonnell AM, Waithman J. Cross-presenting XCR1+ dendritic cells as targets for cancer immunotherapy. Cells. 2020;9(3):E565. doi: 10.3390/cells9030565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veglia F, Gabrilovich DI. Dendritic cells in cancer: the role revisited. Curr Opin Immunol. 2017;45:43–51. doi: 10.1016/j.coi.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broz ML, et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 2014;26(5):638–652. doi: 10.1016/j.ccell.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salmon H, et al. Expansion and activation of CD103(+) dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity. 2016;44(4):924–938. doi: 10.1016/j.immuni.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sánchez-Paulete AR, et al. Cancer immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 2016;6(1):71–79. doi: 10.1158/2159-8290.CD-15-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711–723.e4. doi: 10.1016/j.ccell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veglia F, et al. Lipid bodies containing oxidatively truncated lipids block antigen cross-presentation by dendritic cells in cancer. Nat Commun. 2017;8(1):2122. doi: 10.1038/s41467-017-02186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramakrishnan R, et al. Oxidized lipids block antigen cross-presentation by dendritic cells in cancer. J Immunol. 2014;192(6):2920–2931. doi: 10.4049/jimmunol.1302801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herber DL, et al. Lipid accumulation and dendritic cell dysfunction in cancer. Nat Med. 2010;16(8):880–886. doi: 10.1038/nm.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cubillos-Ruiz JR, et al. ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell. 2015;161(7):1527–1538. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19(2):108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Khami AA, et al. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology. 2017;6(10):e1344804. doi: 10.1080/2162402X.2017.1344804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Khami AA, Rodriguez PC, Ochoa AC. Energy metabolic pathways control the fate and function of myeloid immune cells. J Leukoc Biol. 2017;102(2):369–380. doi: 10.1189/jlb.1VMR1216-535R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veglia F, et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 2019;569(7754):73–78. doi: 10.1038/s41586-019-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohamed E, Al-Khami AA, Rodriguez PC. The cellular metabolic landscape in the tumor milieu regulates the activity of myeloid infiltrates. Cell Mol Immunol. 2018;15(5):421–427. doi: 10.1038/s41423-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol. 2012;22(4):275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez PC, Ochoa AC, Al-Khami AA. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front Immunol. 2017;8:93. doi: 10.3389/fimmu.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinski P. Regulation of immune responses by prostaglandin E2. J Immunol. 2012;188(1):21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205(10):2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condamine T, et al. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J Clin Invest. 2014;124(6):2626–2639. doi: 10.1172/JCI74056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He YM, et al. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat Med. 2018;24(2):224–231. doi: 10.1038/nm.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savina A, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126(1):205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Gajewski TF, Corrales L, Williams J, Horton B, Sivan A, Spranger S. Cancer immunotherapy targets based on understanding the T cell-inflamed versus non-T cell-inflamed tumor microenvironment. Adv Exp Med Biol. 2017;1036:19–31. doi: 10.1007/978-3-319-67577-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37(3):208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu T, Gabrilovich DI. Molecular pathways: tumor-infiltrating myeloid cells and reactive oxygen species in regulation of tumor microenvironment. Clin Cancer Res. 2012;18(18):4877–4882. doi: 10.1158/1078-0432.CCR-11-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Nefedova Y, Lei A, Gabrilovich D. Neutrophils and PMN-MDSC: their biological role and interaction with stromal cells. Semin Immunol. 2018;35:19–28. doi: 10.1016/j.smim.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77(5):598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 32.Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10(7):1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 33.Odobasic D, et al. Neutrophil myeloperoxidase regulates T-cell-driven tissue inflammation in mice by inhibiting dendritic cell function. Blood. 2013;121(20):4195–4204. doi: 10.1182/blood-2012-09-456483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.