Abstract
Platinum-based chemotherapy in combination with immune-checkpoint inhibitors is the current standard of care for patients with advanced lung adenocarcinoma (LUAD). However, tumor progression evolves in most cases. Therefore, predictive biomarkers are needed for better patient stratification and for the identification of new therapeutic strategies, including enhancing the efficacy of chemotoxic agents. Here, we hypothesized that discoidin domain receptor 1 (DDR1) may be both a predictive factor for chemoresistance in patients with LUAD and a potential target positively selected in resistant cells. By using biopsies from patients with LUAD, KRAS-mutant LUAD cell lines, and in vivo genetically engineered KRAS-driven mouse models, we evaluated the role of DDR1 in the context of chemotherapy treatment. We found that DDR1 is upregulated during chemotherapy both in vitro and in vivo. Moreover, analysis of a cohort of patients with LUAD suggested that high DDR1 levels in pretreatment biopsies correlated with poor response to chemotherapy. Additionally, we showed that combining DDR1 inhibition with chemotherapy prompted a synergistic therapeutic effect and enhanced cell death of KRAS-mutant tumors in vivo. Collectively, this study suggests a potential role for DDR1 as both a predictive and prognostic biomarker, potentially improving the chemotherapy response of patients with LUAD.
Keywords: Oncology, Therapeutics
Keywords: Drug therapy, Lung cancer, Molecular biology
Discoidin Domain Receptor 1 (DDR1) expression predicts response to chemotherapy and its inhibition boosts chemosensitivity in lung adenocarcinoma.
Introduction
Lung cancer is the leading cause of cancer mortality worldwide (1). KRAS mutations are the most common drivers of lung adenocarcinoma (LUAD), the major subtype of non–small cell lung cancers (NSCLCs) (2). KRAS mutations are associated with poor response to anti-cancer drug treatments (3–5). Furthermore, additional genetic alterations often coexisting with oncogenic KRAS, such as inactivating mutations in STK11 or KEAP1, enhance the chemoresistant phenotype (6). Mechanistically, chemoresistance may be a consequence of: (a) drug inactivation, (b) reduced drug uptake, (c) increased drug efflux, or (d) enhanced repair of treatment-induced DNA lesions by DNA repair mechanisms (7). Several mediators have been identified for KRAS-dependent chemoresistance, including the transcription factor NRF2 (8) and telomerase activity (9). Potentially druggable vulnerabilities representing new space for therapeutic intervention, such as XPO1 (10), also have been identified. Therefore, considering the limited efficacy of chemotherapy in this clinical setting, an urgent need exists for identification of additional druggable targets.
The discoidin domain receptors, DDR1 and DDR2, belong to the family of receptor tyrosine kinases and regulate cell proliferation, survival, migration, and differentiation in mammalian tissues by interacting with several types of collagen (11, 12). In different human cancers, such as breast, lung and hepatocellular carcinoma, aberrant DDR1 and DDR2 signaling has been observed to be implicated in tumor development and cancer progression (13–16). Moreover, DDR1 promotes local invasion and lung colonization through STAT3 activity, implicating a role for DDR1 in defining the metastatic niche (17). Notably, DDR1 expression has been reported to decrease sensitivity to chemotherapy and induce survival signals, which may lead to cancer relapse (18–21). Recent studies demonstrated that miR-199a-3p–dependent DDR1 expression confers malignant cisplatin resistance phenotype to ovarian cancer (22). Similarly, DDR1 promotes lung cancer chemoresistance in a TM4SF1-dependent manner (23). DDR1 has been identified as a component of a 7-gene group of cancer-associated factors that is maintained after chemotherapy treatment, essential for cell line survival, and increased in drug-resistant cancer stem cells (24). This body of collective evidence suggests that DDR1 is involved in the molecular responses associated with resistance to chemotherapy; however, a direct relationship between abnormal activity of DDR1 and lung cancer chemoresistance has not yet been described. Here, we found that DDR1 expression correlates with response to platinum-based chemotherapy in LUAD. Furthermore, DDR1 inhibition synergistically enhances chemotherapy efficacy in KRAS-mutant LUAD in vivo.
Results
DDR1 expression increases upon treatment with genotoxic agents in LUAD cell lines.
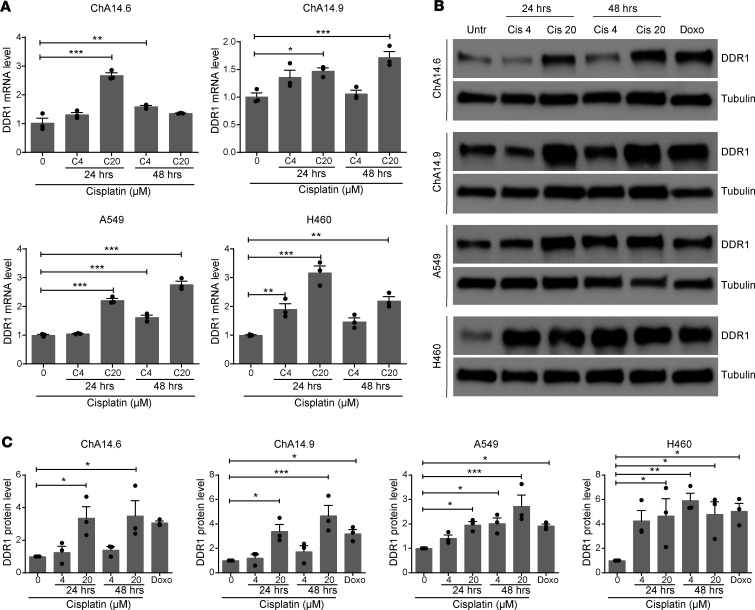
It has been previously reported that DDR1 expression is enhanced by exposure to DNA-damaging agents in various mammalian cell types to convey prosurvival functions (22, 25, 26). However, it is currently unknown whether platinum-based chemotherapy, the conventional treatment option for KRAS-mutant LUAD, also elicits a similar response resulting in DDR1 upregulation. To evaluate this possibility, we exposed both mouse and human KRAS-driven LUAD cell lines to various cisplatin concentrations and exposure times. We used KRasG12V LUAD cell lines previously generated in our laboratory (27) together with commercially available human counterparts. Interestingly, DDR1 levels increased in a dose- and/or time-dependent fashion as measured by both q-PCR and Western blotting to a similar extent than that caused by the classical DNA-damaging agent doxorubicin (Figure 1).
Figure 1. Induction of DDR1 expression in lung adenocarcinoma (LUAD) cell lines following cisplatin treatment.
Murine (ChA14.6 and ChA14.9) and human (A549 and H460) KRAS-mutant lung adenocarcinoma (LUAD) cell lines were treated with cisplatin (4 and 20 μM) and assessed for DDR1 expression either by qPCR (A) or by Western blot (B) after 24 or 48 hours as indicated. Extracts from untreated control cells (Untr) were used as controls. α-Tubulin was used as loading control. Cells were treated with 1 μM doxorubicin during 24 hours as a positive genotoxic control. (C) Quantification of 3 independent Western blot experiments normalized to untreated controls as shown in B. Data were analyzed using 1-way ANOVA followed by Dunnett’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as the mean ± SEM of 3 independent experiments.
DDR1 inactivation in patient-derived xenograft cell lines increases apoptosis and improves the efficacy of chemotherapy.
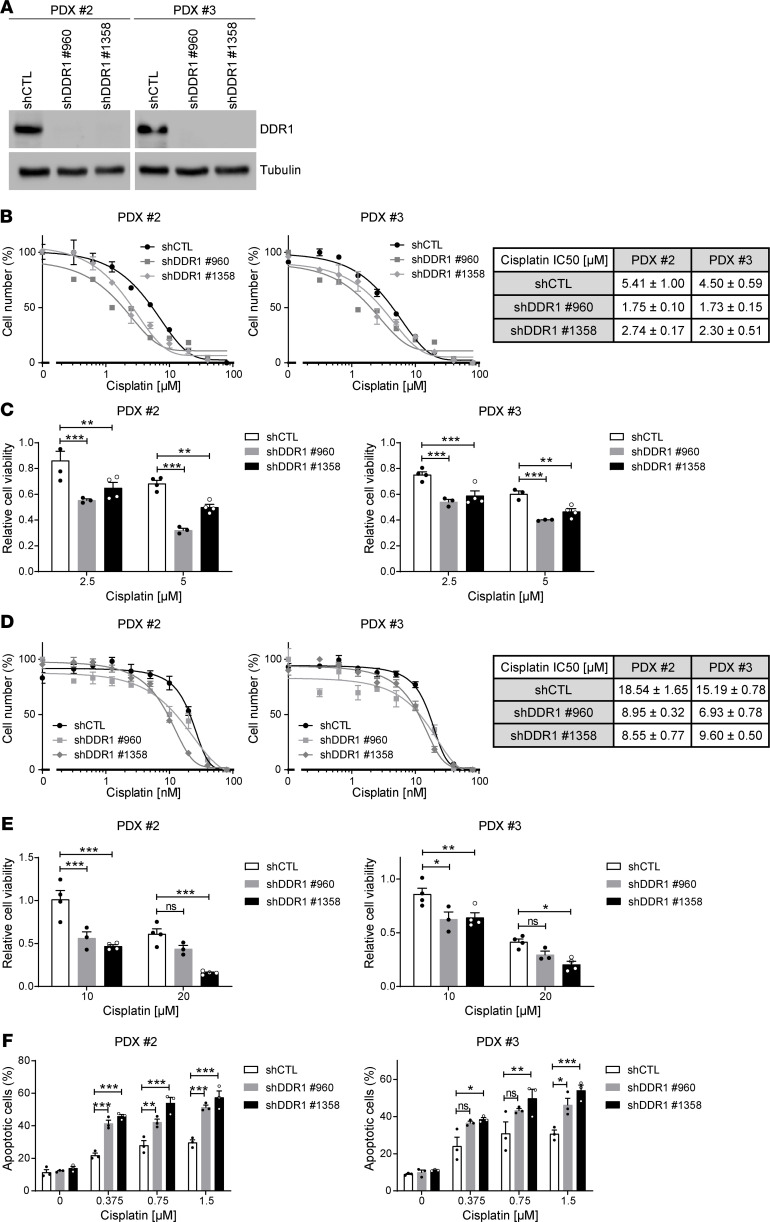
We assessed whether tampering with DDR1 expression concomitantly with chemotherapy could improve treatment efficacy. To this end, patient-derived xenograft (PDX) cell lines harboring KRAS mutations (14) were used to test whether DDR1 knockdown could synergize with cisplatin to enhance cell death. Two independent shRNA sequences were used to efficiently downregulate DDR1 levels (Figure 2A). When grown in 2D conditions, DDR1 knockdown in both PDX cell lines resulted in further sensitization to cisplatin treatment as illustrated by lower IC50 values and reduced cell numbers (Figure 2, B and C). Similar results were obtained in 3D conditions (Figure 2, D and E). Decreased cell numbers could be a direct consequence of apoptotic induction as demonstrated by annexin V/PI detection in dual-treated cells when compared with either sh-DDR1 or cisplatin alone (Figure 2F).
Figure 2. Increased sensitivity to cisplatin in patient-derived xenograft (PDX) lung adenocarcinoma cell lines following DDR1 knockdown.
(A) Western blot validation of DDR1 knockdown in 2 patient-derived xenograft (PDX) cell lines (2 and 3) treated with 2 independent shRNAs (960 and 1358) against DDR1 in parallel with a shRNA control. α-Tubulin was used as loading control. Representative Western blots of 2 independent experiments are shown. (B) Comparison of IC50 values upon 72 hours cisplatin treatment between control and DDR1-knocked down PDX cell lines maintained in 2D growth conditions. Data are shown as the mean ± SEM of 3 independent experiments. The table shows decreased IC50 values (mean ± SD, n = 3) following DDR1 knockdown. (C) Cell number quantification following treatment with the indicated concentrations of cisplatin for 72 hours in control and DDR1-knocked down PDX cell lines maintained in 2D growth conditions. Data were analyzed using 2-way ANOVA followed by Bonferroni’s multiple comparison test. **P value < 0.01, ***P value < 0.001. Data are shown as the mean ± SEM of at least 3 independent experiments. (D) Comparison of IC50 values upon 72 hours cisplatin treatment between control and DDR1-knocked down PDX cell lines maintained in 3D growth conditions. Data are shown as the mean ± SEM of 3 independent experiments. The table shows decreased IC50 values (mean ± SD, n = 3) following DDR1 knockdown. (E) Cell number quantification following treatment with the indicated concentrations of cisplatin for 72 hours in control and DDR1-knocked down PDX cell lines maintained in 3D growth conditions. All values were normalized to their respective untreated controls. Data were analyzed using 2-way ANOVA followed by Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as the mean ± SEM of at least 3 independent experiments. (F) Quantification of apoptosis following treatment with the indicated concentrations of cisplatin for 72 hours in control and DDR1-knocked down PDX cell lines maintained in 2D growth conditions. Data were analyzed using 2-way ANOVA followed by Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as the mean ± SEM of 3 independent experiments. P value ns, nonsignificant.
Association between DDR1 expression and response to therapy in patients with LUAD.
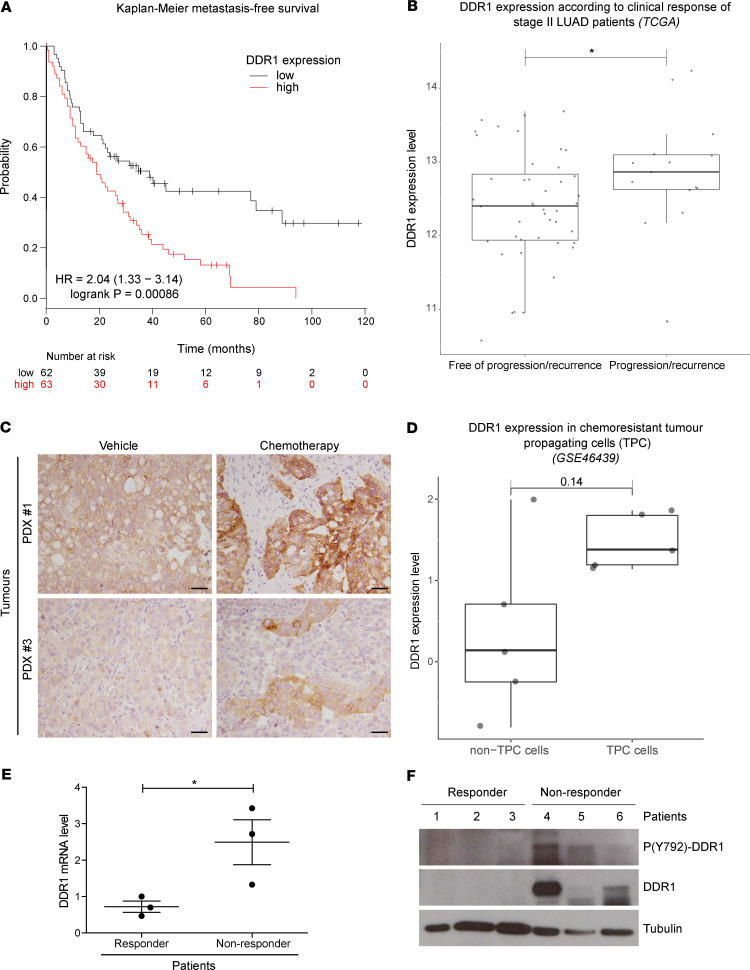
We next evaluated whether the increased response to chemotherapy upon DDR1 knockdown observed in vitro could be relevant in a clinical setting. We surveyed the online Kaplan-Meier plotter platform (https://kmplot.com/analysis/) (28) data sets in search of potential correlations between DDR1 mRNA levels and response to chemotherapy. We next evaluated DDR1 effect on the first progression output after chemotherapy in a cohort of 125 NSCLC patients for which gene expression data were available. Using the median cutoff, we observed that DDR1 overexpression is prognostic of significantly (log-rank, P = 8.6 × 10–4) shorter survival in this cohort (Figure 3A). To further confirm this analysis, we accessed publicly available data from The Cancer Genome Atlas (TCGA) database. We focused our analysis on stage II disease because it is a homogeneous patient cohort that is uniformly treated with surgery followed by adjuvant chemotherapy. Therefore, we hypothesized that any potential contribution of DDR1 in response to chemotherapy was more likely to be detected in this group compared with other stages, such as IIIA, in which treatment tends to be multimodal and more heterogeneous. To further emphasize the importance of chemotherapy treatment, those patients in the TCGA stage II cohort harboring oncogenic drivers, who could have been subjected to targeted treatments, were also excluded from the analysis (see Methods for details). Following this rationale, we retained 59 stage II patients with clinical profiles compatible with a combination of surgery and chemotherapy treatment. We then compared DDR1 expression between the 2 cohorts according to their primary response to treatment. As shown in Figure 3B, higher DDR1 levels are associated with disease progression (Wilcoxon’s test, P < 0.05) as compared with those patients annotated as progression free.
Figure 3. Lung cancer patients with high DDR1 expression display decreased response to chemotherapy and metastasis-free survival.
(A) Kaplan-Meier metastasis-free survival estimates according to DDR1 levels in lung cancer patients subjected to chemotherapy. Data were obtained from www.kmplot.com/lung (28). (B) Clinical response of patients with stage II LUAD from the TCGA database harboring non-druggable oncogenic drivers. Results are plotted based on DDR1 levels (y axis) and indicated along the x axis as patients free of recurrence (n = 44) vs. recurrence (n = 15). Wilcoxon’s test was used to analyze statistical significance. *P < 0.05. (C) Crl:NU-Foxn1nu mice implanted with KRAS-mutant PDX were treated with either vehicle or standard chemotherapy based on cisplatin (3 mg/kg) and paclitaxel (20 mg/kg) administered i.p. every 5 days for 3 weeks (n = 6). After necropsy, tumor samples were fixed and analyzed for DDR1 expression by immunostaining. Clones showing high DDR1 expression are observed in the chemotherapy-treated tumors. Scale bar: 50 μm. (D) Differential DDR1 expression in chemoresistant tumor propagating cells (TPCs) vs. the tumor bulk population (non-TPC). Gene expression data was obtained from GSE46439 (29). Wilcoxon’s test P value is indicated above the box plots. (E and F) Samples from 6 patients with LUAD with lymph node metastasis were classified, following treatment, according to the persistence (nonresponders, n = 3) or absence (responders, n = 3) of the initial lymph node metastasis. Overall DDR1 expression and the activating phosphorylation (Y-792) were evaluated in these samples by qPCR (E) or by Western blot (F). GAPDH was used as loading control. High DDR1 expression shows significant association with poor clinical response. Data were analyzed using unpaired Student’s t test. *P < 0.05. Data are shown as the mean ± SEM.
These analyses indicate that either overall DDR1 expression before treatment and/or DDR1 induction during chemotherapy may predict the clinical outcome. To test this hypothesis, we used an orthotopic PDX model that we previously developed by implanting and expanding patient biopsies directly into the lungs of immunodeficient mice (14). Human biopsies were engrafted in the lungs of Crl:NU-Foxn1nu mice and randomly allocated into 2 cohorts receiving either vehicle or chemotherapy treatment. Mice were sacrificed at the completion of the 4-week protocol, and tumor samples were assessed by immunostaining to detect DDR1 expression in situ. As expected, chemotherapy treatment induced tumor shrinkage (data not shown). Remarkably, we observed that high DDR1 expressing areas were enriched in the residual tumor mass following treatment (Figure 3C). This observation suggests that DDR1 expression correlates with LUAD persistence following chemotherapy. To reinforce this concept, we took advantage of a previously reported gene signature obtained from tumor-propagating cells (TPCs). This is a chemoresistant population that persists after repeated cisplatin treatment, acting as a cell reservoir responsible for disease relapse in mouse LUAD (29). Interestingly, DDR1 levels appeared consistently elevated in TPC samples when compared with the tumor bulk population (Figure 3D and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137869DS1).
Finally, mRNA and protein material samples were taken from a cohort of patients with LUAD with lymph node metastasis before receiving platinum-based therapy. Following treatment, patients were classified as responders or nonresponders based on the disappearance and/or persistence of their lymph node metastases, respectively. We assessed DDR1 expression by q-PCR demonstrating a 3-fold increase in DDR1 expression, when evaluated in pretreatment biopsies from nonresponder patients, when compared with those that subsequently responded to treatment (Figure 3E). This observation was further confirmed by Western blot in 3 patient samples from each cohort. Total DDR1 as well as the activating DDR1 Y-792 phosphorylation were elevated in pretreatment biopsies from the nonresponder patients (Figure 3F). Together, these results suggest that increased DDR1 activity correlates with poor response of patients with LUAD to chemotherapy and, therefore, could potentially serve as a predictive biomarker of treatment efficacy.
Synergistic effect of DDR1 inhibition and chemotherapy in KRasG12V-driven lung cancer in vivo.
We next verified the synergistic effect of DDR1 inhibition and cisplatin in a genetically engineered KRasG12V-inducible mouse model. We therefore assessed whether DDR1 inhibition could synergize with cisplatin/paclitaxel chemotherapy in inducing cell death, which has previously been reported to be an effective therapeutic strategy in endogenous KRAS-driven cancer (13, 14). We chose cisplatin/paclitaxel as an adapted regimen of carboplatin plus paclitaxel, which is a universally accepted regimen in NSCLC (30). Likewise, to pharmacologically inhibit DDR1 we selected the compound 7rh, which is a well-characterized inhibitor previously validated for in vivo use (14). The combination of cisplatin and 7rh enhanced cell death of murine KRasG12V cell lines in vitro when compared with either single treatment alone (Supplemental Figure 2).
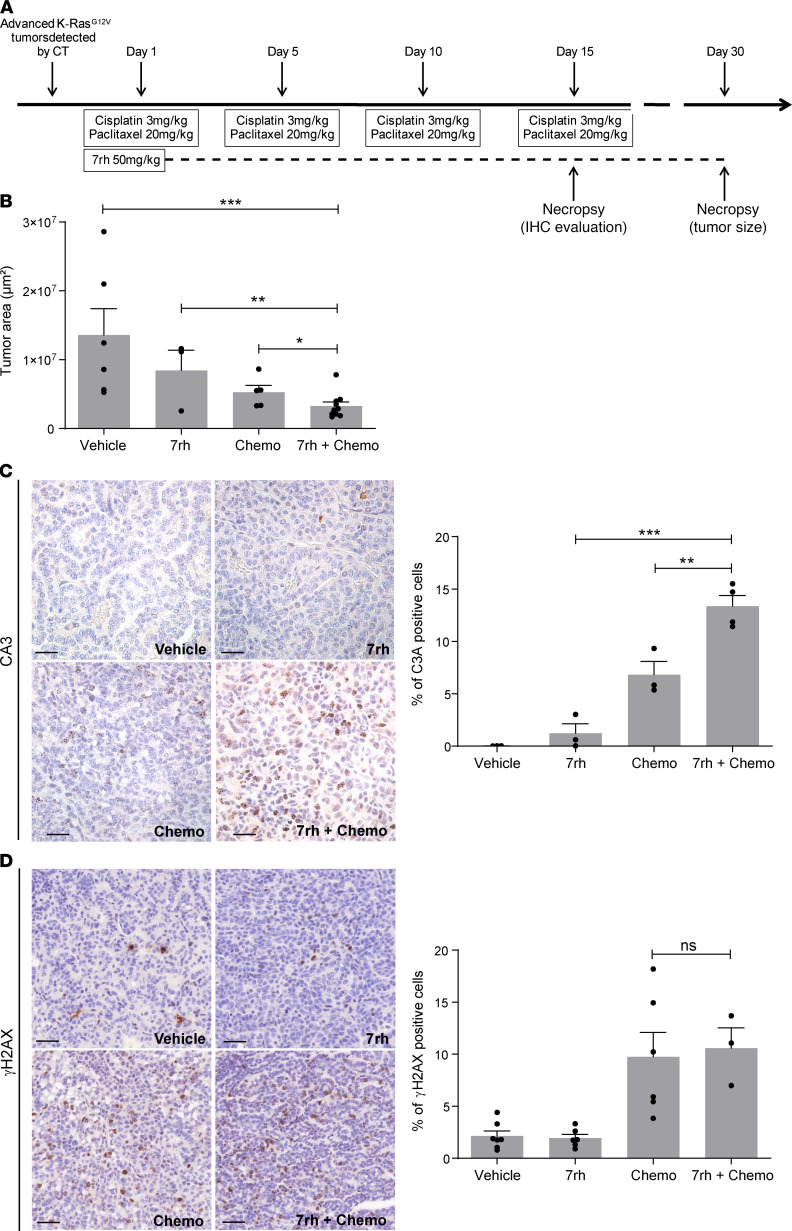
Intratracheal infection of KRasLSLG12Vgeo mice with Cre-recombinase adenoviruses (Ad-Cre) induces the expression of KRasG12V in a limited number of lung cells upon Cre-mediated recombination of the floxed stop cassette inserted within the KRas locus (31). A subset of these KRasG12V-expressing cells progress to LUAD within 8–10 months. Mice bearing KRasG12V-driven LUAD were randomized and treated with either vehicle, the specific DDR1 inhibitor 7rh (32), cisplatin/paclitaxel, or a combination of 7rh+cisplatin/paclitaxel (protocol summarized in Figure 4A). Treatment efficacy was evaluated in lung sections by a quantification of the tumor area showing enhanced tumor shrinkage upon combined 7rh/chemotherapy when compared with either single treatment alone (Figure 4B). Interestingly, we observed a significant increase in apoptosis in tumors treated with the combination therapy in comparison with those subjected to individual treatments (Figure 4C). This increase in cell death upon DDR1 inhibition could be attributed to reduced cell survival or to a previously undescribed effect of DDR1 inhibition in DNA repair resulting in higher DNA-damage burden. To evaluate this possibility, we quantified the presence of the DNA damage marker phosphorylated γ-Histone H2AX (γH2AX) on tumor sections. Interestingly, quantification of γHA2X positive staining was similar in both cisplatin/paclitaxel and 7rh+cisplatin/paclitaxel, indicating that the overall levels of DNA damage induced by cisplatin/paclitaxel in these tumors were not affected by DDR1 inhibition. These results indicate that the increased efficacy of the 7rh+cisplatin/paclitaxel treatment is likely attributed to decreased tumor cell survival as opposed to inefficient DNA repair caused by DDR1 inhibition (Figure 4D).
Figure 4. Treatment with a DDR1 specific inhibitor enhances the efficacy of chemotherapy (chemo) in endogenous KRasG12V-driven lung adenocarcinoma.
(A) Schematic representation of the in vivo protocol used in KRasLSLG12Vgeo mice. (B) Tumor area was calculated on sections obtained from KRasG12V tumors (n = 3–10 tumors per group) following the indicated treatments. Data were analyzed using 1-way ANOVA followed by Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001. Data are shown as the mean ± SEM. (C) Left: representative immunostaining of the apoptotic marker active caspase-3 (C3A) in sections obtained from KRasG12V tumors (n = 6 mice per group) following the indicated treatments. Scale bar: 50 μm. Right: quantification of C3A+ cells. Data were analyzed using 1-way ANOVA followed by Bonferroni’s multiple comparison test. **P < 0.01, ***P < 0.001. Data are shown as the mean ± SEM. (D) Left: representative immunostaining of the DNA damage marker phosphorylated γ-Histone H2AX (γH2AX) in sections of KRasG12V tumors (n = 6 mice per group) following the indicated treatments. Scale bar: 50 μm. Right: quantification of γH2AX+ cells. Data were analyzed using 1-way ANOVA followed by Bonferroni’s multiple comparison test. P value ns, nonsignificant. Data are shown as the mean ± SEM.
Discussion
First-line therapy for advanced KRAS-mutant LUAD consists of platinum-based chemotherapy in combination with pembrolizumab or pembrolizumab alone in patients whose tumors have PD-L1 expression ≥50% (33). In this sense, patients with PD-L1–positive advanced NSCLC harboring KRAS mutations and treated with frontline pembrolizumab in the Keynote-042 were more likely to have a higher overall response rate and longer progression-free survival and overall survival than patients with KRAS WT (34, 35). Patients with advanced LUAD benefited from pembrolizumab combined with carboplatin and pemetrexed regardless of KRAS mutation status (36, 37). Two recent phase I clinical trials (NCT03785249 and NCT03600883) with direct G12C inhibitors showed promising results in this patient subgroup that accounts for approximately one-half of KRAS mutant LUAD (38, 39). However, there is an urgent need to improve treatment of KRAS mutant patients, particularly for advanced-stage disease. Although chemotherapy plus pembrolizumab has yielded promising results, standard chemotherapy also has a high rate of toxicities, and, more important, chemoresistance and relapses invariably occur.
An increasing body of evidence suggests that DDR1 expression is correlated with poor prognosis in different tumor types, including lung (13, 40, 41), is involved in metastatic dissemination (15), and is a suitable target for cancer treatment (13, 42, 43). Notably, DDR1 expression has been linked to decreased sensitivity to chemotherapy (19, 22) and drug resistance (21, 23, 24); however, in vivo DDR1 involvement in KRAS-mutant lung cancer cell persistence upon treatment has not been reported (reviewed in ref. 44).
Our results show a rapid induction of DDR1 expression upon exposure to cisplatin treatment in both murine and human cell lines, suggesting a role for DDR1 in implementing fast adaptive responses to cytotoxic agents (Figure 1). Nevertheless, we cannot exclude that preexisting high DDR1-expressing cells could possess an intrinsic resistance to chemotherapy, which might select for focal viable tumor areas with high DDR1 levels after treatment (Figure 2). Therefore, high DDR1 expression levels in platinum-resistant cells could be attributed to either active DDR1 upregulation and/or to selection of preexisting clones with higher DDR1 baseline expression. Intriguingly, we previously identified a subpopulation of KRasG12V-expressing lung cells in vivo that were committed to an aggressive transcriptional signature as an early event following mutant KRas expression (14): the top hit in this signature was DDR1. Whether high DDR1 expression is a cause or a consequence of selection of aggressive KRas-mutant clones in vivo, and whether this is directly related to intrinsic resistance to chemotherapy remains to be investigated.
To evaluate a potential direct relationship between DDR1 expression and response to therapy in patients with LUAD, we surveyed publicly available gene expression data sets in search of correlations between DDR1 abundance and response to chemotherapy. This was further supported by using mRNA and protein material from our own cohort of LUAD biopsies obtained before platinum-based therapy. Together, these analyses supported the notion that higher DDR1 expression and activity in patients correlated with poor response to chemotherapy (Figure 3). Furthermore, our results obtained both in vitro and in vivo strongly suggest that DDR1 could also be considered as a new predictive biomarker of patient response to chemotherapy. Future studies considering paired pre- and posttreatment biopsies are warranted to further confirm DDR1 as a predictive biomarker in this clinical setting.
We have found substantial variability in DDR1 expression among patients with LUAD reaching up to 10-fold difference when comparing low and high patients (Supplemental Figure 3), suggesting that these differences could be used to stratify patients before chemotherapy. In this context, DDR1 overexpression was not correlated with any particular KRAS oncogenic mutations nor with coexisting alterations in genes known to enhance chemoresistance such as STK11 or KEAP1 (data not shown). This indicates that DDR1 might be an independent prognostic marker of chemoresistance. Although the orthotopic PDX models used in this study are p53-deficient (14), we still detected DDR1 induction upon chemotherapy treatment (Figure 3). In agreement, chemoresistant tumor propagating cells (TPC), which display high levels of DDR1 expression (Figure 3D and Supplemental Figure 1), were identified in p53-deficient LUAD mouse models (29). Additionally, a recent study identified a small subset of genes, including DDR1, which are positively selected upon chemotherapy treatment but unrelated to either proliferation or lineage-associated mutations including p53 (24). In sum, these findings indicate that p53 status is not correlated with DDR1-dependent response to chemotherapy.
We also believe that the results presented here with endogenous KRas-mutant LUAD are proof of the synergistic effect between DDR1 inhibition and chemotherapy. This combination treatment elicited a significant increase in apoptosis in endogenous tumors when compared with either treatment alone (Figure 4). Recently, DDR1 inhibition in a mouse model of pancreatic ductal adenocarcinoma increased tumor response to standard-of-care chemotherapy regimens (45). Similarly, DDR1 inhibition sensitized glioblastoma cells to radio- and chemotherapy resulting in prolonged patient survival (46). Therefore, in addition to LUAD, DDR1-associated chemoresistance has been observed in other tumors. Indeed, DDR1, together with other genes implicated in extracellular matrix remodeling, define a small group of cancer-associated factors that is maintained after chemotherapy in multiple cancer types. In this context, DDR1 is essential for cell line survival and elevated in drug-resistant stem-like cancer cells (24).
To speculate on the applicability of our findings to human tumors, DDR1 inhibition could be achieved with dasatinib, an FDA-approved small-molecule tyrosine kinase inhibitor that was initially developed as a dual ABL-SRC inhibitor and was later found to also display nanomolar inhibitory activity against DDR1 (47). An alternative option could be nilotinib, also FDA-approved and recently shown to reduce colorectal cancer invasion and metastasis by inhibiting DDR1 kinase activity (48).
In conclusion, we propose that stratification based on DDR1 expression might identify responsive patients to the current standard-of-care treatment combining immune checkpoint inhibitors and chemotherapy. Whether DDR1 inhibition might enhance this therapeutic response, resulting in a novel strategy to overcome chemoresistance in KRAS-driven patients with LUAD, will require further investigation.
Methods
Cell lines and drug treatments.
Murine lung cancer cell lines derived from K-RasLSLG12Vgeo mice (ChA14.6 and ChA14.9), human lung cancer cell lines derived from KRAS-mutant PDX (PDX 2 and 3) as previously described (14), and human KRAS-mutant lung cancer cell lines (A549 and H460, purchased from ATCC) were maintained in DMEM containing 10% FBS. Cells were seeded and activated with 50 μg/mL Collagen IV from human placenta (MilliporeSigma, C7521) before adding the corresponding drugs. Cisplatin (MilliporeSigma, P4394) and Doxorubicin (Selleckchem, S1208) were dissolved in 0.9% NaCl and DMSO, respectively, and used at the indicated concentrations. Cell viability was assessed by MTT proliferation assay (Roche). For 3D growth conditions, cells were cultured in ultra-low attachment plates (Corning). For 3D conditions, cell viability was assessed by CellTiter-Glo luminescent viability assay (Promega). For in vivo treatments, the 7rh inhibitor (32) was resuspended in 1% carboxymethyl cellulose, 0.25% Tween 80, and administered daily by oral-gavage at a dose of 50 mg/kg. 7rh was provided by Ke Ding (State Key Laboratory of Respiratory Diseases, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences).
Standard chemotherapy based on cisplatin (3 mg/kg) and paclitaxel (20 mg/kg) was administered i.p. every 5 days. Control mice were treated with the vehicle following identical procedure. Mice were sacrificed within 4–8 hours after the last treatment, and samples were obtained for immunohistochemical analysis.
Mice and patient-derived orthoxenografts.
The KRas+/LSLG12Vgeo strain has been previously described (49). Crl:NU-Foxn1nu mice were purchased from Charles River. Mice were genotyped by Transnetyx (Cordova). Primary tumors were obtained from Bellvitge Hospital (HUB) and Catalan Institute of Oncology (ICO). Tumors were aseptically isolated and placed at room temperature in DMEM supplemented with 10% FBS plus 50 U/mL penicillin and 50 mg/mL streptomycin. The tumors were implanted in Crl:NU-Foxn1nu mice following previously reported procedures (27).
Tumor induction.
Tumors were induced in 8- to 12-week-old mice by single intratracheal Ad-Cre instillation with 1 × 106 pfu/mouse virus after anesthesia (i.p. injection of ketamine 75 mg/kg, xylazine 12 mg/kg) as previously reported (50). A balanced number of both male and female mice was used for each experiment.
Antibodies for in vitro studies.
The antibodies used for Western blotting included those raised against: DDR1 (Cell Signaling, 5583), phospho-DDR1 Y-792 (Cell Signaling, 11994), α-Tubulin (Cell Signaling, 2144), and GAPDH (MilliporeSigma, G8795).
Immunohistochemistry and digital image quantification.
Lung lobes containing either endogenous tumors or implanted PDX biopsies were fixed in 10% buffered formalin (MilliporeSigma) and embedded in paraffin. Antibodies used for immunostaining included those raised against: DDR1 (Cell Signaling, 5583), Caspase3A (R&D Systems, AF835), and phospho-γH2AX (Millipore Sigma, JBW301). Digital images of IHC-stained slides were obtained at 40x magnification (0.12 μm/ pixel) using a whole slide scanner (Mirax scan, Zeiss) fitted with a x40/0.95 Plan Apochromat objective lens (Zeiss). For automated image classification and quantification, the main areas were highlighted according to the tumor morphological differences. These image areas were used as input parameters for the Axiovision 4.8 pattern recognition training software (Zeiss) and analyzed by noise reduction method, threshold, and filter conditions as measurement parameters.
Real-time quantitative PCR.
Total cellular RNA (1 μg), extracted by Quick-RNA MiniPrep plus Kit (Zymo Research), was reverse-transcribed by random primers using One script cDNA Synthesis Kit (Abm), and the reverse transcription reaction was then subjected to PCR amplification using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). PCR signals were recorded on a CFX384 Touch Real-Time PCR System (Bio-Rad) and analyzed using the Bio-Rad CFX Manager software. Primer sets included human DDR1-Fwd (5′-AAGGGACATTTTGATCCTGCC-3′), human DDR1-Rv (5′-CCTTGGGAAACACCGACCC-3′), human HPRT-Fwd (5′-TGACACTGGCAAAACAATGCA-3′), human HPRT-Rv (5′-GGTCCTTTTCACCAGCAAGCT-3′), murine DDR1-Fwd (5′-ATGCTGACATGAAGGGACATTT-3′), murine DDR1-Rv (5′-GGTGTAGCCTACGAAGGTCCA-3′), murine GAPDH-Fwd (5′-AGGTTGTCTCCTGCGACTTCA-3′), and murine GAPDH-Rv (5′-GGTGGTCCAGGGTTTCTTACTC-3′).
DDR1 shRNA-mediated knockdown.
For knockdown assays cells were infected using lentiviral particles obtained using MISSION shRNAs directed against human DDR1, shDDR1 960 (Sigma ref. TRCN0000010094), and shDDR1 1358 (Sigma ref. TRCN0000010084). Lentiviral particles expressing a Non-Target shRNA vector (MilliporeSigma) were used as a negative control.
Patient cohort.
Primary tumors were obtained at Hôpital Arnaud de Villeneuve (Centre Hospitalo-Universitaire de Montpellier, France) following ethical and legal protection guidelines of human subjects, including informed consent. All patients were diagnosed as LUAD and received at least 3 cycles of platinum-based chemotherapy. Patient response to treatment was classified as responders or nonresponders, depending on the absence and/or persistence of the initial lymph-node metastasis, respectively.
External data sets— patient selection criteria.
Using the online KM plotter database, we evaluated a cohort of 125 chemotherapy-treated NSCLC patients, with available data regarding gene-expression and first-progression survival. Of the 125 patients, 106 were diagnosed with squamous cell carcinoma and 19 with LUAD. Kaplan-Meier analysis was performed splitting DDR1 expression using the median value.
For the analysis of the TCGA data set, we focused on patients with stage II LUAD because we hypothesized this was the cohort in which the chemotherapy clinical benefit could be more easily measured. Among 117 stage II LUAD cases with available RNA-Seq data, we first withdrew 41 patients from the study owing to incomplete clinical annotations regarding the primary outcome after treatment. We then removed 17 additional patients who harbored driver oncogenes for which a targeted treatment might have been available at the time of diagnosis (i.e., EGFR, ALK, ERBB2, MET, ROS1, and RIT1). We analyzed DDR1 expression in the remaining 59 stage II patients.
Gene expression analysis.
GSE46439 normalized microarray data were retrieved from the GEO database, and the 450 differentially expressed genes identified by the authors were used to compute Spearman’s correlation between samples. All replicates generated by the authors were used in the analysis, with the exception of replicate 5 from the non-TPC samples. We discarded this replicate because it showed a high correlation with both groups (TPC and non-TPC) as shown in Supplemental Figure 1. DDR1 expression distribution was assessed in the 10 remaining samples.
To analyze DDR1 expression in TCGA patients we used the RSEM-normalized RNA-seq data provided by the TCGA consortium. The difference between groups (responder/nonresponders, Figure 3B, and TPC/non-TPC, Figure 3D) has been evaluated by performing a Wilcoxon rank-sum test.
Statistics.
All experiments were performed as several independent biological replicates. All results are reported as mean ± SD or SEM, as indicated in figure legends. Two group comparisons were performed using unpaired 2-tailed Student’s t test. When an experiment required comparisons between more than 2 groups, statistical analysis was performed using 1- or 2-way ANOVA depending on the number of grouping factors. Dunnett’s test was applied for simple comparisons, whereas Bonferroni’s tests were used for multiple comparisons. A P value less than 0.05 was considered significant. Tumor bearing-mice were randomized before drug treatment. No statistical method was used to predetermine sample size.
Study approval.
All animal experiments were approved by the Ethical Committee of the CNIO (no. PA61-2010, PA30-2011) and the Bellvitge Biomedical Research Institute (IDIBELL, no. AAALAC-1155) and performed in accordance with the guidelines stated in the International Guiding Principles for Biomedical Research Involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). Primary tumors were obtained from Bellvitge Hospital (HUB) and Catalan Institute of Oncology (ICO) upon approval by the Ethical Committee (CEIC Bellvitge Hospital no. PR265/13 and PR036/14) and Hôpital Arnaud de Villeneuve (Centre Hospitalo-Universitaire de Montpellier, France) with approval by the Ethical Committee (Comité de Protection des Personnes Sud Méditerranée I no. RO-2016/33) following ethical and legal protection guidelines of human subjects. Specific informed consent for tumor implantation in mice was obtained from all patients, and the study received the approval of the IDIBELL and the Hôpital Arnaud de Villeneuve Ethics Committees.
Author contributions
MJN, EN, DS, and CA designed the research studies. MJN, CT, BD, AL, SSJ, NC, BT, VPM, MF, MH, and CA conducted the experiments. ED, HW, and MM acquired the data. MJN, ED, CT, HW, MH, MM, AM, DS, and CA analyzed the data. AV, MH, PAJ, JLP, AM, and MB provided the reagents. MJN, AM, EN, DS, and CA wrote the manuscript.
Supplementary Material
Acknowledgments
The results shown here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). This work was supported by grants from Fondation ARC pour la recherche contre le cancer (PJA 20161204784), Region Nouvelle Aquitaine (2017-1R30212), Siric-BRIO and a Senior Chair Idex-Université de Bordeaux (2016-0396) to DS. CA’s laboratory is supported by the Giovanni Armenise–Harvard Foundation and the Lung Cancer Research Foundation (LCRF). CA was also supported by a fellowship from the Spanish Association Against Cancer (AECC). MJN was supported by a bourse d’excellence de la Fédération Wallonie-Bruxelles (WBI) and a postdoctoral fellowship from Fondation ARC pour la recherche contre le cancer. EN received support from the Acció instrumental d’intensificació de professionals de la salut (SLT006/17/00127) of the Department of Health of the Government of Catalonia and from the Spanish Society of Medical Oncology (SEOM) Foundation. We thank CERCA Program/Generalitat de Catalunya for their institutional support and grant 2017SGR448.
Version 1. 08/06/2020
Electronic publication
Footnotes
Conflict of interest: MB reports a research contract from Pfizer and Eli Lilly and a paid consultancy from amcure. AV is a cofounder of the spin-off company Xenopat. EN reports research funding from Pfizer and Roche. PAJ has received consulting fees from AstraZeneca, Boehringer-Ingelheim, Pfizer, Roche/Genentech, Eli Lilly and Company, ACEA Biosciences, Araxes Pharma, Ignyta, Mirati Therapeutics, Daiichi-Sankyo, Takeda Oncology, Novartis, Voronoi, SFJ Pharmaceuticals, Biocartis, Sanofi Oncology, and LOXO Oncology; receives postmarketing royalties from DFCI-owned intellectual property on EGFR mutations licensed to Lab Corp; has sponsored research agreements with AstraZeneca, Daiichi-Sankyo, Boehringer Ingelheim, PUMA, Eli Lilly and Company, Astellas Pharmaceuticals, Revolution Medicines, and Takeda Oncology; and has stock ownership in Gatekeeper Pharmaceuticals and LOXO Oncology.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(15):e137869.https://doi.org/10.1172/jci.insight.137869.
Contributor Information
Marie-Julie Nokin, Email: mj.nokin@iecb.u-bordeaux.fr.
Elodie Darbo, Email: elodie.darbo@u-bordeaux.fr.
Camille Travert, Email: travertcam@gmail.com.
Benjamin Drogat, Email: bdrogat@gmail.com.
Aurélie Lacouture, Email: aurelie.lacouture@gmail.com.
Sonia San José, Email: s.san-jose@iecb.u-bordeaux.fr.
Nuria Cabrera, Email: ncabrera@srv.cnio.es.
Béatrice Turcq, Email: beatrice.turcq@u-bordeaux.fr.
Mattia Falcone, Email: Mattia.Falcone@glasgow.ac.uk.
Alberto Villanueva, Email: avillanueva@iconcologia.net.
Haiyun Wang, Email: wanghaiyun@tongji.edu.cn.
Michael Herfs, Email: m.herfs@uliege.be.
Miguel Mosteiro, Email: mmosteiro@bellvitgehospital.cat.
Jean-Louis Pujol, Email: jl-pujol@chu-montpellier.fr.
Antonio Maraver, Email: antonio.maraver@inserm.fr.
Mariano Barbacid, Email: mbarbacid@cnio.es.
Ernest Nadal, Email: esnadal@iconcologia.net.
David Santamaría, Email: d.santamaria@iecb.u-bordeaux.fr.
Chiara Ambrogio, Email: chiara.ambrogio@unito.it.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Karachaliou N, et al. KRAS mutations in lung cancer. Clin Lung Cancer. 2013;14(3):205–214. doi: 10.1016/j.cllc.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Réjiba S, Wack S, Aprahamian M, Hajri A. K-ras oncogene silencing strategy reduces tumor growth and enhances gemcitabine chemotherapy efficacy for pancreatic cancer treatment. Cancer Sci. 2007;98(7):1128–1136. doi: 10.1111/j.1349-7006.2007.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol. 2013;31(8):1112–1121. doi: 10.1200/JCO.2012.43.0454. [DOI] [PubMed] [Google Scholar]
- 5.Marabese M, et al. KRAS mutations affect prognosis of non-small-cell lung cancer patients treated with first-line platinum containing chemotherapy. Oncotarget. 2015;6(32):34014–34022. doi: 10.18632/oncotarget.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goeman F, et al. Mutations in the KEAP1-NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J Thorac Oncol. 2019;14(11):1924–1934. doi: 10.1016/j.jtho.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Oliver TG, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24(8):837–852. doi: 10.1101/gad.1897010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao S, et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res. 2014;74(24):7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, et al. Kras mutations increase telomerase activity and targeting telomerase is a promising therapeutic strategy for Kras-mutant NSCLC. Oncotarget. 2017;8(1):179–190. doi: 10.18632/oncotarget.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature. 2016;538(7623):114–117. doi: 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitinger B. Discoidin domain receptor functions in physiological and pathological conditions. Int Rev Cell Mol Biol. 2014;310:39–87. doi: 10.1016/B978-0-12-800180-6.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valiathan RR, Marco M, Leitinger B, Kleer CG, Fridman R. Discoidin domain receptor tyrosine kinases: new players in cancer progression. Cancer Metastasis Rev. 2012;31(1–2):295–321. doi: 10.1007/s10555-012-9346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valencia K, et al. Inhibition of collagen receptor discoidin domain receptor-1 (DDR1) reduces cell survival, homing, and colonization in lung cancer bone metastasis. Clin Cancer Res. 2012;18(4):969–980. doi: 10.1158/1078-0432.CCR-11-1686. [DOI] [PubMed] [Google Scholar]
- 14.Ambrogio C, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med. 2016;22(3):270–277. doi: 10.1038/nm.4041. [DOI] [PubMed] [Google Scholar]
- 15.Gao H, et al. Multi-organ Site Metastatic Reactivation Mediated by Non-canonical Discoidin Domain Receptor 1 Signaling. Cell. 2016;166(1):47–62. doi: 10.1016/j.cell.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuge R, et al. Silencing of discoidin domain receptor-1 (DDR1) concurrently inhibits multiple steps of metastasis cascade in gastric cancer. Transl Oncol. 2018;11(3):575–584. doi: 10.1016/j.tranon.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YC, et al. Collagen-rich airway smooth muscle cells are a metastatic niche for tumor colonization in the lung. Nat Commun. 2019;10(1):2131. doi: 10.1038/s41467-019-09878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ongusaha PP, et al. p53 induction and activation of DDR1 kinase counteract p53-mediated apoptosis and influence p53 regulation through a positive feedback loop. EMBO J. 2003;22(6):1289–1301. doi: 10.1093/emboj/cdg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cader FZ, et al. The EBV oncogene LMP1 protects lymphoma cells from cell death through the collagen-mediated activation of DDR1. Blood. 2013;122(26):4237–4245. doi: 10.1182/blood-2013-04-499004. [DOI] [PubMed] [Google Scholar]
- 20.Kim HG, Hwang SY, Aaronson SA, Mandinova A, Lee SW. DDR1 receptor tyrosine kinase promotes prosurvival pathway through Notch1 activation. J Biol Chem. 2017;292(17):7162. doi: 10.1074/jbc.A111.236612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai SL, et al. Collagen induces a more proliferative, migratory and chemoresistant phenotype in head and neck cancer via DDR1. Cancers (Basel) 2019;11(11):E1766. doi: 10.3390/cancers11111766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Y, et al. Suppressing miR-199a-3p by promoter methylation contributes to tumor aggressiveness and cisplatin resistance of ovarian cancer through promoting DDR1 expression. J Ovarian Res. 2017;10(1):50. doi: 10.1186/s13048-017-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye L, et al. Transmembrane-4 L-six family member-1 (TM4SF1) promotes non-small cell lung cancer proliferation, invasion and chemo-resistance through regulating the DDR1/Akt/ERK-mTOR axis. Respir Res. 2019;20(1):106. doi: 10.1186/s12931-019-1071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selfors LM, Stover DG, Harris IS, Brugge JS, Coloff JL. Identification of cancer genes that are independent of dominant proliferation and lineage programs. Proc Natl Acad Sci U S A. 2017;114(52):E11276–E11284. doi: 10.1073/pnas.1714877115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbone A, Gloghini A. Activated DDR1 increases RS cell survival. Blood. 2013;122(26):4152–4154. doi: 10.1182/blood-2013-11-534123. [DOI] [PubMed] [Google Scholar]
- 26.Guo J, et al. 3D culture enhances chemoresistance of ALL Jurkat cell line by increasing DDR1 expression. Exp Ther Med. 2019;17(3):1593–1600. doi: 10.3892/etm.2019.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambrogio C, et al. Modeling lung cancer evolution and preclinical response by orthotopic mouse allografts. Cancer Res. 2014;74(21):5978–5988. doi: 10.1158/0008-5472.CAN-14-1606. [DOI] [PubMed] [Google Scholar]
- 28.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell. 2013;24(1):59–74. doi: 10.1016/j.ccr.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joerger M, et al. Open-label, randomized study of individualized, pharmacokinetically (PK)-guided dosing of paclitaxel combined with carboplatin or cisplatin in patients with advanced non-small-cell lung cancer (NSCLC) Ann Oncol. 2016;27(10):1895–1902. doi: 10.1093/annonc/mdw290. [DOI] [PubMed] [Google Scholar]
- 31.Puyol M, et al. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18(1):63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Gao M, et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. J Med Chem. 2013;56(8):3281–3295. doi: 10.1021/jm301824k. [DOI] [PubMed] [Google Scholar]
- 33.Planchard D, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(suppl 4):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 34.Herbst RS, et al. Use of archival versus newly collected tumor samples for assessing PD-L1 expression and overall survival: an updated analysis of KEYNOTE-010 trial. Ann Oncol. 2019;30(2):281–289. doi: 10.1093/annonc/mdy545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mok TSK, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 2019;393(10183):1819–1830. doi: 10.1016/S0140-6736(18)32409-7. [DOI] [PubMed] [Google Scholar]
- 36.Gadgeel SM, et al. Pembrolizumab and platinum-based chemotherapy as first-line therapy for advanced non-small-cell lung cancer: Phase 1 cohorts from the KEYNOTE-021 study. Lung Cancer. 2018;125:273–281. doi: 10.1016/j.lungcan.2018.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazieres J, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30(8):1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fakih M, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRAS G12C inhibitor, in advanced solid tumors. J Clin Oncol. 2019;37(suppl_15):3003 [Google Scholar]
- 39.Hallin J, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10(1):54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinzelmann-Schwarz VA, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10(13):4427–4436. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- 41.Miao L, et al. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung cancer and promotes cell invasion via epithelial-to-mesenchymal transition. Med Oncol. 2013;30(3):626. doi: 10.1007/s12032-013-0626-4. [DOI] [PubMed] [Google Scholar]
- 42.Villalba M, et al. Identification of a novel synthetic lethal vulnerability in non-small cell lung cancer by co-targeting TMPRSS4 and DDR1. Sci Rep. 2019;9(1):15400. doi: 10.1038/s41598-019-51066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrogio C, et al. KRAS-driven lung adenocarcinoma: combined DDR1/Notch inhibition as an effective therapy. ESMO Open. 2016;1(5):e000076. doi: 10.1136/esmoopen-2016-000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambrogio C, Darbo E, Lee SW, Santamaría D. A putative role for Discoidin Domain Receptor 1 in cancer chemoresistance. Cell Adh Migr. 2018;12(4):394–397. doi: 10.1080/19336918.2018.1445954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguilera KY, et al. Inhibition of discoidin domain receptor 1 reduces collagen-mediated tumorigenicity in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2017;16(11):2473–2485. doi: 10.1158/1535-7163.MCT-16-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vehlow A, et al. Interaction of discoidin domain receptor 1 with a 14-3-3-Beclin-1-Akt1 complex modulates glioblastoma therapy sensitivity. Cell Rep. 2019;26(13):3672–3683.e7. doi: 10.1016/j.celrep.2019.02.096. [DOI] [PubMed] [Google Scholar]
- 47.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 48.Jeitany M, et al. Inhibition of DDR1-BCR signalling by nilotinib as a new therapeutic strategy for metastatic colorectal cancer. EMBO Mol Med. 2018;10(4):e7918. doi: 10.15252/emmm.201707918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4(2):111–120. doi: 10.1016/S1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 50.Blasco RB, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19(5):652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.