Abstract
Pancreatic ductal adenocarcinoma (PDAC) is characterized by a relative paucity of cancer cells that are surrounded by an abundance of nontumor cells and extracellular matrix, known as stroma. The interaction between stroma and cancer cells contributes to poor outcome, but how proteins from these individual compartments drive aggressive tumor behavior is not known. Here, we report the proteomic analysis of laser-capture microdissected (LCM) PDAC samples. We isolated stroma, tumor, and bulk samples from a cohort with long- and short-term survivors. Compartment-specific proteins were measured by mass spectrometry, yielding what we believe to be the largest PDAC proteome landscape to date. These analyses revealed that, in bulk analysis, tumor-derived proteins were typically masked and that LCM was required to reveal biology and prognostic markers. We validated tumor CALB2 and stromal COL11A1 expression as compartment-specific prognostic markers. We identified and functionally addressed the contributions of the tumor cell receptor EPHA2 to tumor cell viability and motility, underscoring the value of compartment-specific protein analysis in PDAC.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Molecular biology, Proteomics
In-depth proteomic analysis of laser-capture isolated pancreatic ductal adenocarcinoma tissue identifies tumor- and stroma-specific prognostic markers.
Introduction
Large-scale omics efforts to identify key mediators of tumor biology have typically focused on the tumor compartment as an entity by itself, and tissue samples for gene expression and genomic analyses are commonly selected based on high tumor purity (1). This bias toward the epithelial compartment understates the influence of the microenvironment and its heterocellular composition (2). Further, by studying tumor tissue in bulk, the composition of specific compartments within the tumor can be masked. Bioinformatics efforts to delineate the compartment-specific expression have overcome this problem partially and increased our understanding of tumor biology (3). However, the inference of cellular composition by bioinformatics remains an estimation that requires validation, for instance, by the analysis of physically separated compartments. Especially in light of current molecular subtyping efforts to improve survival prediction and achieve a personalized treatment schedule, the characterization of the specific expression profiles of tumor cells and their microenvironment is key to identify patient groups more effectively and to discover therapeutic targets.
The relevance of the tumor-stroma interaction is of particular importance in tumors that are characterized by high stromal content, such as pancreatic ductal adenocarcinoma (PDAC). PDAC typically consists of small tumor islands surrounded by abundant extracellular matrix (ECM) and stromal cells, which together constitute the vast majority of the tumor bulk (4). This stroma is known to drive multiple protumorigenic features in PDAC tumor cells; it stimulates proliferation and metastatic growth, creates an immune-suppressive environment (5), and activates signaling via mechanobiology (6). However, stromal depletion strategies had disappointing clinical results, suggesting that PDAC stroma also harbors tumor-restraining properties that suggest that a complex stromal plasticity exists (7–9).
PDAC responds poorly to systemic treatment, and only 9% of all patients diagnosed with PDAC survive more than 5 years (10). New chemotherapeutic regimens have improved the survival rate by several months (11, 12), and, hopefully, (neo)adjuvant multimodal approaches will improve survival further (13, 14). However, a better understanding of this tumor type is essential for the development of more effective, targeted therapeutic approaches and to select patients for such therapies. Several gene expression–based classification efforts have delineated the heterogeneity that is thought to contribute to the typical poor outcomes (15, 16), but most studies have largely ignored the separate stromal and epithelial contributions. Moffitt et al. (17) established molecular PDAC subtypes specific to the epithelial and stromal compartment by using an in silico deconvolution method on the transcriptomic landscape of bulk tumor tissue. Recently, Maurer et al. (18) performed in-depth transcriptomics of laser-capture microdissection (LCM) of PDAC, validating stromal and epithelial subtypes. These data provided evidence of the differential expression and pathways in both tumor cells and stroma and their divergent effects on prognosis. This compartment-specific complexity in PDAC warrants further understanding of each contribution to the disease outcome.
To date, no large-scale proteomic studies have been performed to identify the tumor- and stroma-specific protein landscapes of PDAC. Importantly, LCM can select compartment-specific areas and highly enrich the analysis for specific cell types (19). So far, LCM has predominantly been coupled with genomic or transcriptomic analysis, whereas proteomic studies of LCM material are rare due to the laborious efforts required to yield enough protein from the samples, especially from the stromal compartment (20). With the recent advances in sensitivity, throughput, and scan rate of liquid chromatography–mass spectrometry–based (LC-MS–based) proteomics (21), coupling LCM with LC-MS has become an attractive approach for identifying the functional proteomic landscape of PDAC and unraveling compartment-specific biology.
We hypothesized that tumor and stromal proteomic landscapes can yield complementary molecular information downstream of the transcriptome. Furthermore, we postulated that these data can be leveraged to dissect compartment-specific biology with prognostic value and to reveal targetable pathways. In this study, we report an extensive data set from LCM PDAC tissue and patient-derived xenografts (PDXs) and identify over 6000 proteins.
Results
Clinicopathological evaluation and depth of analysis.
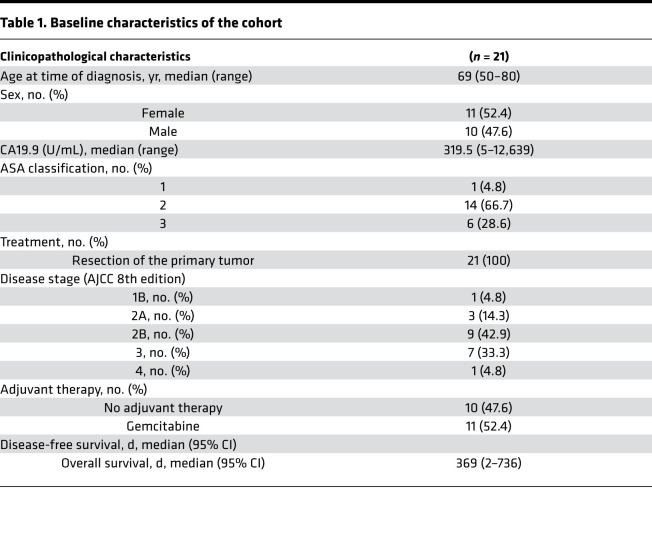
Of the 39 evaluated primary tumor samples, 16 were selected for LCM after confirmation of PDAC histology, with selection based on tissue quantity, tissue quality, and a tumor percentage of at least 5%. Additionally, bulk tumor analysis was performed on 11 samples matched with the LCM samples as well as on 5 unmatched bulk tumor samples. Baseline clinical characteristics of the total group (n = 21) are shown in Table 1 (additional information is provided in Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138290DS1). Three patients showed no sign of recurrence at last follow-up visit. The median overall survival (OS) of the cohort was 358 days. Age, sex, disease stage, and nodal involvement showed no statistically significant association with OS in this surgical cohort, likely due to small group sizes. Eleven patients received adjuvant systemic therapy, which associated with improved OS (log-rank test, P = 0.04). Tumor purities ranged from 5% to 70% (Supplemental Figure 1A), reflecting the known abundance of, and variance in, stromal content in PDAC.
Table 1. Baseline characteristics of the cohort.
A visualization of the workflow is shown in Figure 1A. Large areas of tumor and adjacent stromal areas were successfully isolated by LCM (Figure 1B). Single-shot LC-MS/MS analysis identified a total of 6214 unique proteins, yielding the deepest proteome data set of PDAC to date to our knowledge. Technical replicates were highly correlated (Supplemental Figure 1B), confirming reproducibility of the workflow. We identified 1866 unique epithelial proteins and 220 unique stromal proteins (Supplemental Figure 1C), indicating that protein expression is most heterogeneous in the tumor cell compartment.
Figure 1. Proteome analysis of PDAC samples.
(A) Flowchart of the experimental set up of the proteome landscape analysis (n = 16 for the LCM analysis, n = 16 bulk tumor analysis). (B) Representative example images of 2 PDAC tumors during the LCM procedure. Slides show before and after tumor microdissection and after stroma microdissection. Original magnification, ×10. (C) Principle component analysis (PCA) of MS/MS data shows separation of the epithelial compartment from stroma (n = 16), tumor (n = 15), and bulk samples (n = 11). (D) Unsupervised hierarchical clustering of all proteins shows compartment-specific expression of all samples (n = 42). Heatmap colors indicate relative expression levels.
Principal component analysis (Figure 1C) confirmed successful separation of the compartments, and this was supported by unsupervised clustering (Figure 1D) and correlation analysis (Supplemental Figure 1D). Interestingly, bulk tumors were more similar to the stromal samples than to the tumor compartment, regardless of tumor purity. Pearson correlation analysis confirmed the significantly higher correlation of bulk samples to compared with that of bulk samples to the tumor compartment (P < 0.0001, Supplemental Figure 1E). This result is in agreement with the high fraction of stroma in these tumors and emphasizes the caveats of bulk tumor analyses. Moreover, interpatient correlation of tumor compartment samples was significantly lower (mean R2 = 0.5731) than interpatient correlation of stromal samples (mean R2 = 0.7377; P < 0.0001, Supplemental Figure 1F), indicating that the tumor compartment is the predominant source of heterogeneity in PDAC. This can partly be explained by the lower protein variety of the stromal compartment, as is shown from the lower number of different proteins expressed in stroma (Supplemental Figure 1C), but we propose that this is caused primarily by the relative genetic instability in the tumor compartment.
Biological characterization of epithelial and stromal proteomes.
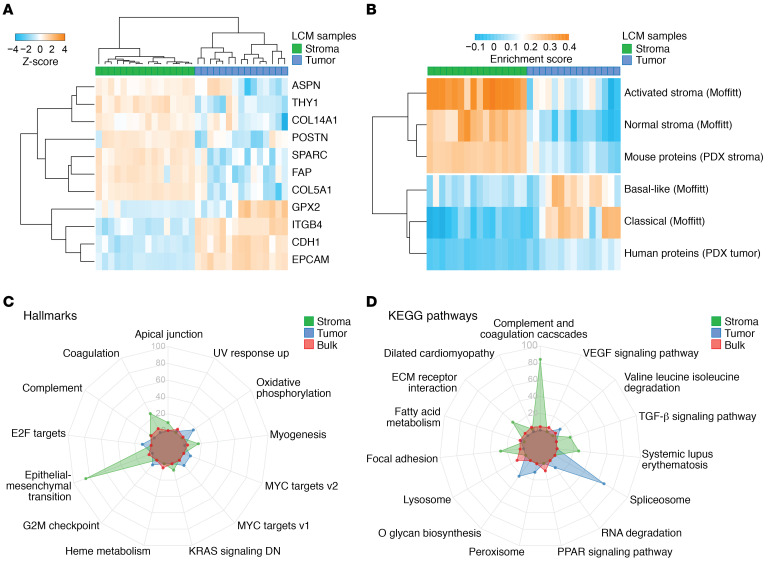
Unsupervised clustering using known markers for epithelial and stromal cells (Figure 2A) confirmed successful separation of compartments. This was also evident from per-sample analysis of selected tumor marker proteins (Supplemental Figure 1G). To further validate our LCM approach and compartment specificity, proteome data from 10 previously established PDAC PDXs (22) were generated. In these models, mouse cells and matrix proteins rapidly replace the human stroma, and the limited homology in amino acid sequence between mouse host and human donor can be leveraged to identify stromal and tumor cell–derived proteins. Species-specific unique proteins were allocated to a human or mouse protein set if they were identified in 5 of 10 samples. This approach yielded 1159 human- and 355 mouse-specific proteins, which we take to represent tumor and stroma proteins, respectively (Supplemental Data File 5). Single-sample gene set enrichment analysis (ssGSEA) showed strong correlation of mouse proteins to the stromal proteins from the LCM approach. Likewise, human proteins associated with the epithelial/tumor LCM compartment, but this enrichment was less evident (Figure 2B). The latter is likely caused by human PDX (tumor cell) proteins that are also expressed in stroma, diluting the relative enrichment. Additionally, ssGSEA of in silico identified stromal and tumor subtypes (17) showed an enrichment pattern confirming compartment specificity. Two tumor samples showed lower enrichment for both previously described Moffitt subtypes (17), implying the existence of tumor cell clones of other subtypes within these tissues or contamination by normal pancreatic tissue.
Figure 2. Successful LCM yields compartment-specific protein identification and biology enrichment.
(A) Unsupervised hierarchical clustering of known tumor and stromal protein markers shows mutually exclusive expression patterns. (B) ssGSEA of previously published and newly created mouse/human PDX tumor and stroma gene sets (17) was performed, showing enrichment in concordance with compartment specificity. Data are shown in heatmap. Each column represents a tumor (n = 15) or stroma sample (n = 16). Each row represents a gene set that was used for enrichment. Colors indicate the enrichment score. (C) Radar plots of GSEA performed on “Hallmarks of cancer” and “KEGG pathways” (D) between sample origin reveal divergent biology of PDAC tumor and stroma, where bulk samples resemble stromal biology. Significant enriched gene sets were identified with GAGE (56), and the top 5 gene sets from each tissue origin with a P value of less than 0.05 are visualized.
Regulation of protein expression downstream of RNA transcription is known to contribute to disparities between transcriptome and proteome data. To assess whether this is the case in models for PDAC, we correlated RNA-Seq data from the PDXs to their proteomics data and found an intermediate correlation of 0.55 between protein and mRNA transcripts, in line with previously reported data (Supplemental Figure 2A) (23). Of note, some proteins showed very high correlation to their transcripts, such as ENO1, while others showed very poor correlation (PSMB1; Supplemental Figure 2, B and C). These findings suggest that a sizeable fraction of proteins are not accurately reflected in transcriptome data and underscore the added value of proteome analysis.
Next, we performed group GSEA of canonical pathways and biological processes. These analyses confirmed the divergent biological functions of tumor and the tumor microenvironment. Global pathway and signature analysis revealed characteristic pathways, such as ECM organization, complement cascade, and epithelial-mesenchymal transition, for the stroma (Figure 2C; Hallmark gene sets). In epithelial samples, enrichment of spliceosome and glycan biosynthesis signatures were found (Figure 2D; KEGG pathway gene sets). These associations confirm the biology known to underlie the epithelial and stromal compartments in PDAC. Moreover, the high enrichment for cancer cell populations also allowed the identification of less-explored biological processes. This includes, for example, oxidative phosphorylation, which was not recognized in bulk human tissue samples that largely reflect stromal GSEA signatures (Figure 2, C and D).
Furthermore, unsupervised clustering on the most variable compartment-specific proteins showed 2 subclasses in the epithelial compartment, (Supplemental Figure 3A), of which cluster 2 showed a trend toward improved OS (Supplemental Figure 3B). Comparison of these groups with existing transcriptome tumor-specific subtypes revealed intermediate concordance (Supplemental Figure 3A), which might be due to sample size, precluding effective unsupervised clustering, or inapplicability of RNA-based classifications on enriched tumor compartment proteome data.
Additionally, a mixture of classical and basal-like protein expression in a single sample was identified in multiple tumors (Figure 2B), highlighting a degree of intratumor heterogeneity that likely results from the existence of populations of cells from different subtypes, as was recently described using gene expression data (24, 25). In the stroma compartment, 2 groups were identified (Supplemental Figure 3A), and these associated with the Moffitt et al. stromal subtypes on the proteome level (17). The normal stroma subgroup showed a trend toward better disease-free survival (DFS, log-rank test, P = 0.081, Supplemental Figure 3C). The activated stromal phenotype was identified more often than normal stroma, as described previously (n = 12 vs. n = 4, respectively) (17). In conclusion, these data reveal a profound heterogeneity in the proteome landscape of PDAC, with complex biological functionality of both tumor and stroma.
Compartment-specific proteins are associated with poor prognosis.
Based on the results above, we hypothesized that LCM can enrich for prognostic proteins previously masked in bulk tissue analysis and that it can uncover tumor biology that underlies the poor disease outcome. To identify prognostic markers, patients with a short survival (n = 6) were compared with patients that survived more than 2 years following resection of the tumor (n = 6). These analyses showed divergent compartment-specific biology associated with disease outcome (Figure 3, A and B). Tumor areas from patients with a poor outcome were enriched for DNA replication and multiple cardiovascular signatures (Figure 3A), which represent proliferation and contractility. Importantly, tumor and stromal areas of patients with poor outcomes both showed enrichment for ECM receptor interaction, emphasizing the interaction of these compartments. Stroma from patients with a worse survival was enriched for focal adhesion pathway genes (Figure 3B), previously described as possible mediators of protumorigenic mechanosignaling (6). The majority of prognostic proteins identified in the stromal compartment by our approach were not specifically expressed in the microenvironment but were also found expressed in tumorous areas. After filtering for compartment specificity, 23 epithelial (Figure 3C) and 5 stromal proteins (Figure 3D) were found to associate with poor prognosis.
Figure 3. Differential expression of short-term versus long-term survival reveals prognostic protein markers.
GSEA of compartment-specific prognostic biology of (A) tumor and (B) stroma compartments. Red, poor prognosis gene sets; blue, good prognosis gene sets. The size of the bar indicates the significant normalized enrichment score (NES). Volcano plots shows differential proteins between poor (n = 6) and good (n = 6) prognosis patients in the tumor (C) and stromal compartment (D) (P < 0.05, limma test). Filtering on compartment specificity (cut-off at significance of P < 0.1, limma test). Black triangles indicate compartment-specific filtering. Colored dots represent significant proteins. (E) Forest plot of meta-analysis of compartment-specific prognostic markers from proteome analysis. Hazard ratio with confidence interval is visualized (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, random effects model). Of the 28 proteins, 17 were validated as poor prognostic markers in transcriptomic data sets.
A meta-analysis based on previously published transcriptomic data sets was implemented for further validation of the identified markers (15–17, 26–28). Of 28 poor prognostic proteins identified, 17 were validated in the meta-analysis (Figure 3E), of which 15 were from the tumor and 2 from the stromal compartment, respectively. Additionally, of the 28 proteins, 16 were previously described in PDAC biology (Table 2), supporting the validity of our approach.
Table 2. Compartment-specific prognostic proteins.
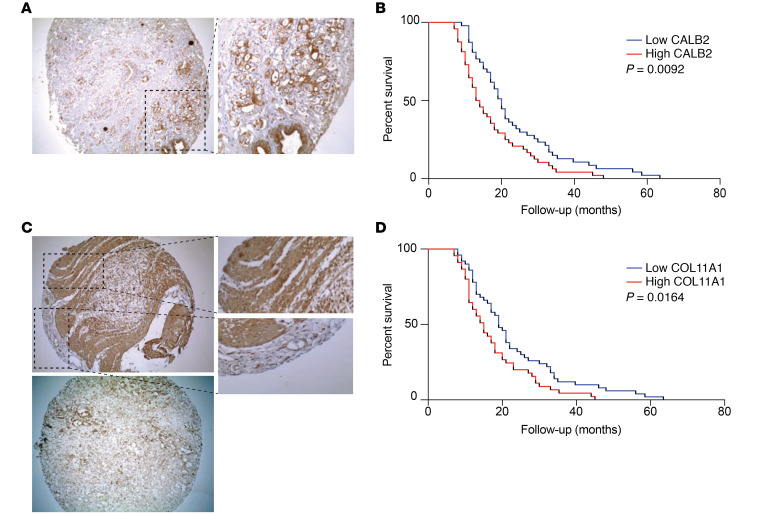
Next, prognostic stromal and epithelial markers were validated by IHC in 2 large independent cohorts of patients with resected PDAC (Supplemental Table 3). The epithelial marker calretinin/calbindin 2 (CALB2) was significantly correlated with OS in both cohorts (Figure 4, A and B; Supplemental Figure 4A; and Supplemental Table 3; P = 0.009 and P = 0.006, respectively). Predicted stromal marker collagen α-1(XI) chain (COL11A1) was significantly correlated with survival in the first independent cohort (Figure 4, C and D; P = 0.016); however, in the second validation cohort, no significance was observed (Supplemental Table 3 and Supplemental Figure 4B). This might be explained by the use of a tissue microarrays (TMAs) with cores selected for high tumor cell content that are therefore less representative of the stroma.
Figure 4. Validation of prognostic proteins CALB2 and COL11A1.
(A) Representative IHC of TMA cores with CALB2-positive staining (original magnification, ×10 [left]; ×40 [right]). (B) Kaplan-Meier survival analysis of high (red, n = 47) or low (blue, n = 48) expression of CALB2 in the first independent cohort shows significant correlation to OS (median OS, 13.5 versus 20 months, P < 0.0092, log-rank test). (C) Representative IHC of TMA cores with CALB2-positive and -negative staining (original magnification, ×10 [left]; ×40 [right]). Additional views of a positive and negative area inside 1 tumor core are highlighted. (D) Kaplan-Meier analysis of high (red, n = 50) or low (blue, n = 45) expression of COL11A1 shows correlation in the first cohort (median OS, 15 versus 19 months, P < 0.016, log-rank test).
Identification of drug targets against PDAC.
PDAC treatment currently depends on combinations of nontargeted cytotoxics (11, 12). The addition of tyrosine kinase inhibitors (TKIs) could expand the horizon for PDAC treatment, but currently available TKIs have not shown sufficient efficacy. We hypothesize that stromal proteins have masked targetable epithelial proteins in bulk proteome analyses, and we performed differential analysis of bulk samples versus the tumor compartment. Focusing on targetable receptor tyrosine kinases (RTKs), we identified 2 targetable RTKs in the LCM tumor cell compartment: epidermal growth factor receptor 1 (EGFR) and ephrin type-A receptor 2 (EPHA2; Figure 5A and Supplemental Figure 5A). EGFR has previously been targeted in an unselected population of patients with PDAC; the results of this study showed limited survival benefit (29). EPHA2 has not been targeted in clinical trials with patients with PDAC, whereas it has shown preclinical potential in modulating the immune evasive characteristic of PDAC (30). Indeed, a panel of PDAC cell lines showed variable EPHA2 expression (Supplemental Figure 5B), representative of the variable EPHA2 expression identified in our tumor compartment (Figure 5A). We selected 2 cell lines, Capan-2 and Hs766t, with high EPHA2 expression and phosphorylation to functionally evaluate EPHA2. Downregulation of EPHA2 by shRNAs (Supplemental Figure 5C) resulted in a reduced proliferation rate in Capan-2 cells (Figure 5B) but not in Hs766t cells (Supplemental Figure 5D), possibly explained by the higher relative phosphorylation of EPHA2 in the Capan-2 cell line (Figure 5D).
Figure 5. EPHA2 phosphorylation in PDAC cells conveys sensitivity to the inhibitor ALW-II-27.
(A) Differential expression of EPHA2 between bulk (n = 16) and isolated epithelial compartments (n = 15) identified EPHA2 as specifically but heterogeneously expressed in the tumor compartment (limma test, corrected for multiple testing, P = 0.05) (mean ± SD). (B) Reduction of EPHA2 expression by shRNA in Capan-2 cells significantly reduced proliferation rate (P < 0.001, unpaired 2-way t test) (mean ± SEM) (n = 7). (C) Dose-response curves of cell lines treated with ALW-II-27. (D) Western blot analysis of cells after treatment with ALW-II-27 shows reduction of EPHA2 expression and phosphorylation of activation site pY-588. See complete unedited blots in the supplemental material. (E) Transwell migration of Capan-2 cells was reduced upon pretreatment with 1 μM ALW-II-27, irrespectively of attractant (EGF, P = 0.0005, FCS P = 0.0029, 2-tailed t test with Welch’s correction) (mean ± SEM) (n = 3).
Recently, ALW-41-27, a EPHA2-specific inhibitor, was identified and was shown to target EPHA2 successfully in preclinical studies in non–small cell lung carcinoma (31). Exposure to this drug resulted in substantial inhibition of proliferation in a dose-dependent manner in PDAC cells (Figure 5C), with the strongest effect again found in Capan-2 cells, which have the highest phosphorylation of this kinase (Figure 5D). Drug treatment resulted in a reduction of EPHA2 phosphorylation on the activating site phospho-Y588 within 2 hours (Figure 5D), confirming reduced activation. After 24 hours, surviving cells expressed less total EPHA2, indicating a possible selection for an EPHA2-negative cell population.
To determine the contributions of EPHA2 activity to tumor cell migration, Transwell migration assays were performed using EGF or FCS as chemoattractants. ALW-41-27 strongly inhibited migration (Figure 5E, P = 0.0005 and P = 0.0029, respectively). Interestingly, EPHA2-silenced cells were highly adherent to cell culture substrates and failed to release during trypsinization (Supplemental Figure 5E), providing further proof for a possible role in attachment and migration of these cells. Given the contributions of tumor cell migration to metastatic disease, these results and those from the proliferation assays suggest that the inhibition of EPHA2 could be very effective against PDAC in a clinical setting as well and underlines the usefulness of the current data set for exploration of PDAC biology.
Discussion
This study reports an in-depth proteomic analysis of LCM PDAC. It disentangles the stromal and tumor cell contributions to the proteome landscape of this aggressive tumor. The analyses performed show that the abundance of stroma in bulk tissue analysis masks important tumor biology and underscores the added value of compartment-specific analysis. We found that protein heterogeneity was more pronounced in tumorous areas as compared with stroma and used this to identify prognostic markers and therapeutic targets.
PDAC is known to harbor relatively small tumor islands in vast areas of stroma, and the biology of both compartments effects multiple oncogenic pathways and thus patient outcome. Despite this knowledge, most efforts to unravel tumor biology have relied on genetic aberrations and transcriptomic networks (15–17, 27). Previous proteomic studies exploring PDAC used bulk tumor samples, performed LCM on very small sample sizes, did not reach the depth of proteome landscape currently described, or did not evaluate the stroma (32, 33). Using our cohort and pipeline, we were able to find a much higher degree of heterogeneity in the tumor compartment compared with the stroma. This can be explained by (a) the larger variety of unique proteins identified in tumor areas and (b) the fact that tumor cells are genetically unstable (compared with stromal cells) and that this is likely reflected in the heterogeneous proteome landscape (34).
Proteins known to associate with previously established RNA-based tumor subtypes (e.g., the Moffitt classification, ref. 17) were identified in our landscape. However, tumors were occasionally found to express proteins from both basal-like and classical subtypes. We propose that this results from the presence of mixed populations within a tumor, as was also found in recent single-cell RNA-Seq analyses of PDAC (25). Importantly, “normal” and “activated” stromal subtypes were fully recapitulated in our data, validating previously described stroma subtypes. Future studies on large cohorts are needed to validate the discriminative power of transcriptomic subtypes in proteome data.
Another proteome approach to evaluate extracellular matrix content is purification of the matrisome, a technique that measures enriched proteins after degradation of cellular content. However, it does not take into account the importance of cellular communication and warrants a large volume of bulk tumor samples, making it less appealing for biopsies or other samples of limited quantity. One study explored the matrisome of PDAC (35) and indicated that prognostic proteins originate mainly from the tumor cells, but this study did not measure contributions from both compartments. Importantly, 4 (AEBP1, C1QTNF5, COL11A1, and MFGE8) of the 5 potential prognostic stromal proteins identified in the current study were significantly upregulated in the PDAC matrisome compared with normal tissue, validating our results on proteome level. Large PDAC transcriptomic (RNA) data sets have been published, and we validated our candidate marker proteins in a meta-analysis of those gene expression data. We confirmed several known prognostic pancreatic markers and validated CALB2 as a poor prognostic tumor marker and COL11A1 as a poor prognostic stromal marker in PDAC. CALB2 has previously been described as a diagnostic marker and tumor-promoting protein in mesothelioma (36). It is associated with poor prognostic basal-like subtypes in other cancers (37). This protein plays an important role in intracellular calcium regulation, and it is necessary for vital cellular functions and cell contractility (38). Its role in cancer has not been fully explored; however, the poor prognostic value could relate to altered mechanobiology known to contribute to PDAC (6). The validated stromal prognostic marker demonstrated in this study, COL11A1, is a recently discovered effector of stromal activation and is suggested to play a pan-cancer role in tumor-stroma interaction (39).
Additionally, the comparison of bulk versus tumor-enriched samples showed the advantage of enrichment in proteomic studies, since these, as do genomic and transcriptomic studies, do not rely on amplification methods. For example, the presence of several kinases was masked in proteomes of bulk tumor. However, by enriching for tumor areas, we uncovered EPHA2 and functionally validated this kinase as therapeutic target in PDAC. shRNA knockdown was effective against cell growth in 1 cell line of 2 tested, which correlated with the phosphorylation of EPHA2. An additional explanation could be that, upon silencing, a bias is introduced for EPHA2-independent clones. A previously published study in which silencing of EPHA2 was achieved by siRNAs showed an antiproliferative effect on PDAC cells upon knockdown (40). Interestingly, EPHA2 was recently identified as a key regulator of the PDAC immune-suppressive microenvironment (30). Experimental work evaluating additional cell lines for the EPHA2-driven effect using inducible knockdown systems would strengthen study results; however, our study did revealed strong tumor-inhibiting and antimigration effects of the EPHA2 inhibitor ALW-41-27 in PDAC cells with high EPHA2 expression. This finding warrants further evaluation of this compound against PDAC.
There are several limitations to our study, the most important being the relatively small sample size. For instance, disease stage did not correlate significantly with survival due to small group size. However, this study remains the largest PDAC proteome landscape to date to our knowledge, and our approach added to the current knowledge on protein localization in PDAC and validated previously described transcriptomic stromal subtypes. In addition, despite the lack of association of outcome with staging, associations of survival with stromal proteome subtypes were strong. The limited sample size might also restrict evaluation of the full heterogeneity present in PDAC. This was apparent from the discordance between our unsupervised clustering analysis and the RNA-based tumor cell–specific classification.
A practical limitation was the surface needed to yield optimal proteome exploration. Adding to this is the fact that proteomics does not incorporate amplification steps, and low abundance proteins can be underrepresented. Therefore, to create the deepest data, identification and exclusion of possible singular tumor cells that may exist in the isolated stromal areas was not performed, which could contaminate the stromal protein landscape to some degree. Despite this limitation, we were able to create a PDAC data set of depth, with very little cross-compartment contamination.
The functional experiments were hampered by the limited number of cell lines tested, and future studies will have to be performed to formally prove the association of EPHA2 levels and phosphorylation with response to EPHA2 inhibitors. Despite this, our comprehensive exploration by IHC and functional investigation of markers/targets underscores the use of our data set for future studies.
In conclusion, we report a large-scale compartment-specific proteome landscape of PDAC. With this data set, future bulk tumor proteome data sets can be deconvolved and annotated. We have shown that our data can be the basis for in vitro analysis of targets of tumor signaling and bidirectional stimulatory communication between PDAC and stromal cells. Moreover, we have shown that prognostic genes can exist in tumor or its microenvironment or can be expressed by both. Our data underline the need to understand the biology of PDAC on multiple levels and warrant future large-scale proteome studies of PDAC.
Methods
Further details are provided in the Supplemental Methods.
Sample collection.
Snap-frozen tumor samples were evaluated for quality and tumor percentage. The workflow is shown in Figure 1A. Sixteen samples were eligible for further LCM analysis and prepared as described previously (41). Additionally, unseparated bulk tumor of 11 matched and 5 unmatched samples was prepared for single-shot MS/MS analysis. Two patients suffered from postoperative complications and were excluded from survival analysis but included in tumor biology analysis.
LCM procedure.
LCM was performed on the Leica LMD7000 instrument to yield a total surface density of 3 × 106 μm2 cells per compartment. Selected areas were captured in 0.1% RapiGest SF Surfactant (Waters) and stored until further use. Samples were sonicated and reduced to a final concentration of 5 mM dithiothreitol (MilliporeSigma) and 15 mM iodoacetamide (MilliporeSigma). Sequencing-grade modified trypsin (Promega) was added overnight to a final concentration of 7 ng/μl. Digestion was stopped with acidification by trifluoroacetic acid (MilliporeSigma). The peptide mixture was centrifuged, and the supernatant was transferred to a glass-lined MS/MS auto sampler vials. Samples were brought to a volume of 20 μl and stored at –80°C until further analysis.
Peptide preparations of PDX models and bulk tumor tissue.
Tissue from 10 previously established PDX models was used for proteome analysis (22). NSG mice were bred in-house. A minimum of 100 mg tissue was digested in lysis buffer (9 M urea, 20 mM HEPES pH 8.0, 1 mM Na3VO4, 2.5 mM Na4P2O7, 1 mM Na2C3H7PO6). 50 μg protein was loaded on a NuPAGE 4%–12% gradient gel (Invitrogen). Proteins were digested and extracted according to our whole-in-gel protocol, as described previously (42). Bulk PDAC tumors were lysed, and in-solution digestion was performed according to the protocol of our laboratories after reduction and alkylation (43).
Nano-LC-MS/MS.
Proteins were identified by nano-LC-MS/MS, as described previously (42). Peptides were separated by an Ultimate 3000 nano-LC-MS/MS system (Dionex LC-Packings). Eluting peptides were ionized into a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Intact masses were measured in the orbitrap using an AGC target value of 3 × 106 charges. The top 15 peptide signals were submitted to MS/MS in a higher-energy collision cell in MS/MS. MS/MS spectra were acquired at resolution 17.500 (at m/z 200) in the orbitrap.
Protein annotation and data analysis.
MS/MS spectra were searched against the Swissprot FASTA file (LCM data: release March 2017; 42,161 entries, canonical and isoforms; proteome profiles from PdX data were searched with both human and mouse fasta files;uniprot_human_referenceproteome_2014_01_NO_fragments_42104entries.fasta, 61,552 entries); Uniprot_Mus_musculus_reference_proteome_2015_06_NO_FRAGMENTS_Canonical and isoforms _34331entries.fasta, 42,296 entries) using MaxQuant 1.5.8.0 (44). Enzyme specificity was set to trypsin, and up to 2 missed cleavages were allowed. Peptide and protein identifications were filtered at an FDR of 1% using the decoy database strategy. The minimal peptide length was 7 amino acids. A match-between-runs setting was implemented for analysis of low abundant proteins in the LCM database. For the PDX data, peptides mapped to solely human proteins were retained and subsequently collapsed in protein groups. Protein compartment specificity was correlated to mouse- and human-specific proteins identified from PDX PDAC tumors (protein had to be uniquely mouse or human specific). The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE (45) partner repository (data set identifiers PXD011289 and PXD017393). Protein and peptide data are available in Supplemental Data Files 1–8.
RNA-Seq of PDX models and correlation with protein expression.
RNA-Seq of PDX was previously performed and described (EMBL-EBI ArrayExpress code E-MTAB-6830) (46). In short, total RNA was extracted from PDX model tissue and amplified with the Total Prep RNA amplification kit (Illumina). RNA was sequenced on the Illumina HiSeq2500. Mapped reads were mapped to both mouse and human genomes. The RNA transcripts mapping solely the human genome were retained for further analysis. RPKM data were log2 transformed and filtered to include transcripts with an average read count ≥1 in all samples, obtaining 14,809 RNA transcripts in total. Human-specific spectral counts were log2 transformed and filtered to include proteins with an average spectral count ≥3 in all samples, obtaining 1705 protein groups in total. Spearman’s correlation was applied to the resulting transcriptome-proteome matched file with 1627 genes (proteins were collapsed at gene level) to evaluate PDX transcriptome and proteome correlation. Methodology was described previously by Wang et al. (23).
In silico validation of prognostic markers.
Data from publicly available transcriptomic (microarray or RNA-Seq) data sets (15–17, 26–28) with survival data were downloaded from GEObase, and each data set was scaled to a mean of 0, with a standard deviation of 1 to allow meta-analysis. Univariate cox proportional hazard regression models were evaluated for genes of interest. The Metafor R package (47) was used to perform meta-analysis validation of identified prognostic markers.
IHC validation on TMAs.
Proteins of interest in both compartments, COL11A1 and CALB2, were evaluated in 2 independent cohorts (n = 95 and n = 95) by staining TMAs as described previously (48). Assessment of IHC staining of COL11A1 was performed in 3 tumor cores per patient containing representatives regions of the desmoplastic reaction. Expression was evaluated in relation to the stromal surface, as described previously (49). IHC of CALB2 was evaluated in the tumorous areas, taking into account positivity and intensity of the staining, as described previously (50).
Cell culture.
PANC1 cells (ATCC) were cultured in RPMI medium (Lonza). Capan-2 cells (ATCC) and Hs766t cells (ATCC) were cultured in DMEM medium (Lonza). Media was supplemented with 10% heat-inactivated fetal calf serum (Biowest) and 1% penicillin-streptomycin (Lonza). An immortalized pancreatic ductal cell line (HPDE, supplied by Ming Tsao, Ontario Cancer Institute, Toronto, Ontario, Canada) (51) was cultured in supplemented KGM medium (Lonza).
Western blot validation of EPHA2 expression.
Cell lysates from PANC1, Hs766t, Capan-2, and HPDE cells were created with diluted ×10 RIPA buffer (Abcam) containing protease and phosphatase inhibitors. 20 μg protein was used for Western blot. Primary antibodies were incubated in 5% BSA (MilliporeSigma) in PBST followed by secondary antibodies in 5% blocking buffer. Visualization was performed by an Uvitec Imaging station (Cleaver Scientific). A list of antibodies is provided in the Supplemental Methods.
Stable EPHA2 knockdown.
Lentiviral plasmids were produced by transfecting HEK293T cells with EPHA2-targeting pLKO.1 constructs (MISSION shRNA Library clone numbers TRCN0000006403 and TRCN0000197131) and a scrambled control (shc002). Transfected supernatant was collected after 48 hours and filtered through a 0.45 μm filter (EMDMillipore). At 30% confluence, PDAC cell lines Capan-2 and Hs766t were transduced and subsequently selected after 48 hours with 2 μg/mL puromycin (MilliporeSigma). Knockdown efficiency was evaluated by Western blot as described above.
In vitro validation of drug target.
Cells were plated in 96-wells plates and allowed to attach overnight. Growth was evaluated over 72 hours with respect to the control at the start of the experiment. To evaluate proliferation, the doubling time was calculated. A specific EPHA2 inhibitor, ALW-II-41-27 (APExBIO), was evaluated for cytotoxic effect. DMSO (MilliporeSigma) was used as control. Effect on cell proliferation was quantified with Sulforhodamine B (MilliporeSigma) protein staining as described previously (52). Migration was evaluated by Transwell migration as described before (53) following 15 minutes pretreatment. The AUC was calculated for each replicate of the migration curve.
Statistics.
Data were analyzed with R (version 3.5.2.). Zeros were imputed based on normal distribution SD of log-transformed intensity data before differential expression analysis. Differential compartment expression was tested by paired statistics analysis (Limma package) (54) and Benjamini-Hochberg corrected for multiple testing. Two patients succumbed to postoperative complication (PDAC10 and PDAC17) and were excluded from the survival analysis. These samples were, however, included in the proteomics and downstream nonclinical analyses. Prognostic proteins were identified by group comparison (short OS, <1 year versus long OS, >2 years). GSEA (55) was performed in R. In vitro experimental comparisons were evaluated by paired or unpaired 2-tailed Student’s t test. Correlation of clinicopathological characteristics and gene expression with DFS/OS were evaluated by Kaplan-Meier and log-rank test. The prognostic value of IHC scoring was tested with uni- and multivariate analysis. Error bars show the mean ± SEM. A P value of < 0.05 was considered statistically significant.
Study approval.
The work described was carried out in accordance with the Declaration of Helsinki. Approval for tissue collection was obtained from the local medical ethical committees at the Amsterdam UMC (protocol 14038 for METC VUmc and 2011_126 and 201_181 for AMC, both Amsterdam, the Netherlands). Ethical approval for the validation cohorts was received from Comitato di Bioetica Azienza Ospedaliero-Universitaria Pisana, University Hospital of Cisanello, Pisa, Italy (protocol 37677; study no. 3909; July 3, 2013)). All patients provided informed consent for participation in the study. Animal work was performed in a previous study according to protocols approved by the animal experiment ethical committee at the Amsterdam UMC (protocol DTB102348, LEX102774).
Author contributions
TYSLL and NF performed LCM. TYSLL, GM, and TVP performed bioinformatic analysis. TYSLL, LLM, NF, and EG were involved with selection of patient material, clinical data collection, and statistical analyses. TYSLL, NF, and NCTVG performed pathological review of LCM material and performed IHC staining. JK, SRP, CRJ, and TVP were responsible for experimental design and mass spectrometry. TYSLL, HWMVL, GK, EG, and MFB were involved with experimental design and manuscript preparation. TYSLL, MFB, and BK performed experimental validation. CRJ, EG, and MFB coordinated and supervised the study. All the authors critically reviewed the manuscript.
Supplementary Material
Acknowledgments
We would like to acknowledge C. Maurer for his help with the meta-analysis of PDAC transcriptomic data, and all the patients participating in our studies that made this research possible. This work was supported by a Cancer Center Amsterdam Alliantie-AIO grant (to EG, MFB, and CRJ), the Bennink Foundation (to GK, EG, and TYSLL), the Dutch Cancer Society (10212 to CRJ, MFB, and EG), and an Italian Association for Cancer Research AIRC/Start-Up grant, Italy (to EG). The Netherlands Organization for Scientific Research (Middelgroot project no. 91116017, to CRJ) is acknowledged for supporting the mass spectrometry infrastructure.
Version 1. 07/07/2020
In-Press Preview
Version 2. 08/06/2020
Electronic publication
Footnotes
Conflict of interest: MFB has received research funding from Celgene and acted as a consultant to Servier. HWMVL has a consultant/advisory role with Bristol-Myers Squibb, Lilly, MSD, Nordic Pharma, Novartis, and Servier. HWMVL received research funding from Bayer, Bristol-Myers Squibb, Celgene, Janssen, Lilly, Nordic Pharma, Philips, Roche, and Servier.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(15):e138290.https://doi.org/10.1172/jci.insight.138290.
Contributor Information
Tessa Y.S. Le Large, Email: t.lelarge@amsterdamumc.nl.
Laura L. Meijer, Email: l.meijer@amsterdamumc.nl.
Nicole C.T. van Grieken, Email: nct.vangrieken@amsterdamumc.nl.
G. Kazemier, Email: g.kazemier@amsterdamumc.nl.
References
- 1.Yadav VK, De S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogeneous tumor tissue samples. Brief Bioinform. 2015;16(2):232–241. doi: 10.1093/bib/bbu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pietras K, Östman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- 3.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson JS, Pirola RC, Apte MV. Stars and stripes in pancreatic cancer: role of stellate cells and stroma in cancer progression. Front Physiol. 2014;5:1–11. doi: 10.3389/fphys.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016;22(8):851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. 2016;22(5):497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhim AD, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steins A, et al. High-grade mesenchymal pancreatic ductal adenocarcinoma drives stromal deactivation through CSF-1. EMBO Rep. 2020;21(5):e48780. doi: 10.15252/embr.201948780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 11.Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conroy T, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364(19):1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 13.Conroy T, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379(25):2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 14.Neoptolemos JP, et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389(10073):1011–1024. doi: 10.1016/S0140-6736(16)32409-6. [DOI] [PubMed] [Google Scholar]
- 15.Collisson E a, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 17.Moffitt RA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47(10):1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurer HC, et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut. 2019;68(6):1034–1043. doi: 10.1136/gutjnl-2018-317706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funel N, Giovannetti E, Pollina LE, del Chiaro M, Mosca F, Boggi UCD. Critical role of laser microdissection for genetic, epigenetic and proteomic analyses in pancreatic cancer. Expert Rev Mol Diagn. 2011;Sep(7):695–701. doi: 10.1586/erm.11.62. [DOI] [PubMed] [Google Scholar]
- 20.Braakman RBH, et al. Optimized nLC-MS workflow for laser capture microdissected breast cancer tissue. J Proteomics. 2012;75(10):2844–2854. doi: 10.1016/j.jprot.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537(7620):347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 22.Damhofer H, et al. Establishment of patient-derived xenograft models and cell lines for malignancies of the upper gastrointestinal tract. J Transl Med. 2015;13(1):1–14. doi: 10.1186/s12967-014-0365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol. 2019;15(2):1–16. doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi A, et al. A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer. 2020;1(1):59–74. doi: 10.1038/s43018-019-0010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter C, et al. Google goes cancer: improving outcome prediction for cancer patients by network-based ranking of marker genes. PLoS Comput Biol. 2012;8(5):e1002511. doi: 10.1371/journal.pcbi.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S, et al. A novel MIF signaling pathway drives the malignant character of pancreatic cancer by targeting NR3C2. Cancer Res. 2016;76(13):3838–3850. doi: 10.1158/0008-5472.CAN-15-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 30.Markosyan N, et al. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2) J Clin Invest. 2019;129(9):3594–3609. doi: 10.1172/JCI127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amato KR, et al. Genetic and pharmacologic inhibition of EPHA2 promotes apoptosis in NSCLC. J Clin Invest. 2014;124(5):2037–2049. doi: 10.1172/JCI72522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takadate T, et al. Nm23/nucleoside diphosphate kinase-A as a potent prognostic marker in invasive pancreatic ductal carcinoma identified by proteomic analysis of laser micro-dissected formalin-fixed paraffin-embedded tissue. Clin Proteomics. 2012;9(1):8. doi: 10.1186/1559-0275-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leca J, et al. Cancer-associated fibroblast-derived annexin A6+ extracellular vesicles support pancreatic cancer aggressiveness. JCI. 2016;126(9):1–17. doi: 10.1172/JCI87734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nones K, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. 2014;135(5):1110–1118. doi: 10.1002/ijc.28765. [DOI] [PubMed] [Google Scholar]
- 35.Tian C, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci U S A. 2019;116(39):19609–19618. doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blum W, Schwaller B. Calretinin is essential for mesothelioma cell growth/survival in vitro: a potential new target for malignant mesothelioma therapy? Int J Cancer. 2013;133(9):2077–2088. doi: 10.1002/ijc.28218. [DOI] [PubMed] [Google Scholar]
- 37.Taliano RJ, et al. Calretinin expression in high-grade invasive ductal carcinoma of the breast is associated with basal-like subtype and unfavorable prognosis. Hum Pathol. 2013;44(12):2743–2750. doi: 10.1016/j.humpath.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui C, Merritt R, Fu L, Pan Z. Targeting calcium signaling in cancer therapy. Acta Pharm Sin B. 2017;7(1):3–17. doi: 10.1016/j.apsb.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia D, et al. A COL11A1-correlated pan-cancer gene signature of activated fibroblasts for the prioritization of therapeutic targets. Cancer Lett. 2016;382(2):203–214. doi: 10.1016/j.canlet.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23(7):1448–1456. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- 41.Caponi S, et al. The good, the bad and the ugly: a tale of miR-101, miR-21 and miR-155 in pancreatic intraductal papillary mucinous neoplasms. Ann Oncol. 2013;24(3):734–741. doi: 10.1093/annonc/mds513. [DOI] [PubMed] [Google Scholar]
- 42.Piersma SR, Warmoes MO, de Wit M, de Reus I, Knol JC, Jiménez CR. Whole gel processing procedure for GeLC-MS/MS based proteomics. Proteome Sci. 2013;11(1):17. doi: 10.1186/1477-5956-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piersma SR, et al. Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines. J Proteomics. 2015;127(Pt B):247–258. doi: 10.1016/j.jprot.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 44.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 45.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dijk F, et al. Unsupervised class discovery in pancreatic ductal adenocarcinoma reveals cell-intrinsic mesenchymal features and high concordance between existing classification systems. Sci Rep. 2019;10(1):337. doi: 10.1038/s41598-019-56826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viechtbauer W. Conducting meta-analyses in R with the metafor. J Stat Softw. 2010;36(3):1–48. [Google Scholar]
- 48.Le Large TYS, et al. Proteomic analysis of gemcitabine-resistant pancreatic cancer cells reveals that microtubule-associated protein 2 upregulation associates with taxane treatment. Ther Adv Med Oncol. 2019;11:1758835919841233. doi: 10.1177/1758835919841233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.García-Pravia C, et al. Overexpression of COL11A1 by cancer-associated fibroblasts: clinical relevance of a stromal marker in pancreatic cancer. PLoS One. 2013;8(10):1–13. doi: 10.1371/journal.pone.0078327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thapa B, et al. Calretinin but not caveolin-1 correlates with tumour histology and survival in malignant mesothelioma. Pathology. 2016;48(7):660–665. doi: 10.1016/j.pathol.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Furukawa T, Duguid W, Rosenberg L, Viallet J, Galloway D, Tsao M. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. Am J Pathol. 1996;148(6):1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 52.Sciarrillo R, et al. Splicing modulation as novel therapeutic strategy against diffuse malignant peritoneal mesothelioma. EBioMedicine. 2019;39:215–225. doi: 10.1016/j.ebiom.2018.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebbing EA, et al. Esophageal adenocarcinoma cells and xenograft tumors exposed to Erb-b2 receptor tyrosine kinase 2 and 3 inhibitors activate transforming growth factor beta signaling, which induces epithelial to mesenchymal transition. Gastroenterology. 2017;153(1):63–76.e14. doi: 10.1053/j.gastro.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:1–17. doi: 10.1186/1471-2105-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P, et al. FHL3 promotes pancreatic cancer invasion and metastasis through preventing the ubiquitination degradation of EMT associated transcription factors. Aging (Albany NY) 2020;12(1):53–69. doi: 10.18632/aging.102564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhasin MK, et al. Meta-analysis of transcriptome data identifies a novel 5-gene pancreatic adenocarcinoma classifier. Oncotarget. 2016;7(17):23263–23281. doi: 10.18632/oncotarget.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hata H, Tatemichi M, Nakadate T. Involvement of annexin A8 in the properties of pancreatic cancer. Mol Carcinog. 2014;53(3):181–189. doi: 10.1002/mc.21961. [DOI] [PubMed] [Google Scholar]
- 60.Iovanna J, Dusetti N. Speeding towards individualized treatment for pancreatic cancer by taking an alternative road. Cancer Lett. 2017;410:63–67. doi: 10.1016/j.canlet.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 61.Castillo J, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29(1):223–229. doi: 10.1093/annonc/mdx542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balasenthil S, et al. A migration signature and plasma biomarker panel for pancreatic adenocarcinoma. Cancer Prev Res (Phila) 2011;4(1):137–149. doi: 10.1158/1940-6207.CAPR-10-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capello M, et al. Carboxylesterase 2 as a determinant of response to irinotecan and neoadjuvant FOLFIRINOX therapy in pancreatic ductal adenocarcinoma. J Natl Cancer Inst. 2015;107(8):djv132. doi: 10.1093/jnci/djv132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ran Y, et al. Derlin-1 is overexpressed on the tumor cell surface and enables antibody-mediated tumor targeting therapy. Clin Cancer Res. 2008;14(20):6538–6545. doi: 10.1158/1078-0432.CCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 65.Kakkar AK, Lemoine NR, Scully MF, Tebbutt S, Williamson RC. Tissue factor expression correlates with histological grade in human pancreatic cancer. Br J Surg. 1995;82(8):1101–1104. doi: 10.1002/bjs.1800820831. [DOI] [PubMed] [Google Scholar]
- 66.Yao H, et al. Glypican-3 and KRT19 are markers associating with metastasis and poor prognosis of pancreatic ductal adenocarcinoma. Cancer Biomarkers. 2016;17(4):397–404. doi: 10.3233/CBM-160655. [DOI] [PubMed] [Google Scholar]
- 67.Yokoyama S, et al. Predicted prognosis of patients with pancreatic cancer by machine learning. Clin Cancer Res. 2020;26(10):2411–2421. doi: 10.1158/1078-0432.CCR-19-1247. [DOI] [PubMed] [Google Scholar]
- 68.Abate-Daga D, et al. A novel chimeric antigen receptor against prostate stem cell antigen mediates tumor destruction in a humanized mouse model of pancreatic cancer. Hum Gene Ther. 2014;25(12):1003–1012. doi: 10.1089/hum.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitsui Y, et al. Upregulation of mobility in pancreatic cancer cells by secreted S100A11 through activation of surrounding fibroblasts. Oncol Res. 2019;27(8):945–956. doi: 10.3727/096504019X15555408784978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharen G, Peng Y, Cheng H, Liu Y, Shi Y, Zhao J. Prognostic value of GLUT-1 expression in pancreatic cancer: results from 538 patients. Oncotarget. 2017;8(12):19760–19767. doi: 10.18632/oncotarget.15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun H, Dai X, Han B. TRIM29 as a novel biomarker in pancreatic adenocarcinoma. Dis Markers. 2014;2014:317817. doi: 10.1155/2014/317817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.