Abstract
Noonan syndrome with multiple lentigines (NSML) is a rare autosomal dominant disorder that presents with cardio-cutaneous-craniofacial defects. Hypertrophic cardiomyopathy (HCM) represents the major life-threatening presentation in NSML. Mutations in the PTPN11 gene that encodes for the protein tyrosine phosphatase (PTP), SHP2, represents the predominant cause of HCM in NSML. NSML-associated PTPN11 mutations render SHP2 catalytically inactive with an “open” conformation. NSML-associated PTPN11 mutations cause hypertyrosyl phosphorylation of the transmembrane glycoprotein, protein zero-related (PZR), resulting in increased SHP2 binding. Here we show that NSML mice harboring a tyrosyl phosphorylation–defective mutant of PZR (NSML/PZRY242F) that is defective for SHP2 binding fail to develop HCM. Enhanced AKT/S6 kinase signaling in heart lysates of NSML mice was reversed in NSML/PZRY242F mice, demonstrating that PZR/SHP2 interactions promote aberrant AKT/S6 kinase activity in NSML. Enhanced PZR tyrosyl phosphorylation in the hearts of NSML mice was found to drive myocardial fibrosis by engaging an Src/NF-κB pathway, resulting in increased activation of IL-6. Increased expression of IL-6 in the hearts of NSML mice was reversed in NSML/PZRY242F mice, and PZRY242F mutant fibroblasts were defective for IL-6 secretion and STAT3-mediated fibrogenesis. These results demonstrate that NSML-associated PTPN11 mutations that induce PZR hypertyrosyl phosphorylation trigger pathophysiological signaling that promotes HCM and cardiac fibrosis.
Keywords: Cardiology
Keywords: Cardiovascular disease, Genetic diseases, Signal transduction
Protein zero-related (PZR) hyper-tyrosylphosphorylation is required for hypertrophic cardiomyopathy in Noonan syndrome with multiple lentigines.
Introduction
Hypertrophic cardiomyopathy (HCM) is a genetic myocardial disorder that is characterized by left ventricular hypertrophy, cardiomyocyte disarray, and myocardial fibrosis (1–3). Although most familial HCM is caused by mutations in genes that encode for sarcomeric proteins, such as myosin subunits, myosin binding proteins, and filament proteins, a variety of disorders also exhibit apparent left ventricular hypertrophy in nonsarcomeric proteins (4). Noonan syndrome with multiple lentigines (NSML, OMIM 151100) is one of the nonsarcomeric forms of HCM showing multiple clinical characteristics, such as growth retardation, dysmorphic facial features, and postdevelopmental abnormalities (5, 6). Approximately 85% of NSML cases exhibit cardiac defects that include HCM and pulmonary stenosis. Up to 85% of patients with NSML have mutations in the PTPN11 gene, which encodes for the SH2 domain–containing protein tyrosine phosphatase 2 (SHP2) (7–9).
SHP2 is a nonreceptor protein tyrosine phosphatase (PTP) that is ubiquitously expressed (7–9). The phosphatase activity of SHP2 is required for the propagation of multiple signaling pathways, including the Ras/MAPK andPI3K/AKT pathways, which are involved in proliferation, differentiation, migration, and development (10–12). The catalytic activity of SHP2 is tightly regulated by intramolecular conformational constraints (13). The “closed” conformation, which is mediated by the interaction between the SH2 and phosphatase domains, is destabilized by the engagement of the N-terminal SH2 domain to phosphotyrosine peptides, resulting in an “open” conformation that renders the catalytic domain substrate accessible (14, 15). NSML-associated SHP2 mutations occur in the PTP domain that result in reduced phosphatase activity and an open conformation (16, 17). The open conformation of NSML-associated SHP2 mutations enhances SH2 domain protein-protein interactions that likely contribute to propagating aberrant signaling that promotes the pathogenesis of NSML (18, 19). However, definitive evidence of such SHP2-mediated SH2 domain engagement in NSML has yet to be established.
We previously reported that PZR is hypertyrosyl phosphorylated in the hearts of NSML mice (20). PZR is a transmembrane glycoprotein comprising an extracellular immunoglobulin-like domain and 2 intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIMs) (21–23). PZR was identified as an SHP2 binding partner, with the SH2 domains of SHP2 binding phosphorylated tyrosine residues (Y241 and Y263) within the ITIM of human PZR (hPZR) (20, 23). Much of the information known about PZR relates to its role in adhesion-mediated cell signaling and migration (24–26). A zebrafish model study revealed that PZR tyrosyl phosphorylation is necessary for convergence and extension cell movements during zebrafish gastrulation (20). We demonstrated NSML-associated SHP2 mutations (SHP2Y279C, NSML-SHP2), which have an “open” conformation, have increased PZR association (20). In order to understand the signaling mechanisms of NSML-associated congenital heart disease, Marin et al. generated a mouse model that expresses knocked in Ptpn11Y279C/+ NSML mutation (referred to herein as NSML mice) (27). These NSML mice exhibit features of the human disease, including those of HCM (27, 28). We found that a low dose of the Src family kinase inhibitor, dasatinib, ameliorated PZR hypertyrosyl phosphorylation and normalized the expression of molecular markers of HCM in the hearts of NSML mice (29). Although our findings suggested the involvement of aberrant PZR/NSML-SHP2 interactions in NSML-associated HCM, it was not definitively addressed as to whether PZR hypertyrosyl phosphorylation and enhanced PZR/SHP2 binding are essential to the development and/or progression of congenital heart disease in general and HCM specifically, in NSML mice.
In this study, we generated a tyrosyl phosphorylation–defective PZR-knockin mutant mouse. Although PZR tyrosyl phosphorylation–defective mice did not exhibit a cardiac phenotype, when intercrossed with NSML mice, PZR hypertyrosyl phosphorylation and enhanced PZR/SHP2 binding were inhibited. Remarkably, NSML mice lacking the ability to generate hypertyrosyl phosphorylated PZR failed to promote enhanced AKT activity in the heart, which correlated with a complete abrogation in the development of HCM. Further, we show that PZR/SHP2 engages NF-κB signaling that stimulates secretion of IL-6 that drives the fibrotic sequelae of HCM. These results demonstrate an essential function for PZR in the development of NSML-associated HCM. Our results provide genetic evidence for targeting PZR tyrosyl phosphorylation as a therapeutic strategy to treat certain forms of RASopathy-associated HCM where PZR is hypertyrosyl phosphorylated.
Results
Generation of NSML mice defective for PZR tyrosyl phosphorylation.
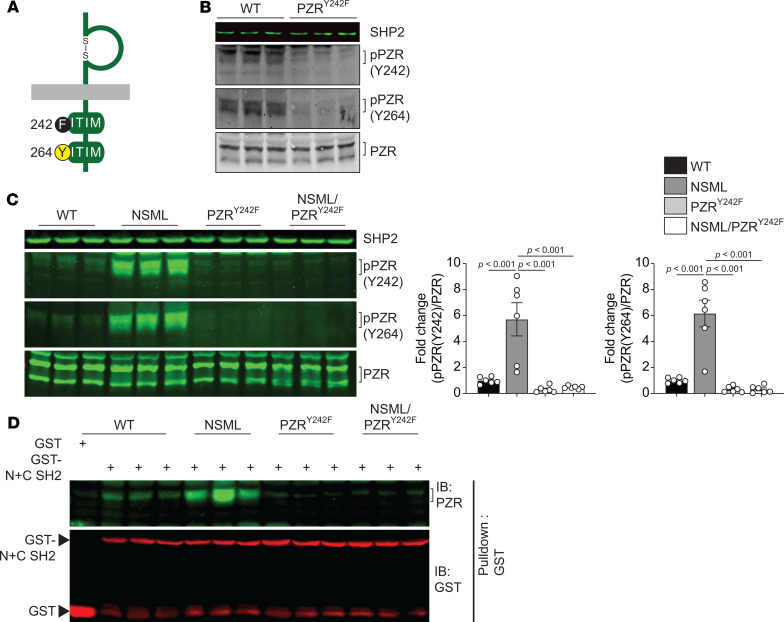
To investigate the role of PZR tyrosyl phosphorylation and hence PZR/SHP2 binding in NSML pathogenesis, we generated a tyrosyl phosphorylation–defective PZR-knockin mutant mouse. Using CRISPR/Cas9, we introduced a tyrosine 242 to phenylalanine mutation in exon 6 of the Mpzl1 gene that encodes for PZR (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.137753DS1). Homozygous PZR-knockin mice (Mpzl1Y242F/Y242F, referred to herein as the PZRY242F mice) were born from heterozygous (Mpzl1+/Y242F) intercrosses. PZRY242F mice were fertile and showed the anticipated ratio of Mendelian offspring (Supplemental Table 1). We confirmed PZR tyrosyl phosphorylation deficiency by immunoblotting with PZR phospho-specific antibodies and found that phosphorylation of both tyrosine resides (Y242 and Y264) was abolished in heart lysates derived from PZRY242F mice (Figure 1B). These results were consistent with previous reports indicating that mutation of one of the ITIM motifs was sufficient to inhibit the phosphorylation of the other (20, 23). PZRY242F mice lacked any major developmental defects and gained weight at a slightly reduced, but significant, rate as compared with WT mice (Supplemental Figure 2A), and there were no differences in the weight of organs, such as lung, liver, spleen, and kidney (Supplemental Figure 2B). Notably, at the age of 20 weeks, PZRY242F mice showed similar heart weight (HW) to body weight (BW) or tibia length (TL) ratios (Supplemental Figure 2C). We also performed echocardiography at 16 weeks of age, and no differences in interventricular septum wall thickness (IVS), left ventricular posterior wall thickness (LVPW), and ejection fraction (EF) in PZRY242F mice as compared with WT mice were observed (Supplemental Table 2). However, analysis of the cardiac fetal gene, Myh7 (myosin heavy chain β); Nppa (atrial natriuretic peptide) and Nppb (brain natriuretic peptide); and fibrotic gene (Col1a) were all reduced in PZRY242F mice as compared with WT mice (Supplemental Figure 3). These data demonstrate that in the context of PZR tyrosyl phosphorylation deficiency alone, mice develop normally without impairment of cardiac functionality.
Figure 1. Generation of PZR tyrosyl phosphorylation–deficient knockin mice.
(A) Schematic of mouse PZRY242F mutation. (B) Heart lysates from WT (Mpzl1+/+) and PZRY242F (Mpzl1Y242F/Y242F+) mice were immunoblotted with anti-SHP2, –p-PZR (Y242), –p-PZR (Y264), and -PZR antibodies (–p, phosphorylated). (C and D) Heart lysates from WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F) and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice were immunoblotted with anti-SHP2, –p-PZR (Y242), –p-PZR (Y264), and -PZR antibodies. The phosphorylation level of PZR was quantitated (n = 6) (C). Heart lysates were incubated with GST-conjugated human SHP2 SH2 domains (GST-N+C SH2). WT lysate was incubated with GST as a negative control. Proteins were affinity purified with glutathione agarose beads and immunoblotted with anti-PZR and -GST antibodies (D). Brackets mark p-PZR and PZR bands. Data represent mean ± SEM. Statistical significance was analyzed by 1-way ANOVA with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
To test the hypothesis that hypertyrosyl phosphorylation of PZR and increased PZR/SHP2 interactions promote HCM in NSML, we introduced the Mpzl1Y242F mutation into NSML (Ptpn11Y279C/+) mice (Supplemental Figure 4). We backcrossed heterozygous Ptpn11Y279C/+ Mpzl1+/Y242F mice in the first founder with PZRY242F mice to generate 4 genotypes: WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F). Analysis of PZR tyrosyl phosphorylation in lysates derived from the hearts of these mice demonstrated that NSML mice exhibited increased PZR tyrosyl phosphorylation as compared with WT mice, as demonstrated previously (20), whereas in NSML/PZRY242F mice PZR tyrosyl phosphorylation at both Y242 and Y264 was undetectable (Figure 1C). We evaluated whether PZR tyrosyl phosphorylation deficiency in NSML mice disrupted PZR/SHP2 binding. Heart lysates from NSML hearts incubated with glutathione-S-transferase–conjugated (GST-conjugated) SH2 domains of SHP2 (GST-SHP2 N+C) showed increased interaction with PZR as compared with WT, and this binding was abrogated in heart lysates prepared from PZRY242F mice. Similarly, heart lysates prepared from NSML/PZRY242F mice also failed to interact with the SHP2 N+C SH2 domains (Figure 1D). These results demonstrate that PZRY242F is phosphorylation defective and fails to bind the SH2 domains of SHP2. Thus, PZRY242F and NSML/PZRY242F mice represent a loss of function for PZR tyrosyl phosphorylation and SHP2 binding.
PZR tyrosyl phosphorylation deficiency rescues HCM in NSML mice.
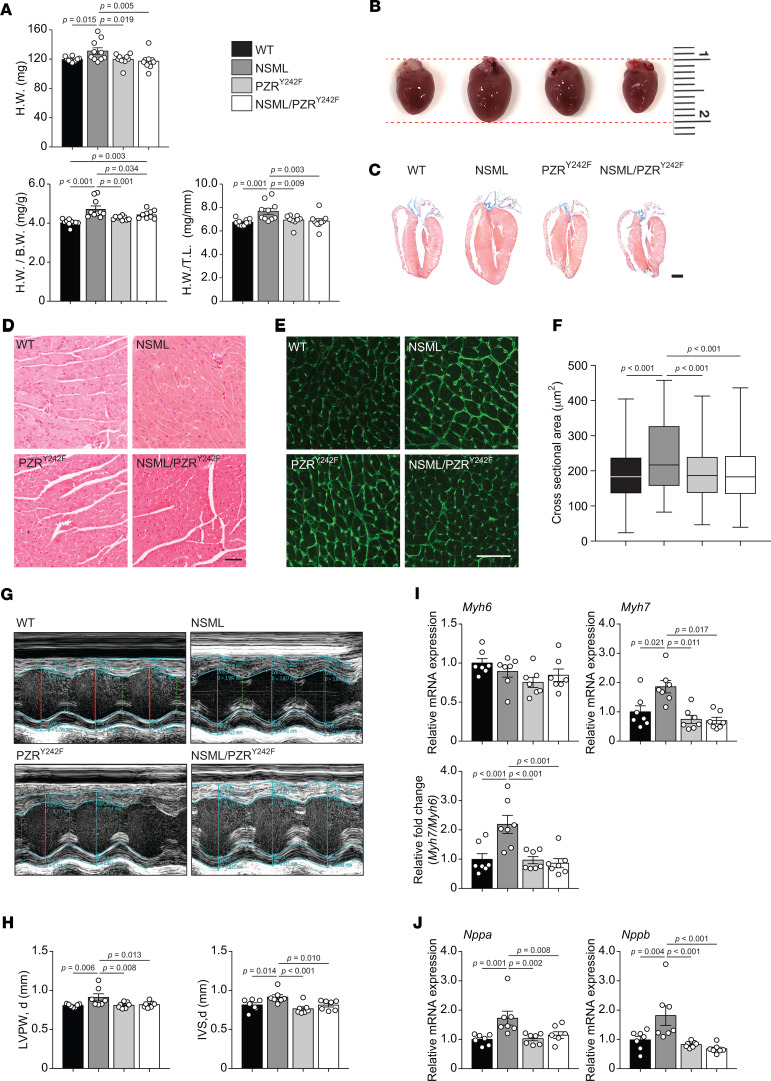
We investigated whether the PZRY242F mutation in NSML mice interferes with the progression of HCM. As previously reported, NSML mice develop HCM by 16 weeks of age (Figure 2, A–C) (27, 28). Although NSML/PZRY242F mice exhibited retarded postdevelopmental growth (Supplemental Figure 5), their cardiac mass and cardiac mass normalized to either body mass or TL was reduced to levels comparable with WT mice (Figure 2, A–C). Morphometric analysis of individual cardiomyocytes from 16-week-old mice demonstrated that the cross-sectional area of cardiomyocytes in NSML mice was significantly increased as compared with WT cardiomyocytes (Figure 2, D–F). In contrast, PZRY242F mutation rescued the NSML-associated cardiomyocyte enlargement (Figure 2, D–F). Additional cardiac parameters were assessed by echocardiography, and these results showed that diastolic LVPW (d) and IVS (d) were significantly reduced in NSML/PZRY242F mice compared with NSML mice (Figure 2, G and H, and Supplemental Table 3). The transition of sarcomeric structural gene expression from myosin heavy chain α (Myh6) to myosin heavy chain β (Myh7) is indicative of cardiomyopathy (30). Whereas Myh7 and the ratio of Myh7 to Myh6 were prominently reexpressed in NSML mice compared with WT mice, this ratio was normalized in NSML/PZRY242F mice (Figure 2I). We also assessed the mRNA expression of clinical markers of cardiomyopathy. Both Nppa and Nppb mRNA expression levels were significantly upregulated in NSML mice as compared with WT mice, while NSML/PZRY242F mice had their levels restored to normal (Figure 2J). Together, these data demonstrate that PZR tyrosyl phosphorylation and, thus, PZR/SHP2 complex formation contribute to the development of HCM in NSML mice.
Figure 2. PZRY242F mutation rescues NSML-associated cardiomyopathies.
(A) Heart weight (H.W.), heart weight to body weight (H.W./B.W.) ratios, and heart weight to tibia length (H.W./T.L.) ratios were measured from 16-week-old WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice (n = 10 for WT and NSML; n = 9 for PZRY242F and NSML/PZRY242F). (B) Gross morphology of hearts from 16-week-old mice. (C) H&E-stained longitudinal sections of hearts from 16-week-old mice (scale bar: 1 mm). (D–F) H&E stain (D) and Alexa Fluor 488–conjugated wheat germ agglutinin (WGA) stain (E) of left ventricles from 16-week-old mice (scale bar: 50 μm). The cross-sectional area of cardiomyocytes of each genotype was quantitated (F). Quantified data are represented as a box-and-whisker plot, with bonds from 25th to 75th percentile, median line, and whiskers ranging from minimum to maximum values (n = 1067 for WT, n = 698 for NSML, n = 1023 for PZRY242F, and n = 1241 for NSML/PZRY242F). (G and H) Representative echocardiographic images of 16-week-old mice (G). LVPW and IVS in diastole (d) were measured from echocardiograms (H) (n = 7 for each group). (I and J) The relative mRNA expression levels of Myh6 and Myh7 and the ratio of Myh7 and Myh6 (I) and Nppa and Nppb (J) in the hearts of 16-week-old mice were measured by quantitative reverse transcription PCR (qRT-PCR) (n = 7 for each group). All data represent mean ± SEM. Statistical significance was analyzed with 1-way ANOVA with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
Effect of PZR tyrosyl phosphorylation on NSML signaling.
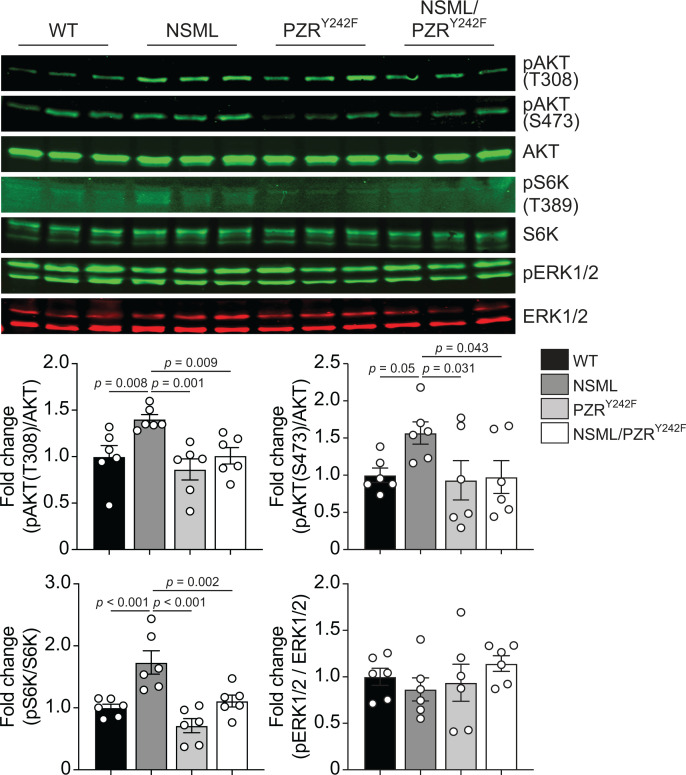
It has been shown that the hearts of NSML mice exhibit HCM because of increased levels of AKT phosphorylation, suggesting that the PI3K/AKT pathway plays a central role in promoting NSML-associated HCM (27, 28, 31). We therefore investigated whether AKT was affected in the hearts of NSML/PZRY242F mice. As previously reported (27), lysates derived from the hearts of NSML mice showed increased levels of AKT and S6 kinase (S6K) phosphorylation as compared with WT mice (Figure 3), whereas heart lysates from NSML/PZRY242F mice failed to show increased AKT and S6K phosphorylation levels as compared with NSML mice (Figure 3). Notably, ERK1/2 phosphorylation levels in heart lysates were comparable across all 4 genotypes (Figure 3). These results demonstrate that PZR tyrosyl phosphorylation, and hence PZR/SHP2 interactions, are required for the hyperactivation of the PI3K/AKT pathway, correlating with the progression of HCM in NSML mice.
Figure 3. PZRY242F mutation reverses NSML signaling in the heart.
Heart lysates from 16-week-old WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice were immunoblotted with anti–p-AKT (T308), –p-AKT (S473), -AKT, –p-S6K (T389), -S6K, –p-ERK1/2, and -ERK1/2 antibodies. The phosphorylation levels of AKT, S6K, and ERK1/2 were quantitated (n = 6). Data represent mean ± SEM, 1-way ANOVA with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
Transcriptome analysis in the hearts of NSML/PZRY242F mice.
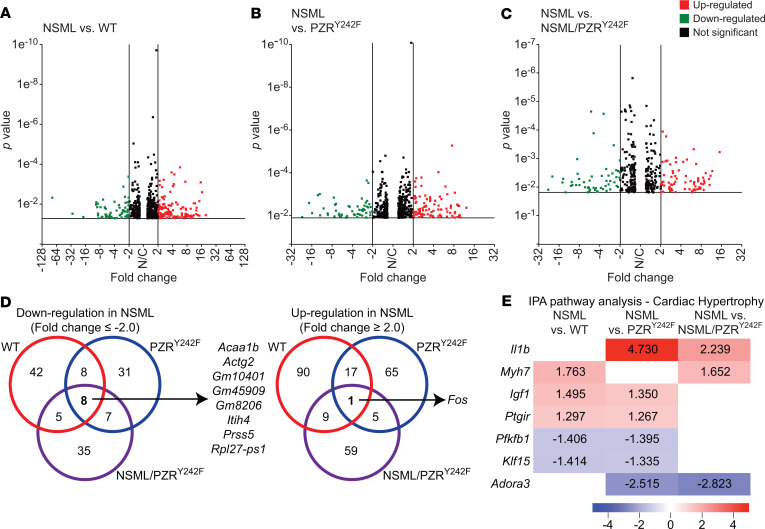
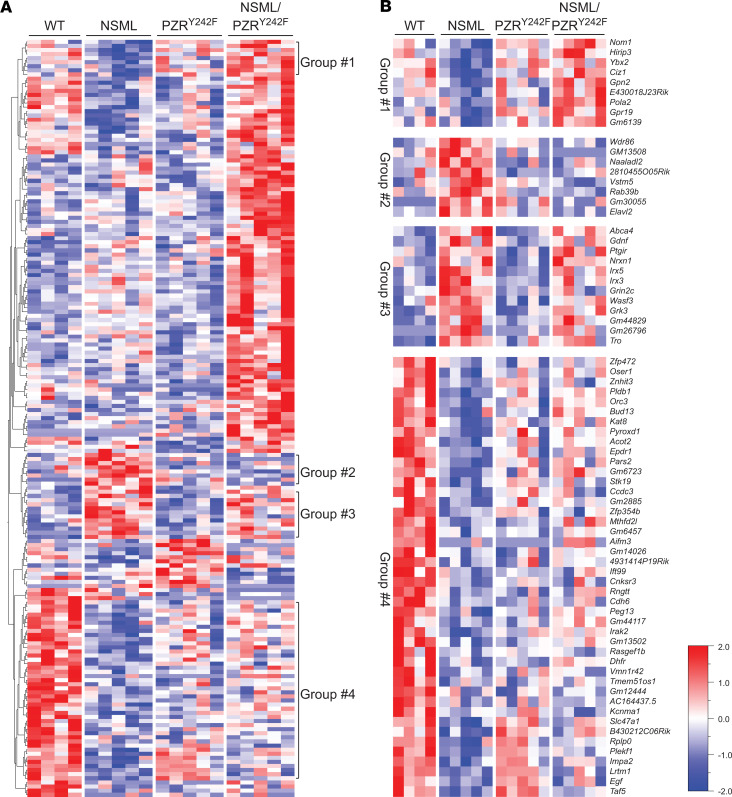
To provide insight into the effects of PZRY242F mutation in NSML-associated gene expression in the heart, whole-transcriptome RNA-sequencing (RNA-Seq) was performed in tissue derived from the hearts of these mice. We generated 22~49 million reads per sample, filtered reads to have high quality scores, and mapped 84.1%–86.3% of those reads to the mouse genome. Principal component analysis showed that samples from the same genotype clustered together, suggesting that transcriptome profiles were different in the 4 genotypes (Supplemental Figure 6). In order to identify candidate genes that are regulated by NSML-SHP2 mutation and/or PZR tyrosyl phosphorylation in the heart, we identified differentially expressed genes (P value < 0.05, fold change ≥ 2.0): 180 genes were found to be differentially expressed between NSML and WT, with 63 downregulated and 117 upregulated in the NSML group, as shown in the volcano plot (Figure 4A). When NSML mice were compared with PZRY242F mice, 142 DEGs were identified, with 54 downregulated and 88 upregulated (Figure 4B); 129 genes were differentially expressed in the NSML compared with NSML/PZRY242F group, in which 55 genes were downregulated and 74 upregulated (Figure 4C). A Venn diagram shows the number of DEGs among the 3 pairwise comparisons (Figure 4D). Interestingly, we found 8 genes (Acaa1b, Actg2, Gm10401, Gm45909, Gm8206, Itih4, Prss5, and Rpl27-ps1) that were commonly downregulated, and 1 gene (Fos) was commonly upregulated in NSML compared with the other 3 groups (Figure 4D). Ingenuity pathway analysis identified cardiac hypertrophy–related genes (P = 0.0187 for NSML vs. WT, and P = 0.0124 for NSML vs. NSML/PZRY242F), such as Myh7 and Igf1, were differentially expressed in the hearts of NSML mice (Figure 4E). The hierarchical clustering of log ratio–transformed gene expression was represented in a heatmap and showed 185 genes that were differentially expressed between 4 genotypes (P < 0.01) (Figure 5A). Further, we identified 4 groups (groups 1, 2, 3, and 4) of genes in the hearts of NSML/PZRY242F mice that were apparently reverted in their expression to WT levels (Figure 5A). Genes identified in groups 1–4 exhibiting revertant behavior (Figure 5B) demonstrate that they are dependent upon PZR tyrosyl phosphorylation and hence represent a transcriptomic “fingerprint” of NSML/PZR target genes that are potentially involved in the progression of HCM.
Figure 4. RNA-Seq transcriptome analysis of NSML and NSML/PZRY242F mice.
Total RNA was isolated from the hearts of 16-week-old mice and RNA-Seq analysis performed. (A–C) Differentially expressed genes (DEGs) of NSML (Ptpn11Y279C/+ Mpzl1+/Y242F) compared with WT (Ptpn11+/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), or NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice are displayed as volcano plots. A cutoff of P ≤ 0.05 and fold change ≤ –2.0 (green) or ≥ 2 (red) was used to determine significance. (D) Venn diagram representation showing the number of significant DEGs in NSML compared with WT, PZRY242F, or NSML/PZRY242F groups. Down- (left) and upregulated (right) genes in NSML are shown. (E) Ingenuity pathway analysis (IPA) performed on statistically significant genes (P < 0.05) for cardiac hypertrophy. Fold changes are shown in the heatmap with values indicated.
Figure 5. Hierarchical clustering analysis of the relative gene expression of NSML and NSML/PZRY242F mice.
(A) Hierarchical clustered heatmap of log2-transformed gene expression in the hearts of WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice (P < 0.01). Each column represents an individual mouse and each row represents a gene. Differential gene expression was shown in the heatmap. (B) Relatively upregulated (group 2 and 3) or downregulated (group 1 and 4) genes in NSML compared with WT, PZRY242F, or NSML/PZRY242F were clustered.
PZR tyrosyl phosphorylation in NSML drives cardiac fibrosis and dysregulated IL-6 secretion.
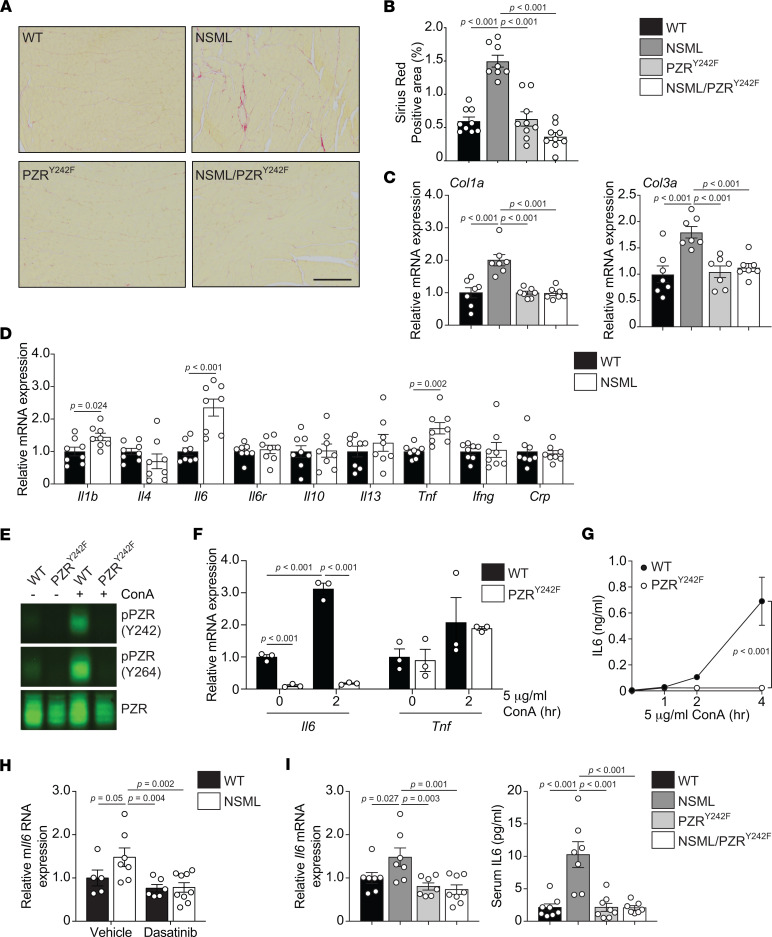
Myocardial fibrosis is one of the clinical sequelae of HCM that contribute to left ventricular dysfunction, heart failure, and sudden cardiac death (1). As reported previously (27, 29), the hearts of NSML mice exhibited myocardial fibrosis as detected by Sirius red staining and increased expression of fibrotic genes (Col1a and Col3a) (Figure 6, A–C). However, hearts of NSML/PZRY242F mice were devoid of myocardial fibrosis, and the expression of fibrotic genes was equivalent to that of WT mice (Figure 6, A–C). These observations suggested that PZR tyrosyl phosphorylation in NSML contributes to myocardial fibrosis. To explore a potential mechanism through which PZR tyrosyl phosphorylation contributes to myocardial fibrosis in NSML mice, we examined the mRNA expression of cytokines in the hearts of NSML mice (Figure 6D). We found that Il1b, Il6, and Tnf mRNA expression levels were significantly upregulated in the hearts of NSML mice as compared with WT mice (Figure 6D). To determine whether the expression of these cytokines was dependent upon PZR tyrosyl phosphorylation, we used mouse embryonic fibroblasts (MEFs) derived from WT and PZRY242F mice. PZR tyrosyl phosphorylation was induced by treating cells with concanavalin A (ConA) (32) (Figure 6E). We found that Il6 and Tnf mRNA expression were significantly increased in WT MEFs by ConA (Figure 6F) but Il1b expression was undetectable. Treatment of PZRY242F MEFs with ConA, which did not induce PZR tyrosyl phosphorylation (Figure 6E), resulted in the diminution of Il6 but not of Tnf mRNA expression (Figure 6F), indicating that the induction of Il6 rather than Tnf mRNA was dependent upon PZR tyrosyl phosphorylation. Furthermore, ConA was capable of inducing IL-6 secretion in WT MEFs; however, in PZRY242F MEFs, IL-6 secretion was undetectable (Figure 6G). Similar results were obtained when PZR tyrosyl phosphorylation was induced by plating cells onto fibronectin (Supplemental Figure 7), further substantiating the interpretation that IL-6 secretion is dependent upon PZR tyrosyl phosphorylation. We also reintroduced PZR WT into PZRY242F MEFs and demonstrated that expression of PZR WT rescues Il6 expression in both basal conditions and upon ConA induction (Supplemental Figure 8). Notably, low-dose dasatinib (0.1 mg/kg/d) treatment from the age of 12 weeks for 4 weeks, which prevents the progression of fibrosis in NSML mice (29), also reduced Il6 mRNA expression in the hearts of NSML mice (Figure 6H). Importantly, the increased levels of Il6 mRNA expression in the heart and increased circulating IL-6 levels in NSML mice were completely restored to WT levels in NSML/PZRY242F mice (Figure 6I). Given the established role of IL-6 in promoting cardiac fibrosis (19, 33, 34), these data demonstrate that PZR tyrosyl phosphorylation in NSML stimulates IL-6 production, which likely contributes to the later development of myocardial fibrosis in NSML mice.
Figure 6. PZR tyrosyl phosphorylation promotes myocardial fibrosis and positively regulates IL-6 expression.
(A–C) Picrosirius red stains of left ventricles from 16-week-old WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice (A) (scale bar: 50 μm). Sirius red–positive areas were quantified (n = 9 for each group; 3 images per mouse, 3 mice per group) (B). The relative mRNA expression levels of Col1a and Col3a in the heart were measured by qRT-PCR (C) (n = 7 for each group). (D) The relative mRNA expression levels of Il1b, Il4, Il6, Il6r, Il10, Il13, Tnf, Ifng, and Crp in the hearts of 16-week-old WT (Ptpn11+/+) and NSML (Ptpn11Y279C/+) mice were measured by qRT-PCR (n = 8 for each group). (E–G) Mouse embryonic fibroblasts (MEFs) from WT (Mpzl1+/+) and PZRY242F (Mpzl1Y242F/Y242F+) mice were serum-starved and stimulated with 5 μg/mL of concanavalin A (ConA) for 2 hours. Whole-cell lysates were immunoblotted with anti–p-PZR (Y242), –p-PZR (Y264), and -PZR antibodies (E). The relative mRNA expression of Il6 and Tnf was measured by qRT-PCR (F) (n = 3). (G) MEFs were serum-starved and stimulated with 5 μg/mL of Con A for 1, 2, and 4 hours. Secreted IL-6 protein levels were measured by ELISA (n = 3 for each group). (H) Low-dose dasatinib (0.1 mg/kg/d, i.p.) was administered to 12-week-old WT (Ptpn11+/+) and NSML (Ptpn11Y279C/+) mice for 4 weeks. The relative mRNA expression of Il6 in the heart was measured by qRT-PCR (n = 5 for vehicle-treated WT, n = 7 for vehicle-treated NSML, n = 6 for dasatinib-treated WT, and n = 9 for dasatinib-treated NSML). (I) The relative mRNA expression level of Il6 in the hearts of 16-week-old WT (Ptpn11+/+ Mpzl1+/Y242F), NSML (Ptpn11Y279C/+ Mpzl1+/Y242F), PZRY242F (Ptpn11+/+ Mpzl1Y242F/Y242F), and NSML/PZRY242F (Ptpn11Y279C/+ Mpzl1Y242F/Y242F) mice were measured by quantitative RT-PCR (n = 7 for WT, NSML, and PZRY242F, and n = 8 for NSML/PZRY242F). Serum was collected from 16-week-old mice, and IL-6 protein levels were measured by ELISA (n = 8 for WT and PZRY242F, n = 7 for NSML and NSML/PZRY242F). All data represent mean ± SEM. Statistical significance was analyzed with 2-tailed Student’s t test (D) or 2-way ANOVA (F–H) or 1-way ANOVA (B, C, and I) with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
PZR tyrosyl phosphorylation activates IL-6–mediated fibrogenesis by coupling to the NF-κB pathway.
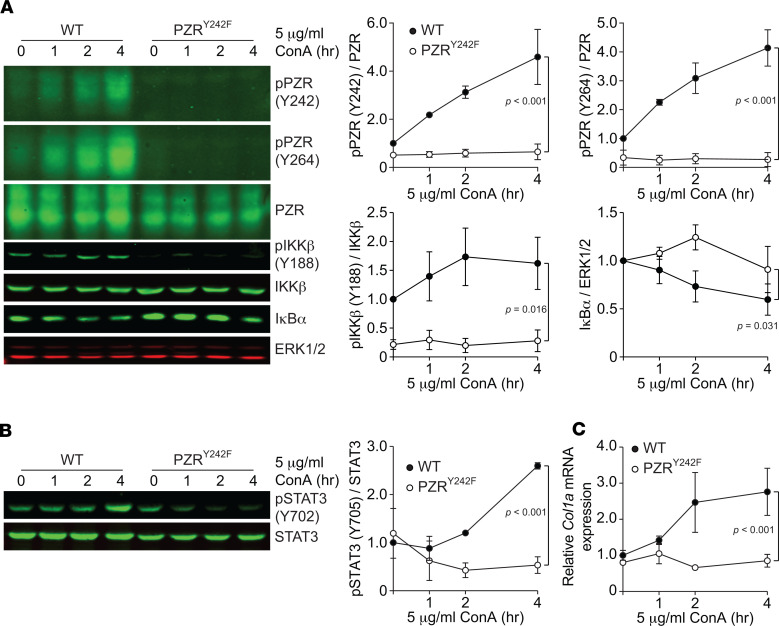
The inhibitor of NF-κB (IκB) kinase (IKK) and its downstream NF-κB signaling pathway are important regulators of inflammation and are implicated in HCM and heart failure (35–39). Il6 is a major NF-κB target gene, and ablation of NF-κB activity ameliorates HCM in a variety of mouse models (35, 36, 39–47). IKK is an enzyme complex that is composed of IKKα, IKKβ, and IKKγ that facilitates NF-κB activation through phosphorylation-mediated protein degradation of IκB (48, 49). c-Src kinase has been shown to phosphorylate and activate IKKβ (50). We previously suggested that PZR hypertyrosyl phosphorylation results in enhanced Src signaling through increased recruitment of SHP2/Src to PZR, thereby driving aberrant signaling in NSML (20, 29). We hypothesized that enhanced PZR/SHP2/Src signaling promotes IKKβ phosphorylation, leading to NF-κB–mediated Il6 expression. To test this, we determined whether NF-κB signaling is dependent on PZR tyrosyl phosphorylation. When WT MEFs were treated with ConA, PZR tyrosyl phosphorylation was induced (Figure 7A). In contrast, ConA-treated PZRY242F MEFs failed to promote tyrosyl phosphorylation of IKKβ (Figure 7A). Consistent with defective IKKβ activation in PZRY242F MEFs, IκB protein levels were increased in these cells as compared with WT MEFs (Figure 7A). These data suggest that increased activation of Il6 gene expression in the hearts of NSML mice is driven by PZR tyrosyl phosphorylation, which activates IKK/NF-κB signaling. To further substantiate this interpretation, we examined the effects of an array of small-molecule inhibitors of the Src and IKK/NF-κB pathway on ConA-induced Il6 mRNA expression in WT MEFs. Consistent with the notion that PZR/SHP2 couples to the NF-κB pathway through c-Src, Src kinase inhibitor (dasatinib), IKKβ inhibitor (IKK-16), NF-κB nuclear transportation inhibitor (JSH23), and NF-κB DNA binding inhibitor (GYY4137) all significantly reduced Il6 mRNA expression and IL-6 secretion in ConA-treated WT MEFs (Supplemental Figure 9, A–D). We also confirmed that these inhibitors ameliorated Il6 mRNA expression in PZRY242F MEFs reexpressing WT PZR (Supplemental Figure 9E). Collectively, these data demonstrate that PZR hypertyrosyl phosphorylation promotes NF-κB signaling, resulting in enhanced IL-6 secretion.
Figure 7. PZR tyrosyl phosphorylation activates the NF-κB pathway and Col1a expression.
MEFs from WT (Mpzl1+/+) and PZRY242F (Mpzl1Y242F/Y242F+) mice were serum-starved and stimulated with 5 μg/mL of ConA for 1, 2, and 4 hours. (A and B) Whole-cell lysates were immunoblotted with anti–p-PZR (Y242), –p-PZR (Y264), -PZR, –p-IKKβ (Y188), -IKKβ, -IκBα, and -ERK1/2 antibodies (A) and anti–p-STAT3 (Y702) and -STAT3 antibodies (B). The phosphorylation levels of PZR, IKKβ, (A) and STAT3 (B) and protein level of IκBα (A) were quantitated (n = 3). (C) Total RNA was isolated and the relative expression of Col1a was measured by qRT-PCR (n = 3). All data represent mean ± SEM. Statistical significance was analyzed with 2-way ANOVA (A–C) with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
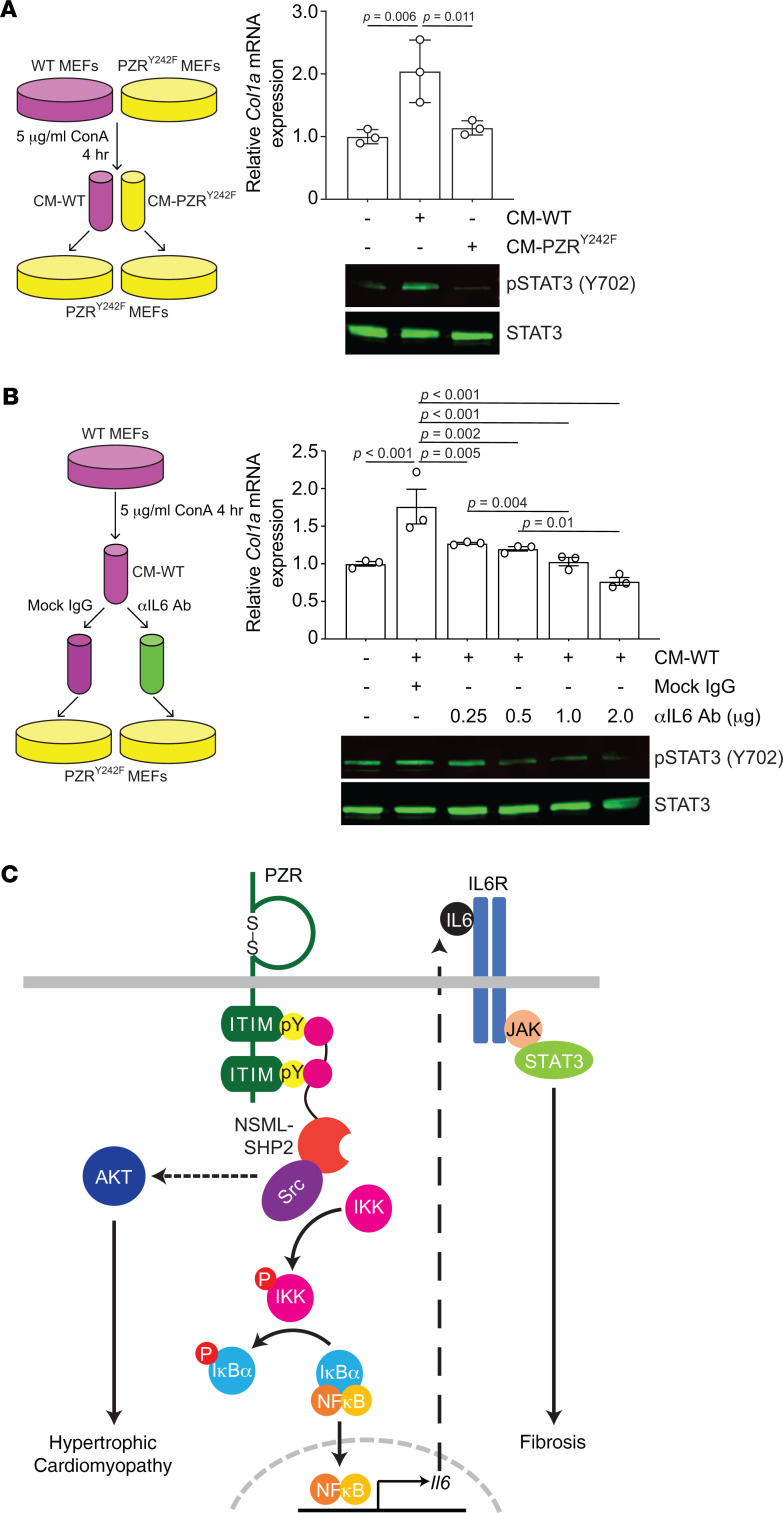
IL-6 activates STAT3, which upregulates fibrotic genes, such as Col1a (51, 52). Because IL-6 secretion was dependent upon PZR tyrosyl phosphorylation (Figure 6, F and G), we hypothesized that STAT3 activity in PZRY242F MEFs would be reduced. Indeed, we found that ConA-induced STAT3 tyrosyl phosphorylation and Col1a expression were dramatically reduced in PZRY242F MEFs as compared with WT MEFs (Figure 7, B and C). These data demonstrate that PZR tyrosyl phosphorylation is responsible for the upregulation of IL-6, which acts in an autocrine manner to induce STAT3 phosphorylation. Furthermore, conditioned medium from ConA-treated WT MEFs (CM-WT) induced STAT3 phosphorylation and Col1a expression in PZRY242F MEFs, which are defective for IL-6 secretion (Figure 8A), but conditioned medium from ConA-treated PZRY242F MEFs (CM-PZRY242F) did not induce STAT3 phosphorylation and Col1a expression (Figure 8A). We confirmed that the effects of the CM-WT on Col1a expression were specifically due to IL-6 by treating the CM-WT with an IL-6–neutralizing antibody followed by assessing Col1a gene expression in PZRY242F MEFs (Figure 8B). These experiments showed that STAT3 phosphorylation and Col1a gene expression were reduced by IL-6–neutralizing antibody treatment in a dose-dependent manner (Figure 8B). These data suggest that PZR hypertyrosyl phosphorylation in the hearts of NSML mice promotes cardiac fibrosis through increased secretion of IL-6 via NF-κB followed by autocrine activation of IL-6/STAT3–mediated fibrotic gene expression.
Figure 8. PZR tyrosyl phosphorylation induces autocrine/paracrine-mediated IL-6 secretion to activate STAT3 phosphorylation and Col1a expression.
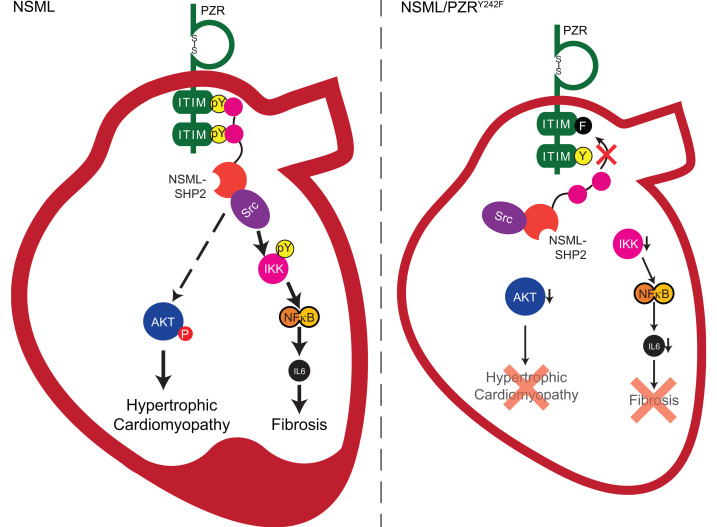
(A) WT (Mpzl1+/+) and PZRY242F (Mpzl1Y242F/Y242F+) MEFs were serum-starved and stimulated with 5 μg/mL of ConA for 4 hours, and medium was collected (conditioned medium, CM). CM from WT MEFs (CM-WT) or PZRY242F MEFs (CM-PZRY242F) was applied to PZRY242F MEFs for 2 hours. Whole-cell lysates were immunoblotted with anti–p-STAT3 (Y702) and -STAT3 antibodies. Total RNA was isolated and the relative expression of Col1a was measured by qRT-PCR (n = 3). (B) CM-WT was incubated with either mock IgG or anti–IL-6 neutralizing antibody and then incubated onto PZRY242F MEFs for 2 hours. Whole-cell lysates were immunoblotted with anti–p-STAT3 (Y702) and -STAT3 antibodies. Total RNA was isolated and the relative expression of Col1a was measured by qRT-PCR (n = 3). (C) Schematic representation of PZR/SHP2 signaling in NSML. All data represent mean ± SEM. Statistical significance was analyzed with 1-way ANOVA with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli correction for multiple comparisons.
Discussion
Previously, we reported that in the hearts of NSML mice the ITIM-containing transmembrane glycoprotein PZR is hypertyrosyl phosphorylated, resulting in enhanced plasma membrane recruitment of SHP2 (20, 29). Although proposed to be involved in congenital heart disease, and possibly HCM in NSML, a causal role of PZR tyrosyl phosphorylation in HCM remained to be formally established. To address the question of whether PZR tyrosyl phosphorylation, and hence SHP2 binding, plays a role in NSML-associated HCM, we generated a knockin mouse harboring a mutant form of PZR (PZRY242F mouse) that is defective for tyrosyl phosphorylation and, thus, SHP2 binding. PZRY242F mice showed no developmental or postdevelopmental defects and exhibited normal postnatal growth. Therefore, tyrosyl phosphorylation of PZR in this context does not appear essential for cardiac development under normal physiological conditions. Whether there are defects in these PZRY242F mice in other tissues cannot be excluded. However, when PZRY242F mice were intercrossed with NSML mice, the development of HCM seen in NSML mice was completely abrogated. These results demonstrate that in the context of an NSML mutation, PZR tyrosyl phosphorylation and SHP2 association play a direct role in promoting HCM. Mechanistically, PZR/SHP2 binding in NSML promotes AKT activity in the hearts of NSML mice (Figure 8C), while PZR/SHP2 binding induces the activation of NF-κB, resulting in the paracrine/autocrine secretion of IL-6 that promotes myocardial interstitial fibrosis (Figure 8C). Thus, NSML-SHP2 mutants engage PZR to activate pathways distinct from, but parallel to, the canonical RASopathy pathway assigned to these mutations.
Multiple signaling pathways have been attributed to the development of HCM, and the interplay of these mechanisms further complicates a clear understanding of how non–sarcomere-driven HCM manifests. Nevertheless, the molecular basis, at least in part, for the development of HCM in NSML mice has been attributed largely to enhanced activation of AKT/mTOR signaling (27, 28, 31, 53). Remarkably, we observed that NSML/PZRY242F mice showed completely normal HW/BW as well as HW/TL as compared with WT mice. Consistent with this, cardiomyocyte size in NSML/PZRY242F mice was completely reverted to WT cardiomyocyte size. These results correlated with the observed increased AKT activity in heart lysates derived from NSML mice as expected, but in NSML/PZRY242F mice, AKT and S6K activity were equivalent to that observed in WT animals. Collectively, these results demonstrate that the observed hypertyrosyl phosphorylation of PZR in the hearts of NSML mice contributes to the hyperactivation of the AKT/mTOR pathway to promote HCM (Figure 8C). The hyperactivation of AKT seen in the hearts of NSML mice that is rescued upon introduction of the PZRY242F allele occurs without appreciable changes in ERK1/2 phosphorylation. This raises the question of the role of ERK1/2 downstream of the NSML-SHP2 mutation in the development of HCM. Our data imply that despite the NSML-SHP2 mutation being inactivating for SHP2 phosphatase activity, there is an evident dispensability for ERK1/2 for the development of NSML-associated HCM. It is important to note that our studies do not define the extent to which PZR hypertyrosyl phosphorylation, AKT and ERK1/2 act in a particular cell type(s) in vivo to promote NSML-mediated HCM. Studies employing cell type–specific PZR tyrosyl phosphorylation–knockin mice will be required to uncover these in vivo details as will extending these studies to assess potential sex differences by examining female mice. Despite the aforementioned limitations, from a molecular standpoint, NSML-SHP2 mutations adopt an open conformation, which is critical for PZR hypertyrosyl phosphorylation and enhanced PZR/SHP2 binding. These results indicate that NSML-SHP2 mutants behave as gain-of-function mutants. We suggest that acquisition of the open SHP2 conformation rendered by the NSML-associated mutations largely defines the molecular basis for NSML, at least for the progression of HCM. However, this interpretation is not mutually exclusive of the possibility that the lower levels of catalytic activity of NSML-associated SHP2 mutations also contribute to the development of HCM through other pathways.
We have previously suggested that low-dose dasatinib could serve as a potential therapeutic for the treatment of HCM in NSML (29). Our results suggest that low-dose dasatinib, which inhibits PZR tyrosyl phosphorylation and prevents the development of HCM in NSML mice, could act by blocking PZR tyrosyl phosphorylation and association with SHP2, and subsequently, AKT activity. Additionally, low-dose dasatinib treatment in NSML mice reduced IL-6 expression levels. Given that other therapeutic strategies against NSML have targeted the AKT/mTOR pathway, although effective in animal models and in a patient (27, 31, 53), toxicity at the effective doses makes this line of therapy prohibitive. If as proposed, enhanced AKT activation can be curtailed through modulating PZR tyrosyl phosphorylation using a low-dose tyrosine kinase inhibitor, such as dasatinib, this strategy might offer an attractive therapeutic path to treat certain NSML cases with HCM with minimum toxicity. PZR is also hypertyrosyl phosphorylated in the hearts of Noonan syndrome mice that also exhibit congenital heart disease phenotypes that are rescuable by low-dose dasatinib treatment (29). Whether PZR hypertyrosyl phosphorylation and increased PZR/SHP2 interactions are also similarly causal in Noonan syndrome–related HCM as well as other forms of HCM, such as that induced by pressure overload, remains to be determined.
Initiation of promiscuous signaling as a result of an SHP2 mutation in NSML that inactivates the phosphatase activity while simultaneously generating an “open” SHP2 conformation creates the formal possibility for both loss-of- and gain-of-function attributes for SHP2 signaling (18). This complexity is further amplified as a result of enhanced upstream SHP2 target binding as observed through an upregulation of PZR/SHP2 interactions in the hearts of NSML mice, which sustains the steady-state lifetime of this and other SHP2 SH2 domain–mediated complexes. Conceivably, these mechanisms are concurrently operable and potentially interoperable, culminating in the dysregulation of multiple downstream pathways that regulate critical gene transcription events that drive congenital heart disease and in particular HCM. Here we define a novel link between PZR hypertyrosyl phosphorylation and downstream signaling to AKT. However, multiple pathways likely contribute to the development of HCM in NSML, and as such, PZR represents one such signaling component in the manifestation of this disease. Given that PZR tyrosyl phosphorylation rescues NSML-mediated HCM, we assessed in an unbiased manner the potential target genes dependent on PZR tyrosyl phosphorylation/SHP2 binding by performing RNA-Seq transcriptome analysis in NSML, PZRY242F, and NSML/PZRY242F mice. A global analysis between these genotypes revealed qualitatively 4 major cohorts of gene sets that were differentially expressed in NSML mice that were reverted back to “normal” in NSML mice expressing the PZR tyrosyl phosphorylation site mutant. These apparent “revertant” groups of genes suggest the existence of an identifiable “fingerprint” of PZR tyrosyl phosphorylation–dependent genes that are dysregulated in the context of the pathophysiological signals driven by the expression of SHP2-associated NSML mutants. Identifying the function of such genes may provide insight into the molecular program governing the initiation and/or maintenance of HCM and possibly other NSML-associated phenotypes. We identified Fos as upregulated by 3–7 fold in NSML mouse hearts, and the upregulation of Fos in the hearts of NSML mice was dependent upon PZR tyrosyl phosphorylation. Provocatively, Fos is an immediate early gene that has been shown to be upregulated in the hearts of patients with HCM (54, 55). Additionally, we identified Grin2c (GluN2C/NR2C), which encodes for the N-methyl-d-aspartate (NMDA) receptor subunit C (19), as an NSML target gene that is reverted to WT levels in NSML/PZRY242F mice. Interestingly, chronic NMDA receptor activation has been associated with cardiac remodeling, cardiac electrical dysfunction, and myocardial fibrosis (19). Given that patients with NSML exhibit electrical conduction disorders of unknown etiology, the identification of potential targets affecting this symptom may be of clinical significance.
A precipitating consequence of HCM is the development of myocardial fibrosis, which underlies the deterioration of cardiac contractility that accompanies heart failure (3, 56, 57). The basis for myocardial fibrosis has been attributed, at least in part, to both local and systemic inflammation caused by increased circulation of proinflammatory cytokines (58). NSML/PZRY242F mice were resistant to the development of myocardial fibrosis consistent with the inhibition of HCM as compared with NSML mice. Although we identified a set of proinflammatory cytokines that were significantly upregulated in NSML mice, only IL-6 expression levels were restored to WT levels in NSML/PZRY242F mice. These observations demonstrate that in NSML mice the PZR/SHP2 interaction is sufficient to promote the upregulation of IL-6 secretion (Figure 6F). The involvement of IL-6 in the development of myocardial fibrosis in HCM is well established (19, 33, 34, 59). IL-6 infusion results in concentric hypertrophy in rats, and IL-6 deletion attenuates left ventricular hypertrophy, myocardial fibrosis, and left ventricular dysfunction in response to pressure overload in mice (34). IL-6 also acts on fibroblasts to promote fibroblast proliferation and collagen production, and as such, the action of IL-6 on fibroblasts is a recognized contributing factor of myocardial fibrosis (34, 60, 61). Our results demonstrating that fibroblasts expressing PZRY242F fail to promote IL-6 expression supports the interpretation that PZR/SHP2 interactions regulate IL-6 expression and secretion. Mechanistically, we demonstrated that PZR/SHP2 interactions activate IL-6 via an Src-dependent pathway that stimulates the IKK enzyme complex, phosphorylation of IκB, and NF-κB nuclear translocation (Figure 8C). Consistent with the notion that this pathway proceeds through an Src-dependent mechanism, low-dose dasatinib treatment of NSML mice restored the elevated Il6 levels to those comparable to WT mice. We further uncovered that the actions of PZR to promote IL-6 secretion in fibroblasts correlated with the upregulation of collagen expression. This was likely a result of IL-6 acting in an autocrine manner on fibroblasts to stimulate STAT3-mediated expression of Col1a1. Although our data showed that at least fibroblasts are a target of IL-6, we cannot exclude other cell types, such as cardiomyocytes, endothelial cells, or hematopoietic cells, that could also be receptive to the supporting auto- or paracrine actions of IL-6 in NSML pathogenesis. Notably, STAT3 has been implicated in HCM (19); at this juncture our data do not exclude a role for STAT3 in HCM of NSML mice.
In summary, our data demonstrate using a PZR tyrosyl phosphorylation–defective mouse model that PZR/SHP2 interactions are critical for the development of NSML-associated HCM. These data identify the NF-κB signaling pathway as a novel PZR/SHP2 downstream target in NSML and suggest that this pathway contributes to the cardiac pathogenesis associated with NSML.
Methods
Generation of Mpzl1Y242F mouse strain.
The Mpzl1Y242F mutation was introduced via CRISPR/Cas9 genome editing essentially as described (62). Potential Cas9 target guide (protospacer) sequences were screened using the MIT CRISPR tool over a genomic sequence spanning 2 potential Tyr codons: 5′-CTCTTCTCCAGGGCCCAGTCATTTACGCACAGTTAGACCACTCTGGCGGACACCACAGCGGCAAGATTAATAAGTCAGAGTCGGTTGTGTATGCGGACATCCGG-3′. The protospacer sequence TCTAACTGTGCGTAAATGAC(PAM TGG) was selected based on its overlap with the Y242 codon, its high specificity score of 81, and the tool’s prediction of no likely off-target sites on chromosome 1. A PCR primer was synthesized containing a T7 promoter, the protospacer, and the 5′ end of the sgRNA scaffold sequence (Supplemental Table 4). The template for sgRNA in vitro transcription was generated by PCR amplification using this primer and the “universal” reverse primer from the pX330 sgRNA expression template (Addgene plasmid 42230) (63). sgRNA was transcribed from the template using the MEGAShortscript kit (Invitrogen, Thermo Fisher Scientific). Cas9 mRNA was similarly transcribed in vitro from the pX330 plasmid using the primers listed in Supplemental Table 4. Transcribed RNAs were purified using MEGAclear kit (Invitrogen, Thermo Fisher Scientific) and eluted using microinjection buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, and 0.25 mM EDTA). Template oligonucleotide (ssODN) containing the specified base pair changes was synthesized by IDT. Components were mixed in injection buffer at a concentration of 30 ng/μL Cas9 mRNA, 15 ng/μL each sgRNA, and 35 ng/μL ssODN; centrifuged at 16,000 g; and microinjected into the cytoplasm of C57BL/6J zygotes. Embryos were transferred to the oviducts of pseudopregnant CD-1 foster females using standard techniques (64). Twenty-one pups were screened for the Mpzl1Y242F mutation, and 4 pups contained the correct base pair (bp) substitutions.
Animal handling.
Mpzl1+/Y242F heterozygotes were crossed with each other, and offspring were genotyped by PCR and digestion with AgeI for the Y242F allele. Ptpn11Y279C/+ male mice were crossed with WT C57BL/6J female, and offspring were genotyped by PCR for the Y279C allele. All mice were maintained on C57BL6/J backgrounds, backcrossed for more than 10 generations, and genotyped by PCR using tail DNA. We used 16- to 20-week-old male mice in this study. Dasatinib was intraperitoneally injected (0.1 mg/kg/d) into male 12-week-old mice for 4 weeks.
Antibodies, chemicals, cell lines, and plasmids.
The following antibodies were used for immunoblotting. Rabbit polyclonal SHP2 (C-18) (catalog sc-280, IB-1:1000) and mouse monoclonal GST (B-14) (catalog sc-138, IB-1:1000) antibodies were from Santa Cruz Biotechnology. Rabbit monoclonal phospho-PZR (Y241; D6F9) (catalog 8131, IB-1:1000), rabbit monoclonal phospho-PZR (Y263; D6A5) (catalog 8088, IB-1:1000), rabbit polyclonal phospho-AKT (S473; catalog 9271, IB-1:1000), rabbit polyclonal phospho-AKT (T308; catalog 9275, IB-1:1000), mouse monoclonal AKT1 (C73H10) (catalog 2938; IB-1:1000), rabbit monoclonal phospho-S6K (T389; 108D2) (catalog 9234; IB-1:1000), rabbit monoclonal S6K (49D7) (catalog 2708; IB-1:1000), rabbit polyclonal phospho-ERK1/2 (T202/Y204; catalog 9101, IB-1:1000), mouse monoclonal ERK (3A7) (catalog 9107, IB-1:1000), rabbit monoclonal PZR (D17B10) (catalog 9893s, IB-1:1000), rabbit monoclonal IKKβ (D30C6) (catalog 8943; IB-1:1000), rabbit monoclonal IκBα (L35A5) (catalog 4814; IB-1:1000), rabbit monoclonal phospho-STAT3 (Y705; D3A7) (catalog 9145; IB-1:1000), and rabbit monoclonal STAT3 (79D7) (catalog 4904; IB-1:1000) antibodies were purchased from Cell Signaling Technology. Rabbit polyclonal phospho-IKKβ (Y188; catalog ab192820, IB-1:1000) was obtained from Abcam. Goat polyclonal IL-6–neutralizing antibody (catalog AB-406-NA) and normal goat IgG control antibody (catalog AB-108-C) were purchased from R&D Systems, Bio-Techne. Rabbit polyclonal PZR antibody (105-6, IB-1:1000) was provided by Z. J. Zhao (21). IL-6 levels in serum and medium were measured using an ELISA kit (mouse IL-6 Quantikine ELISA; R&D Systems, Bio-Techne, catalog M6000B). Dasatinib (catalog 1586-100) was purchased from Biovision. IKK16 (catalog 13313), JSH-23 (catalog 10536), and GYY4137 (catalog 13345) were from Cayman Chemical. MEFs were isolated from WT and PZRY242F mice. Cells were grown in growth medium (DMEM supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum) in a 5% CO2 incubator at 37°C. The mammalian plasmid expressing human PZR (pCDNA3-hPZR-WT) was described previously (24) and was transfected into MEFs using the Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific).
Immunoblotting.
Cells or heart tissue was lysed on ice in lysis buffer (25 mM Tris-HCl at pH 7.4, 136 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1% Nonidet P-40, 1 mM Na3VO4, 10 mM NaF, 1 mM benzamidine, 1 mM PMSF, 1 μg/mL pepstatin A, 5 μg/mL aprotinin, and 5 μg/mL leupeptin). Cell or tissue lysates were incubated at 4°C for 30 minutes and clarified by centrifugation at 20,000 g at 4°C for 10 minutes. Protein concentration was determined using the BCA reagent according to the manufacturer’s instructions (23225; Thermo Fisher Scientific). Total lysates were subjected to SDS-PAGE and immunoblotting. The sites of antibody binding were visualized and were quantified using the Odyssey CLx Imaging System (LI-COR Biosciences).
In vitro GST affinity precipitation assay.
Recombinant GST-SH2 domains of human SHP2 protein were purified from bacterial cells [pGEX-2TK SHP2 N+C, BL21(DE3)]. Affinity precipitation assays were carried out in 1 mg of heart lysates with GST-SH2 of SHP2 protein overnight at 4°C. SH2 domain–bound PZR proteins were affinity purified by BSA-coated GST-Sepharose 4B (17-0756-01; GE Healthcare) for 1 hour at 4°C. The interaction between SH2 domains of SHP2 and PZR was examined by immunoblotting with anti-PZR antibodies.
Histology.
Hearts were isolated, fixed in 4% paraformaldehyde in phosphate-buffered saline, processed for paraffin sections, and stained with H&E, Sirius red, or Alexa Fluor 488–conjugated WGA (Molecular Probes, Thermo Fisher Scientific; W11261). Tissue images were obtained under bright-field microscopy or fluorescence microscopy (Axiovert 200M; Carl Zeiss).
Echocardiography.
Transthoracic echocardiography was performed by the mouse ultrasound imaging core (MUSIC) at Yale Cardiovascular Research Center. Mice were kept anesthetized with 1.5% isoflurane supplied by a nose cone connected to the isoflurane vaporizer while maintaining physiological temperature. Standardized cardiac views were obtained with a high-resolution ultrasound system (Vevo 2100, VisualSonics) equipped with an ultrahigh-frequency (40 MHz) linear array transducer. Left ventricular (LV) EF, LV end-diastolic and end-systolic volumes, and LV wall thickness were measured offline using Vevo Lab software (version 3.1.0, VisualSonics).
RNA extraction and quantitative real-time PCR analysis.
RNA was isolated from mouse hearts using an RNeasy kit (4104; QIAGEN) according to the manufacturer’s instructions. A total of 1 μg RNA was reverse-transcribed to generate cDNA using a reverse transcriptase PCR kit (4368814; Applied Biosystems, Thermo Fisher Scientific). Real-time quantitative PCR was carried out in triplicate using the 7500 Fast real-time PCR system and PowerUp SYBR Green Master Mix (A25742; Applied Biosystems, Thermo Fisher Scientific) with primer pairs listed in Supplemental Table 5. All relative gene expression levels were analyzed using the ΔΔCT method and normalized to 18S rRNA expression.
RNA-Seq analysis.
Total RNA was isolated from the hearts of mice using an RNeasy kit (QIAGEN) according to the manufacturer’s instructions. RNA quality was determined by estimating the A260/A280 and A260/A230 ratios by NanoDrop (Thermo Fisher Scientific). RNA integrity was determined by running an Agilent Bioanalyzer gel, which measures the ratio of the ribosomal peaks. mRNA was purified from approximately 50 ng of total RNA with oligo-dT beads and sheared by incubation at 94°C in the presence of Mg2+ (KAPA mRNA HyperPrep). Following first-strand synthesis with random primers, second strand synthesis and A-tailing were performed with dUTP to generate strand-specific sequencing libraries. Adapter ligation with 3′-dTMP overhangs were ligated to library insert fragments. Library fragments carrying the appropriate adapter sequences at both ends were amplified using high-fidelity, low-bias PCR. Strands marked with dUTP were not amplified. Indexed libraries that met appropriate cutoffs for both were quantified by qRT-PCR using a commercially available kit (KAPA Biosystems), and insert size distribution was determined with the LabChip GX or Agilent Bioanalyzer. Samples with a yield of at least 0.5 ng/μL were used for sequencing. Sample concentrations were normalized to 1.2 nM and loaded onto an Illumina NovaSeq flow cell at a concentration that yielded 25 million passing filter clusters per sample. Samples were sequenced using 100-bp paired-end sequencing on an Illumina NovaSeq according to Illumina protocols. The 10-bp dual index was read during additional sequencing reads that automatically followed the completion of read 1. Data generated during sequencing runs were simultaneously transferred to the Yale Center for Genome Analysis (YCGA) high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time. Signal intensities were converted to individual base calls during a run using the system’s Real Time Analysis software. Base calls were transferred from the machine’s dedicated personal computer to the Yale high-performance computing cluster via a 1 Gigabit network mount for downstream analysis.
Analysis of RNA-Seq data and pathway analysis.
The RNA-Seq statistical analysis was performed using Partek Flow Genomic Analysis software build version 8.0.19.1125 (Partek Inc.). Paired-end reads were trimmed and aligned to the Genome Reference Consortium Mouse Build 38 (mm10) with the STAR alignment tool (ver. 2.6.1d). Total counts per gene were quantified and normalized to identify differentially expressed genes (DEGs). Lists of DEGs or transcripts were generated by DESeq2. Qlucore Omics Explorer 3.5 (Qlucore AB) was used for principal component analysis of log2-transformed global expression values, heatmap generation, and hierarchical clustering. DEGs used in pathway analysis were determined by using a filtering criterion of fold change ≤ –1.2 or ≥ 1.2 (P ≤ 0.05), Benjamini-Hochberg multiple testing correction P value. Ingenuity Pathway Analysis (QIAGEN) software (ver. 10-14) was used to identify top upstream regulators, top canonical pathways, diseases, and functions overrepresented in the DEGs.
Statistics.
No statistical methods were used to predetermine sample size. The number of samples (n) used in each experiment is shown. All in vitro experiments were performed at least 3 times independently. Sample size for animal studies was not estimated and randomization was not applied. The investigators were blinded during echocardiography experiments, histological analyses, and outcome assessment. Statistical analysis and graphing were performed using GraphPad Prism 8 software. We did not estimate variations in the data. The variances were similar between the groups that were statistically compared. All data represent the means ± SEM. For P value determinations, we used 2-tailed Student’s t test or 1- or 2-way ANOVA with 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli for multiple comparisons. P < 0.05 was considered statistically significant.
Study approval.
Animals were housed and cared for in facilities run by the Division of Animal Care and were routinely monitored by the veterinary staff. Animal handling was approved by Yale University Institutional Animal Care and Use Committee.
Author contributions
JSY and AMB conceptualized the research, designed the experiments, and wrote the manuscript. JSY performed the majority of the experiments and analyzed data. JSY and SP conducted RNA-Seq data analysis. JSY and LE conducted histological experiments and analyzed data.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 HL134166 to AMB. We thank Timothy Nottoli at the Yale Genome Editing Center for assistance with the generation of the PZRY242F mice. The MUSIC and the YCGA were supported by S10RR023602 and UM1 HG006504, respectively. We thank Lei Zhang for providing technical support.
Version 1. 06/25/2020
In-Press Preview
Version 2. 08/06/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(15):e137753.https://doi.org/10.1172/jci.insight.137753.
Contributor Information
Jae-Sung Yi, Email: jae-sung.yi@yale.edu.
Anton M. Bennett, Email: anton.bennett@yale.edu.
References
- 1.Varnava AM, Elliott PM, Sharma S, McKenna WJ, Davies MJ. Hypertrophic cardiomyopathy: the interrelation of disarray, fibrosis, and small vessel disease. Heart. 2000;84(5):476–482. doi: 10.1136/heart.84.5.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 3.Elliott P, McKenna WJ. Hypertrophic cardiomyopathy. Lancet. 2004;363(9424):1881–1891. doi: 10.1016/S0140-6736(04)16358-7. [DOI] [PubMed] [Google Scholar]
- 4.Marian AJ. Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest. 2010;40(4):360–369. doi: 10.1111/j.1365-2362.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-Quintana E, Rodríguez-González F. Leopard syndrome: clinical features and gene mutations. Mol Syndromol. 2012;3(4):145–157. doi: 10.1159/000342251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkozy A, Digilio MC, Dallapiccola B. Leopard syndrome. Orphanet J Rare Dis. 2008;3:13. doi: 10.1186/1750-1172-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carcavilla A, et al. LEOPARD syndrome: a variant of Noonan syndrome strongly associated with hypertrophic cardiomyopathy. Rev Esp Cardiol (Engl Ed) 2013;66(5):350–356. doi: 10.1016/j.recesp.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29(4):465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 9.Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78(2):279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tajan M, de Rocca Serra A, Valet P, Edouard T, Yart A. SHP2 sails from physiology to pathology. Eur J Med Genet. 2015;58(10):509–525. doi: 10.1016/j.ejmg.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochem J. 2007;402(1):1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, et al. An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Dev Cell. 2006;10(3):317–327. doi: 10.1016/j.devcel.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92(4):441–450. doi: 10.1016/S0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 14.Lechleider RJ, et al. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor. J Biol Chem. 1993;268(29):21478–21481. [PubMed] [Google Scholar]
- 15.Mohi MG, et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7(2):179–191. doi: 10.1016/j.ccr.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Qiu W, et al. Structural insights into Noonan/LEOPARD syndrome-related mutants of protein-tyrosine phosphatase SHP2 (PTPN11) BMC Struct Biol. 2014;14:10. doi: 10.1186/1472-6807-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281(10):6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 18.Yu ZH, et al. Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry. 2014;53(25):4136–4151. doi: 10.1021/bi5002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L, et al. Deletion of interleukin-6 attenuates pressure overload-induced left ventricular hypertrophy and dysfunction. Circ Res. 2016;118(12):1918–1929. doi: 10.1161/CIRCRESAHA.116.308688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paardekooper Overman J, et al. PZR coordinates Shp2 Noonan and LEOPARD syndrome signaling in zebrafish and mice. Mol Cell Biol. 2014;34(15):2874–2889. doi: 10.1128/MCB.00135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao ZJ, Zhao R. Purification and cloning of PZR, a binding protein and putative physiological substrate of tyrosine phosphatase SHP-2. J Biol Chem. 1998;273(45):29367–29372. doi: 10.1074/jbc.273.45.29367. [DOI] [PubMed] [Google Scholar]
- 22.Zhao R, Fu X, Teng L, Li Q, Zhao ZJ. Blocking the function of tyrosine phosphatase SHP-2 by targeting its Src homology 2 domains. J Biol Chem. 2003;278(44):42893–42898. doi: 10.1074/jbc.M306136200. [DOI] [PubMed] [Google Scholar]
- 23.Zhao R, Zhao ZJ. Dissecting the interaction of SHP-2 with PZR, an immunoglobulin family protein containing immunoreceptor tyrosine-based inhibitory motifs. J Biol Chem. 2000;275(8):5453–5459. doi: 10.1074/jbc.275.8.5453. [DOI] [PubMed] [Google Scholar]
- 24.Eminaga S, Bennett AM. Noonan syndrome-associated SHP-2/Ptpn11 mutants enhance SIRPalpha and PZR tyrosyl phosphorylation and promote adhesion-mediated ERK activation. J Biol Chem. 2008;283(22):15328–15338. doi: 10.1074/jbc.M801382200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia D, et al. Amplification of MPZL1/PZR promotes tumor cell migration through Src-mediated phosphorylation of cortactin in hepatocellular carcinoma. Cell Res. 2014;24(2):204–217. doi: 10.1038/cr.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roubelakis MG, Martin-Rendon E, Tsaknakis G, Stavropoulos A, Watt SM. The murine ortholog of the SHP-2 binding molecule, PZR accelerates cell migration on fibronectin and is expressed in early embryo formation. J Cell Biochem. 2007;102(4):955–969. doi: 10.1002/jcb.21334. [DOI] [PubMed] [Google Scholar]
- 27.Marin TM, et al. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121(3):1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauriol J, et al. Developmental SHP2 dysfunction underlies cardiac hypertrophy in Noonan syndrome with multiple lentigines. J Clin Invest. 2016;126(8):2989–3005. doi: 10.1172/JCI80396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi JS, et al. Low-dose dasatinib rescues cardiac function in Noonan syndrome. JCI Insight. 2016;1(20):e90220. doi: 10.1172/jci.insight.90220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita H, Seidman J, Seidman CE. Genetic causes of human heart failure. J Clin Invest. 2005;115(3):518–526. doi: 10.1172/JCI24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Chandrasekhar V, Abbadessa G, Yu Y, Schwartz B, Kontaridis MI. In vivo efficacy of the AKT inhibitor ARQ 092 in Noonan Syndrome with multiple lentigines-associated hypertrophic cardiomyopathy. PLoS One. 2017;12(6):e0178905. doi: 10.1371/journal.pone.0178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao R, Guerrah A, Tang H, Zhao ZJ. Cell surface glycoprotein PZR is a major mediator of concanavalin A-induced cell signaling. J Biol Chem. 2002;277(10):7882–7888. doi: 10.1074/jbc.M111914200. [DOI] [PubMed] [Google Scholar]
- 33.Fredj S, Bescond J, Louault C, Delwail A, Lecron JC, Potreau D. Role of interleukin-6 in cardiomyocyte/cardiac fibroblast interactions during myocyte hypertrophy and fibroblast proliferation. J Cell Physiol. 2005;204(2):428–436. doi: 10.1002/jcp.20307. [DOI] [PubMed] [Google Scholar]
- 34.Meléndez GC, McLarty JL, Levick SP, Du Y, Janicki JS, Brower GL. Interleukin 6 mediates myocardial fibrosis, concentric hypertrophy, and diastolic dysfunction in rats. Hypertension. 2010;56(2):225–231. doi: 10.1161/HYPERTENSIONAHA.109.148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown M, et al. Cardiac-specific blockade of NF-kappaB in cardiac pathophysiology: differences between acute and chronic stimuli in vivo. Am J Physiol Heart Circ Physiol. 2005;289(1):H466–H476. doi: 10.1152/ajpheart.00170.2004. [DOI] [PubMed] [Google Scholar]
- 36.Dawn B, et al. Cardiac-specific abrogation of NF- kappa B activation in mice by transdominant expression of a mutant I kappa B alpha. J Mol Cell Cardiol. 2001;33(1):161–173. doi: 10.1006/jmcc.2000.1291. [DOI] [PubMed] [Google Scholar]
- 37.Jones WK, Brown M, Ren X, He S, McGuinness M. NF-kappaB as an integrator of diverse signaling pathways: the heart of myocardial signaling? Cardiovasc Toxicol. 2003;3(3):229–254. doi: 10.1385/CT:3:3:229. [DOI] [PubMed] [Google Scholar]
- 38.Maier HJ, et al. Cardiomyocyte-specific IκB kinase (IKK)/NF-κB activation induces reversible inflammatory cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2012;109(29):11794–11799. doi: 10.1073/pnas.1116584109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young D, Popovic ZB, Jones WK, Gupta S. Blockade of NF-kappaB using IkappaB alpha dominant-negative mice ameliorates cardiac hypertrophy in myotrophin-overexpressed transgenic mice. J Mol Biol. 2008;381(3):559–568. doi: 10.1016/j.jmb.2008.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontes JA, Rose NR, Čiháková D. The varying faces of IL-6: from cardiac protection to cardiac failure. Cytokine. 2015;74(1):62–68. doi: 10.1016/j.cyto.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gupta, S, Sen, S. Role of the NF-κB signaling cascade and NF-κB-targeted genes in failing human hearts. J Mol Med (Berl). 2005;83(12):4. Retracted in: J Mol Med (Berl). 2020;98(613) [Google Scholar]
- 42.Wakatsuki S, et al. A novel IKK inhibitor suppresses heart failure and chronic remodeling after myocardial ischemia via MMP alteration. Expert Opin Ther Targets. 2008;12(12):1469–1476. doi: 10.1517/14728220802551140. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, et al. IκB kinase inhibitor attenuates sepsis-induced cardiac dysfunction in CKD. J Am Soc Nephrol. 2017;28(1):94–105. doi: 10.1681/ASN.2015060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higuchi Y, et al. Cardioprotection afforded by NF-kappaB ablation is associated with activation of Akt in mice overexpressing TNF-alpha. Am J Physiol Heart Circ Physiol. 2006;290(2):H590–H598. doi: 10.1152/ajpheart.00379.2005. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura N, et al. Blockade of NF-kappaB improves cardiac function and survival without affecting inflammation in TNF-alpha-induced cardiomyopathy. Cardiovasc Res. 2005;66(3):520–529. doi: 10.1016/j.cardiores.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Kawano S. Blockade of NF-kappaB improves cardiac function and survival after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;291(3):H1337–1344. doi: 10.1152/ajpheart.01175.2005. [DOI] [PubMed] [Google Scholar]
- 47.Timmers L, et al. Targeted deletion of nuclear factor kappaB p50 enhances cardiac remodeling and dysfunction following myocardial infarction. Circ Res. 2009;104(5):699–706. doi: 10.1161/CIRCRESAHA.108.189746. [DOI] [PubMed] [Google Scholar]
- 48.Rothwarf DM, Karin M. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci STKE. 1999;1999(5):RE1. doi: 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- 49.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 50.Huang WC, Chen JJ, Chen CC. c-Src-dependent tyrosine phosphorylation of IKKbeta is involved in tumor necrosis factor-alpha-induced intercellular adhesion molecule-1 expression. J Biol Chem. 2003;278(11):9944–9952. doi: 10.1074/jbc.M208521200. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar S, Vellaichamy E, Young D, Sen S. Influence of cytokines and growth factors in ANG II-mediated collagen upregulation by fibroblasts in rats: role of myocytes. Am J Physiol Heart Circ Physiol. 2004;287(1):H107–H117. doi: 10.1152/ajpheart.00763.2003. [DOI] [PubMed] [Google Scholar]
- 52.Mir SA, Chatterjee A, Mitra A, Pathak K, Mahata SK, Sarkar S. Inhibition of signal transducer and activator of transcription 3 (STAT3) attenuates interleukin-6 (IL-6)-induced collagen synthesis and resultant hypertrophy in rat heart. J Biol Chem. 2012;287(4):2666–2677. doi: 10.1074/jbc.M111.246173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hahn A, et al. Rapidly progressive hypertrophic cardiomyopathy in an infant with Noonan syndrome with multiple lentigines: palliative treatment with a rapamycin analog. Am J Med Genet A. 2015;167A(4):744–751. doi: 10.1002/ajmg.a.36982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hengstenberg C, Maisch B. Increased nuclear proto-oncogene expression in hypertrophic cardiomyopathy. Cardioscience. 1993;4(1):15–20. [PubMed] [Google Scholar]
- 55.Kroumpouzou E, Gomatos IP, Kataki A, Karayannis M, Dangas GD, Toutouzas P. Common pathways for primary hypertrophic and dilated cardiomyopathy. Hybrid Hybridomics. 2003;22(1):41–45. doi: 10.1089/153685903321538071. [DOI] [PubMed] [Google Scholar]
- 56.Shirani J, Pick R, Roberts WC, Maron BJ. Morphology and significance of the left ventricular collagen network in young patients with hypertrophic cardiomyopathy and sudden cardiac death. J Am Coll Cardiol. 2000;35(1):36–44. doi: 10.1016/S0735-1097(99)00492-1. [DOI] [PubMed] [Google Scholar]
- 57.Factor SM, Butany J, Sole MJ, Wigle ED, Williams WC, Rojkind M. Pathologic fibrosis and matrix connective tissue in the subaortic myocardium of patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 1991;17(6):1343–1351. doi: 10.1016/S0735-1097(10)80145-7. [DOI] [PubMed] [Google Scholar]
- 58.Fang L, Ellims AH, Beale AL, Taylor AJ, Murphy A, Dart AM. Systemic inflammation is associated with myocardial fibrosis, diastolic dysfunction, and cardiac hypertrophy in patients with hypertrophic cardiomyopathy. Am J Transl Res. 2017;9(11):5063–5073. [PMC free article] [PubMed] [Google Scholar]
- 59.Yin F, et al. Interleukin-6 family of cytokines mediates isoproterenol-induced delayed STAT3 activation in mouse heart. J Biol Chem. 2003;278(23):21070–21075. doi: 10.1074/jbc.M211028200. [DOI] [PubMed] [Google Scholar]
- 60.Duncan MR, Berman B. Stimulation of collagen and glycosaminoglycan production in cultured human adult dermal fibroblasts by recombinant human interleukin 6. J Invest Dermatol. 1991;97(4):686–692. doi: 10.1111/1523-1747.ep12483971. [DOI] [PubMed] [Google Scholar]
- 61.Wang JH, et al. Hypoxia-stimulated cardiac fibroblast production of IL-6 promotes myocardial fibrosis via the TGF-β1 signaling pathway. Lab Invest. 2016;96(8):839–852. doi: 10.1038/labinvest.2016.65. [DOI] [PubMed] [Google Scholar]
- 62.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nagy, A, Gertsenstein, M, Vintersten, K, Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press;2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.