Abstract
Objectives
To further reveal the phylogenetic evolution and molecular characteristics of the whole genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) based on a large number of genomes and provide a basis for the prevention and treatment of SARS-CoV-2.
Methods
Various evolution analysis methods were employed.
Results
The estimated ratio of the rates of non-synonymous to synonymous changes (Ka/Ks) of SARS-CoV-2 was 1.008 or 1.094 based on 622 or 3624 SARS-CoV-2 genomes and nine key specific sites of high linkage, and four major haplotypes were found: H1, H2, H3 and H4. The results of Ka/Ks, detected population size and development trends of each major haplotype showed that H3 and H4 subgroups were going through a purify evolution and almost disappeared after detection, indicating that they might have existed for a long time. The H1 and H2 subgroups were going through a near neutral or neutral evolution and globally increased with time, and the frequency of H1 was generally high in Europe and correlated with the death rate (r >0.37), suggesting that these two haplotypes might relate to the infectivity or pathogenicity of SARS-CoV-2.
Conclusions
Several key specific sites and haplotypes related to the infectivity or pathogenicity of SARS-CoV-2, and the possible earlier origin time and place of SARS-CoV-2 were indicated based on the evolution and epidemiology of 16,373 SARS-CoV-2 genomes.
Keywords: SARS-CoV-2, Evolution, Classification, Haplotype
Introduction
The global outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently and increasingly being recognized as a serious global public health concern. To date, seven types of coronaviruses that can infect humans have been found. Four coronaviruses of them could cause a cold, including hCoV-229E, hCoV-NL63, hCoV-OC43, and hCoV-HKU1, while the other three viruses usually cause mild to severe respiratory diseases, including: severe acute respiratory syndrome coronavirus (SARS-CoV); Middle East respiratory syndrome coronavirus (MERS-CoV); and SARS-CoV-2. SARS-CoV-2 has particularly shown a greater adaptation to the human host compared with the other coronaviruses and the other reference hosts (Dilucca et al., 2020). The total number of SARS-CoV and MERS-CoV infections are 8069 and 2,494, with the reproduction number (R0) fluctuating from 2.5 to 3.9 and 0.3–0.8, respectively. However, SARS-CoV-2 had infected 2,471,136 people in 212 countries by 22 April 2020 (WHO, 2020), with the basic R0 ranging from 1.4 to 6.49 (Liu et al., 2020). Among these three typical coronaviruses, MERS-CoV has the highest death rate of 34.40%, SARS-CoV has a modest death rate of 9.59% and SARS-CoV-2 has a death rate of about 6.99% and 7.69% globally and in Wuhan, respectively. However, some European countries have quite high death rates – such as Belgium, Italy, United Kingdom, Netherlands, Spain, and France – which have reached rates of 14.95%, 13.39%, 13.48%, 11.61%, 10.42%, and 13.60%, respectively, according to data from 22 April 2020. Except for the shortage of medical supplies and aging, it is unclear whether there is a virus mutation effect in these countries with such significantly increased death rates.
It has been reported that SARS-CoV-2 belongs to β-coronavirus, and is mainly transmitted by the respiratory tract, which belongs to the same subgenus (SarbeCoVirus) as SARS-CoV (Lu et al., 2020). Through analyzing the genome and structure of SARS-CoV-2, its receptor-binding domain (RBD) has been found to bind with angiotensin-converting enzyme 2 (ACE2), which is also one of the receptors for binding SARS-CoV (Wrapp et al., 2020). Some previous genomic studies have shown that SARS-CoV-2 is similar to certain bat viruses (RaTG13, with the whole genome homology of 96.2% (Zhou et al., 2020)) and Malayan pangolin coronaviruses (GD/P1L and GDP2S, with the whole genome homology of 92.4% (Lam et al., 2020)). Several possible origins of SARS-CoV-2 have been speculated based on the spike protein characteristics (cleavage sites or the RBD) (Lam et al., 2020, Zhang and Holmes, 2020, Zhou et al., 2020). In particular, the RBD of SARS-CoV-2 exhibits 97.4% amino acid sequence similarity to that of Guangdong pangolin coronaviruses, even though it is most closely related to bat coronavirus RaTG13 at the whole genome level. However, it is not enough to present genome-wide evolution by a single gene or local evolution of RBD, and it remains uncertain whether bats or pangolins play an important role in the zoonotic origin of SARS-CoV-2 (Andersen et al., 2020). Since there is little known about SARS-CoV-2 epidemic trends, origins and whether it has significant variation that affects its phenotype, this study integrated various evolution analysis methods to further reveal the phylogenetic evolution and molecular characteristics of the whole genome of SARS-CoV-2 based on a large number of genomes and to provide some basis for the prevention and treatment of SARS-CoV-2.
Materials and methods
Genome sequences
The complete genome sequences of SARS-CoV-2 were downloaded from the China National Center for Bioinformation (https://www.gisaid.org/) by 22 March 2020. The sequences were filtered out according to the following criteria: (1) sequences with ambiguous time; (2) low-quality sequences, which contained the counts of >15 unknown bases and >50 degenerate bases (https://bigd.big.ac.cn/ncov/release_genome); (3) sequences with 100% similarity were removed to leave the unique one. Finally, 624 high-quality genomes with precise collection times were selected and aligned using MAFFT v7 (Katoh and Standley, 2013) with automatic parameters. The genome sequences of seven SARS-CoV and 475 MERS-CoV were also downloaded from the National Center for Biotechnological Information (NCBI) (https://www.ncbi.nlm.nih.gov/), and the MERS-CoV dataset, which includes samples collected from both humans and camels. In addition, for further exploring evolution and molecular characteristics of SARS-CoV-2 based on the larger amount of genomic data, validation datasets of the genome sequences were redownloaded from GISAID by 06 April 2020 and 10 June 2020, resulting in 3624 and 16373 sequences, respectively.
Estimate of evolution rate and the time to the most recent common ancestor for SARS-CoV, MERS-CoV and SARS-CoV-2
The average rates of non-synonymous (Ka), rates of synonymous (Ks) and ratio of the rates of non-synonymous to synonymous changes (Ka/Ks) for all coding sequences were calculated using KaKs_Calculator v1.2 (Zhang et al., 2006), and the substitution rate and tMRCA were estimated using BEAST v2.6.2 (Bouckaert et al., 2019). The temporal signal with root-to-tip divergence was visualized in TempEst v1.5.3 (Rambaut et al., 2016) using a Maximum Likelihood (ML) whole genome tree with bootstrap value as input. For SARS-CoV and SARS-CoV-2, a strict molecular clock and Coalescent Exponential Population Model were selected. For MERS-CoV, a relaxed molecular clock and Birth Death Skyline Serial Cond Root Model were selected. The tip dates were used and the HKY was chosen as the site substitution model in all these analyses. The Markov Chain Monte Carlo (MCMC) chain length was set to 10,000,000 steps sampling after every 1000 steps. The output was examined in Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer/).
Variants calling of SARS-CoV-2 genome sequences
Each genome sequence was aligned to the reference genome (NC_045512.2) using bowtie2 (Langmead and Salzberg, 2012) with default parameters, and variants were called by samtools (sort; mpileup -gf) and bcftoots (call -vm). The merge Variant Call Format (VCF) files were created by bgzip and bcftools (merge--missing-to-ref) (Li, 2011, Li et al., 2009).
Phylogenetic tree construction and virus isolates clustering for SARS-CoV-2
After alignment of 624 high-quality SARS-CoV-2 genomes and manually deleting two highly divergent genomes (EPI_ISL_415710 and EPI_ISL_414690) according to the first constructed phylogenetic tree, the aligned dataset of 622 sequences was phylogenetically analyzed. The Smart Model Selection (SMS) method was used to select GTR + G as the base substitution model (Lefort et al., 2017). PhyML 3.1 (Guindon et al., 2010) and MEGA (Kumar et al., 2018) were used to construct the no-root phylogenetic tree by the ML method with the bootstrap value of 100. The online tool iTOL (Letunic and Bork, 2019) was used to visualized the phylogenetic tree. The clusters were defined by the shape of the phylogenetic tree.
Detection of specific sites from each Cluster
Information (ID, countries/regions and collection times) and variants (NC_045512.2 as reference genome) of each genome from each Cluster were extracted. The allele frequency and nucleotide divergency (pi) for each site in the virus population of each Cluster were measured by vcftools (Danecek et al., 2020). The Fst were also calculated by vcftools (Danecek, et al. 2020) to assess the diversity between the Clusters. Sites with high levels of Fst together with different major alleles in each Cluster were filtered as the specific sites. Principal Component Analysis (PCA) was analyzed by the GCTA v1.93.1beta (Yang et al., 2011) with the specific sites and all SNV datasets.
Linkage analysis of specific sites and characteristics of major haplotype subgroups
The linkage disequilibrium of the specific sites was analyzed by haploview (Barrett et al., 2005), and the statistics of the haplotype of the specific sites for each Cluster or country were used with in-house perl script.
Phylogenetic network of haplotype subgroups
The Templeton, Crandall and Sing (TCS) network is constructed using an agglomerative approach, where clusters are progressively combined with one or more connecting edges (Cotten et al., 2014), and the minimum spanning network (MSN) networks contain all edges that appear in a minimum spanning tree (Leigh et al., 2015). Hence, in order to estimate genealogical relationships of haplotype groups, the phylogenetic networks were inferred by PopART package v1.7.2 (Leigh et al., 2015) using the TCS method and MSN, respectively.
Frequencies of specific sites or haplotypes and correlation with death rate
The frequencies of specific sites for each country were calculated. The death rate was estimated with total deaths/confirmed cases based on data from Johns Hopkins resources on 12 May 2020 (https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6).The correlation coefficient between death rate and frequencies of specific site or haplotype in different countries was calculated using the Pearson method.
Results
Genome sequences
A total of 1053 genomic sequences were found by 22 March 2020. According to the filter criteria, 37 sequences with ambiguous time, 314 with low quality and 78 with similarity of 100% were removed for further analysis. A total of 624 sequences were obtained to perform multiple sequences alignment. Two highly divergent sequences (EPI_ ISL_414690, EPI_ISL_415710), according to the first constructed phylogenetic tree, were also filtered out (Table S1). The remaining 622 sequences were used to reconstruct a phylogenetic tree. In addition, a total of 3624 and 16,373 genomic sequences were redownloaded by 06 April 2020 and 10 May 2020, respectively, to further explore the evolution and molecular characteristics of SARS-CoV-2.
Estimate of evolution rate and the time to the most recent common ancestor for SARS-CoV, MERS-CoV and SARS-CoV-2
The average Ka/Ks for all the coding sequences of 622 genome sequences ranging from 26 December 2019 to 18 March 2020 was closer to 1 (1.008), indicating that the genome was going through a neutral evolution. The Ka/Ks of SARS-CoV and MERS-CoV were also reevaluated through the whole period, and it was found that the ratio was smaller than SARS-CoV-2 (Table 1 ). To estimate the more credible Ka/Ks for SARS-CoV-2, it was recalculated using 3624 redownloaded genome sequences ranging from 26 December 2019 to 18 March 2020. As a result, the average Ka/Ks was 1.094 (Table 1), which was almost same with the above result.
Table 1.
Statistics of Ka, Ks and Ka/Ks ratios for all coding regions of the SARS-CoV, MERS-CoV and SARS-CoV-2 genome sequences.
| Ka (mean) 10−3 | s.e. (Ka) 10−3 | Ks (mean) 10−3 | s.e. (Ks) 10−3 | Ka/Ks (mean) | s.e. (Ka/Ks) | Hypothesis | p-value* 622 genomes | p-value* 3624 genomes | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV | 0.985 | 0.018 | 1.310 | 0.049 | 0.760 | 0.038 | Ka/Ks (SARS-CoV < SARS-CoV-2) | 0.139 | 0.084 | ||
| MERS-CoV | 1.319 | 0.040 | 4.887 | 0.096 | 0.260 | 0.003 | Ka/Ks (MERS-CoV < SARS-CoV-2) | 0.000 | 0.000 | ||
| SARS-CoV-2 | 622 genomes | 0.231 | 0.004 | 0.265 | 0.005 | 1.008 | 0.020 | ||||
| 3624 genomes | 0.287 | 0.002 | 0.298 | 0.002 | 1.094 | 0.010 | |||||
Note: *One-sided Mann-Whitney U test for the means of two independent samples.
The temporal signal was assessed using TempEst v1.5.3 (Rambaut et al., 2016). All three datasets exhibited a positive correlation between root-to-tip divergence and sample collecting time (Figure S1), so that they were suitable for molecular clock analysis in BEAST (Bouckaert et al., 2019, Rambaut et al., 2016). The substitution rate of SARS-CoV-2 genome was estimated to be 1.601 × 10−3 (95% CI 1.418–1.796 × 10-3, Table 2 , Figure S2A) substitution/site/year, which was in the same order of magnitude as SARS-CoV and MERS-CoV. The tMRCA was inferred in late September 2019 (95% CI 08 August–26 October 2019, Table 2, Figure S2B), which was about two months before the early cases of SARS-CoV-2 (Huang et al., 2020).
Table 2.
Substitution rate and tMRCA estimated by BEAST v2.6.2.
| substitution rate (10−3) | 95% CI (10−3) | tMRCA | 95% CI | references | |
|---|---|---|---|---|---|
| SARS-CoV | 1.050 | 0.489, 1.654 | 28 June 2002 | 19 January 2019, 03 November 2002 | this study |
| – | 0.800, 2.380 | spring of 2002 | – | (Zhao, et al., 2004) | |
| MERS-CoV | 1.516 | 1.392, 1.632 | 07 June 2012 | 04 June 2012, 09 June 2012 | this study |
| 1.120 | 0.876, 1.370 | March 2012 | December 2011, June 2012 | (Cotten, et al., 2014) | |
| SARS-CoV-2 | 1.601 | 1.418, 1.796 | 27 September 2019 | 28 October 2019, 26 October 2019 | this study |
Phylogenetic tree and clusters of SARS-CoV-2
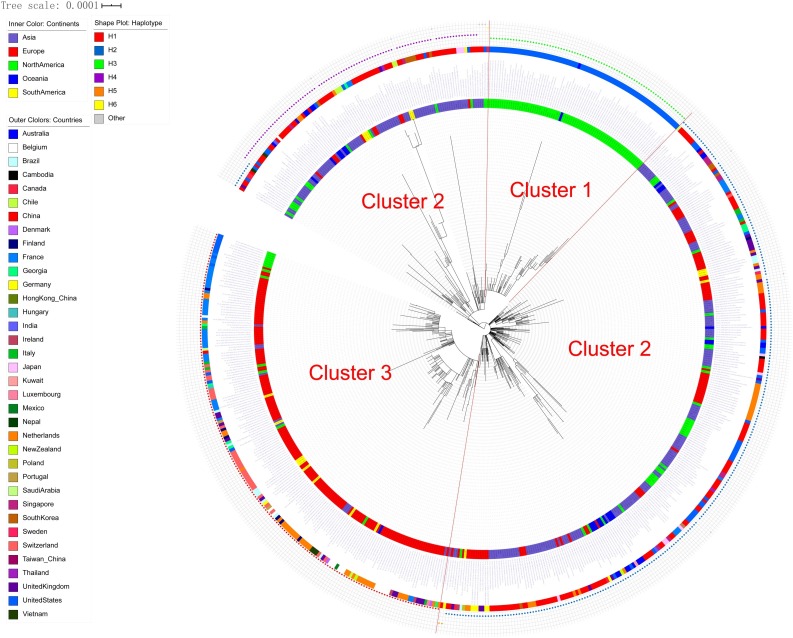
The no-root phylogenetic trees constructed by the ML method with PhyML 3.1 and MEGA are shown in Fig. 1 and Figure S3. According to the shape of the phylogenetic trees, 622 sequences were divided into three clusters (Fig. 1): Cluster 1, including 76 sequences mainly from North America; Cluster 2, including 367 sequences from all regions of the world; and Cluster 3, including 179 sequences mainly from Europe (Table S2).
Fig. 1.
Phylogenetic tree and clusters of 622 SARS-CoV-2 genomes.
The 622 sequences were clustered into three clusters: Cluster 1 was mainly from North America, Cluster 2 was from regions all over the world, and Cluster 3 was mainly from Europe.
The specific sites of each Cluster
The Fst and population frequency of a total of nine sites (NC_045512.2 as reference genome) were detected (Table 3 , Table S3). Three (C17747 T, A17858 G and C18060 T) were the specific sites of Cluster 1, and four (C241 T, C3037 T, C14408 T, and A23403 G) were the specific sites of Cluster 3. Notably, C241 T was located in the 5′-UTR region and the others were located in coding regions (six in ofr1ab gene, one in S gene and one in ORF8 gene). Five of them were missense variants, including C14408 T, C17747 T and A17858 G in ofr1ab gene, A23403 G in S gene, and T28144C in ORF8 gene. The PCA results showed that these nine specific sites could clearly separate the three Clusters, while all SNV dataset could not clearly separate Cluster 1 and Cluster 2 (Figure S4), which further suggests that these nine specific sites were the key sites for separating the three Clusters.
Table 3.
Information of the nine specific sites in each Cluster.
| Pos | Ref | Alt |
Cluster 1 |
Cluster 2 |
Cluster 3 |
Fst | Gene region | Mutation type | Protein changed | codon changed | Impact | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pi | Major allele | frequency | pi | Major allele | frequency | pi | Major allele | frequency | |||||||||
| 241 | C | T | 0.0000 | C | 1.0000 | 0.0109 | C | 0.9945 | 0.0000 | T | 1.0000 | 0.9912 | 5′UTR | upstream | NA | NA | Modifier |
| 3037 | C | T | 0.0000 | C | 1.0000 | 0.0109 | C | 0.9945 | 0.0000 | T | 1.0000 | 0.9912 | gene-orf1ab | synonymous | 924F | 2772ttC > ttT | Low |
| 8,782 | C | T | 0.0000 | T | 1.0000 | 0.3863 | C | 0.7390 | 0.0000 | C | 1.0000 | 0.5821 | gene-orf1ab | synonymous | 2839S | 8517agC > agT | Low |
| 14,408 | C | T | 0.0000 | C | 1.0000 | 0.0055 | C | 0.9973 | 0.0000 | T | 1.0000 | 0.9956 | gene-orf1ab | missense | 4715 P > L | 14144cCt > cTt | Moderate |
| 17,747 | C | T | 0.0735 | T | 0.9620 | 0.0000 | C | 1.0000 | 0.0000 | C | 1.0000 | 0.9752 | gene-orf1ab | missense | 5828 P > L | 17483cCt > cTt | Moderate |
| 17,858 | A | G | 0.0497 | G | 0.9747 | 0.0000 | A | 1.0000 | 0.0000 | A | 1.0000 | 0.9836 | gene-orf1ab | missense | 5865Y > C | 17594tAt > tGt | Moderate |
| 18,060 | C | T | 0.0000 | T | 1.0000 | 0.0164 | C | 0.9918 | 0.0000 | C | 1.0000 | 0.9761 | gene-orf1ab | synonymous | 5932 L | 17796ctC > ctT | Low |
| 23,403 | A | G | 0.0000 | A | 1.0000 | 0.0164 | A | 0.9918 | 0.0000 | G | 1.0000 | 0.9869 | gene-S | missense | 614D > G | 1841gAt > gGt | Moderate |
| 28,144 | T | C | 0.0000 | C | 1.0000 | 0.3915 | T | 0.7335 | 0.0000 | T | 1.0000 | 0.5785 | gene-ORF8 | missense | 84 L > S | 251tTa > tCa | Moderate |
Linkage of specific sites
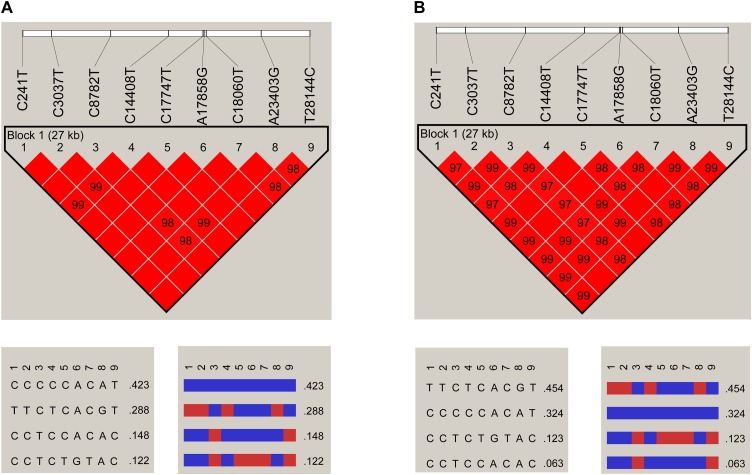
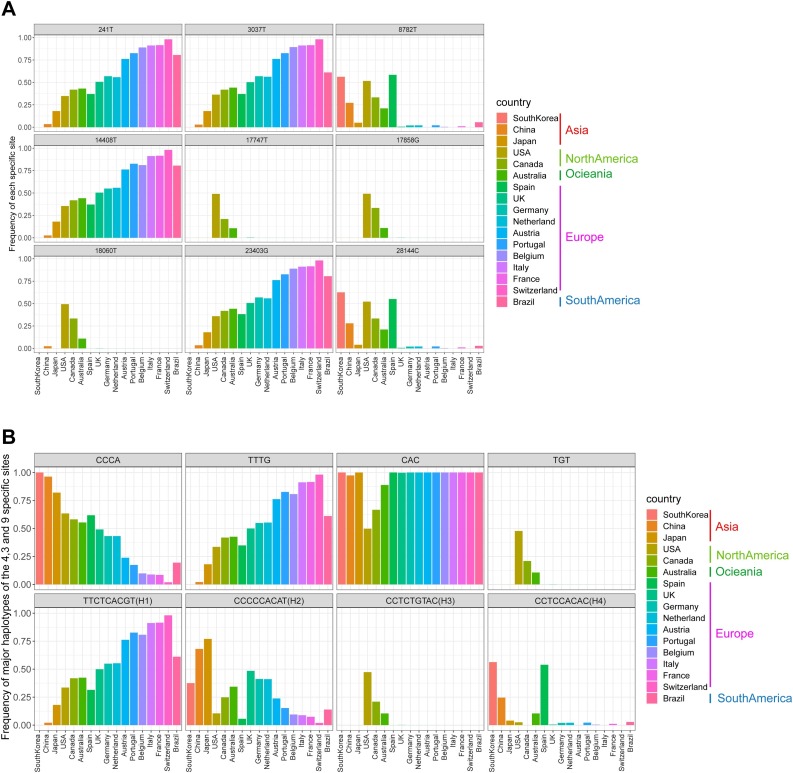
It was found that the nine specific sites were highly linked based on 622 genome sequences (Fig. 2 A), then a further linkage analysis was carried out using the 3624 genome sequences (Fig. 2B). As a result, for the 3624 genome sequences, three specific sites in Cluster 1 were almost completely linked, and haplotypes CAC and TGT accounted for 98.65% of all the three site haplotypes. The same phenomenon was also found in four specific sites of Cluster 3, and haplotypes CCCA and TTTG accounted for 97.68% of all four site haplotypes. Intriguingly, the nine specific sites were still highly linked, and four haplotypes – TTCTCACGT (H1), CCCCCACAT (H2), CCTCTGTAC (H3), and CCTCCACAC (H4) – accounted for 95.89% of all the nine site haplotypes. H1 and H3 had completely different bases at the nine specific sites. The frequencies of each site and major haplotype for each country are shown in Fig. 3 and Table S4. The data showed that the haplotype TTTG of the four specific sites in Cluster 3 currently exist globally, and still exhibit high frequencies in most European countries but are quite low in Asian countries. The haplotype TGT of the three specific sites in Cluster 1 exist almost only in North America and Australia. For the nine specific sites, most countries have two or three major haplotypes, except America and Australia, which have all four major haplotypes with relative higher frequencies.
Fig. 2.
Linkage disequilibrium plot of haplotypes of the nine specific sites.
A. The plot for 622 genome sequences; B. The plot for 3624 genome sequences.
Fig. 3.
The frequencies of both the nine specific sites and haplotypes.
The frequencies of the nine specific sites (A) and haplotypes (B) in each country for 3624 genomes.
Characteristics and epidemic trends of major haplotype subgroups
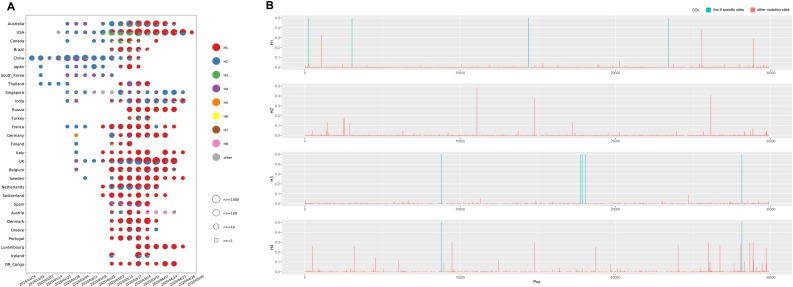
All haplotypes of nine specific sites for 3624 or 16,373 genomes and the numbers of them are shown in Table S5. Four major haplotypes – H1, H2, H3, and H4 – three minor haplotypes – H5, H7 and H8 – close to H1 and one minor haplotype – H6 – close to H3 were found in both datasets. The numbers of these haplotypes for 16,373 genomes with clear collection dates detected in each country in chronological order are shown in Fig. 4 A. As show in these results, the H2 and H4 subgroups have existed for a long time (24 December 2019 to 28 April 2020), and the former had a far greater detected population size. The H3 subgroup almost disappeared after detection (18 February–28 April 2020), while the H1 subgroup globally increased with time (18 February–05 May 2020), indicating that the H1 subgroup has adapted to the human hosts and undergoing an adaptive growth period worldwide. However, due to the nonrandom sampling in the early phase (only patients with recent travel to Wuhan were detected), some earlier cases of H3 may have been lost, which could be indicated by the high proportion of the H3 subgroup during 18 February–10 March 2020.
Fig. 4.
The characteristics of haplotype subgroups.
A. The numbers of haplotypes of the nine specific sites for 16,373 genomes with clear collection data detected in each country in chronological order; B. The whole genome mutations in each major haplotype subgroup.
The H3 and H4 subgroups had a lower Ka/Ks ratio (about 0.7–0.8) than that of the H1 and H2 subgroups (about 1.1–1.3) in 3624 or 16,373 genomes among the four major subgroups (Table 4 ), suggesting that the H3 and H4 subgroups might be going through a purifying evolution and have existed for a long time, while the H1 and H2 subgroups might be going through a near neutral or neutral evolution, which was consistent with the above phenomenon that only H1 and H2 subgroups have been spreading around the world over time. From the whole genome mutations in each major haplotype subgroup (Fig. 4B, Table S6), it was found that, except for the nine specific sites, there were no common mutations with frequencies >0.05 in all four subgroups but one at the position of 14805 exists between H2 and H4 subgroups.
Table 4.
Statistics of Ka, Ks and Ka/Ks ratios for all coding regions of each four major haplotype subgroup with 3624 and 16,373 genomes, respectively.
| Ka (mean) 10−3 | s.e. (Ka)10−3 | Ks (mean) 10−3 | s.e. (Ks)10−3 | Ka/Ks (mean) | s.e. (Ka/Ks) | Hypothesis | p-value* | ||
|---|---|---|---|---|---|---|---|---|---|
| 3624 genomes | H1 | 0.510 | 0.057 | 0.498 | 0.056 | 1.099 | 0.010 | Ka/Ks (H3 < H1) | 0.000 |
| H2 | 0.347 | 0.042 | 0.334 | 0.044 | 1.268 | 0.023 | Ka/Ks (H3 < H2) | 0.000 | |
| H3 | 0.298 | 0.007 | 0.397 | 0.009 | 0.795 | 0.010 | – | – | |
| H4 | 0.334 | 0.023 | 0.414 | 0.019 | 0.796 | 0.019 | – | – | |
| 16,373 genomes | H1 | 0.416 | 0.011 | 0.384 | 0.011 | 1.178 | 0.005 | Ka/Ks (H3 < H1) | 0.000 |
| H2 | 0.332 | 0.014 | 0.316 | 0.014 | 1.224 | 0.012 | Ka/Ks (H3 < H2) | 0.000 | |
| H3 | 0.305 | 0.004 | 0.401 | 0.005 | 0.809 | 0.007 | – | – | |
| H4 | 0.354 | 0.010 | 0.477 | 0.009 | 0.749 | 0.010 | – | – | |
Note: *One-sided Mann-Whitney U test for the means of two independent samples.
Phylogenetic network of haplotype subgroups
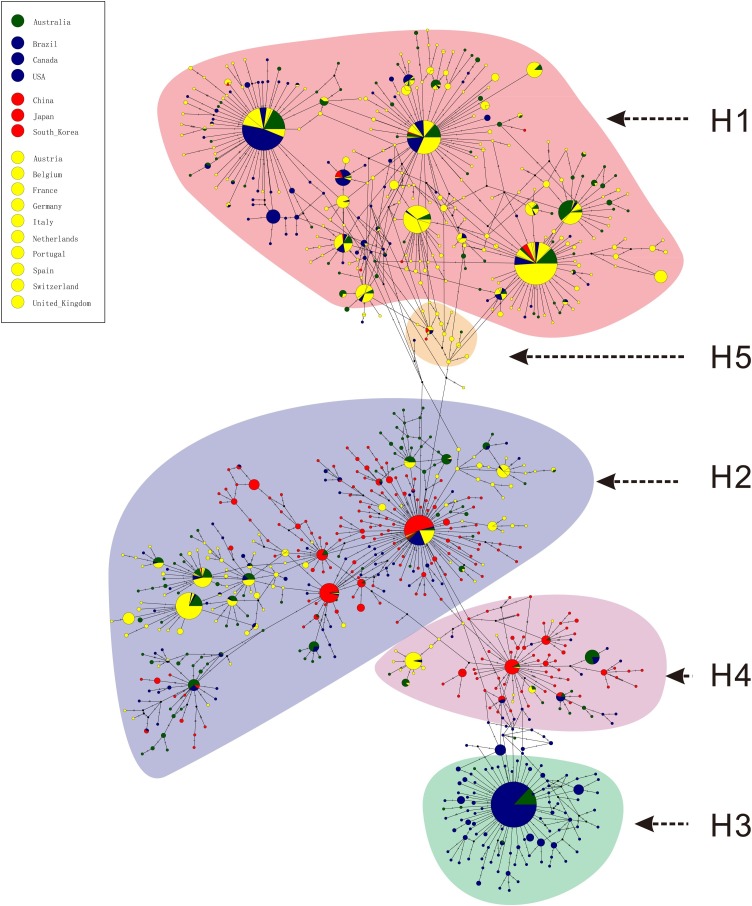
Phylogenetic networks were inferred with 697 mutations from 3624 genome datasets, and the network structures of TCS and MSN were similar. The major haplotype subgroups H2 and H4 were in the middle of the network, while H1 and H3 were in the end nodes of the network (Fig. 5 ). According to the phylogenetic networks, four hypotheses were proposed: (1) the ancestral haplotypes evolved in four different directions to obtain H1, H2, H3, and H4, respectively, or evolved in two or more different directions to obtain two or more major haplotypes and then evolved into the other major haplotype(s); (2) H2 or H4 evolved in different directions and finally generated H1 and H3; (3) H1 evolved in one direction to generate H2 and H4, and then evolved into H3; (4) H3 evolved in one direction to generate H4 and H2, and then evolved into H1. The first hypothesis cannot be excluded based on the present data; however, according to the Ka/Ks, detected population size and development trends in chronological order of each major haplotype subgroup, if there are evolutionary relationships among the four major haplotypes, it is speculated that the most likely evolution hypothesis is that H3 and H4 are the earliest haplotypes, which have gradually eliminated with selection, while H2 is the transitional haplotype in the evolution process, and H1 may be the finally fixed haplotype.
Fig. 5.
Phylogenetic network of haplotype subgroups for 3624 genomes.
The network was inferred by POPART using the TCS method. Each colored vertex represents a haplotype, with different colors indicating the different sampling areas. Hatch marks along the edge indicate the number of mutations. Small black circles within the network indicate unsampled haplotypes. H1-H5 subgroups are pointed out according to haplotypes of the nine specific sites, and other small subgroups are not especially pointed out.
Correlation analysis of specific sites with death rate and infectivity
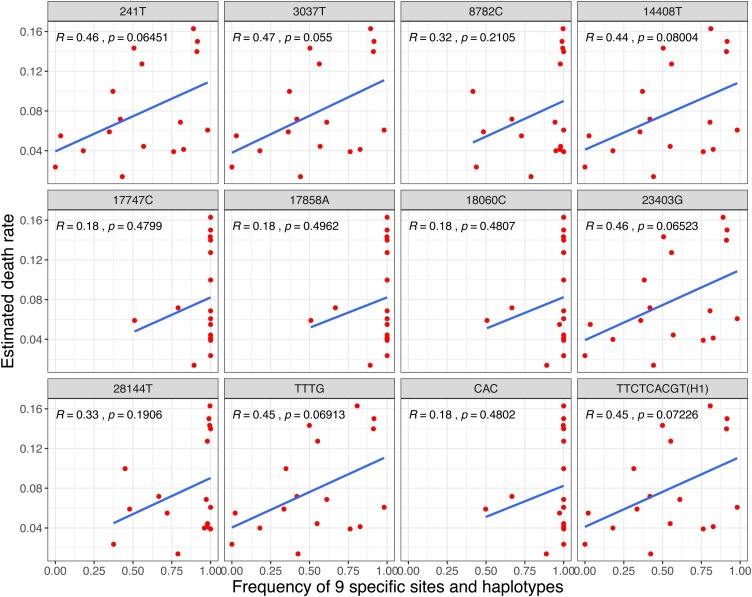
To explore the relationship between death rate and the nine specific sites, the Pearson method was used to calculate the correlation coefficient between death rate and frequency of each specific site or major haplotype in 17 countries with 3624 genomes in the early stages. As a result, all r values of 241 T, 3037 T, 14,408 T, 23,403 G and haplotype TTTG and H1 were >0.4 (Fig. 6 , Table S4). The correlation coefficient with 16,373 genomes in 30 countries was also evaluated, and the r values of haplotype TTTG and H1 were still >0.37 (Table S4). Integrated with their high frequencies in most European countries, these finding indicate that the four sites – C241 T, C3037 T, C14408 T, and A23403 G – and haplotype TTTG and H1 might be related to the pathogenicity of SARS-CoV-2.
Fig. 6.
The correlation between death rates and frequencies of both the nine specific sites and haplotypes.
To explore the relationship between infectivity and the nine specific sites, the population size of the major haplotypes was used to deduce the possible specific sites related to infectivity. It was assumed that these major haplotypes in each country were subject to similar virus transmission and control patterns, while the population sizes of H1 and H2 subgroups were far greater than those of H3 and H4 subgroups (Fig. 4A, Table S4). Thus, the common different specific sites of H1 and H2 subgroups with H3 and H4 subgroups, C8782 T and T28144C, might be related to the infectivity of SARS-CoV-2, and the viruses with C8782 and T28144 might be more infectious than those viruses with 8782 T and 28144C.
Discussion
SARS-CoV-2 poses a great threat to the production, living and survival of human beings (Guo et al., 2020). With a further outbreak of SARS-CoV-2 in the world, comprehensive understanding on the evolution and molecular characteristics of SARS-CoV-2 based on a large number of genome sequences will enable the world to better deal with the challenges brought about by SARS-CoV-2.
Exploring the evolution rate, tMRCA and phylogenetic tree of SARS-CoV-2 would help to better understand the virus (Yuen et al., 2020). The average Ka/Ks for all the coding sequences of 622 and 3624 SARS-CoV-2 genomes was 1.008 and 1.094, which was higher than those of SARS-CoV and MERS-CoV, indicating that the SARS-CoV-2 is going through a neutral evolution. Interestingly, it was also found that the subgroups of different haplotypes – H1, H2, H3, and H4 – seemed to undergo the different evolutionary patterns according to their Ka/Ks. The H3 subgroup disappeared soon after detection (18 February–28 April 2020, Fig. 4A), while the H1 subgroup increased globally with time. These characteristics of evolution and change should be considered in developing therapeutic drugs and vaccines. The tMRCA of SARS-CoV-2 was inferred in late September 2019 (95% CI 28 August–26 October 2019), about two months before the early cases of SARS-CoV-2 (Huang et al., 2020). The tMRCA of SARS-CoV and MERS-CoV was also estimated with the same methods; both were about 3 months later than the corresponding tMRCAs estimated by previous studies (Cotten et al., 2014, Zhao et al., 2004). A recent study used the TreeDater method to estimate tMRCA for >7000 SARS-CoV-2 genomes and indicated the tMRCA of SARS-CoV-2 was around 06 October–11 December 2019, which was in broad agreement with six previous studies all performed on no more than 120 early SARS-CoV-2 genomes with the BEAST method (van Dorp et al., 2020). The current study chose the most common method – BEAST – to estimate the tMRCA of SARS-CoV-2 based on 622 genomes, and the results of SARS-CoV and MERS-CoV are consistent with the previous studies, indicating that the estimated tMRCA in the present study is reliable.
A recent study clustered 160 SARS-CoV-2 whole-genome sequences into A, B and C groups through a phylogenetic network analysis by taking bat RaTG13 as a root (Forster et al., 2020). The clustering result was similar to the current study: both the samples in Cluster A and current Cluster 1 were mainly from the United States; the samples in Cluster C and current Cluster 3 were mainly from European countries, while Cluster B and current Cluster 2 were mainly from China and other regions. It is interesting that the markers C8782 T and T28144C, which were discovered by Yu et al. (Yu et al., 2020), in Cluster B were also found in the current study, but the other markers in Cluster A (T29095C) and Cluster C (G26144 T) were not significant in the current study, which may have been caused by different sample sizes and different constructing methods of the phylogenetic tree. Based on the base substitution model, the ML method avoids the possible "long-branch attraction" problem in the maximum parsimony method and is faster than the Bayesian method (Holder and Lewis, 2003), indicating that it could be used as a reliable method for phylogenetic analysis. Some studies used the genome of bat SARS-like-CoV (Zhang et al., 2020a), RaTG13 (Zhang et al., 2020b) or MT019529 (https://bigd.big.ac.cn/ncov/tree) as the root of the phylogenetic tree. There is no obvious evidence showing that SARS-CoV-2 is from the bat coronavirus, even though the identity between SARS-CoV-2 and RaTG13 is up to 96.2% (Zhou et al., 2020). In the current study, the tMRCA of SARS-CoV-2 was inferred in late September 2019, which indicated that there might have been an earlier SARS-CoV-2 strain that was not found. In the case of unclear sources of SARS-CoV-2 and high homology of its genomes (>99.9% homology), it may be inappropriate to identify the evolutionary characteristics inside the genomes by taking bat SARS-like-CoV, RaTG13 or MT019529 as a root. Therefore, the ML method was used in the current study to construct a no-root tree to obtain the reliable clusters with different characteristics. Based on the no-root tree, nine specific sites were identified of high linkage that successfully played a decisive role in the classification of clusters.
Among the nine specific sites, eight of them are located in coding regions (six in ofr1ab gene, one in S gene and one in ORF8 gene), and five of them are missense variants, including C14408 T, C17747 T and A17858 G in ofr1ab gene, A23403 G in S gene, and C28144 T in ORF8 gene. This study found that the four specific sites – C241 T, C3037 T, C14408 T, and A23403 G – in Cluster 3 were almost completely linked, and the frequency of haplotype TTTG was generally high in European countries and correlated with death rates (r >0.37) based on 3624 or 16,373 SARS-CoV-2 genomes, which provides a new perspective on the reasons for the relatively high death rate in Europe, and provides a new opportunity in designing new vaccine and drug development for SARS-CoV-2. Two possible specific sites – C8782 T and T28144C – related to the infectivity of SARS-CoV-2 were also shown in the present study, which would provide a basis for SARS-CoV-2 epidemiology. Among these genes with specific sites, Orf1ab comprises two partially overlapping open reading frames (orf): orf1a and orf1b. It is a proteolytic cleaved into 16 non-structural proteins (nsp), including nsp1 (suppress antiviral host response), nsp9 (RNA/DNA binding activity), nsp12 (RNA-dependent RNA polymerase), nsp13 (helicase), and others (Chan et al., 2020), indicating the vital role of it in transcription, replication, innate immune responses, and virulence (Graham et al., 2008). C14408 T, with high frequencies of T in European countries, was located at the nsp12 region, indicating that this missense variant might influence the role of RNA polymerase. Spike glycoprotein, the largest structural protein on the surface of coronaviruses, comprises S1 and S2 subunits mediating binding of the receptor on the host cell surface and fusing membranes, respectively (Li, 2016). It has been reported that the S protein of SARS-CoV-2 can bind ACE2 with higher binding affinity than that of SARS (Wrapp et al., 2020, Zhou et al., 2020). Recently, several in vitro studies (Daniloski et al., 2020, Hu et al., 2020) posted on preprint showed that A23403 G in S gene (D614 G mutation in S protein) could promote virus entry into the host cells and enhance the infectivity of host cells by 2–7 times, which have partially verified the current conclusions.
It seems to take a long time to finally fix mutations according to the mutation frequency of each subgroup. For example, H2 and H4 subgroups, which were detected for >4 months from 24 December 2019–05 May 2020 (Fig. 4A), have more mutations with higher frequencies, but the highest mutation frequency is 0.486 at the position of 11,083 (Fig. 4B, Table S6). From these phenomena, it can be inferred that it takes a long time for the specific sites of each major subgroup to be fixed, but it may be faster if the early population is smaller. In addition, there is also the possibility that an ancestor strain evolved in four directions by directly obtaining the specific mutations and produced the four current major haplotypes, so the evolution time for obtaining the four major haplotypes may have been shorter, which seems to be consistent with the phenomenon that the four major haplotypes were detected in 2 months (Fig. 4A). However, if there is an evolutionary relationship among the major haplotypes of SARS-CoV-2, it would have been difficult to complete the evolution among the four major haplotypes within 2 months (24 December 2019– 18 February 2020, Fig. 4A) at the current evolution rate of each major haplotype population (Table 4). Therefore, it is speculated that the transformation among the four major haplotypes may have been completed for a long time, and not have been detected. It is interesting that only the United States and Australia, among 29 countries, have all of four major haplotypes with relatively higher frequencies (Fig. 4A, Table S4), which indicates that the two countries are the most likely places where the virus appeared earlier, based on the present data.
Conclusion and prospective
The Ka/Ks ratio and tMRCA of SARS-CoV-2 indicate that SARS-CoV-2 might have completed the selection pressure of cross-host evolution in earlier stages and currently be going through a neutral evolution. Nine specific sites with high linkage were found to play a decisive role in the classification of clusters. Several key specific sites and haplotypes related to infectivity or pathogenicity of SARS-CoV-2 as well as the possible earlier origin time and place of SARS-CoV-2 were indicated based on the evolution and epidemiology of 16,373 SARS-CoV-2 genomes. The relationship between the key specific sites or haplotype TTTG (or H1) and the infectivity or pathogenicity of SARS-CoV-2 needs to be further verified by clinical samples or virus infectivity and virulence test experiments. Given the different evolution patterns of different haplotypes subgroups, the evolution and changes should be considered in the development of therapeutic drugs and vaccines.
Authors’ contributions
HD conceived the study. YM, YB, DJ, JL, and XC carried out the data analysis and wrote the manuscript. MH, SL, and ZC collected data and revised the manuscript. XW attended the discussions. HD and YM supervised the whole work and revised the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not required.
Footnotes
Electronic supplementary tables are available online at https://figshare.com/articles/dataset/SARS-CoV-2_genomes_evolution/12366449
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was supported by the National Key R&D Program of China [2018YFC0910201], the K ey R&D Program of Guangdong Province [2019B020226001], the S cience and the Technology Planning Project of Guangzhou [201704020176 and 2020Q-P013]. Additionally, we thank Dr. Li Junhua for his participation in the discussion and revision of the paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.08.066.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nature Med. 2020 doi: 10.1038/s41591-020-0820-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Jf, Kok Kh, Zhu Z. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Watson S.J., Zumla A.I. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio. 2014;5 doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G. The variant call format and VCFtools. Bioinformatics (Oxford, England) 2020;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Guo X., Sanjana N.E. The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. bioRxiv. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon Usage and Phenotypic Divergences of SARS-CoV-2 Genes. Viruses. 2020;12 doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. PNAS. 2020 doi: 10.1073/pnas.2004999117202004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133:88–100. doi: 10.1016/j.virusres.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder M., Lewis P.O. Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Hu J., He C.-L., Gao Q.-Z. The C.-L. mutation of SARS-CoV-2 spike protein enhances viral infectivity and decreases neutralization sensitivity to individual convalescent ser. bioRxiv. 2020 doi: 10.1101/2020.06.20.161323. [DOI] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H.C. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Longueville J.E., Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J.W., Bryant D., Nakagawa S. Popart: full‐feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gkz239. W256-W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England) 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklov J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020:27. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genetics Evol. 2020 doi: 10.1016/j.meegid.2020.104351104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO; 2020. Coronavirus disease 2019 (COVID-19) Situation Report – 93. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.B., Tang G.D., Zhang L., Corlett R.T. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2 / HCoV-19) using whole genomic data. Zool Res. 2020;41:247–257. doi: 10.24272/j.issn.2095-8137.2020.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K.-S., Ye Z.-W., Fung S.-Y., Chan C.-P., Jin D.-Y. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- Zhang L., Shen F.-m, Chen F., Lin Z. Origin and Evolution of the 2019 Novel Coronavirus. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X.Q., Wang J., Wong G.K., Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Li H., Wu X. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.