Highlights
-
•
Fecal SARS-CoV-2 was relevant to milder condition and better radiological recovery.
-
•
The first attempt of using a survival analysis for SARS-CoV-2 RNA duration.
-
•
SARS-CoV-2 RNA survival in feces was significantly longer than in oropharyngeal swabs.
-
•
In spite of the negative oropharyngeal swabs, Fecal viral load easily rebounded.
Keywords: COVID-19, Fecal specimens, Viral load, Survival analysis
Abstract
Background
To investigate the clinical significance, viral shedding duration and viral load dynamics of positive fecal SARS-CoV-2 signals in COVID-19.
Methods
COVID-19 patients were included. SARS-CoV-2 RNA was tested in stool and respiratory specimens until two sequential negative results were obtained. Clinical, laboratory and imaging data were recorded.
Results
Of the 69 COVID-19 patients, 20 (28.99 %) had positive fecal viral tests who were younger, had lower C-reactive protein (CRP) and fibrinogen (FIB) levels on admission (all P < 0.05), and showed more improvement and less progression on chest CT during recovery. The median duration of positive viral signals was significantly longer in stool samples than in respiratory samples (P < 0.05). In spite of the negative oropharyngeal swabs, eleven patients were tested positive for viral RNA in stool specimens, with their fecal SARS-CoV-2 RNA Ct (cycle threshold) values reaching 25–27. 6 of these 11 patients' Ct values rebounded.
Conclusion
SARS-CoV-2 RNA in stool specimens was associated with a milder condition and better recovery of chest CT results while the median duration of SARS-CoV-2 RNA persistence was significantly longer in fecal samples than in oropharyngeal swabs. The fecal viral load easily reached a high level and rebounded even though respiratory signals became negative.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19) and has become a global pandemic. Until July 11, 2020, the virus had spread to 200 countries, with more than 12 million people infected and over 550,000 deaths globally. SARS-CoV-2 belongs to beta-coronavirus genus lineage B and is phylogenetically closely related to bat SARS-like coronaviruses (Chan et al., 2020). Infection with SARS-CoV-2 causes a spectrum of respiratory symptoms, including severe acute respiratory syndrome. COVID-19 spreads mainly via respiratory droplets and human-to-human contact (Li et al., 2020; Chen et al., 2020a; Zhou et al., 2020a; Wang et al., 2020). SARS-CoV-2 RNA has been reported to be detected in stool specimens from patients with COVID-19 (Chen et al., 2020b; Xie et al., 2020). Furthermore, not only SARS-CoV-2 RNA but also intracellular staining of the viral nucleocapsid protein was found in gastric, duodenal and rectal epithelia (Lamers et al., 2020; Zhou et al., 2020b; Zang et al., 2020). Importantly, viral host receptor angiotensin converting enzyme 2 was positivity expressed in gastrointestinal epithelial cells (Xiao et al., 2020a). These reports demonstrate that SARS-CoV-2 infection might occur in the gastrointestinal tract, which may result in transmission by the fecal-oral route. Most importantly, the risk of aerosol or contact transmission associated with fecal samples warrants more attention. Notably, clearance of viral RNA from patients’ stool specimens requires more time than clearance from oropharyngeal samples (Ling et al., 2020; Wu et al., 2020a). The dynamic changes in the viral load of prolonged SARS-CoV-2 RNA in stool specimens are not clear. In addition to the epidemiology, COVID-19 patients with gastrointestinal symptoms are more likely to test positive for viral RNA in stool, which is considered to be associated with prolonged symptoms (Wei et al., 2020; Han et al., 2020a). However, whether SARS-CoV-2 RNA in stool specimens has a strong relationship with the condition and prognosis of COVID-19 is unclear.
To further investigate the clinical significance, viral shedding duration and viral load dynamics of SARS-CoV-2 RNA in stool specimens, we compared the characteristics and prognoses of COVID-19 patients with positive and negative fecal viral tests, conducted a survival analysis of viral shedding durations in fecal and respiratory samples, and analyzed dynamic changes in fecal viral loads.
2. Methods
2.1. Patients
From February 6, 2020, to February 22, 2020, we enrolled 69 patients with laboratory-confirmed SARS-CoV-2 infection who were admitted to the Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University in Oujiangkou district. The diagnosis was based on the guidelines issued by the National Health Commission of the People’s Republic of China. The present study was performed in accordance with the Helsinki Declaration and was approved by the Ethics Committees of the Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University.
2.2. Real-time RT-PCR
Real-time reverse-transcriptase polymerase chain reaction (RT-PCR) was performed at the Wenzhou Municipal Center for Disease Prevention and Control (CDC) until two sequential negative results were obtained. Two sets of primers and probe sets were used for two target genes separately (i.e, open reading frame 1ab[ORF1ab] and nucleocapsid protein [N]) according to the protocol issued by the National Institute for Viral Disease Control and Prevention in China (National Institute for Viral Disease Control and Prevention, 2020). The primer sequences were as follows: ORF1ab, CCCTGTGGGTTTTACACTTA A and CCCTGTGGGTTTTACACTTAA; N, GGGGAACTTCTCCTGCTAGAAT and CAGACATTT TGCTCTCAAGCTG. The Probe sets were as follows: ORF1ab, 5ʹ-VIC-CCGTCTGCGGTATG TGGAAAGGTTATGG-BHQ1-3ʹ; N, 5ʹ-FAM- TTGCTGCTGCTTGACAGATT-TAMRA-3ʹ. If two targets tested positive by specific RT-PCR, the patients would be considered to be laboratory-confirmed.
Negative: no Ct value or Ct = 40.
Positive: a Ct value < 37.
A Ct value between 37–40 is indeterminate. It is required confirmation by repeating. If, when repeated, the Ct value is < 40 with a obvious amplification peak, the sample is positive, otherwise, it is negative.
2.3. Data collection
The date of disease onset was defined as the day of the onset of symptoms. Laboratory and chest CT examinations were conducted on admission and at the time of the last negative respiratory test and fecal test at least. Epidemiological data, clinical data, laboratory results and radiological findings were recorded, including sex, age, exposure history, familial aggregation, chronic medical illnesses, symptoms, neutrophil counts, leukocyte counts, lymphocyte counts, C-reactive protein (CRP) levels, albumin (ALB) levels, alanine transaminase (ALT) levels, fibrinogen (FIB) levels, D-dimer levels and chest CT results. The dynamic cycle threshold (Ct) values of the ORF1ab and N genes in fecal samples were recorded after the viral RNA test in oropharyngeal swabs became negative. Ct values are inversely related to the viral RNA load, with Ct values of 30.76, 27.67, 24.56, and 21.48 corresponding to 1.5 × 104, 1.5 × 105, 1.5 × 106, and 1.5 × 107 copies per milliliter, respectively. All data were reviewed by two physicians.
2.4. Statistical analysis
Continuous variables are presented as the medians and interquartile ranges (IQRs). Nonparametric comparative tests were used for continuous data. Categorical variables are presented as the number and percentages. Survival analysis was performed with the Kaplan-Meier method. All statistical analyses were performed with SPSS software (version 22.0, SPSS, Inc.). P < 0.05 was considered significant.
3. Results
3.1. Patient characteristics
From February 6, 2020, to February 22, 2020, a total of 69 patients with laboratory-confirmed SARS-CoV-2 infection were included, with 29 (42.03 %) females and 40 (57.97 %) males. The characteristics of patients are shown in Table 1 .The median age was 49 years (IQR, 39–55.5), with ages ranging from 10 to 77 years. 20(28.99 %) of the 69 patients tested positive for viral RNA in fecal samples. The median age was 43 years (IQR, 31.25–51.0) among the patients with positive fecal samples, which was significantly younger than the patients with negative fecal samples (P = 0.003). Of the 20 patients with positive fecal viral tests, 11 (55 %) patients had negative results in stool samples later than in oropharyngeal swabs. In addition, 2 (10 %) of the 20 patients had first-time positive signals in fecal samples after the respiratory samples became negative.
Table 1.
Characteristics of patients with fecal positive and negative SARS-CoV-2 RNA tests.
| Parameters | Patients with positive fecal tests (n = 20) | Patients with negative fecal tests (n = 49) | P value |
|---|---|---|---|
| Age(yr) | 43 (31.25–51.0) | 52 (42–62.5) | 0.003 |
| Sex | |||
| Male |
13 (65 %) | 27 (55.10 %) | 0.45 |
| Female |
7 (35 %) | 22 (44.90 %) | |
| Exposure history | |||
| Stay in Hubei Province | 5 (25 %) | 7 (14.29 %) | 0.287 |
| Contact with people from Hubei Province | 5 (25 %) | 8 (16.31 %) | 0.403 |
| Familial aggregation | 11 (55 %) | 19 (38.78 %) | 0.217 |
| Chronic medical illness | |||
| Diabetes mellitus | 1 (5 %) | 10 (20.4 %) | 0.112 |
| Hypertension |
3 (15 %) | 10 (20.4 %) | 0.602 |
| Treatment Glucocorticoids |
|||
| Glucocorticoids Antibiotics |
1 (5 %) | 10 (20.4 %) | 0.112 |
| Antibiotics |
5 (25 %) | 14 (28.6 %) | 0.763 |
| Arbidol |
16 (80 %) | 39 (79.6 %) | 0.969 |
| Recombinant Human Interferon α2b Spray | 11 (55 %) | 3 (65.3 %) | 0.423 |
| Chloroquine Phosphate | 1 (5 %) | 0 | 0.115 |
Data are presented as interquartile ranges (IQRs), numbers and percentages.
3.2. Symptoms and laboratory and radiological findings on admission
The comparison of clinical characteristics and laboratory and radiological findings between patients with positive and negative fecal viral tests is shown in Table 2 . Fever was more common in patients with negative fecal viral tests than in patients with positive viral tests (58 % vs 22 %, P = 0.039). Six (10.53 %) of the 49 patients with negative fecal viral tests presented breathlessness, which was not observed in the patients with positive fecal viral tests. Patients with positive fecal viral tests had significantly lower CRP and FIB levels than patients with negative fecal viral tests [2.935 (0.90–8.77) vs 13.9 (4.29–43.03 mg/L, P = 0.001); 4.02 (3.505–4.85) vs 5.15 (3.56–5.905), P < 0.001]. In order to exclude the influence of age and gender, Generalized Linear Model was used for analysis. After correcting for effects of age and sex, CRP and FIB levels were still significantly lower in fecal RNA positive patients(CRP: Waldchi-square=5.681, P=0.017; FIB: Wald chi-square=11.746, P=0.001).
Table 2.
Symptoms, laboratory and chest CT results of patients with fecal positive and negative SARS-CoV-2 RNA tests on admission.
| Parameters | Patients with positive fecal tests (n = 20) | Patients with negative fecal tests (n = 49) | P value |
|---|---|---|---|
| Symptoms | |||
| Fever |
10 (50.00 %) | 37 (75.51 %) | 0.039 |
| Dry cough | 9 (45.00 %) | 20 (40.82 %) | 0.749 |
| Cough with sputum | 6 (30.00 %) | 21 (42.86 %) | 0.321 |
| Muscle ache | 2 (10.00 %) | 13 (26.53 %) | 0.235 |
| Sore throat | 4 (20.00 %) | 5 (10.20 %) | 0.483 |
| Breathlessness | 0 | 6 (12.24 %) | 0.243 |
| Chest tightness | 5 (25.00 %) | 8 (16.33 %) | 0.619 |
| Fatigue | 4 (20.00 %) | 13 (26.53 %) | 0.568 |
| Headache | 2 (10.00 %) | 6 (12.24 %) | 1 |
| Nausea and vomiting | 1 (5.00 %) | 7 (14.29 %) | 0.497 |
| Diarrhea | 5 (25.00 %) | 7 (14.29 %) | 0.474 |
| Laboratory results | |||
| CRP, 0∼8 mg/L | 2.935 (0.90–8.77) | 13.9 (4.29–43.03) | 0.001 |
| >8 | 6 (30 %) | 32 (65.31 %) | 0.007 |
| Leukocyte counts4 −10 × 10^9/L |
4.925 (3.9–5.62) | 4.89 (4.35–5.95) | 0.579 |
| >10 | 0 | 2 (4.08 %) | 0.9 |
| Neutrophil counts 1.8-6.3 × 10^9/L |
2.85 (2.055–3.48) | 3.13 (2.50–4.11) | 0.203 |
| >6.3 | 1 (5 %) | 3 (6.12 %) | 1 |
| Lymphocyte counts 1.1–3.2 × 10^9/L |
1.50 (1.18–1.70) | 1.34 (1.01–1.44) | 0.12 |
| <1.1 | 3 (15 %) | 17 (34.69 %) | 0.102 |
| ALT, 0–69 IU/L | 33.5 (24.5–51.5) | 29 (21.0–38.0) | 0.309 |
| >69 | 0 | 5 (10.20 %) | 0.331 |
| ALB, 35−55 g/L | 39.45 (38.425–41.35) | 38.5 (35.1–39.9) | 0.071 |
| <35 | 1 (5 %) | 10 (20.41 %) | 0.221 |
| FIB, 2.00–4.00 g/L | 4.02 (3.505–4.85) | 5.15 (3.56–5.905) | <0.001 |
| >4 | 10 (50 %) | 45 (91.84 %) | <0.001 |
| D-dimer, 0-0.5 μg/L | 0.33 (0.22-0.5225) | 0.43 (0.32-0.69) | 0.136 |
| >0.5 | 5 (25 %) | 22 (44.90) | 0.124 |
| Chest CT | |||
| Unilateral | 4 (20 %) | 6 (12.24 %) | 0.169 |
| Bilateral lung nvolvement | 15 (75 %) | 42 (85.71) | |
| No abnormal density shadow | 1 (5 %) | 1 (2.04 %) | |
Data are presented as interquartile ranges (IQRs), numbers and percentages.
CRP, C-reactive protein; ALT, alanine transaminase; ALB, albumin; FIB, Fibrinogen.
3.3. Survival analysis of viral shedding duration
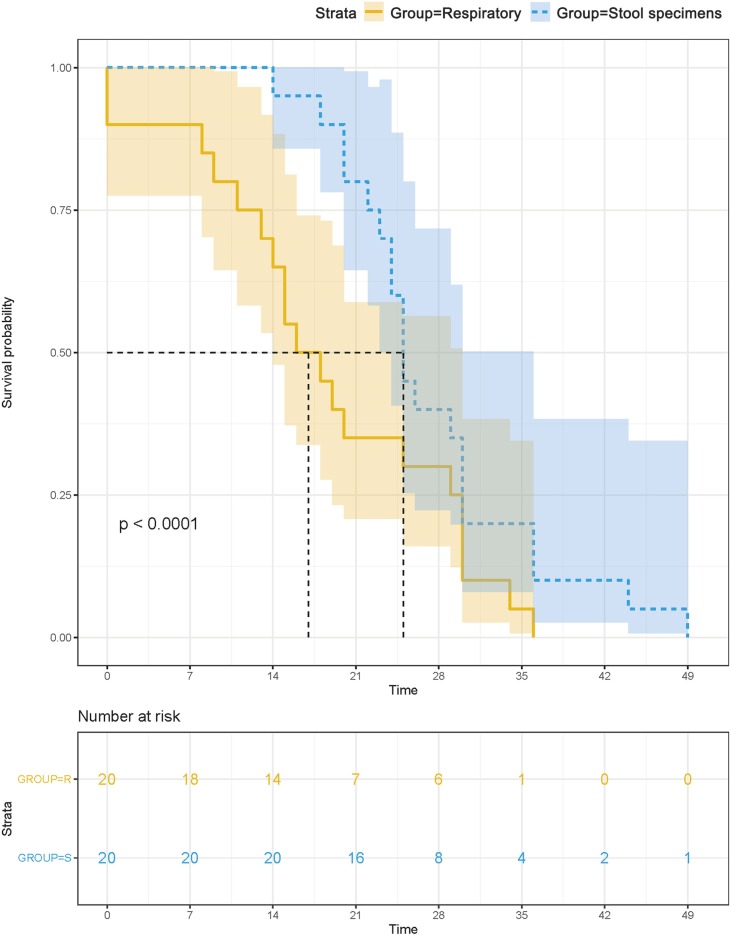
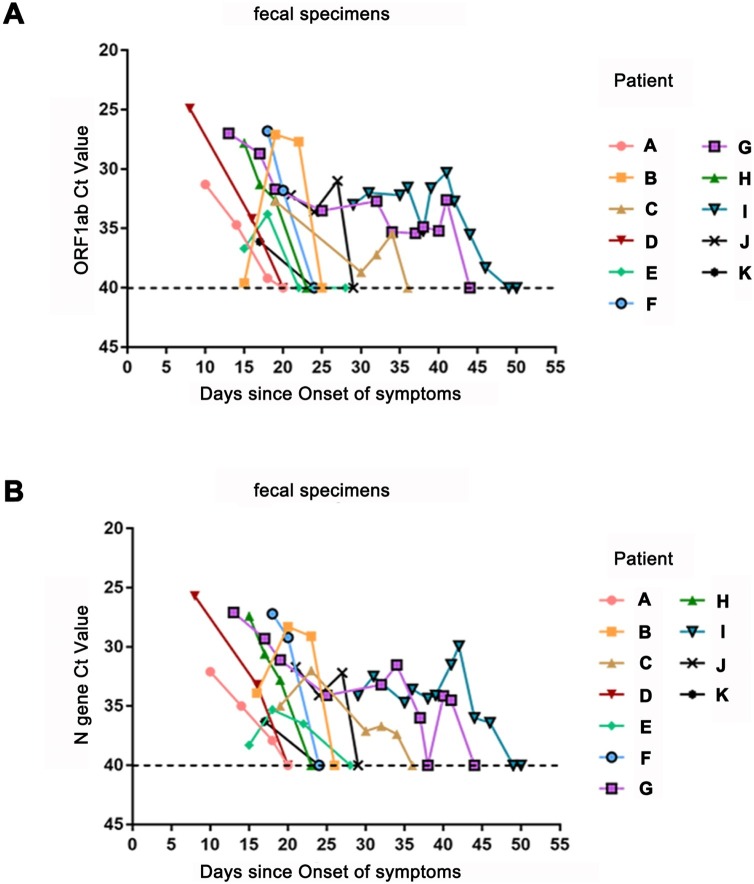
Survival analysis showed no significant difference in the duration of positive signals for SARS-CoV-2 RNA in respiratory samples between patients with negative and positive fecal viral tests (P > 0.05). The median survival of positive viral signals was significantly longer in fecal samples than that in oropharyngeal swabs (P < 0.001, Fig. 1 ). The median durations of positive signals for SARS-CoV-2 RNA were 25 days in fecal samples (95 % CI, 23.55–26.45) and 16 days in oropharyngeal swabs (95 % CI, 9.43–22.57) after symptom onset; therefore, the duration was approximately 9 days longer in fecal samples than in oropharyngeal swabs. In particular, positive fecal signals persisted for 49 days in patient I after symptom onset and for 33 days in patient G after viral tests in oropharyngeal swabs became negative (Fig. 2 ).
Fig. 1.
Survival analysis of positive SARS-CoV-2 signals in stool and respiratory specimens.
The median survival of positive viral signals was significantly longer in fecal samples than that in oropharyngeal swabs.
Fig. 2.
Viral load of SARS-CoV-2 detected in stool specimens after respiratory specimens became negative.
Panel A shows the Ct values for ORF1b from RT-PCR of specimens obtained from 11 patients, with a longer duration of positive signals in stool specimens than in respiratory specimens. Panel B shows the Ct values for the N gene from RT-PCR of specimens obtained from 11 patients, with a longer duration of positive fecal signals after respiratory specimens became negative.
3.4. Fecal viral load dynamics after respiratory signals became negative
To further understand the reason for the persistent positive fecal signals in 11 patients despite negative respiratory signals, Ct values, which are inversely related to the viral load, were recorded for fecal samples after the viral RNA test in oropharyngeal swabs became negative. As shown in Fig. 2, the Ct values in fecal samples reached high levels of 25–27 within 8–18 days even though they became negative in oropharyngeal swabs. The viral loads in fecal samples from five patients (namely, patients A, D, F, K and H) gradually decreased to negative values. However, the viral loads in fecal samples from the other 6 patients (namely, patients B, C, E, G, I and J) rebounded and lasted longer. Interestingly, the viral loads in both patient G and patient I showed second peaks on the 40th day after symptom onset. Moreover, patient I, who had persistent positive SARS-CoV-2 RNA signals in his fecal samples, had a negative outcome after 3 days of treatment of chloroquine phosphate.
3.5. Comparison of prognoses between patients with positive and negative fecal tests
The symptoms and laboratory and radiological findings of patients with either positive or negative SARS-CoV-2 RNA tests in stool samples at the time of the last negative respiratory sample are listed in Table 3 . We found that most of the symptoms and laboratory findings did not differ between patients with positive and negative fecal viral tests. Chest CT results showed heterogeneity of the lesions in different lobes of the lung. The most frequently involved lobe on admission and at the time of the last negative respiratory sample was the right lower lobe in patients with positive and negative signals in stool samples. Although most involved lobes showing glass ground opacity and consolidation shadows on CT scans improved, Patients with positive fecal viral tests had more complete absorption and no lesion progression in each lobe compared with patients with positive fecal viral tests (15 % vs 2.04 %, 5 % vs 2.04 %, 10 % vs 2.04 %, 15 % vs 6.12 %, 5 % vs 2.04 %; 0 % vs 2.04 %, 4.08 %, 2.04 %, 2.04 %, 4.08 %).
Table 3.
Symptoms, laboratory and chest CT results of patients with fecal positive and negative SARS-CoV-2 RNA tests at the time of the last negative respiratory sample.
| Parameters | Patients with positive fecal tests (n = 20) | Patients with negative fecal tests (n = 49) | P value |
|---|---|---|---|
| Symptoms | |||
| Fever | 1 (5 %) | 1 (2.04 %) | |
| Dry cough | 1 (5 %) | 8 (16.33 %) | |
| Cough with sputum | 0 | 0 | |
| Muscle ache | 0 | 0 | |
| Sore throat | 0 | 0 | |
| Breathlessness | 0 | 0 | |
| Chest tightness | 0 | 0 | |
| Fatigue | 0 | 1 (2.04 %) | |
| Headache | 0 | 0 | |
| Nausea and vomiting | 0 | 1 (2.04 %) | |
| Diarrhea | 1 (5 %) | 1 (2.04 %) | |
| Laboratory results | |||
| CRP,0∼8 mg/L | 0.57 (0.22–1.725) | 1.14 (0.43–2.95) | 0.074 |
| >8 | 1 (5 %) | 6 (12.24 %) | 0.642 |
| Leukocyte counts4 −10 × 10^9/L |
4.67 (3.98–6.07) | 5.4 (4.53–6.81) | 0.08 |
| >10 | 0 | 2 (4.08 %) | 0.9 |
| Neutrophil counts 1.8-6.3 × 10^9/L |
2.82 (1.68–3.54) | 3.33 (2.57–4.67) | 0.031 |
| >6.3 | 0 | 3 (6.12 %) | 0.631 |
| Lymphocyte counts 1.1–3.2 × 10^9/L |
1.74 (1.5–1.89) | 1.53 (1.21–1.95) | 0.146 |
| <1.1 | 1 (5 %) | 6 (12.24 %) | 0.642 |
| ALT, 0–69IU/L | 35.0(23.0–47.75) | 33.0(25.0–50.50) | 0.548 |
| >69 | 1 (5 %) | 9 (18.37 %) | 0.292 |
| ALB, 35−55 g/L | 42.3 (38.625–43.65) | 39.3 (36.85–41.45) | 0.010 |
| <35 | 1 (5 %) | 5 (10.20 %) | 0.822 |
| Involved lobes (chest CT) | |||
| Right upper lobe | 8 (40 %) | 29 (59.18 %) | |
| Right middle lobe | 11 (55 %) | 35 (71.43 %) | |
| Right lower lobe | 14 (70 %) | 44 (89.80 %) | |
| Left upper lobe | 10 (50 %) | 33 (67.35 %) | |
| Left lower lobe | 13 (65 %) | 39 (79.59 %) | |
| Bilateral lung involvement | 13 (65 %) | 41 (83.67 %) | |
| Percentage of improvement (chest CT) | |||
| Right upper lobe | 8 (40 %) | 26 (53.14 %) | |
| Right middle lobe | 10 (50 %) | 31 (63.27 %) | |
| Right lower lobe | 14 (70 %) | 41 (83.67 %) | |
| Left upper lobe | 10 (50 %) | 31 (63.27 %) | |
| Left lower lobe | 13 (65 %) | 35 (71.43 %) | |
| Percentage of progress (chest CT) | |||
| Right upper lobe | 0 | 1 (2.04 %) | |
| Right middle lobe | 0 | 2 (4.08 %) | |
| Right lower lobe | 0 | 1 (2.04 %) | |
| Left upper lobe | 0 | 0 | |
| Left lower lobe | 0 | 2 (4.08 %) | |
| complete absorbed lobes (chest CT) | |||
| Right upper lobe | 3 (15 %) | 1 (2.04 %) | |
| Right middle lobe | 1 (5 %) | 1 (2.04 %) | |
| Right lower lobe | 2 (10 %) | 1 (2.04 %) | |
| Left upper lobe | 3 (15 %) | 3 (6.12 %) | |
| Left lower lobe | 1 (5 %) | 1 (2.04 %) | |
Data are presented as interquartile ranges (IQRs), numbers and percentages.
CRP, C-reactive protein; ALT, alanine transaminase; ALB, albumin.
3.6. Prognoses of patients with longer durations of fecal SARS-CoV-2 persistence
Clinical data, laboratory results and imaging examinations were compared at the times of the last negative viral respiratory tests and fecal tests in the 11 patients with a longer duration of positive fecal signals than positive respiratory signals. No significant symptoms were noted by the time of the negative viral RNA test in oropharyngeal swabs except for patient A who had a mild cough and diarrhea. No significant differences in leukocyte counts, lymphocyte counts, CRP levels, ALT levels or ALB levels by the time of the negative viral RNA test were found between oropharyngeal swabs and fecal samples (P > 0.0.5). No lesion progression was observed in any lobes of the 11 patients at the time of the last negative viral fecal tests, and imaging findings had mostly improved compared with those at the time of the last negative viral respiratory tests.
4. Discussion
This study showed that 20 (28.99 %) of 69 patients had positive fecal viral tests. These patients were younger, had lower CRP and FIB levels on admission, and showed better recovery of chest CT results, with no differences in gastrointestinal symptoms and the viral shedding duration in respiratory samples compared with patients with negative fecal tests. The median duration of SARS-CoV-2 RNA persistence was significantly longer in fecal samples than in oropharyngeal swabs. The viral load of SARS-CoV-2 easily reached a high level and rebounded even though respiratory viral tests became negative.
Previous reports showed significantly different positivity rates of SARS-CoV-2 RNA in fecal samples. Our study showed that 20 (28.99 %) of 69 patients had positive fecal viral tests, which is higher than the rate in the report from Wu et al. (2020b). Two studies showed that 8 of 9 patients and 10 of 10 patients had positive SARS-CoV-2 RNA signals in fecal samples (Zang et al., 2020; Lo et al., 2020). The different rates of positive RNA tests may be due to the small sample sizes, discontinuous detection and different timing of testing. Importantly, inconsistent with the positive COV-2 RNA signals in oropharyngeal swabs, the presence of the positive RNA signals from fecal samples may lagged behind that for the signals from oropharyngeal swabs. Wu reported that the first-time positive SARS-CoV-2 RNA signals were detected in fecal samples from partial patients with COVID-19 two weeks after the respiratory samples became negative (Wu et al., 2020a). Similarly, our study found that 2 patients had first-time positive results presented in fecal samples even though respiratory signals became negative.
The incidence of diarrhea differs in recent reports. Cheung KS et al. reported that 17.6 % of patients with COVID-19 had gastrointestinal symptoms based on an analysis of data from a Hong Kong cohort of patients with COVID-19 and a meta-analysis of findings from publications (Cheung et al., 2020). In our study, 5 (25.00 %) patients had diarrhea, and only 1 (5.00 %) patient had nausea and vomiting among 20 patients with positive fecal viral tests, and no significant differences were observed between patients with positive and negative fecal viral tests. Lin L et al. reported that the presence of SARS-CoV-2 RNA in feces does not necessarily indicate more severe gastrointestinal symptoms (Lin et al., 2020). Possible reasons for digestive symptoms in SARS-nCoV-2 infection include direct injury to the gastrointestinal tract, a simple manifestation of viremia, and systemic inflammation, which are difficult to distinguish (Aroniadis et al., 2020). Whether the presence of SARS-CoV-2 RNA in feces is associated with direct injury to the gastrointestinal tract is worth exploring.
Furthermore, our present study revealed that fewer patients with positive fecal viral tests experienced fever, and none experienced breathlessness. Likewise, a younger onset age and lower CRP and FIB levels were observed in patients with positive signals in fecal samples, as well as less comorbidity and a higher level of ALB. Lower lymphocyte counts, ALT levels, and D-dimer levels were detected in patients with positive fecal viral tests on admission and at the time of the last negative oropharyngeal swab test. Coagulation function is considered a potential risk factor for a poor prognosis and death in patients with COVID-19 (Connors and Levy, 2020; Tang et al., 2020). The above data imply that patients with positive signals in fecal samples might have a milder condition of COVID-19.
Our study presented the first attempt of using a survival analysis to investigate the duration of SARS-CoV-2 RNA persistence in both respiratory and fecal specimens. Survival analysis showed no differences in the duration of SARS-CoV-2 in respiratory samples between patients with positive and negative fecal tests. The median duration of positive SARS-CoV-2 signals was significantly longer in fecal specimens than in respiratory specimens by approximately 9 days. The median duration of positive fecal signals for SARS-CoV-2 RNA was 25 (95 % CI, 23.55–26.45) days, which is similar to that reported by Guangzhou (Wu et al., 2020a) but longer than that reported by Ling Y et al. who reported a short observation period (Ling et al., 2020). Previous studies have demonstrated that SARS-CoV-2 and MERS-CoV were both viable for days in an environment with low temperature and humidity, leading to environmental contamination and even fecal–oral transmission (Wang et al., 2005; Chan et al., 2011; van Doremalen et al., 2013). Moreover, SARS-CoV-2 has greater transmissibility than other coronaviruses (Goh et al., 2020). Notably, Luo reported a cluster spreading event that occurred as a result of people bathing in a public bath center where the temperature and humidity were high (Luo et al., 2020). Although knowledge of the viability of SARS-CoV-2 remains to be clarified, the possibility of SARS-CoV-2 transmission from stool should be regarded as a dissemination route for COVID-19, especially in some areas with poor sanitation (Yeo et al., 2020).
To further understand the reason for the persistent positive fecal signals in 11 patients despite negative respiratory signals, Ct values, which are inversely related to the viral load, were recorded for fecal samples after the viral RNA test in oropharyngeal swabs became negative. The Ct values in fecal samples reached peaks of 25–27 within 8–18 days of symptom onset and rebounded more by than half after oropharyngeal swabs became negative. In particular, the peak was maintained even approximately 40 days after the onset of symptoms in two cases. Studies have reported that the clearance of the virus in salivary and pharyngeal samples peaked in the first week after the onset of symptoms when infectious viruses are readily isolated from respiratory specimens but not from stool specimens (To et al., 2020; Wölfel et al., 2020). In addition, Xiao F et al. reported that alive virus was isolated from stool specimen of a patient with COVID-19 on the 29th day after the onset of symptoms (Xiao et al., 2020b). An other study have reported that when a stool sample from a COVID-19 patient was used to inoculate naive ferrets they could isolate SARS-CoV-2 from the animals (Jeong et al., 2020). These results indicated that the clearance of the virus in stool might occur later than in the respiratory tract. It is possible that the virus in the digestive tract might remain alive for a long time, but requires further studies.
Since no differences in the duration of SARS-CoV-2 RNA persistence in respiratory samples were found between with positive and negative fecal tests, comparisons of symptoms and laboratory and radiological findings between with the two groups at the time of the last respiratory tests were discussed. We found that patients with positive fecal viral tests had higher proportions of complete absorption and no lesion progression in each lobe but no significant differences in most of the symptoms and laboratory findings compared with patients with negative fecal viral tests at the time of the last respiratory negative tests. In addition, Zheng S et al. reported that the viral shedding duration in respiratory samples was associated with disease severity (Zheng et al., 2020). These findings suggest that SARS-CoV-2 RNA in stool specimens is associated with better recovery of chest CT results.
Last, clinical data and laboratory and radiological findings were compared at the times of the last negative respiratory viral tests and fecal tests in the 11 patients with a longer duration of SARS-CoV-2 in fecal samples than in respiratory samples. We found no significant differences in clinical symptoms, leukocyte counts, lymphocyte counts, CRP levels, ALB levels or ALT levels, but differences were identified in the CT results between the times of the last negative respiratory viral tests and fecal tests. Lesions mostly improved in all lobes of the 11 patients at the time of the last negative viral fecal tests. Han X et al. reported that most patients with SARS-CoV-2 infection showed initial radiographic deterioration, with the worst manifestations in the second week after admission, followed by radiographic improvement in the third and fourth weeks (Han et al., 2020b). Therefore, it is possible that COVID-19 patients with negative RNA signals in the respiratory tract but with positive RNA signals in the digestive tract may still transmit the virus.
Several limitations exist in this study. First, the sample size was small. Second, the sample size of COVID-19 patients with positive fecal viral tests was relatively smaller than the sample of patients with negative respiratory viral tests (20 vs 49).
5. Conclusion
SARS-CoV-2 RNA in stool specimens SARS-CoV-2 RNA in stool specimens does not necessarily indicate more severe condition and poor recovery. The median duration of SARS-CoV-2 RNA persistence was significantly longer in fecal samples than in oropharyngeal swabs. The viral load of SARS-CoV-2 in stool specimens easily reached a high level and rebounded even though respiratory signals became negative. Fecal testing for SARS-CoV-2 RNA is crucial for informing the precautions necessary to prevent transmission in patients with SARS-CoV-2 infection.
Funding source
This work was supported by the Zhejiang Provincial Program for the Cultivation of High-level Innovative Health Talents.
CRediT authorship contribution statement
Xiaoming Wang: Investigation, Formal analysis, Writing - original draft. Jingwei Zheng: Formal analysis. Lei Guo: Investigation. Hao Yao: Investigation. Lingya Wang: Investigation. XiaoDong Xia: Conceptualization, Writing - original draft, Writing - review & editing. Weixi Zhang: Conceptualization, Writing - original draft, Writing - review & editing, Project administration.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
We thank the patients, the nurses and physicians who provided care for the patients, and the investigators at the Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198147.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Aroniadis O.C., DiMaio C.J., Dixon R.E. Current knowledge and research priorities in the digestive manifestations of COVID-19. Clin. Gastroenterol. Hepatol. 2020;18(8):1682–1684. doi: 10.1016/j.cgh.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.H., Peiris J.S., Lam S.Y. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011;2011 doi: 10.1155/2011/734690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China : a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Lan Y., Yuan X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infect. 2020;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F., Chan P.P. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135(23):2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G.K.-M., Dunker A.K., Foster J.A. Rigidity of the outer shell predicted by a protein intrinsic disorder model sheds light on the COVID-19 (Wuhan-2019-nCoV) infectivity. Biomolecules. 2020;10(2):331. doi: 10.3390/biom10020331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Duan C., Zhang S. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am. J. Gastroenterol. 2020;115(June (6)):916–923. doi: 10.14309/ajg.0000000000000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Cao Y., Jiang N. Novel coronavirus pneumonia (COVID-19) progression course in 17 discharged patients: comparison of clinical and thin-section CT features during recovery. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa271. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.W., Kim S.M., Kim H.S. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.07.020. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. (Engl.) 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I.L., Lio C.F., Cheong H.H. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16(10):1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Yao L., Zhang L. Possible transmission of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a Public Bath Center in Huai’an, Jiangsu Province, China. JAMA Netw. Open. 2020;3(3) doi: 10.1001/jamanetworkopen.2020.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.htmlNational Institute for Viral Disease Control and Prevention. Novel Primers and Probes for Detection of Novel Coronavirus 2019. January 21, 2020. (accessed March 12, 2020; in Chinese).
- Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Munster V.J. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18:20590. doi: 10.2807/1560-7917.es2013.18.38.20590. [DOI] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J., Guo T. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th hospital of the Chinese people’s liberation army. Water Sci. Technol. 2005;52:213–221. [PubMed] [Google Scholar]
- Wei X.S., Wang X., Niu Y.R. Diarrhea is associated with prolonged symptoms and viral carriage in COVID-19. Clin. Gastroenterol. Hepatol. 2020;18(8):1753–1759. doi: 10.1016/j.cgh.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Liu J., Li S. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med. Infect. Dis. 2020;18(April) doi: 10.1016/j.tmaid.2020.101673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Jiang L., Huang G. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int. J. Infect. Dis. 2020;93:264–267. doi: 10.1016/j.ijid.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26(7):1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.