Abstract
The most basic form of locomotion in limbed vertebrates consists of alternating activities of the flexor and extensor muscles within each limb coupled with left/right limb alternation. Although larval zebrafish are not limbed, their pectoral fin movements exhibit the following fundamental aspects of this basic movement: abductor/adductor alternation (corresponding to flexor/extensor alternation) and left/right fin alternation. Because of the simplicity of their movements and the compact neural organization of their spinal cords, zebrafish can serve as a good model to identify the neuronal networks of the central pattern generator (CPG) that controls rhythmic appendage movements. Here, we set out to investigate neuronal circuits underlying rhythmic pectoral fin movements in larval zebrafish, using transgenic fish that specifically express GFP in abductor or adductor motor neurons (MNs) and candidate CPG neurons. First, we showed that spiking activities of abductor and adductor MNs were essentially alternating. Second, both abductor and adductor MNs received rhythmic excitatory and inhibitory synaptic inputs in their active and inactive phases, respectively, indicating that the MN spiking activities are controlled in a push-pull manner. Further, we obtained the following evidence that dmrt3a-expressing commissural inhibitory neurons are involved in regulating the activities of abductor MNs: (1) strong inhibitory synaptic connections were found from dmrt3a neurons to abductor MNs; and (2) ablation of dmrt3a neurons shifted the spike timing of abductor MNs. Thus, in this simple system of abductor/adductor alternation, the last-order inhibitory inputs originating from the contralaterally located neurons play an important role in controlling the firing timings of MNs.
SIGNIFICANCE STATEMENT Pectoral fin movements in larval zebrafish exhibit fundamental aspects of basic rhythmic appendage movement: alternation of the abductor and adductor (corresponding to flexor–extensor alternation) coupled with left–right alternation. We set out to investigate the neuronal circuits underlying rhythmic pectoral fin movements in larval zebrafish. We showed that both abductor and adductor MNs received rhythmic excitatory and inhibitory synaptic inputs in their active and inactive phases, respectively. This indicates that MN activities are controlled in a push-pull manner. We further obtained evidence that dmrt3a-expressing commissural inhibitory neurons exert an inhibitory effect on abductor MNs. The current study marks the first step toward the identification of central pattern generator organization for rhythmic fin movements.
Keywords: CPG, dmrt3, pectoral fin, rhythmic movement, spinal cord, zebrafish
Introduction
In land-dwelling limbed vertebrates, the most basic form of locomotion involves alternating activities both of the flexor and extensor muscles within the limb and of the muscles on the two sides of the body (Grillner, 1975). Pectoral and pelvic fins in fish are homologs of forelimbs and hindlimbs, respectively. Evolutionally, alternating limb/fin movements can be traced back to cartilaginous fish: some skate species engage in walking-like movements using their pelvic fins, suggesting a very ancient origin of the neural substrate for alternating appendage movements (Jung et al., 2018).
Neuronal circuits involved in controlling rhythmic limb movements have been investigated for more than a century (Brown, 1911). Studies have shown that the precise timings and patterns of muscle movements are generated by the activity of neuron assemblies in the spinal cord that are known as central pattern generators (CPGs; Grillner, 2003; Kiehn, 2006). The advent of molecular genetic techniques, which enable us to label particular types of neurons (and to manipulate their activities) in mice, has greatly advanced our knowledge of the CPG constituent neurons (Goulding and Pfaff, 2005; Goulding, 2009; Arber, 2012; Kiehn, 2016). Nevertheless, the organizational details of spinal CPG in mammals are not yet completely understood.
Two main obstacles make it difficult to study the spinal CPG in mammals. One is the complexity of mammalian movements, and the other is the complexity of the mammalian nervous system. Mammalian limbs are composed of many flexor and extensor muscles, each of which contracts with a slightly different timing during locomotion, making the analysis of motor outputs complicated. Additionally, the mammalian spinal cord is large in size, making recordings (either electrophysiological or calcium imaging) of CPG-constituent neuron activities difficult.
Research would be made easier by the development of a simple and compact model organism that exhibits certain fundamental aspects of rhythmic appendage movements. Pectoral fin movements in larval zebrafish could serve as such a model. The structure of the pectoral fin in larval zebrafish is very simple: there are only two types of muscles, the abductor and adductor (Thorsen and Hale, 2005). Yet, the movements of this fin fulfil all the hallmarks of rhythmic limb movements: alternation of the abductor and adductor (corresponding to flexor–extensor alternation) coupled with left–right alternation (Thorsen et al., 2004). The spinal cords of larval zebrafish are also small in size, enabling physiological recordings of fin motor neurons (MNs; and, potentially, CPG interneurons) during fictive rhythmic movements (Green and Hale, 2012). An additional advantage of the zebrafish as a model is that it is amenable to molecular genetics. Particular classes of candidate CPG neurons can be identified by the expression patterns of developmentally regulated transcription factors, allowing researchers to perform fine resolution analyses (Kimura and Higashijima, 2019; Satou et al., 2020). Because the expression patterns of transcription factors are largely conserved across all vertebrate species, analyses of CPG organization for rhythmic paired fin movements in fish could provide insight into the evolutionary origins of rhythmic limb movements (Jung et al., 2018).
Here, we set out to investigate neuronal circuits underlying rhythmic pectoral fin movements in larval zebrafish. We first showed that both abductor and adductor MNs received rhythmic excitatory synaptic inputs in their active phase and inhibitory inputs in their inactive phase, consistent with previous observations during scratching in turtles (Robertson and Stein, 1988) and walking in mammals (Shefchyk and Jordan, 1985; Cazalets et al., 1996; Hochman and Schmidt, 1998; Endo and Kiehn, 2008). We then sought the source neurons that provide synaptic inputs to fin MNs and identified dmrt3a (zebrafish homolog of mammalian dmrt3)-expressing neurons as possible candidates. In mammals, dmrt3 neurons have been implicated in commissural inhibitory pathways in limb movements (Andersson et al., 2012; Perry et al., 2019). Our results show that dmrt3a-expressing commissural inhibitory neurons exert an inhibitory effect on abductor MNs. The current study marks the first step toward the identification of CPG organization for rhythmic pectoral fin movements.
Materials and Methods
Animals.
Zebrafish adults, embryos, and larvae were maintained at 28.5°C. Experiments were performed at room temperature (23–28°C). All procedures were performed in compliance with the guidelines approved by the animal care and use committees of the National Institutes of Natural Sciences. Animals were staged according to days postfertilization (dpf).
The following transgenic fish strains were used in this study: Tg[zCREST2-hsp70:GFP] (Uemura et al., 2005), Tg[UAS:Kaede], Tg[UAS:GFP] (Kimura et al., 2013), Tg[UAS:RFP] (Asakawa et al., 2008), Tg[hspzGFFgDMC26A] (found among collections of the enhancer trap lines in the Kawakami laboratory; Asakawa et al., 2008), Tg[dmrt3a:GFP], Tg[dmrt3a:loxP-DsRed-loxP-DTA], Tg[glyt2:loxP-DsRed-loxP-GFP] (Satou et al., 2020), Tg[zCREST2-hsp70:Gal4], Tg[hoxa4a:Cre], Tg[drmt3a:tTA], Tg[tetO:Dendra2], Tg[tetO:ChR], and Tg[lhx1a:GFP] (this study). For the generation of Tg[zCREST2-hsp70:Gal4], GFP in the construct of zCREST2-hsp70:GFP (Uemura et al., 2005) was replaced with Gal4FF (Asakawa et al., 2008). For the generation of Tg[hoxa4a:Cre], the enhancer sequence of the hoxa4a gene (Kimura and Higashijima, 2019) and the hsp70 promoter Cre-mCherry-NLS (Satou et al., 2012) were used. The tetO and tTA sequences that were used to generate Tg[drmt3a:tTA], Tg[tetO:ChR] were described previously (Natsubori et al., 2017). A channelrhodopsin (ChR) sequence used in Tg[tetO:ChR] was ChR-WideReceiver (Wang et al., 2009). Tg[zCREST2-hsp70:Gal4], Tg[hoxa4a:Cre], Tg[tetO:Dendra2], and Tg[tetO:ChR] transgenic fish were generated using Tol2-based transgenesis (Urasaki et al., 2006). Tg[dmrt3a:tTA] and Tg[lhx1a:GFP] transgenic fish were generated using the CRISPR/Cas9-mediated knock-in method with the hsp70 promoter (Kimura et al., 2014).
Behavioral analyses.
Behavioral experiments were performed essentially as described previously (Satou et al., 2009; Kimura and Higashijima, 2019) using 3 dpf (range, 3.1–3.5 dpf) larvae. Sequential images of swimming or rhythmic fin movements were captured at 1000 frames/s with a high-speed camera (FASTCAM-ultima1024, Photron). Fish were filmed from their dorsal side. We analyzed swimming or rhythmic fin movements that occurred spontaneously.
To determine the frequency of the rhythmic fin movements, the start of abduction (left or right fin) was used as the standard time point. The cycle period of each cycle of the movement was then determined, and the frequency was calculated as the inverse of the cycle period. For image analysis of fin–body coordination, the start of abduction was again used as the reference time point. Swim cycles whose frequencies were between 28 and 34 Hz (representing ∼75% of all cycles) were included in our analysis. To extract the body contour of each fish, a small sample of points in the eye region that were visible at the lowest pixel intensity were identified in image frames and used to define a scanning line that was orthogonal to the line between the eyes. At intervals of eight pixels along the line between the eyes, pixel intensities within the arc-shaped region were sampled and the borders of left/right sides were determined by finding the change points for the variance in their intensities. This procedure was repeatedly applied with shifted sampling arc positions to reduce the proportion of erroneous body-border detections. Segments of cubic spline functions were then obtained by applying a smoothing spline interpolation method to the pairs of border positions (i.e., along the midline). This curve describing body shape was fitted to a straight line (linear regression). During the line fitting, disturbances in tail–tip fluctuations were excluded from the anterior half of the curve. The output is treated as the regression line. The farthest protruding point of the body was determined using the midline and the regression line described above.
Confocal imaging.
GFP-, Kaede-, or Dendra2-labeled neurons were imaged in the living fish by confocal microscopy (SP8 Leica; or FV1200, Olympus). Photoconversion of Kaede in fin MNs was performed using a 405 nm laser line. For the expression of Kaede in abductor MNs, compound transgenic fish of Tg[zCREST2-hsp70:Gal4] and Tg[UAS:Kaede] were used. For the expression of Kaede in adductor MNs, compound transgenic fish of Tg[hspzGFFgDMC26A] and Tg[UAS:Kaede] were used. Photoconversion of Dendra2 in dmrt3a neurons in the compound transgenic fish of Tg[dmrt3a:tTA] and Tg[tetO:Dendra2] was performed according to the primed photoconversion technique using 488 and 635 nm laser lines (Taniguchi et al., 2017). After photoconversion of Kaede or Dendra2 proteins in cell bodies, samples were kept for 2–3 h to ensure that all photoconverted Kaede or Dendra2 protein would be distributed to the axons. Then, confocal imaging was performed.
For the morphologic analysis of dmrt3a neurons using Dendra2, a specialized class of commissural inhibitory neurons (CoLo neurons) that are exclusively involved in escape behaviors (Satou et al., 2009) were excluded based on their distinctive morphology characterized by a spherical soma and thick short axons.
Electrophysiology.
In vivo loose-path or whole-cell recordings and ventral root (VR) recordings were performed as described previously (Kimura and Higashijima, 2019; Satou et al., 2020) with some modifications. Recordings were conducted using 3 dpf (range, 3.1–3.5 dpf) larvae. Larvae were immobilized by soaking in the neuromuscular blocker d-tubocurarine (0.1 mg/ml in distilled water) for 5–15 min, then pinned through the notochord to a SYLGARD-coated, glass-bottomed dish with short pieces of fine tungsten pins. Animals were then covered with an extracellular recording solution that contained the following (in mm): 134 NaCl, 2.9 KCl, 1.2 MgCl2, 2.1 CaCl2, 10 HEPES, 0.01 d-tubocurarine, and 10 glucose, adjusted to pH 7.8 with NaOH. The skin covering the rostral part of the body was removed with a pair of forceps. Then, muscle fibers of muscle segment 4 were carefully removed manually with a tungsten needle. For all electrophysiology experiments, the preparations were observed using a water-immersion objective (40×, numerical aperture 0.80; Olympus) on an upright microscope (BX51WI, Olympus) fitted with differential interference contrast optics. Spinal neurons located at muscle segment 4 were targeted for loose-patch or whole-cell recordings. We used the following transgenic fish for targeted recordings: Tg[zCREST2-hsp70:GFP] for abductor MNs, Tg[hspzGFFgDMC26A], and Tg[UAS:GFP] for adductor MNs; and Tg[dmrt3a:GFP] for dmrt3a neurons. We analyzed fictive swimming that occurred spontaneously. VR recordings of axial nerves were made at the boundary between muscle segments 9 and 10. Electrodes for VR recordings (tip diameter, 30–50 µm) and loose-patch recordings (resistance, 9–12 MΩ) were filled with the extracellular recording solution. Patch electrodes (resistance, 11–15 MΩ) were filled with intracellular solution. The intracellular solution for current-clamp recording contained the following (in mm): 119 K-gluconate, 6 KCl, 2 MgCl2, 10 HEPES, 10 EGTA, and 4 Na2 ATP at 290 mOsm, adjusted to pH 7.2 with KOH. The intracellular solution for voltage-clamp recording contained the following (in mm): 140 CsMeSO4, 1 QX314-Cl, 1 TEA-Cl, 3 MgCl2, 10 HEPES, 1 EGTA, and 4 Na2-ATP, adjusted to pH 7.2 with CsOH. Before performing voltage-clamp recordings from abductor or adductor MNs, we always made loose-patch recordings from the same neurons to examine the firing patterns of the neurons. When we measured excitatory currents, cells were held at −74 mV. When we measured inhibitory currents, cells were held at +10 mV. These values represent the calculated chloride ion and cation reversal potentials, respectively. Values were corrected for a calculated junction potential of −10 mV (pClamp 10, Molecular Devices). Electrophysiological recordings were generated using MultiClamp700B amplifiers and digitized with Digidata1440A digitizer (Molecular Devices).
For ChR experiments, Tg[drmt3a:tTA]; Tg[tetO:ChR] fish were used to express ChR in drmt3a neurons. The fish were crossed to either Tg[zCREST2-hsp70:GFP] (for targeting abductor MNs) or Tg[hspzGFFgDMC26A]; Tg[UAS:GFP] (for targeting adductor MNs). A 20 ms pulse of blue light illumination (power, ∼2 mW/mm2) was applied over an area covering muscle segments 3.5–5.5. Blue light illumination was applied at least 10 times for each cell. Postsynaptic currents were monitored at whole-cell voltage-clamp configuration with the holding potential set to +10 mV. If any IPSC was observed within 40 ms (i.e., during the blue light illumination period plus an additional 20 ms), the IPSCs were judged to be evoked by ChR-expressing neurons. The probability of responses was defined as the number of positive trials per all trials. The amplitude of the IPSC was measured as the peak of the current.
Data analysis.
Electrophysiological data were analyzed with DataView (software by William Heitler, University of St. Andrews, St. Andrews, UK) and Excel (Microsoft). VR recordings were rectified and smoothened. To detect each instance of VR activity, a threshold value was set by visual inspection. For the phase analysis, the middle time point of a VR activity was assigned a phase value of 0, and that of the next VR activity was assigned a phase value of 1. In determining the frequency of swimming, the interval between time points 0 and 1 was defined as the cycle period. The swimming frequency was the inverse of the cycle period.
For the data analyses of fin MNs or dmrt3a neurons (circular plot analyses and current analyses in voltage-clamp recordings), we only included data obtained from the cells that fired relatively consistently in each cycle (i.e., those that exhibited a spike in more than ∼60% of cycles). Circular plot analysis was performed essentially as described previously (Kjaerulff and Kiehn, 1996) to provide a statistical measure of the coupling between neuronal firing and the phase of VR bursts. Spikes that occurred during smooth slow swimming (28–34 or 29–35 Hz in the case of wild-type fish and 29–35 Hz in the case of dmrt3a-DTA fish, representing ∼45, ∼40, and ∼45% of the total cycles, respectively) were subject to analysis. For each of the recorded cells, 30 spikes that fulfilled the criterion described above were randomly selected by a computer, and the phase values of these selected spikes were determined and plotted in a circle. For voltage-clamp analyses, cycles occurring during smooth slow swimming (described above) were subject to analysis. Each cycle was binned into 200 segments. In each segment, the averaged value of the currents was calculated and plotted. For each cell, 20 traces were made. In our presentations of population data, each cell is represented by the mean of its 20 traces.
The IPSC response delay time from the spike in a dmrt3a neuron to that in its potential target neuron on the contralateral side was presumed to be 1.5 ms (1 ms of conduction time and 0.5 ms of synaptic transmission time). We used 1 ms as the conduction time for the following reason. The conduction velocity of an action potential in the axon was assumed to be 0.2 m/s (Menelaou and McLean, 2019). The length from the soma to the target was assumed to be 200 µm [100 µm for crossing the spinal cord and 100 µm for running on the contralateral side (∼1 segment)]. The 1.5 ms corresponds to the value of 0.05/cycle (one cycle, 0–1).
Statistics.
Results are presented as the mean ± SD. Statistical significance was assessed using the t test or Mann–Whitney U test.
Results
Coordinated rhythmic pectoral fin movement during slow swimming in 3 dpf larvae
It has been demonstrated that larval zebrafish at 5 dpf show rhythmic pectoral fin movements (alternation of the abductor and adductor coupled with left–right alternation) associated with body undulation during slow swimming (Thorsen et al., 2004). In the current study, we chose to use younger animals, namely 3 dpf larvae, since the duration of swimming bouts tends to be longer in 3 dpf larvae (typically, longer than 1 s and consisting of >30 swim cycles) than in 5 dpf larvae (typically, several hundred milliseconds and consisting of ∼10 swim cycles), making physiological analyses easier. Because pectoral fin movements in 3 dpf larvae have not been described previously, we first performed high-speed camera filming of the movements. As Figure 1A shows, the movements are essentially the same as those described in 5 dpf larvae: alternated fin movements are coupled with undulation movements of the body. Frequencies of the rhythmic fin movements are summarized in Figure 1B, showing that the most common frequency was ∼30 Hz (mean, 31.1 ± 2.5 Hz). Coordination of the fin and body movements appeared to be stereotyped among cycles in each fish, and across fish, such that similar body shape appeared in a particular phase of the fin movement cycle (e.g., at the time points of 0 and 32 ms; these time points correspond to near-maximum abduction of the right fin). We quantified this by examining the body shape at the time point when the right fin started to abduct. The farthest protruding point of the body (Fig. 1C, circle) tended to occur at a similar body position across swim cycles (Fig. 1D,E; position value, 0.81 ± 0.016 along the body length; 15 swim cycles). The values were very similar in all seven fish examined (Fig. 1F; 0.82 ± 0.010). These results indicate that the rhythmic pectoral fin movements are highly coordinated with the body movements.
Figure 1.
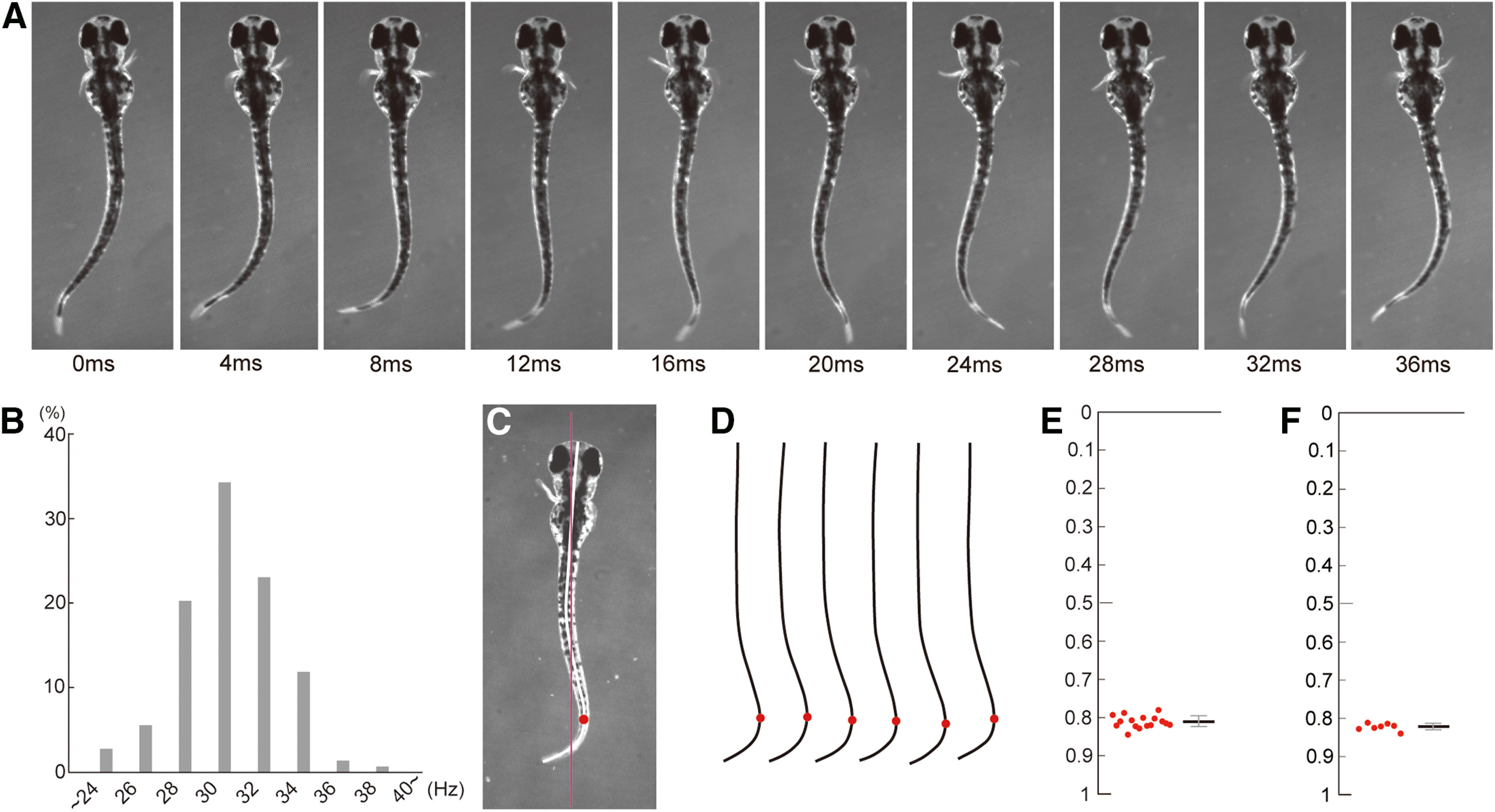
Rhythmic pectoral fin movements in 3 dpf larvae. A, A typical slow swimming of larval zebrafish. The fins alternate rhythmically in conjunction with the body undulation. B, Histogram of swimming frequency (143 cycles from 11 fish). C, Example body shape at the time point when the right fin started to abduct. The red line shows the regression line for the rostral half of the body. The midline of the body is indicated by the thick white line. The red circle shows the farthest protruding point from the regression line. D, Midlines of the body in six swim cycles are aligned. E, Plots of the farthest protruding point in the larva at the time point when the right fin started to abduct (15 swim cycles). F, Same as in E, but the averaged values obtained from seven fish are plotted.
Genetic visualization of abductor and adductor MNs
A critical experimental requirement in the current study was to distinguish abductor and adductor MNs. For the identification of abductor MNs, we used Tg[zCREST2-hsp70:GFP], in which GFP is known to be expressed in abductor MNs (Uemura et al., 2005; Green and Hale, 2012). Figure 2, A1 (lateral view) and A2 (dorsal view), shows images of these transgenic fish. From the dorsal view, GFP-labeled motor axons that innervate to the lateral side of the fin, where the abductor muscles are located, are clearly visible (Fig. 2A2). For the identification of adductor MNs, we searched for an adductor-specific to MN-specific line. In a collection of enhancer/gene trap lines (Asakawa et al., 2008), we found the enhancer trap line Tg[hspzGFFgDMC26A], in which Gal4 is expressed in abductor MNs. Figure 2, B1 (lateral view) and B2 (dorsal view), shows images of the compound transgenic fish of Tg[hspzGFFgDMC26A] and Tg[UAS:GFP]. From the dorsal view, GFP-labeled motor axons that innervate to the medial side of the fin, where the adductor muscles are located, are clearly visible (Fig. 2B2). Somata of the fin MNs were located in the rostral spinal cord around the location near muscle segments 2–5 (Fig. 2A1,B1).
Figure 2.
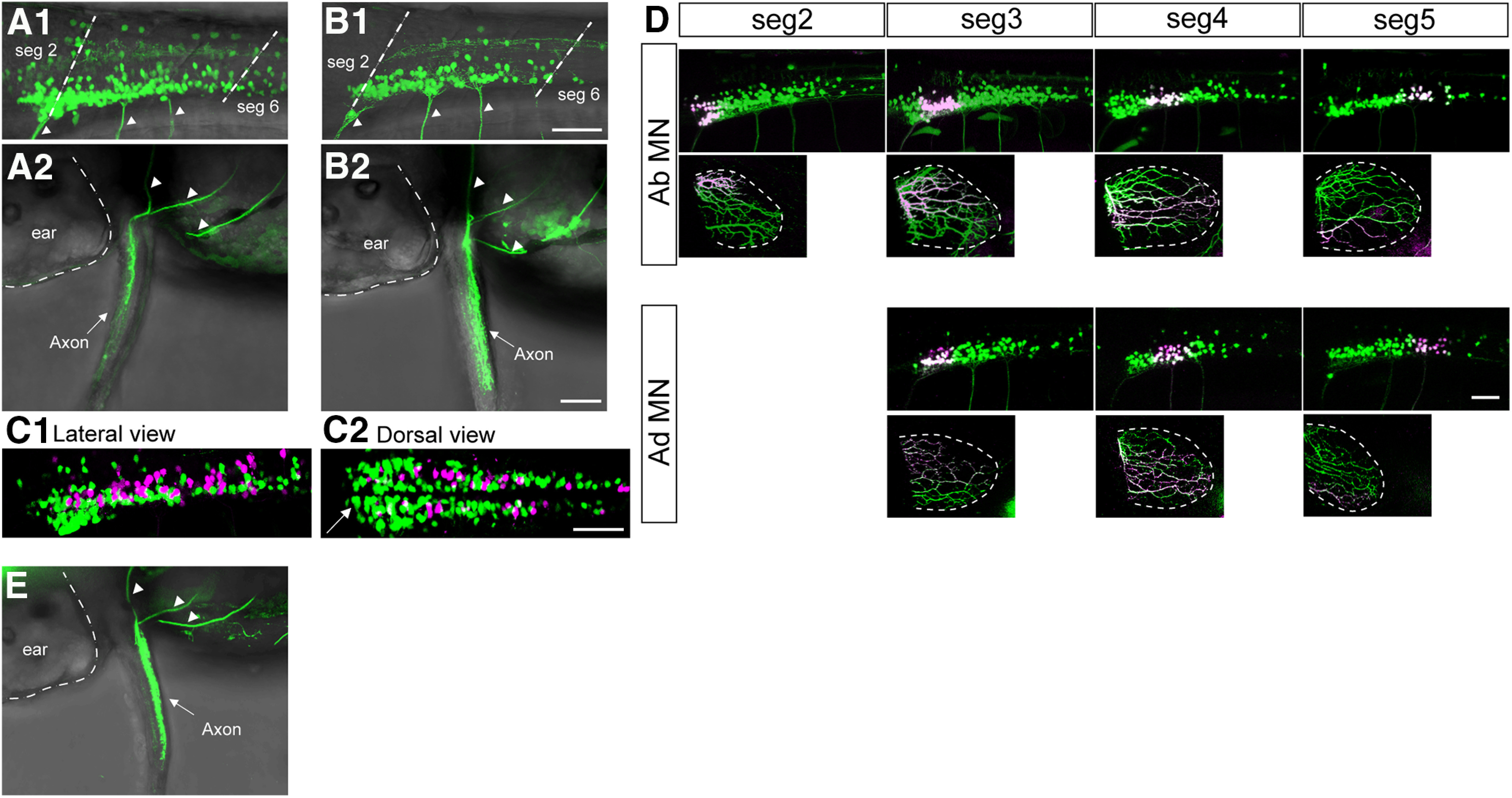
Live visualization of abductor and adductor MNs. A1, A2, Stacked image of confocal optical sections of Tg[zCREST2-hsp70:GFP]. A1 is a lateral view of the rostral spinal cord. A2 is a dorsal view of a fin. Arrowheads indicate axons extending into the fin. B1, B2, Same as A1 and A2, but of compound transgenic fish of Tg[hspzGFFgDMC26A] and Tg[UAS:GFP]. C1, C2, Stacked image of confocal optical sections of the triple transgenic fish of Tg[zCREST2-hsp70:GFP], Tg[hspzGFFgDMC26A], and Tg[UAS:RFP] (C1, lateral view; C2, dorsal view) The arrow shows that the majority of labeled neurons in the most rostral region are abductor MNs (green). D, The results of Kaede photoconversion experiments. In the top half of each panel, Kaede is expressed in abductor MNs. In the lower half of each panel, Kaede is expressed in adductor MNs. For each type of MN, the top panels show the lateral view of the spinal cord, while the bottom panels show the lateral view of the fin. E, Dorsal view of confocal stacked image of Tg[lhx1a:GFP]. Arrowheads indicate axons extending into the fin. Scale bar, 50 µm.
A previous study has suggested that abductor and adductor MN somata are intermingled in 5 dpf larvae (Thorsen and Hale, 2007). We examined this possibility in 3 dpf larvae. Figure 2C shows images of the triple transgenic fish of Tg[zCREST2-hsp70:GFP], Tg[hspzGFFgDMC26A], and Tg[UAS:RFP], in which abductor and adductor MNs were labeled by GFP and RFP, respectively. From both the lateral view (Fig. 2C1) and the dorsal view (Fig. 2C2), there is no clear separation of the two MN populations. In the rostral-most part of the fin MN residing area, the majority of the neurons were abductor MNs (Fig. 2C2, arrow), indicating that there was some bias in the distribution along the rostrocaudal dimension.
We examined the topographic relation between MN somata and the innervation area as described previously (Thorsen and Hale, 2007). For this purpose, the photoconvertible protein Kaede was expressed in abductor or adductor MNs using the Gal4-UAS system. Green Kaede proteins in the MN somata in a restricted location were photoconverted to red. Then, red Kaede distribution was investigated in the axons in the fin. We found a clear topographic relationship between the somata location and the innervation area (Fig. 2D). Thus, our results are consistent with the previous results (rostrocaudal dimension of the cell body location is converted to the dorsoventral dimension of the axonal targets in the fin) and showed that the topographic relationship applies to both abductor and adductor MNs.
Activities of abductor and adductor MNs during fictive swimming
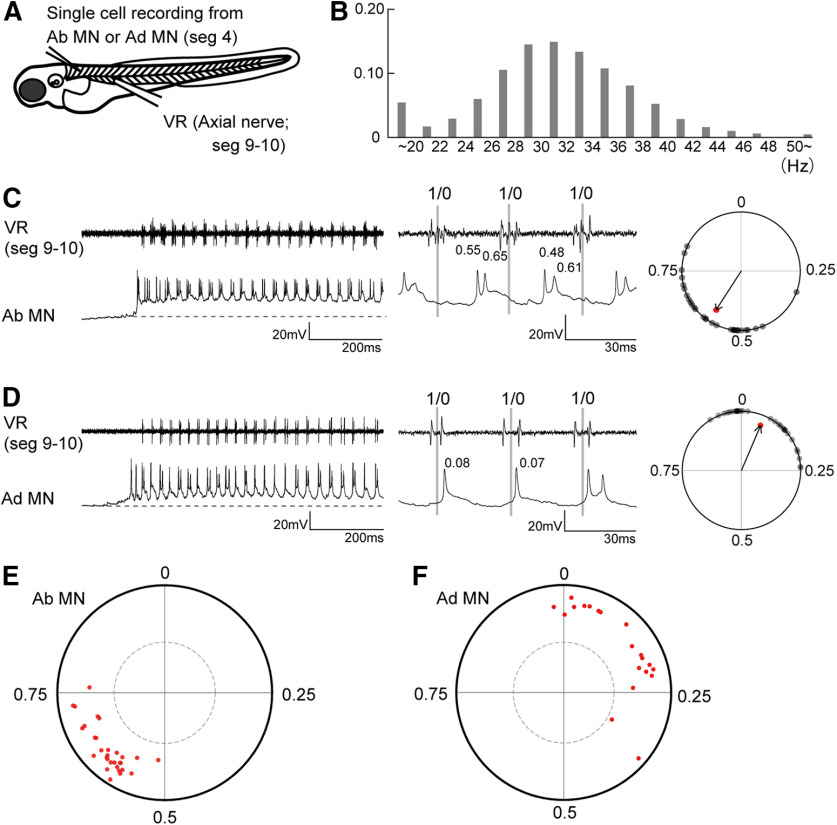
Next, we examined firing activities of abductor and adductor MNs during fictive swimming. Figure 3A shows a schematic of the electrophysiological experiment. Firing activities of abductor or adductor MNs located in segment 4 (MNs that innervate the middle region of the fin along the dorsoventral dimension; Fig. 2D) were examined either by loose-patch or whole-cell current-clamp recordings. Simultaneously, activities of axial MN axons that ran along the border between muscle segments 9 and 10 were examined (VR recordings). Because fin and body movements are highly coordinated and stereotyped during slow swimming (Figs. 1D,E), we used the axial VR activity as a reference timing for our phase analysis of fin MN spiking activities (this applies throughout the present study).
Figure 3.
Firing patterns of abductor and adductor MNs during fictive swimming. A, A schematic illustration of the simultaneous recordings of an abductor or adductor MN (loose patch or whole cell) and VR. B, Histogram of fictive swimming frequency (2920 cycles from seven fish). C, An example of the recording (whole cell current clamp) from the abductor MN. The dotted line indicates the resting membrane potential. The middle panel shows a close-up view of three swim cycles. For the phase analysis of spike timings, the middle time point of a VR activity was assigned a phase value of 0, and that of the next VR activity was assigned a phase value of 1. The right panel shows a circular plot of the spikes relative to VR activity during fictive swimming. D, Same as C, but for the adductor MN. E, Circular plot showing spike timing of abductor MNs (n = 30). Dotted circle line marks the 5% significance level. F, Circular plot showing spike timing of adductor MNs (n = 20).
We first investigated the frequency distribution of fictive swimming. Figure 3B shows that the most common frequency was ∼30 Hz (mean, 30.8 ± 7.8 Hz), which is quite similar to that observed during actual swimming (31.1 ± 2.5 Hz; Fig. 1B).
Figure 3, C and D (left panels), shows examples of whole-cell recordings of abductor and adductor MNs, respectively. Both MNs exhibited rhythmic spiking activities that were coordinated with VR activities. We performed phase analyses of these spikes. The middle time point of one VR burst was set as time 0, and that of the next cycle was set as time 1. The timing of each spike was represented as its phase in the cycle (Fig. 3C,D, middle panels). The phase values of 30 randomly selected spikes are plotted in the circles (Fig. 3C, right, black dots), and the average value was used as the vector (Fig. 3C, right, red arrows). In this circular plot analysis (Kjaerulff and Kiehn, 1996), the direction of the vector shows the mean of the phase value, whereas the length of the vector shows the strength of the rhythmicity.
Figure 3, E and F, shows population data for abductor and adductor MNs, respectively (each dot represents the tip of the vector for the neuron examined). Phase values of abductor MNs are clustered around the timing between 0.5 and 0.75 (mean, 0.63 ± 0.06). Phase values of adductor MNs are more broadly distributed around the timing between 0 and 0.35 (mean, 0.14 ± 0.11). Despite the broader phase distribution of adductor–MN vectors, the mean lengths of the vectors are not significantly different between abductor MNs and adductor MNs (abductor MNs, 0.79 ± 0.08; adductor MNs, 0.79 ± 0.09; p = 0.72, t test). For all of the MNs examined, the vectors exceed significance (Fig. 3E,F, dotted circle), indicating that each MN, regardless of type, fired in a highly rhythmic manner. The phase values of abductor MNs (0.5–0.75) and those of adductor MNs (0–0.35) are nonoverlapping and approximately opposite each other. This indicates that abductor and adductor MNs are essentially alternating, a finding that is consistent with the behavioral analysis.
Voltage-clamp recordings from abductor and adductor MNs during fictive swimming
Next, we performed voltage-clamp recordings from abductor and adductor MNs to obtain insights into the synaptic inputs underlying the rhythmic firings of the MNs. Our experimental design was as follows. First, the firing pattern of an MN was examined using loose-patch recording. Then, the electrode was changed for whole-cell voltage-clamp recording, and the same cell was targeted (note that firing pattern could not be examined with the intracellular solution for voltage-clamp recording). During voltage-clamp recording, the recorded MN was held at two different holding potentials, as follows: −74 mV for the examination of the excitatory current and +10 mV for the examination of the inhibitory current.
Figure 4, A1 (loose patch), A2 (voltage clamp with the holding potential at −74 mV), and A3 (voltage-clamp with the holding potential at +10 mV), shows a representative example of the series of experiments conducted on each abductor MN. All the traces show rhythmicity in conjunction with VR activity. The excitatory inputs (Fig. 4A2, red trace) consisted of the following two components: tonic and phasic (Fig. 4A2, right-most edge). During swimming, the tonic component was maintained at a near-constant value. On top of this tonic component, the MN received rhythmic phasic excitation. The inhibitory inputs are composed of phasic inputs only.
Figure 4.
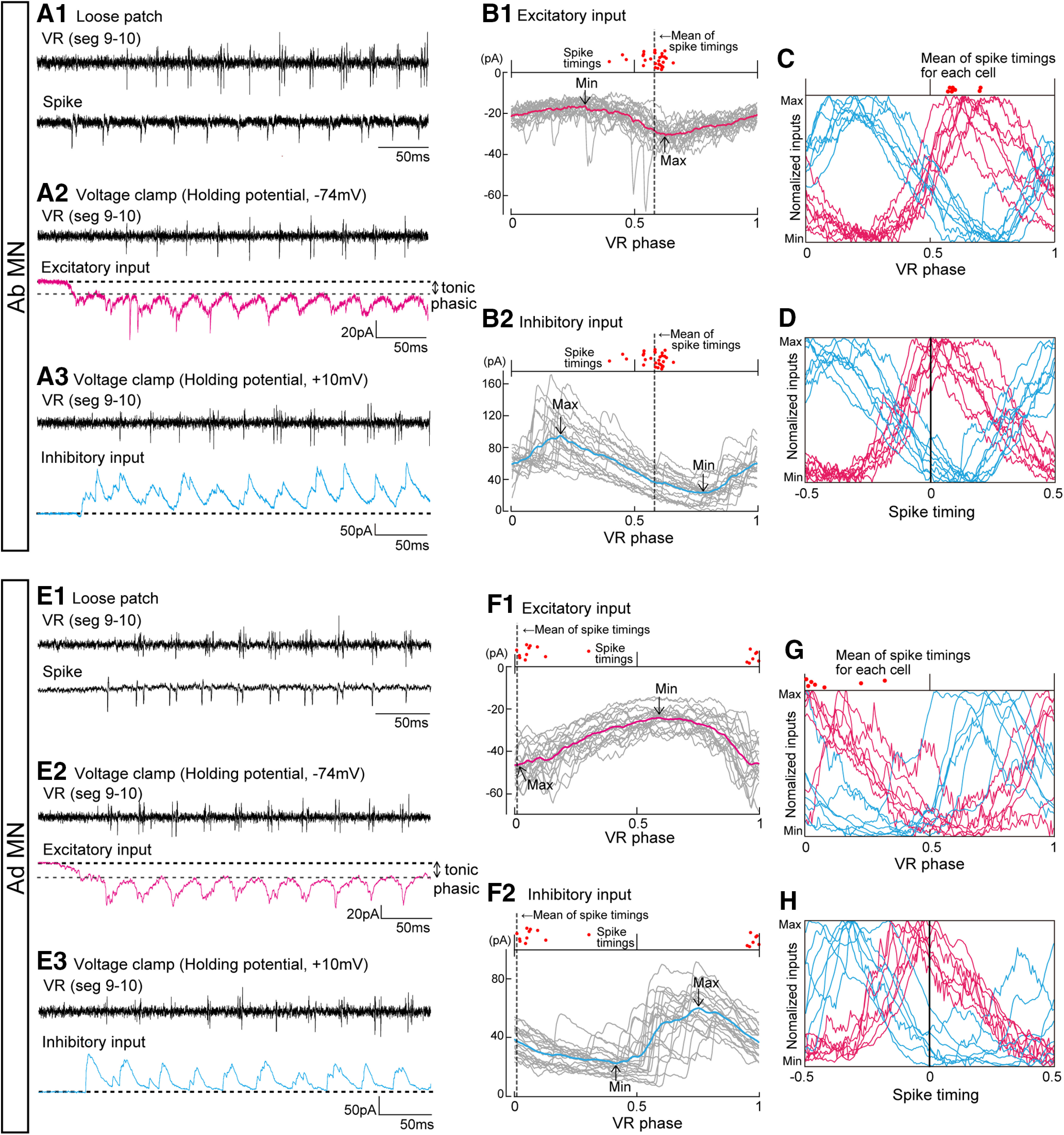
Voltage-clamp recordings from abductor and adductor MNs during fictive swimming. A1, An example of the loose-patch recordings from the abductor MN. A2, Voltage-clamp recording of the same cell with the holding potential at −74 mV. A3, Voltage-clamp recording of the same cell with the holding potential at +10 mV. B1, Twenty traces of voltage-clamp (holding potential at −74 mV) recording from the same cell shown in A are aligned by swim cycles. The average is shown in red. Spike timings (n = 30) of the same cell are shown at the top of the graph (red dots). Mean timing of the spikes is indicated as a dotted line. B2, Same as B1, but with the holding potential at +10 mV. C, Averaged voltage-clamp traces for eight abductor MNs. The vertical axis is normalized to the maximum and minimum values for each trace. Averages of the spike timings of the cells are shown at top of the graph (red dots). D, Same as C, but each trace is aligned with the average spike timing of each cell. E1–E3, Same as A1, A2, and A3, but for the adductor MN. F1, Same as B1 (holding potential at −74 mV), but for the adductor MN (the same cell for E1, E2 and E3). F2, Same as F1, but with the holding potential at +10 mV. G, Same as C, but in adductor MNs (n = 7). H, Same as G, but each trace is aligned with the average spike timing of each cell.
For more quantitative phase analyses, excitatory or inhibitory inputs were aligned by cycles, and 20 traces were superimposed (Fig. 4B1,B2, gray traces). Then, the average of the traces was calculated (Fig. 4B1, excitatory inputs, red trace, B2 for inhibitory inputs, blue trace). Spike timings (30 spikes) of the MNs recorded in loose-patch recordings are indicated by dots at the top of the graphs. On the graph of the excitatory input (Fig. 4B1), the mean spike timing (dotted line) is located near the peak of the inward currents (Max). On the graph of the inhibitory inputs (Fig. 4B2), the mean spike timing is located in the region where the large outward currents are mostly over, and the current value is approaching the trough (Min). Figure 4C shows population data (n = 8). Each trace, which is normalized to Min and Max, represents one cell. The mean spike timing of each MN is shown at the top of the graph (red dots). As shown in the example data (Fig. 4B1,B2), the mean spike timing tends to be located (1) near the peak of the excitatory inputs and (2) around the region where the value of inhibitory inputs approaches the trough. This trend is more clearly seen when the traces of individual cells are aligned with the mean spike timing of the cells (Fig. 4D).
Figure 4E1–E3 shows a representative example of the series of experiments conducted on adductor MNs. Essentially, the same phenomena that were observed in the abductor MNs (Fig. 4A1–A3) were observed in the adductor MNs as well. In Figure 4, F1 and F2, the traces of excitatory (Fig. 4F1) or inhibitory (Fig. 4F2) inputs are aligned per cycle. The mean spike timing (dotted line) is located near the peak of the excitatory inputs and around the region where the value of inhibitory inputs is approaching the trough. Figure 4G shows population data (n = 7). Compared with the corresponding data in abductor MNs (Fig. 4C), the traces are rather scattered in the VR phase-aligned graph. This is likely because of the fact that the spike timings of adductor MNs are less tightly clustered than those of abductor MNs (Fig. 3E,F, dispersed red dots at the top of the graph). When traces are aligned with the mean spike timings of the individual cells (Fig. 4H), the same trends observed in abductor MNs are apparent: namely, spike timing tends to be located (1) near the peak of the excitatory inputs and (2) around the region where the value of inhibitory inputs is approaching the trough.
Together, the results indicate that both abductor and adductor MNs received rhythmic excitatory synaptic inputs in their active phase and inhibitory inputs in their inactive phase. This is consistent with the idea that the spike timings of MNs are determined by push-pull control originating from excitatory and inhibitory inputs (Endo and Kiehn, 2008; Johnson et al., 2012).
dmrt3a neurons are candidates that provide inhibition onto abductor MNs
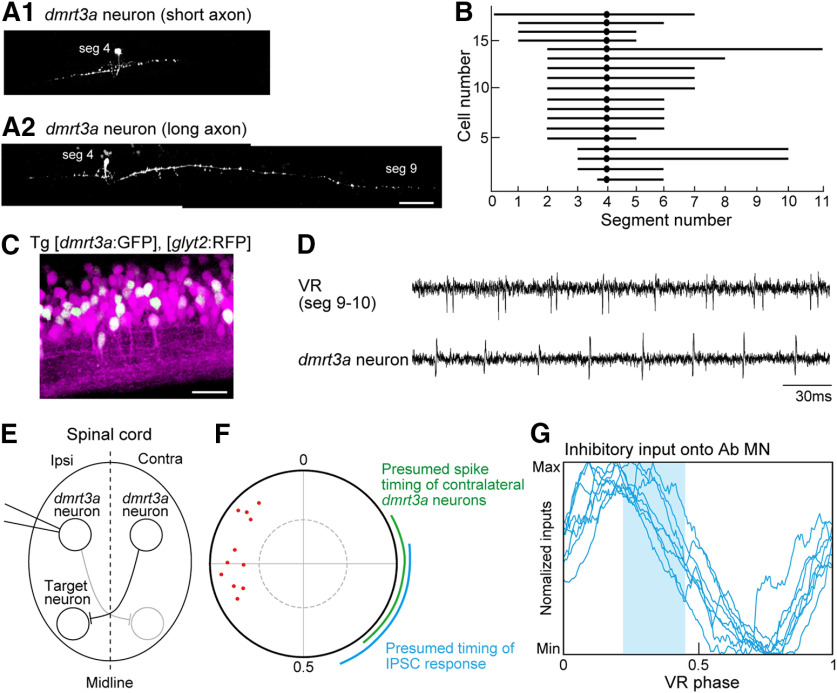
We then sought to identify the upstream sources of interneurons that provide synaptic inputs onto fin MNs by the following strategy. Using a collection of transgenic fish (Satou et al., 2013), we examined the firing patterns of each of several classes of interneurons. Then, we asked whether the phase of their spikes could be correlated with the synaptic inputs onto fin MNs. If a tight correlation was observed, that class of interneurons was treated as a candidate source of synaptic inputs onto fin MNs. We focused on abductor MNs because the synaptic inputs onto abductor MNs are well clustered in the VR phase-aligned graph (Fig. 4, compare C, G), making the exploration of candidate neurons easier. Through our exploration, we found that neurons expressing dmrt3a may convey inhibition to abductor MNs. Below, before presenting the electrophysiological data, we first describe what is known about dmrt3a neurons to date and provide the results of our morphologic analysis of drmt3a neurons in fin MN residing areas.
In the mid-trunk, we have recently shown that (1) dmrt3a neurons are commissural inhibitory neurons, and that (2) they play a role in providing mid-cycle inhibition to contralateral axial MNs and CPG interneurons during fictive swimming, thereby preventing cocontraction of the left and right sides of the body (Satou et al., 2020). It has also been shown that dmrt3a mutant larvae exhibit impaired swimming with decreased velocity and acceleration (Del Pozo et al., 2020). We investigated the morphology of dmrt3a neurons in the fin MN residing area (near muscle segment 4), and found that all were commissural neurons, as in the trunk. Two examples of the axonal morphology are shown in Figure 5, A1 and A2. The summary plot of the axonal lengths is shown in Figure 5B: the average axonal length in the fin MN residing area was 5.9 ± 1.7 segments, which was significantly shorter than that in the trunk (8.1 ± 3.2 segments; Satou et al., 2020; p = 0.011, t test). As in the trunk, dmrt3a neurons are positive for glyt2 (Fig. 5C), indicating that they are glycinergic inhibitory neurons.
Figure 5.
Properties of dmrt3a neurons in the fin region containing MNs. A1, A2, Two examples of the morphology of dmrt3a neurons. Scale bar, 50 µm. B, Ball and stick plots of the somata locations and axon projection distances of dmrt3a neurons. C, Stacked image of confocal optical sections of the compound transgenic fish of Tg[dmrt3a:GFP] and Tg[glyt2:loxP-DsRed-loxP-GFP] (Tg[glyt2:RFP]). Lateral view of the spinal cord around muscle segments 4–5. Scale bar, 20 µm. D, An example of loose-patch recordings from a dmrt3a neuron. E, Schematic representation of the recording of a dmrt3a neuron on the ipsilateral side, a dmrt3a neuron on the contralateral side, and its potential synaptic target neuron on the ipsilateral side. F, Circular plot showing spike timing of dmrt3a neurons (n = 11). The green arc shows the presumed spike timing of contralateral dmrt3a neurons. The blue arc shows the presumed timing of the synaptic transmission from the dmrt3a neurons. G, The presumed timing of the synaptic transmission from dmrt3a neurons (shaded in blue) is imposed on the normalized traces of inhibitory currents for abductor MNs (the same data are shown in Fig. 4C).
We now present our electrophysiological data on dmrt3a neurons in the rostral spinal cord during fictive swimming. Figure 5D shows a representative example. The dmrt3a neurons show rhythmic spiking activities. The circular plot of the population data shows that spiking activities of dmrt3a neurons are clustered at ∼0.67–0.90 (Fig. 5F). Given that dmrt3a neurons are commissural neurons, their synaptic target neurons receive inputs from the dmrt3a neurons located on the contralateral side (Fig. 5E). This means that we need to consider the firing timings of contralaterally located dmrt3a neurons. Considering the symmetry of the spinal cord, the timings are presumed to be 180° opposite each other (Fig. 5F, green arc; timing values, 0.17–0.40). Assuming that the delay of the IPSC response from the spike is ∼1.5 ms (1 ms of conduction time; 0.5 ms of synaptic transmission time; see Materials and Methods), the timing of the IPSC response is presumed to be 0.22–0.45 (Fig. 5F, blue arc). This timing corresponds to the late phase of the inhibition received by the abductor MNs (Fig. 5G), raising the possibility that dmrt3a neurons participate in conveying this inhibition to abductor MNs in accordance with this timing.
dmrt3a neurons make synaptic connections onto abductor MNs
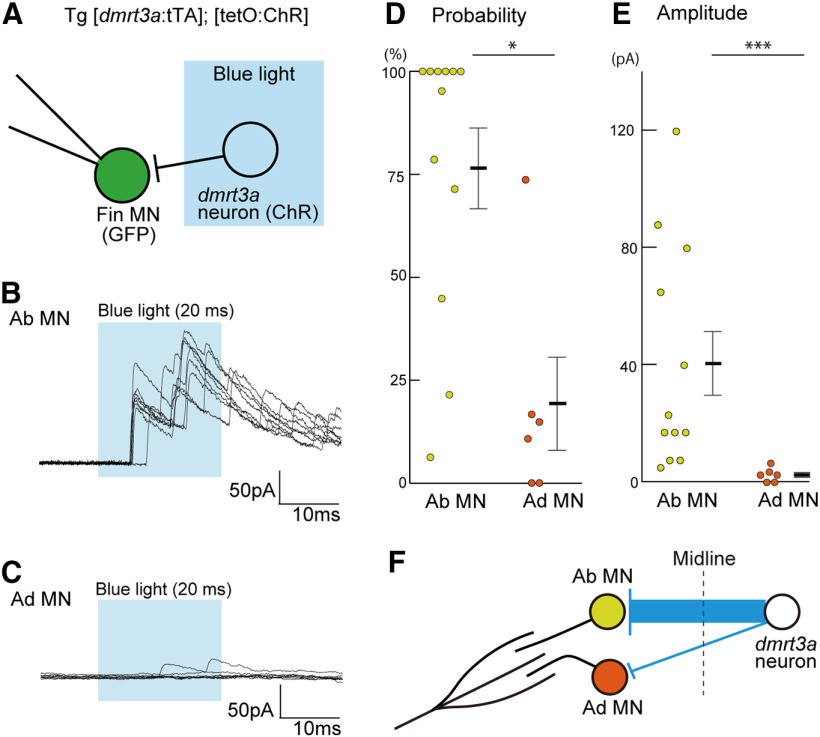
We expected that dmrt3a neurons would make synaptic inhibitory connections onto the contralateral abductor MNs. To test this hypothesis, we prepared animals that expressed ChR in dmrt3a neurons using the Tet system. We then applied brief photostimulation to elicit action potentials in ChR-expressing neurons, while performing voltage-clamp recordings in fin MNs to examine connectivity (Fig. 6A). Holding potential was set at +10 mV to reveal inhibitory currents.
Figure 6.
dmrt3a neurons form strong inhibitory synaptic connections onto abductor MNs. A, Experimental scheme. ChR was expressed in dmrt3a neurons using the Tet system. Whole-cell voltage-clamp recording with the holding potential at +10 mV was made for an abductor or adductor MN. A 20 ms pulse of blue light illumination was applied near the region surrounding the recorded neuron (covering muscle segments 3.5–5.5). B, An example whole-cell recording from an abductor MN. Eight traces are superimposed. C, Same as B, but for adductor MNs. D, Connection probability from dmrt3a neurons to abductor or adductor MNs. Each circle represents one cell (n = 12 for abductor MNs; n = 6 for adductor MNs). *p < 0.05 (Mann–Whitney U test, p = 0.013). E, Amplitude of the peak currents recorded in abductor or adductor MNs. The amplitude presented is the mean of the peak currents in the trials for each cell. Each circle represents one cell (n = 12 for abductor MNs; n = 6 for adductor MNs). ***p < 0.005 (Mann–Whitney U test, p = 0.0017). F, A schematic illustration of the connections from dmrt3a neurons to abductor or adductor MNs.
We obtained recordings from 12 abductor MNs and 6 adductor MNs. In the experiments for the abductor MNs, large inhibitory currents were observed in most cases on photostimulation (Fig. 6B). By contrast, small or negligible inhibitory currents were observed in the case of adductor MNs (Fig. 6C). Figure 6, D and E, shows population data. In both connection probability and current amplitude, there were significant differences between abductor and adductor MNs. These results indicate that dmrt3a neurons make strong synaptic connections onto abductor MNs (Fig. 6F). Synaptic connections onto adductor MNs, on the other hand, are very sparse or weak if they are formed at all (Fig. 6F).
Genetic ablation of dmrt3a neurons in the rostral spinal cord impairs firing patterns of abductor MNs
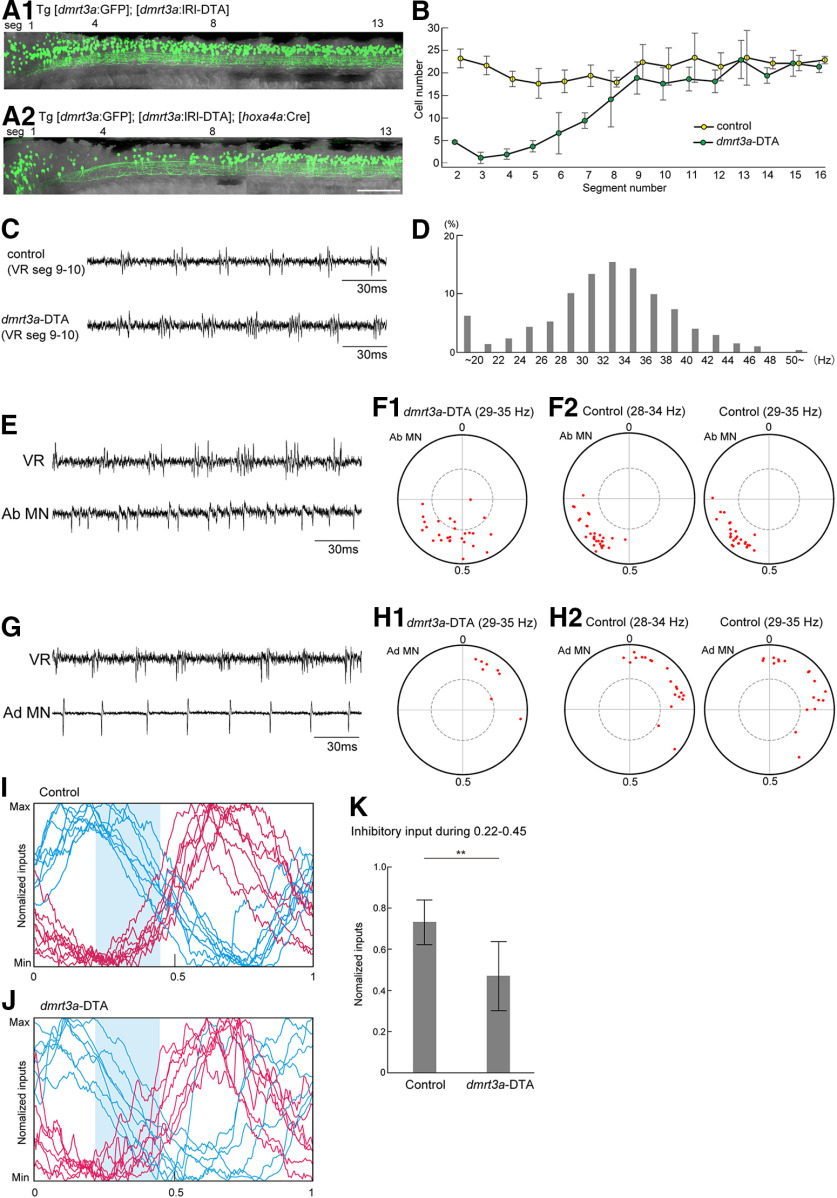
The results we have described are consistent with the idea that dmrt3a neurons are involved in conveying inhibition to abductor MNs on the contralateral side. The timing of this possible inhibition corresponds to the late phase of the inhibition received by abductor MNs (Fig. 5G). This would imply that the loss of dmrt3a neurons should suppress inhibition in this phase, leading to earlier and prolonged firings of abductor MNs. We tested this hypothesis by selective ablation of dmrt3a neurons in the rostral spinal cord by crossing Tg[dmrt3a:loxP-DsRed-loxP-DTA (lRl-DTA)] (DTA, diphtheria toxin A subunit) to Tg[hoxa4a:Cre], in which Cre is mainly expressed in the rostral spinal cord. Successful ablation of dmrt3a neurons (loss of GFP neurons) in the rostral spinal cord in these compound fish was verified using a triple transgenic fish (Tg[dmrt3a:lRl-DTA], Tg[hoxa4a:Cre], and Tg[dmrt3a:GFP] fish; Fig. 7, compare A1 and A2). Examination of the number of dmrt3a neurons in each segment (Fig. 7B) shows that the loss of dmrt3a neurons is severe in the rostral spinal cord (around segments 2–7). dmrt3a neurons in segments 8–12 were mildly affected, while dmrt3a neurons in the more caudal region (caudal to segment 13) were hardly affected.
Figure 7.
Genetic ablation of dmrt3a neurons in the rostral spinal cord alters the spiking patterns of abductor MNs. A1, A2, Fluorescent images (green channel) of compound transgenic fish of Tg[dmrt3a:GFP] and Tg[dmrt3a:lRl-DTA] without (A1) or with (A2) the cross to Tg[hoxa4a:Cre]. In the presence of Tg[hoxa4a:Cre], Cre-mediated recombination occurred in the rostral spinal cord, leading to the expression of DTA. This resulted in ablation of dmrt3a (GFP-positive) neurons in the rostral spinal cord (A2). Scale bar, 100 µm. B, Numbers of dmrt3a neurons in each hemisegment in control and dmrt3a-DTA fish (n = 4 for each fish). C, VR recordings from control and dmrt3a-DTA fish. D, Histogram of fictive swimming frequency (1864 cycles from four fish) in dmrt3a-DTA fish. E, An example of the loose-patch recordings from an abductor MNs in dmrt3a-DTA fish. F1, Circular plot showing spike timing of abductor MNs in dmrt3a-DTA fish (n = 27). Swim cycles of 29–35 Hz were subject to the analysis. F2, Circular plot showing spike timing of abductor MNs in control fish. In the left panel, swim cycles of 28–34 Hz were subject to the analysis (n = 30). The same data are shown in Figure 3E. In the right panel, swim cycles of 29–35 Hz were subject to the analysis (n = 27). G, An example loose-patch recording from an adductor MN in a dmrt3a-DTA fish. H1, Same as F1, but for adductor MNs (n = 8). H2, Same as F2, but for adductor MNs. Left, n = 20; right, n = 18. I, Voltage-clamp recordings from abductor MNs in control fish (the same data are shown in Fig. 4C). Shaded area represents the timing of 0.22–0.45. J, The same as I, but for dmrt3a-DTA fish. K, Normalized inhibitory inputs in the timing of 0.22–0.45 in control and dmrt3a-DTA fish. **p < 0.01 (t test, p = 0.0028).
In our previous study, in which almost the entire population of dmrt3a neurons in the spinal cord was ablated, rhythmic fictive swimming was often disorganized (Satou et al., 2020). In the DTA fish used in the current study (Fig. 7A2; called dmrt3a-DTA, hereafter), however, rhythmic swimming remained almost normal (Fig. 7C). Figure 7D shows the histogram of fictive swimming frequency in dmrt3a-DTA fish. Compared with the wild type (Fig. 3B), swimming frequency was slightly higher in these fish (30.8 ± 7.8 for control vs 32.0 ± 8.1 for dmrt3a-DTA).
We examined firing patterns of abductor and adductor MNs in dmrt3a-DTA fish. Figure 7, E and G, shows representative examples of the recordings. Figure 7, F and H, shows circular plots of the MNs recorded. For dmrt3a-DTA fish, spikes that occurred during swim cycles of the most common swimming frequency (29–35 Hz) were analyzed (Fig. 7F1,H1). For control fish, swim cycles in the following two swimming frequency ranges were analyzed: (1) the most common swimming frequency (28–34 Hz; Fig. 7F2,H2, left panels; these are the same data shown as in Fig. 3E,F); and (2) the swimming frequency at which the analyses in dmrt3a-DTA fish were performed (29–35 Hz; Fig. 7F2,H2, right panels). Swim cycles in the latter frequency range were analyzed to exclude the possibility that the observed phenotypes (described below) were because of a difference in swimming frequency.
In the case of abductor MNs, regular rhythmic firing patterns are often disorganized in dmrt3a-DTA fish (Fig. 7E). Vectors in the circular plot were broadly dispersed (Fig. 7, compare F1, F2). The average length of the vectors was significantly shorter than that of control fish (0.62 ± 0.17 for dmrt3a-DTA vs 0.79 ± 0.08 for control of 28–34 Hz, p = 1.9 × 10−5, t test; 0.79 ± 0.07 for control of 29–35 Hz, p = 1.3 × 10−5, t test). In dmrt3a-DTA fish, several vectors did not reach significance level (Fig. 7F1, red dots within the dotted circles). These results indicate that phase-locked firings of abductor MNs were disorganized in dmrt3a-DTA fish. The averaged direction of the vectors was shifted toward an earlier timing (0.54 ± 0.11 for dmrt3a-DTA, 0.63 ± 0.06 for control at 28–34 Hz; 0.60 ± 0.09 for control at 29–35 Hz). Collectively, the results are consistent with the prediction: firings of abductor MNs occur earlier in a cycle with prolonged duration of the active phase.
In adductor MNs, by contrast, rhythmic firing patterns were mostly maintained in dmrt3a-DTA fish (Fig. 7G). The distribution of the vectors in the circular plot was not severely disorganized (Fig. 7H1,H2). The average length of the vectors did not change significantly (0.76 ± 0.13 for dmrt3a-DTA vs 0.79 ± 0.09 for control at 28–34 Hz, p = 0.46, t test; 0.77 ± 0.09 for control at 29–35 Hz, p = 0.92, t test). The averaged direction of the vectors was likewise not markedly different (0.12 ± 0.08 for dmrt3a-DTA, 0.14 ± 0.11 for control at 28–34 Hz; 0.15 ± 0.13 for control at 29–35 Hz). These results indicate that the firing patterns of adductor MNs were less strongly affected than those of abductor MNs in dmrt3a-DTA fish.
We further performed voltage-clamp recordings of abductor MNs in dmrt3a-DTA fish to directly show that late-phase inhibition was reduced. The comparison between Figure 7, I (the summary data from control fish; the same as in Fig. 4C) and J (the data from dmrt3a-DTA fish) suggests that late-phase (0.22–0.45; shaded area) inhibition was reduced in dmrt3a-DTA fish. To quantify these differences, the average amplitude of inhibition at this phase (0.22–0.45) was normalized to the maximum amplitude of inhibition. As shown in Figure 7K, the inhibitory inputs at the timing of 0.22–0.45 were significantly reduced in dmrt3a-DTA fish.
Discussion
In this study, we set out to investigate neuronal circuits underlying rhythmic pectoral fin movements in larval zebrafish. We showed that both abductor and adductor MNs received rhythmic excitatory synaptic inputs in their active phase and inhibitory inputs in their inactive phase. This indicates that MN activities are controlled in a push-pull manner. We also obtained evidence strongly suggesting that inhibition originating from dmrt3a-expressing commissural inhibitory neurons plays an important role in regulating abductor MN activities. The current study marks the first step toward the identification of CPG organization for rhythmic pectoral fin movements.
Topographic relationships between MN position and innervation area
Our Kaede photoconversion experiments revealed topographic innervation of fin MNs such that the rostrocaudal dimension of cell body locations is converted to the dorsoventral dimension in the innervation area of the fin. This may be important for enabling fine-tuned control of fin movements in the late stage of life. In the early larval stage (3–5 dpf), rhythmic movements of the pectoral fins appear to consist of simple abduction and adduction. A primary role of this movement in early-stage larvae may be to assist in cutaneous respiration by stirring the water (Green et al., 2011). In the later stages (7 dpf and onward), pectoral fin movements have been shown to generate lift (force that moves the fish upward) during swimming (Ehrlich and Schoppik, 2019), suggesting the capacity for fine-tuned control of the fin (e.g., twisting movements). To produce such sophisticated movements, the dorsal (i.e., the leading edge in adults) and ventral (i.e., the trailing edge in adults) edges of the fin need to be contracted according to different timings. Topographic innervation may play an important role in enabling such fine control of the fins by independently regulating the activities of rostrally located and caudally located MNs.
Properties of firing patterns of abductor and adductor MNs
Abductor and adductor MN activities are essentially alternated during rhythmic fin movements. This is analogous to the alternated activities seen in flexor and extensor MNs during mammalian walking (Kiehn, 2016) or scratching in turtles (Stein, 2010), but the fin MNs exhibit a much simpler organization, with only two types of MNs, each of which innervates either the flexor and extensor muscles. Spiking activities of adductor MNs are more broadly distributed than those of abductor MNs (Fig. 3E,F). At each cell level, however, the strength of the rhythmicity (i.e., the length of the vectors in the circular plot) in adductor MNs is comparable to that in abductor MNs. This indicates that preferred firing timings (but not strength of the rhythmicity) are more broadly distributed in adductor MNs. The differences in spike timing among adductor MNs are likely because of the differences in the timing of synaptic inputs to each cell (Fig. 4I,H). This suggests that upstream interneurons controlling adductor MN firings are more heterogeneous than those controlling abductor MN firings. It should be noted that our recordings were taken from fin MNs located exclusively in segment 4 and that, therefore, MN location could not have been the main cause of heterogeneity. Nevertheless, adductor MNs in segment 4 have wider innervation fields than abductor MNs have, which introduces the possibility that this projection difference may have resulted in a somewhat wider distribution of spike timing among adductor MNs.
Synaptic inputs to abductor and adductor MNs
Our voltage-clamp recordings from fin MNs showed that these MNs receive rhythmic excitation and inhibition. For both abductor and adductor MNs, excitatory synaptic inputs peaked in the active phase and inhibitory inputs peaked in the inactive phase (Fig. 4D,H). This finding strongly suggests that firings of the abductor and adductor MNs are controlled by an alternating combination of excitatory and inhibitory inputs (i.e., push-pull control; Johnson et al., 2012). This is consistent with previous observations during scratching in turtles (Robertson and Stein, 1988) and walking in mammals (Shefchyk and Jordan, 1985; Cazalets et al., 1996; Hochman and Schmidt, 1998; Endo and Kiehn, 2008).
Crossed inhibition from dmrt3a neurons is involved in controlling spike timings of abductor MNs on the contralateral side
Neurons expressing dmrt3a are commissural inhibitory neurons. Firing timings of dmrt3a neurons on the contralateral side matched the late phase of inhibitory inputs received by abductor MNs, raising the possibility that dmrt3a neurons contribute to the inhibition during the corresponding timing. If this is the case, we can make the following predictions: (1) there are inhibitory synaptic connections between dmrt3a neurons and abductor MNs; and (2) the ablation of dmrt3a neurons leads to earlier and prolonged firings of abductor MNs. As described below, our experimental results support these hypotheses.
ChR-mediated photostimulation experiments showed that dmrt3a neurons form strong, presumably monosynaptic, inhibitory synaptic connections onto abductor MNs. Ablation of dmrt3a neurons in the rostral spinal cord resulted in earlier activations of abductor MNs in swim cycles. In addition to this shift in spike timing, the strength of rhythmicity (length of vectors in a circular plot) was degraded (Fig. 7F1). This finding suggests that firings of abductor MNs are prolonged in dmrt3a-DTA fish. We also directly measured the inhibitory currents from abductor MNs in dmrt3a-DTA fish by performing voltage-clamp recordings. Our result showed that the inhibitory inputs onto abductor MNs were significantly reduced during the expected timing (Fig. 7K). In summary, all the results are consistent with our prediction.
In the classic half-center model, reciprocal inhibition between the flexor module and the extensor module within a limb is considered to play the central role in controlling flexor/extensor alternation (Brown, 1911). According to this model, one can imagine that the interneurons controlling the firing timings of flexor and extensor MNs would be confined to the corresponding half of the spinal cord. Contrary to that expectation, however, it has been shown that neuronal circuits on the contralateral side are required to generate proper activities of the flexor and extensor MNs during scratching movements in turtles (note that this is a unilateral movement; Stein et al., 1995). This led the authors to propose that the neuronal circuits controlling the precise firing timings of flexor and extensor MNs are bilaterally distributed. Our results are compatible with that conclusion, and further show that this bilateral distribution reaches to the lowest level of CPG (the last order interneurons) in zebrafish fin motor system. We expect that a similar configuration is present in the CPG controlling walking in mammals.
Alternation of the left and right fins is the only gait that early larval zebrafish exhibit. Older zebrafish, however, can exhibit synchronous gait, as can the adults of many other fish species (Hale et al., 2006; Ehrlich and Schoppik, 2019). Further study is needed to reveal whether or how drmt3a neurons are involved in synchronous gait.
dmrt3a neurons in axial and pectoral fin movements
We have previously shown that dmrt3a neurons are involved in axial swimming movements by providing mid-cycle inhibition to the contralateral side (Satou et al., 2020). Ablation of dmrt3a neurons throughout the spinal cord resulted in swimming patterns that were often disorganized, with frequent coactivation of the left and right axial MNs. In this study, we restricted the neuronal ablation area to the rostral spinal cord only to minimize the effects on swimming for our phenotypic analyses of fin movements.
Currently, it is unclear whether dmrt3a neurons controlling abductor MN activities are also involved in the regulation of axial MN activities. Our morphologic analysis of dmrt3a neurons in the fin region where MNs reside showed that the average axonal length of dmrt3a neurons is shorter in this region than in the mid-trunk. Some dmrt3a neurons have very short axons (Fig. 5A1). This suggests that these dmrt3a neurons may be dedicated to the control of fin movements.
Evolutionary considerations
dmrt3a is a zebrafish homolog of mammalian dmrt3. The basic features of dmrt3 neurons are conserved across vertebrate species. dmrt3 neurons in mammals are predominantly commissural and inhibitory, and have been implicated in left–right coordination, specifically, in providing inhibition to CPG and MNs (Andersson et al., 2012; Vallstedt and Kullander, 2013; Perry et al., 2019). Detailed characterization of synaptic targets, however, has not been achieved.
The present study has shown that dmrt3a neurons form strong inhibitory synaptic connections onto abductor MNs. The observed gene expression pattern suggests that abductor MNs correspond to the MNs innervating muscles that are derived from ventrally located muscle precursors in the fin/limb, while adductor MNs correspond to the MNs innervating muscles that are derived from dorsally located muscle precursors [one example of this evidence is that lhx1a in zebrafish is expressed in adductor MNs (Fig. 2E), while lhx1 in mammals is expressed in the MNs that innervate muscles derived from dorsal precursors in mammals]. Ventrally and dorsally located muscle precursors in mammals predominantly produce flexors and extensors, respectively (Jung et al., 2018). It would be interesting to ask whether dmrt3a neurons in mammals preferentially innervate flexor-innervating MNs and control their firing timings during walking.
Footnotes
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and from National BioResource Project (NBRP) and NBRP/Fundamental Technologies Upgrading Program from the Japan Agency for Medical Research and Development. We thank Dr. Kamikouchi for continuous encouragement and discussion. We also thank the Spectrography and Bioimaging Facility of the National Institute for Basic Biology Core Research Facilities for providing technical support.
The authors declare no competing financial interests.
References
- Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin C-J, Patra K, Arnason T, Wellbring L, Hjälm G, Imsland F, Petersen JL, McCue ME, Mickelson JR, Cothran G, Ahituv N, Roepstorff L, Mikko S, Vallstedt A, Lindgren G, et al. (2012) Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488:642–646. 10.1038/nature11399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S. (2012) Motor circuits in action: specification, connectivity, and function. Neuron 74:975–989. 10.1016/j.neuron.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K (2008) Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A 105:1255–1260. 10.1073/pnas.0704963105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TG. (1911) The intrinsic factors in the act of progression in the mammal. Proc Royal Acad Sci 84:308–319. [Google Scholar]
- Cazalets JR, Borde M, Clarac F (1996) The synaptic drive from the spinal locomotor network to motoneurons in the newborn rat. J Neurosci 16:298–306. 10.1523/JNEUROSCI.16-01-00298.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pozo A, Manuel R, Iglesias Gonzalez AB, Koning HK, Habicher J, Zhang H, Allalou A, Kullander K, Boije H (2020) Behavioral characterization of dmrt3a mutant zebrafish reveals crucial aspects of vertebrate locomotion through phenotypes related to acceleration. eNeuro 7:ENEURO.0047-20.2020 10.1523/ENEURO.0047-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich DE, Schoppik D (2019) A primal role for the vestibular sense in the development of coordinated locomotion. eLife 8:e45839 10.7554/eLife.45839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Kiehn O (2008) Asymmetric operation of the locomotor central pattern generator in the neonatal mouse spinal cord. J Neurophysiol 100:3043–3054. 10.1152/jn.90729.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M. (2009) Circuits controlling vertebrate locomotion: moving in a new direction. Nat Rev Neurosci 10:507–518. 10.1038/nrn2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M, Pfaff SL (2005) Development of circuits that generate simple rhythmic behaviors in vertebrates. Curr Opin Neurobiol 15:14–20. 10.1016/j.conb.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Green MH, Hale ME (2012) Activity of pectoral fin motoneurons during two swimming gaits in the larval zebrafish (Danio rerio) and localization of upstream circuit elements. J Neurophysiol 108:3393–3402. 10.1152/jn.00623.2012 [DOI] [PubMed] [Google Scholar]
- Green MH, Ho RK, Hale ME (2011) Movement and function of the pectoral fins of the larval zebrafish (Danio rerio) during slow swimming. J Exp Biol 214:3111–3123. 10.1242/jeb.057497 [DOI] [PubMed] [Google Scholar]
- Grillner S. (1975) Locomotion in vertebrates: central mechanisms and reflex interaction. Physiol Rev 55:247–304. 10.1152/physrev.1975.55.2.247 [DOI] [PubMed] [Google Scholar]
- Grillner S. (2003) The motor infrastructure: from ion channels to neuronal networks. Nat Rev Neurosci 4:573–586. 10.1038/nrn1137 [DOI] [PubMed] [Google Scholar]
- Hale ME, Day RD, Thorsen DH, Westneat MW (2006) Pectoral fin coordination and gait transitions in steadily swimming juvenile reef fishes. J Exp Biol 209:3708–3718. 10.1242/jeb.02449 [DOI] [PubMed] [Google Scholar]
- Hochman S, Schmidt BJ (1998) Whole cell recordings of lumbar motoneurons during locomotor-like activity in the in vitro neonatal rat spinal cord. J Neurophysiol 79:743–752. 10.1152/jn.1998.79.2.743 [DOI] [PubMed] [Google Scholar]
- Johnson MD, Hyngstrom AS, Manuel M, Heckman CJ (2012) Push-pull control of motor output. J Neurosci 32:4592–4599. 10.1523/JNEUROSCI.4709-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Baek M, D'Elia KP, Boisvert C, Currie PD, Tay B-H, Venkatesh B, Brown SM, Heguy A, Schoppik D, Dasen JS (2018) The ancient origins of neural substrates for land walking. Cell 172:667–682.e15. 10.1016/j.cell.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehn O. (2006) Locomotor circuits in the mammalian spinal cord. Annu Rev Neurosci 29:279–306. 10.1146/annurev.neuro.29.051605.112910 [DOI] [PubMed] [Google Scholar]
- Kiehn O. (2016) Decoding the organization of spinal circuits that control locomotion. Nat Rev Neurosci 17:224–238. 10.1038/nrn.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Higashijima S-i (2019) Regulation of locomotor speed and selection of active sets of neurons by V1 neurons. Nat Commun 10:2268. 10.1038/s41467-019-09871-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Satou C, Fujioka S, Shoji W, Umeda K, Ishizuka T, Yawo H, Higashijima SI (2013) Hindbrain V2a neurons in the excitation of spinal locomotor circuits during zebrafish swimming. Curr Biol 23:843–849. 10.1016/j.cub.2013.03.066 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S-i (2014) Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep 4:6545. 10.1038/srep06545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff O, Kiehn O (1996) Distribution of networks generating and coordinating locomotor activity in the neonatal rat spinal cord in vitro: a lesion study. J Neurosci 16:5777–5794. 10.1523/JNEUROSCI.16-18-05777.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menelaou E, McLean DL (2019) Hierarchical control of locomotion by distinct types of spinal V2a interneurons in zebrafish. Nat Commun 10:4197. 10.1038/s41467-019-12240-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsubori A, Tsutsui-Kimura I, Nishida H, Bouchekioua Y, Sekiya H, Uchigashima M, Watanabe M, de Kerchove d'Exaerde A, Mimura M, Takata N, Tanaka KF (2017) Ventrolateral striatal medium spiny neurons positively regulate food-incentive, goal-directed behavior independently of D1 and D2 selectivity. J Neurosci 37:2723–2733. 10.1523/JNEUROSCI.3377-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S, Larhammar M, Vieillard J, Nagaraja C, Hilscher MM, Tafreshiha A, Rofo F, Caixeta FV, Kullander K (2019) Characterization of Dmrt3-derived neurons suggest a role within locomotor circuits. J Neurosci 39:1771–1782. 10.1523/JNEUROSCI.0326-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GA, Stein PS (1988) Synaptic control of hindlimb motoneurones during three forms of the fictive scratch reflex in the turtle. J Physiol 404:101–128. 10.1113/jphysiol.1988.sp017281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Kohashi T, Horikawa K, Takeda H, Oda Y, Higashijima S (2009) Functional role of a specialized class of spinal commissural inhibitory neurons during fast escapes in zebrafish. J Neurosci 29:6780–6793. 10.1523/JNEUROSCI.0801-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Higashijima S (2012) Generation of multiple classes of V0 neurons in zebrafish spinal cord: progenitor heterogeneity and temporal control of neuronal diversity. J Neurosci 32:1771–1783. 10.1523/JNEUROSCI.5500-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Hirata H, Suster ML, Kawakami K, Higashijima S (2013) Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development 140:3927–3931. 10.1242/dev.099531 [DOI] [PubMed] [Google Scholar]
- Satou C, Sugioka T, Uemura Y, Shimazaki T, Zmarz P, Kimura Y, Higashijima SI (2020) Functional diversity of glycinergic commissural inhibitory neurons in larval zebrafish. Cell Rep 30:3036–3050.e4. 10.1016/j.celrep.2020.02.015 [DOI] [PubMed] [Google Scholar]
- Shefchyk SJ, Jordan LM (1985) Excitatory and inhibitory postsynaptic potentials in alpha-motoneurons produced during fictive locomotion by stimulation of the mesencephalic locomotor region. J Neurophysiol 53:1345–1355. 10.1152/jn.1985.53.6.1345 [DOI] [PubMed] [Google Scholar]
- Stein PS. (2010) Alternation of agonists and antagonists during turtle hindlimb motor rhythms. Ann N Y Acad Sci 1198:105–118. 10.1111/j.1749-6632.2010.05500.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein PS, Victor JC, Field EC, Currie SN (1995) Bilateral control of hindlimb scratching in the spinal turtle: contralateral spinal circuitry contributes to the normal ipsilateral motor pattern of fictive rostral scratching. J Neurosci 15:4343–4355. 10.1523/JNEUROSCI.15-06-04343.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi A, Kimura Y, Mori I, Nonaka S, Higashijima SI (2017) Axially-confined in vivo single-cell labeling by primed conversion using blue and red lasers with conventional confocal microscopes. Dev Growth Differ 59:741–748. 10.1111/dgd.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen DH, Hale ME (2005) Development of zebrafish (Danio rerio) pectoral fin musculature. J Morphol 266:241–255. 10.1002/jmor.10374 [DOI] [PubMed] [Google Scholar]
- Thorsen DH, Hale ME (2007) Neural development of the zebrafish (Danio rerio) pectoral fin. J Comp Neurol 504:168–184. 10.1002/cne.21425 [DOI] [PubMed] [Google Scholar]
- Thorsen DH, Cassidy JJ, Hale ME (2004) Swimming of larval zebrafish: fin-axis coordination and implications for function and neural control. J Exp Biol 207:4175–4183. 10.1242/jeb.01285 [DOI] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H (2005) Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol 278:587–606. 10.1016/j.ydbio.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174:639–649. 10.1534/genetics.106.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallstedt A, Kullander K (2013) Dorsally derived spinal interneurons in locomotor circuits. Ann N Y Acad Sci 1279:32–42. 10.1111/j.1749-6632.2012.06801.x [DOI] [PubMed] [Google Scholar]
- Wang H, Sugiyama Y, Hikima T, Sugano E, Tomita H, Takahashi T, Ishizuka T, Yawo H (2009) Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J Biol Chem 284:5685–5696. 10.1074/jbc.M807632200 [DOI] [PubMed] [Google Scholar]