Abstract
Background
Neuroblastoma (NB) is a common malignant tumor of the sympathetic nervous system, mainly disturbing children. Long non-coding RNAs (lncRNAs) serving as promising cancer biomarkers have been well recognized. Our study intends to explore the functions of lncRNA X–inactive specific transcript (DLX6-AS1) in NB and provide a potential action mechanism.
Methods
The expression of DLX6-AS1, miR-506-3p and signal transducer and activator of transcription 2 (STAT2) was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation was assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and colony formation assay. Cell cycle distribution was determined by flow cytometry assay. The protein level of cell cycle-related markers and STAT2 was detected by Western blot. Glycolysis progress was evaluated according to glucose consumption, lactate production and ATP level. The target genes were predicted by the online database Starbase3.0 and verified by dual-luciferase reporter assay.
Results
DLX6-AS1 expression was highly elevated in NB tissues and cells. DLX6-AS1 deficiency inhibited NB cell proliferation, cell cycle and glycolysis in vitro. MiR-506-3p was a target of DLX6-AS1, and miR-506-3p absence partly reversed the effects of DLX6-AS1 deficiency. Besides, STAT2 was targeted by miR-506-3p, and its expression was regulated by DLX6-AS1 through miR-506-3p. MiR-506-3p restoration also inhibited NB cell malignant behaviors, and STAT2 overexpression partially abolished the role of miR-506-3p restoration. Moreover, DLX6-AS1 deficiency weakened tumor growth in vivo.
Conclusion
DLX6-AS1 regulated cell proliferation, cell cycle and glycolysis in vitro and tumor growth in vivo to promote the development of NB by upregulating STAT2 via targeting miR-506-3p.
Keywords: DLX6-AS1, miR-506-3p, STAT2, neuroblastoma
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in childhood of neural crest origin,1 accounting for approximately 7% of childhood malignant tumors and 15% of childhood cancer mortality.2 About 60% of NB patients are diagnosed with a high risk of metastases, and these patients remain a therapeutic challenge for pediatric oncologists.3 Despite the fact that intensive treatment with multiple therapies has developed, including radiation, surgery and chemotherapy, the overall survival rate of high-risk patients is still less than 40%.4 NB usually cannot be removed and requires intensive multimodal treatment for children with NB over the age of 18 months. However, children with NB younger than 18 months can be cured with moderate-intensity chemotherapy, and it is possible to regress spontaneously.5,6 Therefore, early diagnosis is of great significance for the treatment of NB, and more specific biomarkers need to be tapped for early diagnosis.
The involvement of long non-coding RNAs (lncRNAs) in cancer initiation or regression has aroused much concern. LncRNAs, over 200 nucleotides, are a class of major non-coding transcripts.7 Until now, abundant lncRNAs have been functionally characterized via gene-specific research, involving multiple biological processes.8 As for the development of NB, several studies documented diagnostic or prognostic lncRNA signatures by RNA sequencing technology.9 Besides, several NB-associated lncRNAs have been functionally identified, such as FOXD3-AS1, HOXD-AS1 and SNHG1.10−12 They are aberrantly expressed in NB and function as tumor suppressors or oncogenes to participate in the pathogenesis of NB. LncRNA Distal-Less Homeobox 6 antisense RNA 1 (DLX6-AS1) is a newly identified oncogene in tumorigenesis of several cancers, including NB.13 However, the functional role of DLX6-AS1 in NB pathogenesis has not been fully elucidated, and the associated mechanism about DLX6-AS1 is limited.
MicroRNAs (miRNAs), 19~22 nucleotides in length, are well-known cellular molecules to mediate gene expression at the post-transcriptional level.14 The implication of miRNAs in cell-specific functions, such as differentiation, division, metastasis and apoptosis, thus taking part in mechanisms of tumor development.15 Several studies maintain that miRNAs are promising candidates of NB biomarkers for non-invasive diagnosis, risk stratification and prognosis monitoring.16 MiR-506-3p acts as a tumor inhibitor that has been extensively studied in common cancers.17,18 Unfortunately, the role of miR-506-3p in NB is obscured, and relevant studies are lacking.
Signal transducer and activator of transcription 2 (STAT2) encodes a protein that induces the transcription of genes in response to the signaling mediation through the interferon pathway.19 STAT2 was proved to harbor oncogenic functions and critically function in cancer progression, including colorectal cancer, glioma, and lung cancer.20–22 Given the critical role of STAT2, it was served as a worthwhile target for cancer therapy. However, the potential role and action mechanisms of STAT2 in NB remain largely unknown.
In our study, we examined the expression of DLX6-AS1 in NB tissues and cells. Besides, the functional role of DLX6-AS1 was investigated in cell proliferation, cycle and glycolysis. Moreover, the relationship among DLX6-AS1, miR-506-3p and STAT2 was determined, thus providing a potential regulatory mechanism of DLX6-AS1 in NB. This paper aimed to obtain several valuable biomarkers for NB treatment.
Materials and Methods
Tissue Collection
Total 31 pairs of NB tissues and adjacent normal tissues were collected from NB patients recruited from Luoyang Central Hospital Affiliated to Zhengzhou University. All excised tissues were immediately dealt with liquid nitrogen and preserved in −80°C conditions. Written informed consent was obtained from each patient or their guardian. This study was conducted with the approval of the Ethics Committee of Luoyang Central Hospital Affiliated to Zhengzhou University. The association between clinical features of NB patients and DLX6-AS1 expression were shown in Table 1.
Table 1.
Association Between Clinical Features and DLX6-AS1 Expression of NB Patients (n=31)
| Parameters | Case | DLX6-AS1 Expressiona | P value | |
|---|---|---|---|---|
| High (n=16) | Low (n=15) | |||
| Age (years) | ||||
| ≤2.5 | 12 | 5 | 7 | 0.3785 |
| >2.5 | 19 | 11 | 8 | |
| Gender | ||||
| Male | 10 | 4 | 6 | 0.3719 |
| Female | 21 | 12 | 9 | |
| INSS stage | ||||
| 1–2 | 18 | 6 | 12 | 0.0166* |
| 3–4S | 13 | 10 | 3 | |
| Lymph node metastasis | ||||
| No | 16 | 5 | 11 | 0.0191* |
| Yes | 15 | 11 | 4 | |
| Differentiation | ||||
| Well | 17 | 12 | 5 | 0.0198* |
| Moderate-poor | 14 | 4 | 10 | |
Notes: *P<0.05. aUsing median expression level of DLX6-AS1 as cutoff.
Cell Culture
NB cell lines, including SK-N-SH and LAN-6, were purchased from BeNa Culture Collection (Beijing, China) and DSMZ (Braunschweig, Germany), respectively. All cells were cultured in 90% Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA) added with 10% fetal bovine serum (FBS; Gibco) at 37°C containing 5% CO2 incubator.
Cell Transfection
Small interference RNA targeting DLX6-AS1 (si-DLX6-AS1: 5ʹ-AAUAAAGAACACUUACACUACUG-3ʹ) together with its negative control (si-NC: 5ʹ- UUCUCCGAACGUGUCACGUTT-3ʹ) were assembled by Ribobio (Guangzhou, China). MiR-506-3p mimics (miR-506-3p; Product ID: miR10002878-1-5), miR-506-3p inhibitors (anti-miR-506-3p; Product ID: miR20002878-1-5) and respective controls, including miR-NC (Product ID: miR1N0000001-1-5) and anti-miR-NC (Product ID: miR2N0000001-1-5) were purchased from Ribobio. The sequences of DLX6-AS1 and STAT2 were amplified and cloned into the pcDNA3.1 overexpression vector, named as pcDNA-DLX6-AS1 and pcDNA-STAT2 (Sangon, Shanghai, China), and pcDNA empty vector was served as the negative control (pcDNA-NC). For stable DLX6-AS1 knockdown, a lentiviral vector containing short hairpin RNA targeting DLX6-AS1 (sh-DLX6-AS1: 5ʹ-GGTTCAGTATAGATTTCTA-3ʹ) and its negative control (sh-NC: 5ʹ- TTCTCCGAACGTGTCACGT-3ʹ) were synthesized by Research-Bio Co., Ltd (Shanghai, China). These oligonucleotides (20 nM) and plasmids (1 µg) were transfected into SK-N-SH and LAN-6 cells (5×104 cells) using Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using the TransZol Kit (TransGen, Beijing, China). Next, reverse transcription PCR was performed using the TransScript First-Strand cDNA Synthesis SuperMix (TransGen) or TransScript miRNA First-Strand cDNA Synthesis SuperMix (TransGen). Subsequently, the TransStart Green qPCR SuperMix was used to conduct the qRT-PCR amplification reaction. The expression data were processed using the 2−ΔΔCT method, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was adopted as the reference. The primer sequences were listed as follows: DLX6-AS1, forward: 5ʹ-AGTTTCTCTCTAGATT-GCCTT-3ʹ and reverse: 5ʹ-ATTGACATGTTAGTGCCCTT-3ʹ; miR-506-3p, forward: 5ʹ-ACACTCATAAGGCACCCTTC-3ʹ and reverse: 5ʹ- TCTACTCAGAAGGGGAGTAC-3ʹ; STAT2, forward: 5ʹ-GCAGCACAATTTG GGAA-3ʹ and reverse: 5ʹ-ACAGGTGTTTCGAGAACTGGC-3ʹ; GAPDH, forward: 5ʹ-AACGTGTCAGTGGTGGACCTG-3ʹ and reverse: 5ʹ- AGTGGGTGTCGCTGTTGAAGT-3ʹ; U6, forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse: 5ʹ-AACGCTTCACGAATTTGCGT −3ʹ. All experiments were repeated in triplicate.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
The MTT Cell proliferation Assay Kit (Beyotime, Shanghai, China) was used to detect cell proliferation capacity. Generally, cells with different transfections were grown in 96-well plates (2000 cells/well) for the desired time (24, 48 and 72 h). Four hours before the end time point of culture, 10 μL MTT solution was added into each well, incubating for another 4 h at 37°C. Afterwards, 100 μL formazan solving liquid was added into each well until the formazan was fully dissolved. The absorbance at 490 nm was recorded using the iMark microplate reader (Bio-Rad, Hercules, CA, USA). All experiments were repeated in triplicate.
Colony Formation Assay
Cells with different transfections were cultured at the logarithmic growth phase and then seeded into 10 cm diameter culture plates at a density of 200 cells/plate. The culture plates were maintained at 37°C containing 5% CO2 incubator. After 12 d, the arisen colonies were fixed using methanol, stained with crystal violet and then counted under a microscope (Olympus, Tokyo, Japan). These experiments were performed in triplicate.
Cell Cycle Detection
Cell cycle was determined through flow cytometry assay using the Cell Cycle Analysis Kit (Beyotime). Briefly, cells with different transfections were collected by trypsinization and fixed with 70% alcohol for 6 h at 4°C. Then, cells were subjected to centrifugation and resuspended with phosphate-buffered saline (PBS). Afterwards, propidium iodide (PI) staining solution containing RNase A was added to stain for 30 min at 37°C away from light on the ice. Finally, cell sorting was executed using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA), and cell distribution in the G1, S and G2/M phases was assessed using CellQuest Pro software (BD Biosciences). These experiments were performed in triplicate.
Western Blot
Western blot was performed to examine the expression of cell cycle-related markers, including Cyclin-dependent kinase 1 (CDK1) and Cyclin D1, and the expression of STAT2. Concisely, total proteins were isolated using the RIPA Lysis Buffer (Beyotime) and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Afterwards, the proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad), followed by non-fat milk. Next, the membranes were probed with the primary antibodies against CDK1 (ab18; Abcam, Cambridge, MA, USA), Cyclin D1 (ab134175; Abcam), STAT2 (ab124283; Abcam), GAPDH (ab9485; Abcam) and secondary antibodies (ab205718, ab205719; Abcam). The protein blots were presented using an enhanced chemiluminescence kit (Beyotime). All experiments were repeated in triplicate.
Glycolysis Detection
Glycolysis was investigated according to glucose consumption, lactate production and ATP level of cells after transfection using Glucose Uptake Assay Kit (Abcam), Lactate Assay Kit-WST (Abcam) and ATP Assay Kit (Abcam) in line with the corresponding protocol. These experiments were performed in triplicate.
Target Gene Prediction and Verification
The target genes of lncRNA or miRNA were predicted using the online bioinformatics tool Starbase3.0 (http://starbase.sysu.edu.cn/).
The targeted relationship was verified by dual-luciferase reporter assay. Briefly, the DLX6-AS1 partial sequences harboring the wild-type binding sites with miR-506-3p were amplified and cloned into pGL4 vector, terming as Luc-DLX6-AS1-WT, and the DLX6-AS1 partial sequences harboring the mutant binding sites with miR-506-3p were also amplified and constructed into pGL4 vector, terming as Luc-DLX6-AS1-MUT. Similarly, Luc-STAT2 3ʹUTR-WT and Luc-STAT2 3ʹUTR-MUT were also assembled. Subsequently, these fusion plasmids together with miR-506-3p or miR-NC were cotransfected into SK-N-SH and LAN-6 cells, respectively. After 48 h, transfected cells were collected and followed by the detection of luciferase activity using the TransDetect Double-Luciferase Reporter Assay Kit (TransGen). All experiments were repeated in triplicate. The primer sequences used for subclone were listed as follows: DLX6-AS1-WT, F, 5ʹ-CTAGCTAGCGGAGATACAAGGTAAACCTGAGAC-3ʹ and R, 5ʹ-CGGATATCAATTATCTGATGTGTATCTCAAGGC-3ʹ; DLX6-AS1-MUT, F, 5ʹ-CTCAAGCAGACTTTGUAACTATGCGAGATAC-3ʹ and R, 5ʹ-GTATCTCGCATAGTTACAAAGTCTGCTTGAG-3ʹ. STAT2 3ʹUTR-WT, F, 5ʹ-CTAGCTAGCATACTGGCATTGGCCCTTGG-3ʹ and R, 5ʹ-CGGATATCGGAACAGGTACAGCCAGCTT-3ʹ; STAT2 3ʹUTR-MUT, F, 5ʹ-GAGCTTTCGATACGAGCCTACCATGT-3ʹ and R, 5ʹ-TGGTAGGCTCGTATCGAAAGCTCTC-3ʹ
Animal Experiment in vivo
Animal experiment was authorized by the Animal Care and Use Committee of Luoyang Central Hospital Affiliated to Zhengzhou University. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. BALB/c nude mice (n=12, male, 4-weeks-old) were purchased from HFK Bioscience Co., Ltd (Beijing, China). SK-N-SH cells (3×106) with the transfection of sh-DLX6-AS1 and sh-NC were subcutaneously inoculated into experimental mice. The mice were averagely divided into two groups: the sh-DLX6-AS1 group and the sh-NC group. Two groups were maintained in the same growth conditions. From 5th d after inoculation, the tumor volume was measured every 5 d according to the method: length × width2 × 0.5. At the end of growth (30 d), all mice were killed, and tumor tissues were removed for the following analysis.
Statistical Analysis
The statistical analysis was performed using SPSS version 21.0 (IBM, Armonk, NY, USA). All data were obtained from at least three repeats and presented as the mean ± standard deviation. Data sets were compared using the Student’s t-test and analysis of variance where appropriate. The correlation between miR-506-3p expression and DLX6-AS1 expression or STAT2 expression was assessed according to Spearman rank correlation. The level of statistical significance set was at P < 0.05.
Result
The Expression of DLX6-AS1 Was Highly Upregulated in NB Tissues and Cell Lines
Firstly, the expression level of DLX6-AS1 was examined to monitor whether DLX6-AS1 was abnormally expressed in NB. Using qRT-PCR, we obtained that the level of DLX6-AS1 was significantly enhanced in NB tissues compared with that in normal tissues (Figure 1A). Consistently, the level of DLX6-AS1 in SK-N-SH and LAN-6 cells was strikingly higher than that in HUVEC cells (Figure 1B). The data hinted that NB development might be associated with dysregulation of DLX6-AS1.
Figure 1.
The abundance of DLX6-AS1 was enhanced in NB tissues and cell lines. (A) The expression of DLX6-AS1 in NB tissues and normal tissues was detected by qRT-PCR. (B) The expression of DLX6-AS1 in NB cells (SK-N-SH and LAN-6) and normal cells (HUVEC) was detected by qRT-PCR. *P < 0.05.
DLX6-AS1 Deficiency Reduced Cell Proliferation, Arrested Cell Cycle and Blocked Glycolysis in NB Cells
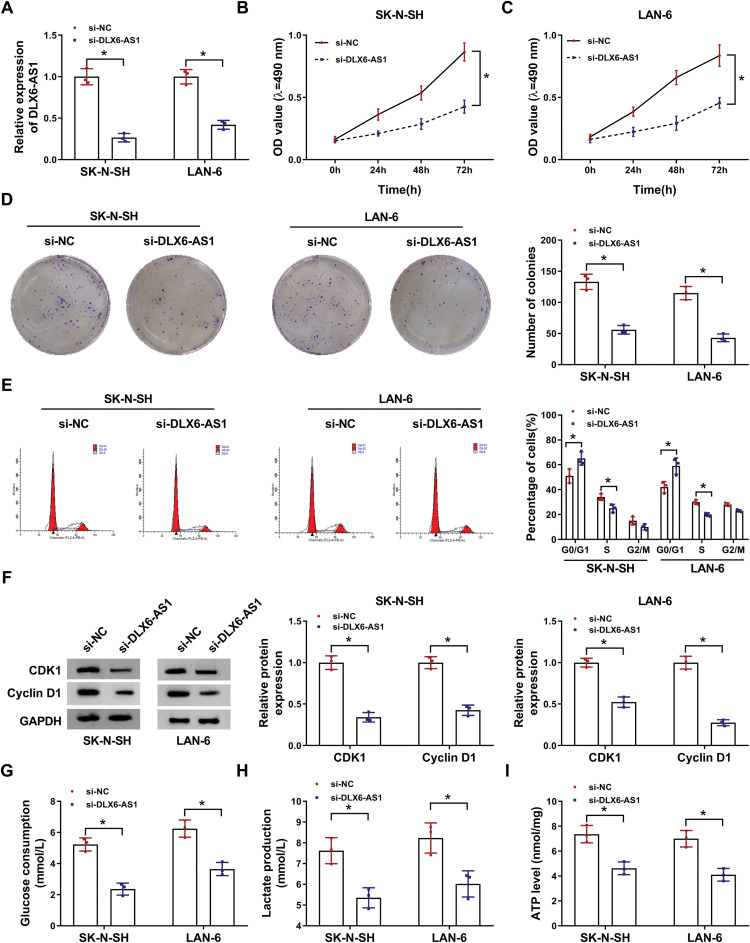
Given that the expression of DLX6-AS1 was enhanced in NB cells, the endogenous level of DLX6-AS1 was decreased to investigate its function in vitro. The expression of DLX6-AS1 in SK-N-SH and LAN-6 cells transfected with si-DLX6-AS1 was notably lower than that transfected with si-NC (Figure 2A). Cell proliferation was assessed by MTT assay and colony formation assay, and the results showed that DLX6-AS1 deficiency diminished the OD value at 490 nm and the number of colonies relative to si-NC in both SK-N-SH and LAN-6 cells (Figure 2B-D). Besides, the analysis of cell cycle distribution presented that DLX6-AS1 deficiency increased the proportion of SK-N-SH and LAN-6 cells in the G0/G1 phase and decreased the proportion in S phase (Figure 2E), indicating that DLX6-AS1 deficiency inhibited the G1/S transition of the cell cycle. In addition, the levels of cell cycle-related proteins were quantified, and the data displayed that DLX6-AS1 deficiency markedly diminished the levels of CDK1 and Cyclin D1 in SK-N-SH and LAN-6 cells (Figure 2F). Moreover, the glycolysis progress was elevated by glucose consumption, lactate production and ATP levels, and all of them were prominently weakened by DLX6-AS1 deficiency in SK-N-SH and LAN-6 cells (Figure 2G-I). These experiments suggested that DLX6-AS1 deficiency blocked the progression of NB in vitro.
Figure 2.
DLX6-AS1 deficiency suppressed NB development in vitro. (A) The expression of DLX6-AS1 in SK-N-SH and LAN-6 with the transfection of si-DLX6-AS1 was checked by qRT-PCR at 48 h post-transfection. (B and C) Cell proliferation at different time points was checked using MTT assay. (D) At 48-h post transfection, colony formation assay was performed to monitor colony growth. (E) Cell cycle was analyzed at 48 h post-transfection using flow cytometry assay. (F) The expression of CDK1 and Cyclin D1 SK-N-SH and LAN-6 with the transfection of si-DLX6-AS1 was checked by Western blot at 48 h post-transfection. (G–I) Glucose production, lactate production and ATP level were examined at 48 h post-transfection to assess glycolysis using the corresponding kit. *P < 0.05.
MiR-506-3p, Downregulated in NB Tissues and Cell Lines, Was a Target of DLX6-AS1
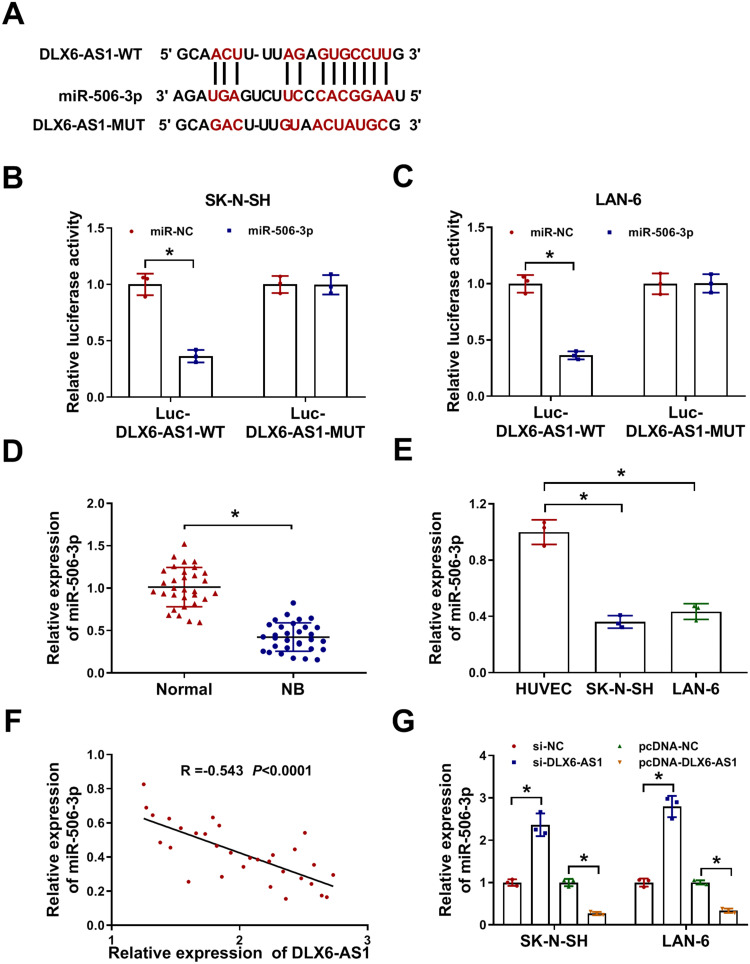
Through the analysis of the online bioinformatics tool Starbase3.0 (http://starbase.sysu.edu.cn/), miR-506-3p was one of the targets of DLX6-AS1. As shown in Figure 3A, there were several binding sites between DLX6-AS1 and miR-506-3p, and the mutant sequence mutated at the binding sites was generated to conduct dual-luciferase reporter assay. The result showed that the luciferase activity in SK-N-SH and LAN-6 cells transfected with miR-506-3p and Luc-DLX6-AS1-WT was significantly reduced relative to miR-NC, while the luciferase activity hardly changed in SK-N-SH and LAN-6 cells transfected with Luc-DLX6-AS1-MUT and miR-506-3p or miR-NC (Figure 3B and C). The expression analysis revealed that the expression of miR-506-3p was remarkably lower in NB tissues and cell lines (SK-N-SH and LAN-6) compared with that in normal tissues and HUVEC cells, respectively (Figure 3D and E). Besides, Spearman correlation analysis presented that miR-506-3p expression was negatively associated with DLX6-AS1 expression in NB tissues (Figure 3F). The expression of miR-506-3p was pronouncedly increased in SK-N-SH and LAN-6 cells with DLX6-AS1 knockdown but decreased with DLX6-AS1 overexpression (Figure 3G). All data manifested that DLX6-AS1 might function in NB by targeting miR-506-3p.
Figure 3.
MiR-506-3p was a target of DLX6-AS1. (A) The binding sites between miR-506-3p and DLX6-AS1 were analyzed by the online tool Starbase3.0. (B and C) Their interaction was verified by dual-luciferase reporter assay. (D and E) The expression of miR-506-3p in NB tissues and cell lines relative to normal tissues and cells was determined using qRT-PCR. (F) The expression correlation was analyzed using Spearman correlation analysis. (G) The expression of miR-506-3p affected by DLX6-AS1 deficiency or overexpression was examined by qRT-PCR at 48 h post-transfection. *P < 0.05.
DLX6-AS1 Deficiency Blocked NB Cell Malignant Behaviors via Enhancing the Expression of miR-506-3p
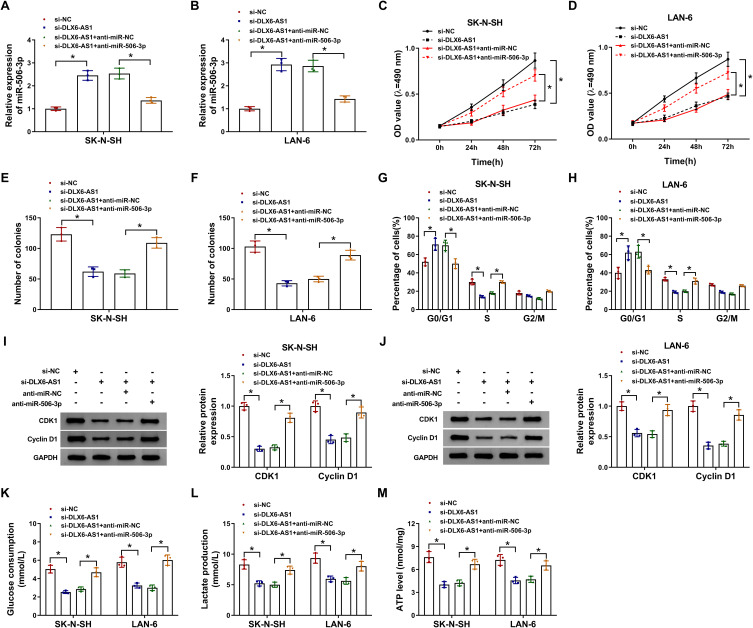
Rescue experiments of miR-506-3p absence were performed in DLX6-AS1 downregulated cells. The expression of miR-506-3p was notably strengthened in SK-N-SH and LAN-6 cells transfected with si-DLX6-AS1 relative to si-NC but impaired in cells transfected with si-DLX6-AS1+anti-miR-506-3p relative to si-DLX6-AS1+anti-miR-NC (Figure 4A and B). Functionally, DLX6-AS1 deficiency-inhibited cell proliferation was partly recovered in SK-N-SH and LAN-6 cells with si-DLX6-AS1+anti-miR-506-3p transfection relative to that with si-DLX6-AS1+anti-miR-NC transfection (Figure 4C and D). Also, DLX6-AS1 deficiency-inhibited colony formation in SK-N-SH and LAN-6 cells was restored by miR-506-3p inhibition (Figure 4E and F). Besides, the inhibited G1/S transition of the cell cycle in cells transfected with si-DLX6-AS1 was partially improved in cells transfected with si-DLX6-AS1+anti-miR-506-3p relative to si-DLX6-AS1+anti-miR-NC (Figure 4G and H). The protein levels of CDK1 and Cyclin D1 depleted in cells with DLX6-AS1 knockdown were substantially reinforced in cells with DLX6-AS1 knockdown plus miR-506-3p inhibition (Figure 4I and J). Moreover, glucose consumption, lactate production and ATP level were blocked in cells transfected with si-DLX6-AS1 but strikingly elevated in cells transfected with si-DLX6-AS1+anti-miR-506-3p (Figure 4K-M). These experiments suggested that miR-506-3p inhibition reversed the role of DLX6-AS1 deficiency.
Figure 4.
DLX6-AS1 targeted miR-506-3p to regulate NB development. SK-N-SH and LAN-6 cells were introduced with si-DLX6-AS1 or si-DLX6-AS1+anti-miR-506-3p, si-NC or si-DLX6-AS1+anti-NC serving as the control. (A and B) The expression of miR-506-3p in transfected cells was checked by qRT-PCR at 48 h post-transfection. (C and D) Cell proliferation was assessed using CCK-8 assay at different time points. (E and F) Colony formation assay was conducted at 48 h post-transfection. (G and H) Cell cycle was examined using flow cytometry assay at 48 h post-transfection. (I and J) The levels of CDK1 and Cyclin D1 at 48 h post-transfection were quantified by Western blot. (K–M) Glucose production, lactate production and ATP level were measured at 48 h post-transfection to assess glycolysis using the corresponding kit. *P < 0.05.
STAT2 Was a Target of miR-506-3p, and DLX6-AS1 Regulated STAT2 Expression by Targeting miR-506-3p
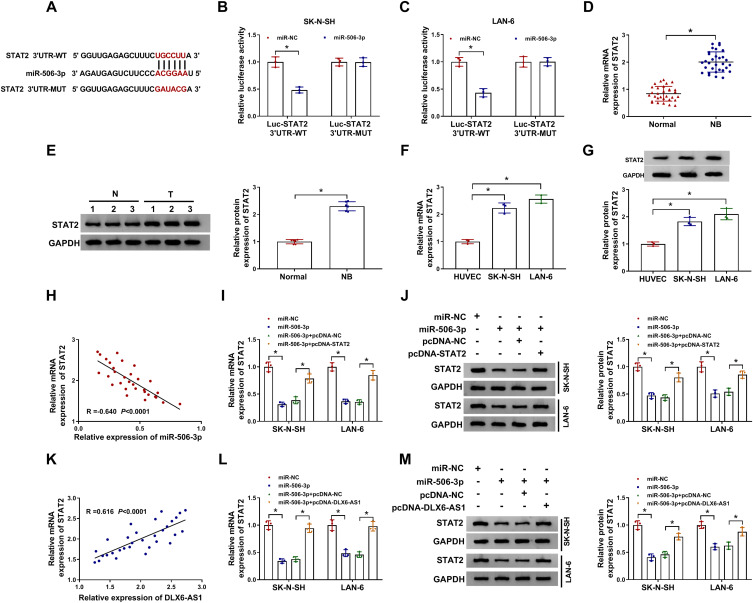
The analysis of the online tool Starbase3.0 revealed that there was a special binding site between miR-506-3p and STAT2 3ʹUTR (Figure 5A), hinting that STAT2 was a potential target of miR-506-3p. Then, this prediction was verified by dual-luciferase reporter assay, and miR-506-3p reintroduction significantly dwindled the luciferase activity in SK-N-SH and LAN-6 cells with the transfection of STAT2 3ʹUTR-WT but not STAT2 3ʹUTR-MUT relative to miR-NC (Figure 5B and C). In addition, the expression of STAT2 in NB tissues was aberrantly higher than that in normal tissues at both mRNA and protein levels (Figure 5D and E). Similarly, the expression of STAT2 in NB cells was also significantly higher than that in HUVEC cells at both mRNA and protein levels (Figure 5F and G). Moreover, the expression of STAT2 was negatively correlated with miR-506-3p expression in NB tissues (Figure 5H), and the expression of STAT2 was notably decreased in NB cells transfected with miR-506-3p relative to miR-NC but substantially recovered in NB cells transfected with miR-506-3p+pcDNA-STAT2 relative to miR-506-3p+pcDNA-NC at both mRNA and protein levels (Figure 5I and J). Furthermore, STAT2 expression was positively linked to DLX6-AS1 expression in NB tissues (Figure 5K), and its expression depleted in NB cells transfected with miR-506-3p was remarkably elevated in cells transfected with miR-506-3p+pcDNA-DLX6-AS1 at both mRNA and protein levels (Figure 5L and M). These analyses demonstrated that the expression of STAT2 was modulated by DLX6-AS1 through miR-506-3p.
Figure 5.
STAT2 was a target of miR-506-3p, and DLX6-AS1 regulated STAT2 expression by targeting miR-506-3p. (A) The potential target relationship between STAT2 and miR-506-3p was predicted by the online tool Starbase3.0. (B and C) The potential target relationship between STAT2 and miR-506-3p was verified by dual-luciferase reporter assay. (D and E) The expression of STAT2 in NB tissues was detected by qRT-PCR and Western blot. (F and G) The expression of STAT2 in NB cells was detected by qRT-PCR and Western blot. (H) The correlation between STAT2 expression and miR-506-3p expression in NB tissues was analyzed according to Spearman correlation analysis. (I and J) The expression of STAT2 in SK-N-SH and LAN-6 cells transfected with miR-506-3p, miR-NC, miR-506-3p+pcDNA-STAT2 or miR-506-3p+pcDNA-NC was detected by qRT-PCR and Western blot at 48 h post-transfection. (K) The correlation between STAT2 expression and DLX6-AS1 expression in NB tissues was analyzed according to Spearman correlation analysis. (L and M) The expression of STAT2 in SK-N-SH and LAN-6 cells transfected with miR-506-3p, miR-NC, miR-506-3p+pcDNA-DLX6-AS1 or miR-506-3p+pcDNA-NC was examined by qRT-PCR and Western blot at 48 h post-transfection. *P < 0.05.
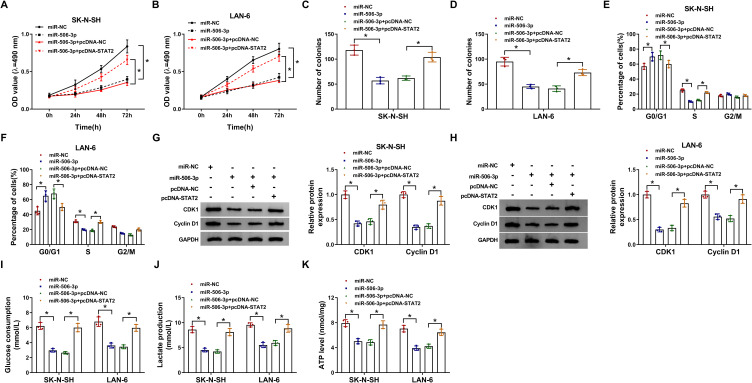
MiR-506-3p Reintroduction Suppressed NB Cell Malignant Behaviors by Diminishing the Expression of STAT2
To explore the function of miR-506-3p and STAT2, SK-N-SH and LAN-6 cells were transfected with miR-506-3p or miR-506-3p+pcDNA-STAT2, miR-NC or miR-506-3p+pcDNA-NC serving as the respective control. Cell proliferation was significantly inhibited in SK-N-SH and LAN-6 cells transfected with miR-506-3p relative to miR-NC but partly recovered in cells transfected with miR-506-3p+pcDNA-STAT2 relative to miR-506-3p+pcDNA-NC (Figure 6A and B). Likewise, the number of colonies was notably reduced in SK-N-SH and LAN-6 cells transfected with miR-506-3p but partly increased in cells transfected with miR-506-3p+pcDNA-STAT2 (Figure 6C and D). Moreover, miR-506-3p reintroduction induced cell cycle arrest at the G1-S phase, while the transfection of miR-506-3p+pcDNA-STAT2 lessened this cell cycle arrest in SK-N-SH and LAN-6 cells (Figure 6E and F). Besides, the expression of CDK1 and Cyclin D1 was strikingly weakened in SK-N-SH and LAN-6 cells transfected with miR-506-3p but substantially recovered in cells transfected with miR-506-3p+pcDNA-STAT2 (Figure 6G and H). Furthermore, glucose consumption, lactate production and ATP level were all impaired by miR-506-3p reintroduction but promoted by combined STAT2 overexpression in SK-N-SH and LAN-6 cells (Figure 6IK). These analyses showed that STAT2 overexpression partly reversed the effects of miR-506-3p reintroduction.
Figure 6.
MiR-506-3p bound to STAT2 to modulate the development of NB. SK-N-SH and LAN-6 cells were transfected with miR-506-3p, miR-NC, miR-506-3p+pcDNA-STAT2 or miR-506-3p+pcDNA-NC. (A and B) Cell proliferation was monitored at different time points using MTT assay. (C and D) Colony formation assay was performed at 48 h post-transfection. (E and F) Cell cycle was assessed by flow cytometry assay at 48 h post-transfection. (G and H) The levels of CDK1 and Cyclin D1 at 48 h post-transfection were quantified by Western blot. (I–K) Glucose production, lactate production and ATP level were checked at 48 h post-transfection to assess glycolysis using the corresponding kit. *P < 0.05.
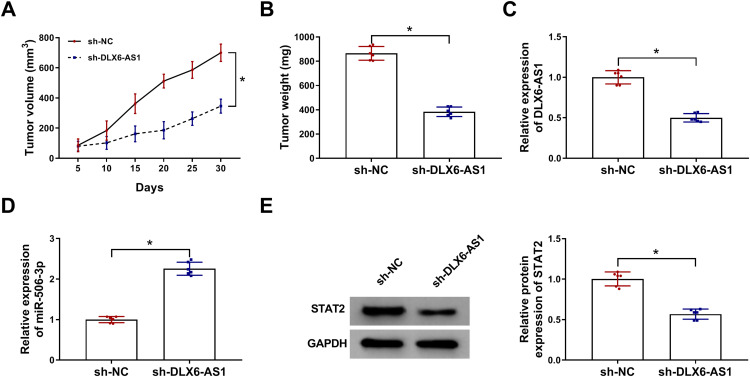
DLX6-AS1 Deficiency Inhibited Tumor Growth in vivo by Regulating miR-506-3p and STAT2 Expression
Additionally, the role of DLX6-AS1 in vivo was also explored. Xenograft model was established, and the record presented that DLX6-AS1 deficiency remarkably weakened tumor volume and tumor weight (Figure 7A and B). The expression of DLX6-AS1, miR-506-3p and STAT2 was detected in excised tumor tissues. Compared with the data in the sh-NC group, the expression of DLX6-AS1 and STAT2 was considerably declined in the sh-DLX6-AS1 group, while the expression of miR-506-3p was noticeably strengthened in the sh-DLX6-AS1 group (Figure 7C–E). We concluded that DLX6-AS1 regulated NB growth in vivo.
Figure 7.
DLX6-AS1 deficiency inhibited NB development in vivo. Experimental mice were inoculated with SK-N-SH cells transfected with sh-DLX6-AS1 or sh-NC. (A) Tumor volume was measured every five days. (B) At 30 d post-inoculation, all tumor tissues were excised to detect tumor weight. (C–E) The expression of DLX6-AS1, miR-506-3p and STAT2 in excised tumor tissues was examined using qRT-PCR or Western blot. *P < 0.05.
Discussion
The exploration of the mechanisms of NB development from the lncRNA landscape contributes to the insights of NB pathogenesis. The present study focused on an aberrantly upregulated lncRNA DLX6-AS1 and functionally discovered that DLX6-AS1 deficiency blocked the progression of NB by suppressing cell proliferation, cycle and glycolysis in vitro and inhibiting tumor growth in vivo. Through bioinformatics analysis and luciferase activity verification, the DLX6-AS1/miR-506-3p/STAT2 regulatory network was constructed to further illustrate the possible action mode of DLX6-AS1 in NB. These results introduced that DLX6-AS1 played a vital role in NB tumorigenesis and supplied a therapeutic approach for the treatment of NB.
DLX6-AS1 was abundantly regulated in colorectal cancer tissues and cells, and its depletion-repressed cell migration and invasion and restrained tumor outgrowth in colorectal cancer.23,24 Besides, DLX6-AS1 was substantially upregulated in non-small cell lung cancer (NSCLC), and DLX6-AS1 silence weakened NSCLC cell proliferation but accelerated cell apoptosis.25 The similar situation of DLX6-AS1 was also involved in renal cell carcinoma.26 These findings emphasize the carcinogenic role of DLX6-AS1 in different cancers. However, the effects of DLX6-AS1 on NB and underlying molecular mechanisms remain insufficient. Limited studies introduced that DLX6-AS1 was abundantly expressed in NB tissues and cell lines, and DLX6-AS1 downregulation-impaired cell proliferation, metastasis and invasion in NB cells.27 Besides, high expression of DLX6-AS1 was associated with advanced stage and poor survival.28 Here, consistent with these previous studies, the aberrant overexpression of DLX6-AS1 was observed in NB tissues and cells. Our loss-function experiments revealed the additional functions of DLX6-AS1 knockdown on cell cycle, glycolysis progression and solid tumor growth, indicating that DLX6-AS1 silence blocked tumor growth and development both in vitro and in vivo.
In general, lncRNAs can act as the decoys of special miRNAs to repress the partial functions of miRNAs.29 Here, miR-506-3p was predicted as one of the targets of DLX6-AS1, which was weakly expressed in NB tissues and negatively correlated with the expression of DLX6-AS1. Rescue experiments presented that miR-506-3p absence reversed the effects of DLX6-AS1 deficiency, indicating that DLX6-AS1 exerted functions by sponging miR-506-3p. A recent study recorded the anti-tumor role of miR-506-3p in NB, and it harbored the idea that miR-506-3p reintroduction sequestered NB cell proliferation, migration and invasion.30 The characteristics of miR-506-3p about tumor suppressor were widely documented in other cancers, including osteosarcoma,31 esophageal squamous cell carcinoma32 and nasopharyngeal carcinoma.33 Similarly, our experimental data displayed that miR-506-3p reintroduction inhibited cell proliferation, cell cycle and glycolysis in NB, hinting that its anti-tumor properties also pervaded in NB.
Further, our analysis proved that miR-506-3p mediated STAT2 expression at the post-transcriptional level. Aberrant expression of STAT2 has been mentioned in dozens of cancers, including solid tumors and hematological malignancy.34,35 STAT2 overexpression contributed to several cellular malignant behaviors, such as migration, invasion, EMT and oncogene expression.36 STAT2 functioned as a target mRNA of miRNAs to participate in tumor development. For example, STAT2 was a target of miR-3908, and STAT2 abolished the partial role of miR-3908 to promote the deterioration of glioblastoma.21 Likewise in NB, miR-653-5p directly bound to STAT2 3ʹUTR, and STAT2 overexpression exerted the same effects as miR-653-5p deficiency, encouraging cell viability, migration and invasion.37 In our study, STAT2 was interacted by miR-506-3p, and its overexpression reversed the effects of miR-506-3p reintroduction. Interestingly, a previous study clarified that DLX6-AS1 regulated lung adenocarcinoma cell proliferation by mediating the JAK/STAT signaling pathway.38 Moreover, another STAT family member, such as STAT3, was also reported to promote tumorigenicity and enhance chemoresistance of NB.39 These data suggested that STAT family played crucial roles in the development of NB. Moreover, our data showed that STAT2 expression was positively correlated with DLX6-AS1 expression in NB tissues, and expression analysis presented that DLX6-AS1 directly regulated STAT2 expression by targeting miR-506-3p.
Conclusion
The expression level of DLX6-AS1 was aberrantly upregulated in NB tissues and cells. DLX6-AS1 deficiency suppressed proliferation, cell cycle and glycolysis of NB cells in vitro and depleted tumor growth in vivo by modulating the expression of miR-506-3p and STAT2. In short, the DLX6-AS1/miR-506-3p/STAT2 regulatory network may be an action mode of DLX6-AS1 that participates in the pathogenesis of NB, and DLX6-AS1 is a promising biomarker for the treatment of NB.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202–2211. doi: 10.1056/NEJMra0804577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stafman LL, Beierle EA. Cell proliferation in neuroblastoma. Cancers (Basel). 2016;8:1. doi: 10.3390/cancers8010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaturvedi NK, McGuire TR, Coulter DW, et al. Improved therapy for neuroblastoma using a combination approach: superior efficacy with vismodegib and topotecan. Oncotarget. 2016;7(12):15215–15229. doi: 10.18632/oncotarget.7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgenstern DA, Baruchel S, Irwin MS. Current and future strategies for relapsed neuroblastoma: challenges on the road to precision therapy. J Pediatr Hematol Oncol. 2013;35(5):337–347. doi: 10.1097/MPH.0b013e318299d637 [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;11(12):704–713. doi: 10.1038/nrclinonc.2014.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diede SJ. Spontaneous regression of metastatic cancer: learning from neuroblastoma. Nat Rev Cancer. 2014;14(2):71–72. doi: 10.1038/nrc3656 [DOI] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu D, Ho SY, Juan HF, Huang HC. High-risk, expression-based prognostic long noncoding RNA signature in neuroblastoma. JNCI Cancer Spectr. 2018;2(2):pky015. doi: 10.1093/jncics/pky015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Li D, Huang D, et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26(3):755–773. doi: 10.1016/j.ymthe.2017.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Yarmishyn AA, Batagov AO, Tan JZ, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahu D, Hsu CL, Lin CC, et al. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7(36):58022–58037. doi: 10.18632/oncotarget.11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsson M, Beck S, Kogner P, Martinsson T, Caren H. Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics. 2016;11(1):74–84. doi: 10.1080/15592294.2016.1138195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravindan N, Subramanian K, Somasundaram DB, Herman TS, Aravindan S. MicroRNAs in neuroblastoma tumorigenesis, therapy resistance, and disease evolution. Cancer Drug Resist. 2019;2:1086–1105. doi: 10.20517/cdr.2019.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galardi A, Colletti M, Businaro P, Quintarelli C, Locatelli F, Di Giannatale A. MicroRNAs in neuroblastoma: biomarkers with therapeutic potential. Curr Med Chem. 2018;25(5):584–600. doi: 10.2174/0929867324666171003120335 [DOI] [PubMed] [Google Scholar]
- 17.Huang B, Liu C, Wu Q, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482(4):828–834. doi: 10.1016/j.bbrc.2016.11.120 [DOI] [PubMed] [Google Scholar]
- 18.Guo X, Xiao H, Guo S, Dong L, Chen J. Identification of breast cancer mechanism based on weighted gene coexpression network analysis. Cancer Gene Ther. 2017;24(8):333–341. doi: 10.1038/cgt.2017.23 [DOI] [PubMed] [Google Scholar]
- 19.Arimoto KI, Lochte S, Stoner SA, et al. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol. 2017;24(3):279–289. doi: 10.1038/nsmb.3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamero AM, Young MR, Mentor-Marcel R, et al. STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev Res (Phila). 2010;3(4):495–504. doi: 10.1158/1940-6207.CAPR-09-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Chen J, Zhang J. AdipoR1-mediated miR-3908 inhibits glioblastoma tumorigenicity through downregulation of STAT2 associated with the AMPK/SIRT1 pathway. Oncol Rep. 2017;37(6):3387–3396. doi: 10.3892/or.2017.5589 [DOI] [PubMed] [Google Scholar]
- 22.Yang M, Chen H, Zhou L, Chen K, Su F. Expression profile and prognostic values of STAT family members in non-small cell lung cancer. Am J Transl Res. 2019;11(8):4866–4880. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou FR, Pan ZP, Shen F, et al. Long noncoding RNA DLX6-AS1 functions as a competing endogenous RNA for miR-577 to promote malignant development of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23(9):3742–3748. doi: 10.26355/eurrev_201905_17800 [DOI] [PubMed] [Google Scholar]
- 24.Zhang JJ, Xu WR, Chen B, et al. The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur Rev Med Pharmacol Sci. 2019;23(19):8321–8331. doi: 10.26355/eurrev_201910_19143 [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Ni R, Wang J, Liu Y. Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed Pharmacother. 2019;109:1851–1859. doi: 10.1016/j.biopha.2018.09.151 [DOI] [PubMed] [Google Scholar]
- 26.Zeng X, Hu Z, Ke X, et al. Long noncoding RNA DLX6-AS1 promotes renal cell carcinoma progression via miR-26a/PTEN axis. Cell Cycle. 2017;16(22):2212–2219. doi: 10.1080/15384101.2017.1361072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang HY, Xing MQ, Guo J, et al. Long noncoding RNA DLX6-AS1 promotes neuroblastoma progression by regulating miR-107/BDNF pathway. Cancer Cell Int. 2019;19:313. doi: 10.1186/s12935-019-0968-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wang S, Yang C. Long non-coding RNA DLX6-AS1 regulates neuroblastoma progression by targeting YAP1 via miR-497-5p. Life Sci. 2020;252:117657. doi: 10.1016/j.lfs.2020.117657 [DOI] [PubMed] [Google Scholar]
- 29.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D, Cao Y, Li J, Xu J, Liu Q, Sun X. miR-506 suppresses neuroblastoma metastasis by targeting ROCK1. Oncol Lett. 2017;13(1):417–422. doi: 10.3892/ol.2016.5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiashi W, Chuang Q, Zhenjun Z, Guangbin W, Bin L, Ming H. MicroRNA-506-3p inhibits osteosarcoma cell proliferation and metastasis by suppressing RAB3D expression. Aging (Albany NY). 2018;10(6):1294–1305. doi: 10.18632/aging.101468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Zhang Z, Yu X, et al. Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene. 2019;38(12):2042–2055. doi: 10.1038/s41388-018-0476-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang TS, Zheng YJ, Wang J, Zhao JY, Yang DK, Liu ZS. MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/beta-catenin signaling pathway by down-regulating LHX2. J Exp Clin Cancer Res. 2019;38(1):97. doi: 10.1186/s13046-019-1023-4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Li S, Sheng B, Zhao M, Shen Q, Zhu H, Zhu X. The prognostic values of signal transducers activators of transcription family in ovarian cancer. Biosci Rep. 2017;37:4. doi: 10.1042/BSR20170650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamauchi H, Sakai I, Narumi H, Takeuchi K, Soga S, Fujita S. Development of interferon-alpha resistant subline from human chronic myelogenous leukemia cell line KT-1. Intern Med. 2001;40(7):607–612. doi: 10.2169/internalmedicine.40.607 [DOI] [PubMed] [Google Scholar]
- 36.Ogony J, Choi HJ, Lui A, Cristofanilli M, Lewis-Wambi J. Interferon-induced transmembrane protein 1 (IFITM1) overexpression enhances the aggressive phenotype of SUM149 inflammatory breast cancer cells in a signal transducer and activator of transcription 2 (STAT2)-dependent manner. Breast Cancer Res. 2016;18(1):25. doi: 10.1186/s13058-016-0683-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chi R, Chen X, Liu M, et al. Role of SNHG7-miR-653-5p-STAT2 feedback loop in regulating neuroblastoma progression. J Cell Physiol. 2019;234(8):13403–13412. doi: 10.1002/jcp.28017 [DOI] [PubMed] [Google Scholar]
- 38.Li J, Li P, Zhao W, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi: 10.1186/s12935-015-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Odate S, Veschi V, Yan S, Lam N, Woessner R, Thiele CJ. STAT3inhibition of with the generation 2.5 antisense oligonucleotide, AZD9150, decreases neuroblastoma tumorigenicity and increases chemosensitivity. Clin Cancer Res. 2017;23(7):1771–1784. doi: 10.1158/1078-0432.CCR-16-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]