Abstract
In this study, we report the molecular diagnosis and retrospective study of porcine circovirus 3 (PCV3) in frozen and formalin-fixed paraffin-embedded swine tissues (FFPE) collected from 1967 to 2018 in southeastern Brazil (Espírito Santo and Rio de Janeiro states). Frozen tissues from 35 pigs and FFPE tissues from 143 pigs were tested by nested PCR, targeting the PCV3 partial capsid gene. Bidirectional sequencing of 16 positive samples was performed, followed by sequence analysis and haplotype networks. A total of 26/178 samples (14.6%) tested positive for PCV3: 14/35 (40%) frozen tissue and 12/143 (8.4%) FFPE tissue. PCV3 was detected in the 1960s, 1970s, 2000s, and 2010s with the characterization of types PCV3a and PCV3b. A star-like distribution was observed in the grid of haplotypes, with a low haplotype diversity and more recent dispersal of the virus. A total of 40% of asymptomatic animals considered fit for slaughter tested positive for PCV3. In conclusion, PCV3 DNA was detected over 51 years of study, prior to initial reports and, so far, the sample detected in 1967 is the oldest partial capsid sequence described. The circulation of two different genotypes was reported, suggesting more than one introduction event of this virus into Brazil. Moreover, taken together, our studies indicated an ancient origin of PCV3 and its circulation in asymptomatic animals in Brazilian herds.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00281-6) contains supplementary material, which is available to authorized users.
Keywords: Sequence analysis, Formalin-fixed paraffin-embedded tissues, Haplotype network
Introduction
In 2015, in the USA, there was an increase in the mortality rate of sows and the presence of mummified fetuses on farms in North Carolina. After metagenomic sequencing, the presence of a new virus, subsequently classified as porcine circovirus 3 (PCV3), was revealed [1]. In 2016, another study detected PCV3 DNA in tissue samples from cases of acute myocarditis and multisystem inflammation where no PCV2 genome was detected [2].
PCV3 is a single stranded DNA non-enveloped virus and since 2017 was formally accepted into the Circoviridae family, with PCV1 and PCV2 [3, 4]. The novel sequences showed less than 70% of identity in the predicted whole genome and capsid protein amino acid (aa) sequence compared with other virus in family [3]. In the last 4 years, PCV3 has been described worldwide in animals affected by different clinical conditions including porcine dermatitis and nephropathy syndrome (PDNS), reproductive, respiratory, gastrointestinal and neurological disorders, multisystemic inflammation, myocarditis, and in subclinical infections [1, 3, 5, 6]. PCV-3 genome has been detected by PCR in oral fluids and nasal swabs as well as in feces, semen, and colostrums and horizontal transmission through direct contact is probably the main route of transmission [3]. Its high occurrence may pose a potential threat to the swine industry worldwide [7].
In Brazil, few studies about the circulation of PCV3 were performed using serum and tissues (lung, pool of lymph nodes, spleen, and mummified fetuses [6, 8, 9]) and data about the introduction of PCV3 in this country, as well as the impact of this new type of circovirus in Brazilian herds, are still unknown. In this study, we report the molecular diagnosis and characterization of PCV3 in frozen and formalin-fixed paraffin-embedded (FFPE) swine tissues collected from 1967 to 2018 (51 years) in the southeast region of Brazil (Espírito Santo and Rio de Janeiro states).
Material and methods
Swine tissues were obtained from two different origins: frozen tissue from 35 slaughtered pigs and FFPE tissue samples from 143 pigs.
The frozen tissue samples (lymph node tissue, lungs, liver, and kidneys) were obtained from asymptomatic animals considered fit for slaughter, during the evisceration process performed in the daily routine of the pig slaughterhouse located in the state of Espírito Santo, Brazil, from June 2017 to May 2018 [10]. The samples were stored in sterile flasks and kept under − 20 °C until processing.
After thawing, fragments of organs from the same animal were cut into smaller fragments using sterile scalpel blades and pooled in a 2.0 mL microtubes prior to tissue digestion processing. The digestion process was performed with lysis buffer (ATL buffer, Qiagen®) and proteinase K, and the DNA extraction process was performed using guanidinium thiocyanate and silica [11].
FFPE tissue (intestine, heart, lymph node, lung, liver, kidneys, brain, spleen, and others) was obtained from the collection of the Geraldo Manhães Carneiro Animal Health Research Center, Pesagro-Rio. The samples from 143 pigs were part of the Pesagro-Rio collection, which was originally used for routine diagnostic purposes over a period of 50 years (1967 to 2017), and come from different regions of the state of Rio de Janeiro and Espírito Santo. Ten-micrometer sections of each sample block were cut from paraffin using sterile scalpel blades, pooled by animal in a 2.0 mL microtubes, homogenated, and dewaxed with deparaffinization solution (QIAGEN, Germany). Samples were digested with lysis buffer (ATL buffer, Qiagen®) and proteinase K. For DNA extraction, a QIAamp® DNA FFPE Tissue Kit (QIAGEN, Germany) was used according to the manufacturer’s instructions.
For PCV3 molecular detection, a 649 bp fragment (1339–1987) of capsid protein gene was amplified [12]. In order to improve the detection, a novel nested PCR primer pair was designed (5’-CCATTGAACGGTGGGGTCAT-3′ and 5’-TGGACCACAAACACTTGGCT-3′) in Prime 3 0.4.0 software [13], targeting a 203 bp internal fragment (1442–1645). The reaction was performed with 11 μl of a commercial master mix, 10 pmol of each PCV3-NF and PCV3-NR primers, and 5 μl of template DNA with a final reaction volume of 20 μl. The thermal cycling conditions were as follows: 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 52 °C for 30 s, 72 °C for 30 s, and 72 °C for 10 min [14]. PCR assay was also performed to identify the swine β-actin gene as an extraction control in frozen and FFPE tissues, with a set of primers that amplified an 850 bp fragment, as described by Hui et al. [15].
Single bidirectional sequencing was performed by Myleus Biotechnology (Belo Horizonte, Brazil) using an ABI3130 automated sequencer (Applied Biosystems, Foster City, CA) with BigDye v3.1. Raw sequences were analyzed by Codon Code aligner (http://www.codoncode.com/aligner/) using the Phred quality score tool and a cutoff of 20 was established. Sequences generated in this study were deposited in the GenBank (accession numbers MN651451–MN65157 and MN65159–MN651467). Multiple alignments of nucleotide sequences (and amino acids deduced from them) and phylogenetic analysis were performed with MEGA 10.0.5 [16] and BEAST v1.7.4—Bayesian Evolutionary Analysis Sampling Trees package [17].
For haplotype networks, the datasets containing partial sequences of the gene encoding the 203 bp for PCV3 capsid protein were used (84 reference sequences and 16 sequences obtained in this study). The information about each sequence (GenBank accession number, year and country of origin) is described in Supplementary File 1. Sequences were converted into the DnsSP5.10.1 program [18] and used for the analysis of median-joining (MJ) using parsimony criterion with the NETWORK 4.5.1.6 program [19].
This study was approved by the Ethics Committee on Animal Use of the Fluminense Federal University (Protocol No. 1013/2018).
Results
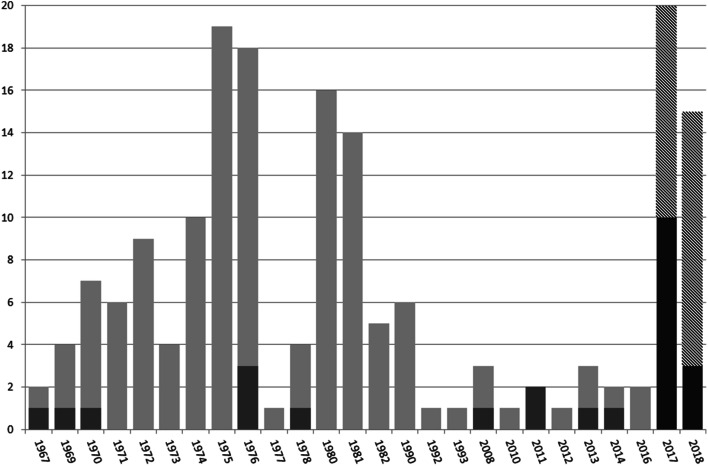
A total of 26/178 samples (14.6%) tested positive for PCV3: 14/35 (40%) frozen tissue and 12/143 (8.4%) FFPE tissues. PCV3 was detected in the 1960s, 1970s, 2000s, and 2010s, with the 1967 sample being the oldest in the world to date. The distribution of samples tested for PCV3 over 51 years is shown in Fig. 1.
Fig. 1.
Distribution of positive and tested samples for PCV3 over 51 years (from 1967 to 2018) using the nested PCR technique. In light gray, FFPE samples tested from 1967 to 2016; in dark gray, positive samples tested from 1967 to 2016; in hatch, frozen samples tested from 2017 to 2018; and in solid black, positive samples tested from 2017 to 2018
Comparing the results of swine β-actin and PCV3 capsid gene PCR from frozen and FFPE samples, a 100% concordance was observed in frozen tissue samples from 2017 to 2018. A total of 18 of the 26 (69%) FFPE PCV3 positive samples also tested positive in swine β-actin PCR detection.
Conventional and nested PCR products were sequenced, and 16 sequences were of sufficient quality for phylogenetic analysis: 4 PCR products (648 bp) and 12 nested PCR products (203 bp). For 4 samples, ES6P/2017, ES8L/2017, ES8P/2017, and ES33L/2018 (648 bp products), based on group-specific marker codons 24, 27, 77, and 150, it was possible to determinate PCV3 types, and PCV3a and PCV3b types were detected [20]. The remaining 12 sequences (203 bp) were not large enough to allow characterization by types of PCV3. Other non-synonymous substitutions (aa 104, 114, 131, 132, 134, 137, 161, 168, 183, and 214) were detected in the PCV3 sequences of this study compared with the reference sequences available on GenBank (Table 1 and Supplementary File 1).
Table 1.
Representation of amino acid substitutions found from the alignment of the partial sequences of the region encoding the PCV3 capsid protein. The classification of types of PCV3 in circulation in the states of Espírito Santo and Rio de Janeiro, Brazil, was carried out according to Fux et al. [20]. Other amino acid changes described in this study are also shown
| Reference sequences | Type | 3 | 24 | 27 | 77 | 104 | 114 | 131 | 132 | 134 | 137 | 150 | 161 | 168 | 183 | 214 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KY354061 | PCV 3a | H | V | K | S | F | A | T | R | V | S | I | Q | R | G | L |
| MK117056 | PCV 3a | . | A | . | . | Y | . | . | . | . | . | . | . | . | . | . |
| MK060073 | PCV 3a | – | A | R | G | Y | . | . | . | . | . | . | . | . | . | – |
| MK060075 | PCV 3a | – | – | – | . | . | . | . | . | . | . | L | . | . | . | – |
| MK117052 | PCV 3b | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| This study | ||||||||||||||||
| ES6P/2017 | PCV 3a | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| ES8L/2017 | PCV 3a | P | . | . | . | . | . | . | . | . | . | . | . | . | . | F |
| ES8P/2017 | PCV 3a | – | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| ES33L/2018 | PCV 3b | – | A | R | . | Y | . | . | . | . | . | . | . | . | . | – |
| RJ15/1969 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| RJ16/1969 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| RJ60/1976 | NA | – | – | – | – | – | – | I | C | G | . | . | . | . | – | – |
| RJ63/1976 | NA | – | – | – | – | – | – | . | C | . | F | . | . | K | – | – |
| RJ31/2008 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| RJ22/2011 | NA | – | – | – | – | – | – | . | . | . | F | . | K | . | – | – |
| RJ30/2011 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| RJ32/2013 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| ES7L/2017 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | R | – |
| ES7P/2017 | NA | – | – | – | – | – | F | . | . | . | . | . | . | . | – | – |
| ES12L/2017 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
| ES19L/2017 | NA | – | – | – | – | – | – | . | . | . | . | . | . | . | – | – |
NA* not applicable; “–” not sequenced; “.” same amino acid
Analysis of partial ORF2 nucleotide sequences showed that sequences from this study share from 96.5 to 100% of identity with reference sequences obtained from GenBank (Table 1 and Supplementary File 1). Phylogenetic analysis of the partial capsid sequences were performed with different molecular clock models (strict and relaxed uncorrelated lognormal), as well as different population dynamics models (constant population size; exponential, expansional and logistic population growth; and the Bayesian Skyline coalescent model) were tested but phylogenetic tree showed low posterior probabilities values in all adopted models. The model with the highest values of posterior probabilities was chosen and the best fitted phylogenetic tree is shown in Supplementary File 2.
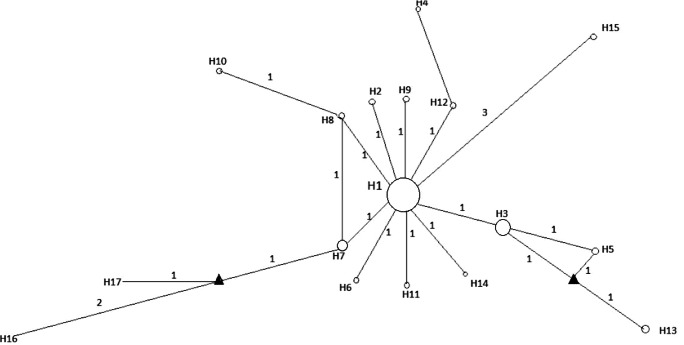
The 101 sequences (16 sequences from this study and 84 reference sequences) generated 17 haplotypes (Fig. 2 and Supplementary File 1). The network revealed a main haplotype occupying a central position in the network, with two medium vectors separating the neighboring haplotypes, ranging from 1 to 3 the number of substitutions (Fig. 2). A star-shaped configuration with the most central haplotype (H1) was observed, represented by samples from the USA, China, Korea, Colombia, Spain, Japan, England, Thailand, Denmark, and those sequences from this study (from Espírito Santo 2017/ 2018 Rio de Janeiro 1969, 2008, and 2001). Sequences from the same year and locality were grouped into different haplotypes, such as samples from Rio de Janeiro 1976 (H15 and H16), Rio de Janeiro 2011 (H1 and H17), and those from Espirito Santo de 2017 (H1 and H7), demonstrating the absence of structure related to geographic origin (Fig. 2).
Fig. 2.
Median-joining haplotype network for PCV3. Circles correspond to haplotypes and the size of the circles is proportional to the frequency of the haplotypes in the network. Triangles correspond to the median vectors. The number of substitutions is represented in the connections between the haplotypes. Numbers close to the lines correspond to the numbers of transitions and crossings between the haplotypes. The three major haplotypes were haplotype H1 includes sequences from USA, China, Brazil, South Korea, Colombia, Denmark, Spain, Japan, UK, and Thailand; haplotype H3 includes sequences from China, Brazil, Korea, USA, and UK; haplotype H7 includes sequences from China and Brazil. The information about each sequence (GenBank accession number, year and country of origin) is described in Supplementary File 1
The greatest distances were observed in haplotypes 15 and 16, separated from the Central Haplotype by 3 transitions, both of which were composed of samples from 1976. Most other haplotypes were unique and distributed around the central, demonstrating the existence of a small genetic distance among them, which was reinforced by the low haplotypic diversity (0.48).
The network did not recover the topology of the Bayesian analysis, which presented two clades supported with a high posterior probability value (1.0), one formed by the samples sequenced in this study from Rio de Janeiro 1970, 1976, 2011, and 2017 and the Espirito Santo 2017, and the other cluster formed by the other samples sequenced in this study together with all samples from other countries (Supplementary File 2).
Discussion
In this study, two sample sources were tested: FFPE samples were obtained from diseased animals that underwent necropsy and histopathological examination, and frozen tissue samples were obtained from clinically healthy slaughtered animals.
The largest number of positive samples was found in frozen tissues obtained in 2017–2018 and similar results were also observed in the swine β-actin PCR detection. This can be explained by the degradation of the genetic material present in the FFPE blocks. One of the main challenges of this study was the use of swine FFPE tissue samples since DNA extraction from ancient FFPE blocks has some critical points that may culminate in PCR amplification failure [21]. In this case, the design of a nested PCR approach based on a small (203 bp) DNA fragment increased PCV3 detection in samples of up to 50 years of stocking time.PCV3 DNA has been detected over the 51 years of study in both sources, and the FFPE sample detected in 1967 (RJ10/1967) is so far the oldest PCV3 positive sample.
PCV3 was first reported in Brazil by Tochetto et al. [6] in swine serum pools collected in 2017. The first retrospective study was performed by Saraiva et al. [8] in Brazil with DNA extracted from swine tissue collected between 2006 and 2007. The detection of PCV3 in Brazil in 1967 (present study), Sweden in 1993 [22], and China and Spain in 1996 [3, 23] indicates that the virus circulated in swine populations decades before initial reports, whether or not it was associated with clinical manifestation.
Identification of PCV3 genotypes combined with amino acid analysis has been reported by Fu et al. [24] and Fux et al. [19]. Fux et al. [20] defined the classification of PCV3 in 2 groups (a and b). To support the definition of these two main groups, they aligned the PCV3 ORFs and identified the group-specific marker codons. Based on this, the strains found here belong to the PCV3a and PCV3b genotypes. The presence of at least two different PCV3 genotypes corroborates the results obtained by Saraiva et al. [8], which indicated more than one event introducing PCV3 to Brazil. These introductions may have occurred through animal trade or through the movement of pigs in reservoirs [25]. The virus was detected in wild boars with high prevalence, suggesting a potential role as a reservoir for domestic pigs [26].
Franzo et al. [27] provided an up-to-date representation of the origin and evolution of PCV3 and showed limited genetic variability and high similarity between recent sequences and those obtained in the early 1990s. Our sequences corroborate their studies, since a high homology was found between the sequences obtained in 1967 and those obtained up to 2018.
Phylogenetic analysis is a powerful tool and is now widely used to investigate the evolution of PCV3, but there is a large number of different inference methods and a lack of uniformity in the use of these methods in different studies. The posterior probability of root and ancestral node localization, described by Franzo et al. [27], was often low, revealing a widely expected uncertainty considering the long period of time between the estimated tMRCA and the oldest available sequences. The same kind of result was found in our studies where the posterior probabilities obtained were also low. Therefore, according to the authors Li et al. [7], accurate phylogenetic trees for the identification of PCV3 genotypes should be constructed using a complete genome, but this were not possible due to DNA fragmentation in ancient FFPE samples.
Haplotype one was the most common and central, suggesting that ancestral sequences are represented in it. This centralized star-shaped structure, with several haplotypes of more recent origin connected by few substitutions, with low genetic diversity and number of medium vectors can also indicate the probable occurrence of a virus expansion event [28].
Our studies have not allowed the association of PCV3 viral DNA detection with the clinical presentation of the disease, but 40% of animals considered fit for slaughter tested positive for PCV3. Our results agree with STADEJEK et al. [29], YE et al. [22], and ZHENG et al. [30], who found a high prevalence of PCV3 in asymptomatic animals. Other authors such as WEN et al. [31] and ARRUDA et al. [32] described an association between the prevalence of PCV3 and various clinical signs of the disease. There is a possibility that PCV3 associated diseases have the same evolution and impact on pig breeding as PCV-2 [32]. The question arises as to whether PCV3 would behave like PCV-2 in the absence of other factors.
Conclusions
PCV3 has circulated in Brazilian herds since 1967 and, so far, this is the oldest partial capsid sequence described. The circulation of two different genotypes was reported, suggesting more than one introduction event of PCV3 in Brazil. Besides, low haplotype diversity and more recent dispersal were observed. Taken together, our studies indicated an ancient origin of PCV3 and its circulation in asymptomatic animals in Brazilian herds.
Electronic supplementary material
(DOCX 19 kb).
(PDF 208 kb).
Acknowledgements
This research was supported by CAPES. We thank the veterinarian Márcio Figueiredo, who made the collection of slaughtered pig samples possible. We also thank the State Center for Research in Animal Health, Agricultural Research Company of the State of Rio de Janeiro-Pesagro-Rio, which allowed access to its collection enabling the execution of this work.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, Byers E, Hause BM. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. 2017;91(1):1879–1816. doi: 10.1128/JVI.01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, Deng X, Resende T, Vannucci F, Delwart E. Detection of a novel circovirus PCV-3 in pigs with cardiac and multi-systemic inflammation. Virol J. 2016;13:184. doi: 10.1186/s12985-016-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klaumann F, Correa-Fiz F, Franzo G, Sibila M, Nunez JI, Segales J. Current knowledge on porcine circovirus 3 (PCV-3): a novel virus with a yet unknown impact on the swine industry. Front Vet Sci. 2018;5:315. doi: 10.3389/fvets.2018.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ICTV Taxonomy history: Porcine circovirus 3. International Committee on Taxonomy of Viruses (ICTV) Publishing.https://talk.ictvonline.org//taxonomy/p/taxonomy-history?taxnode_id=201855768. Accessed 04 April 2020
- 5.Zhao D, Wang X, Gao Q, Huan C, Wang W, Gao S, Liu X. Retrospective survey and phylogenetic analysis of porcine circovirus type 3 in Jiangsu province, China, 2008 to 2017. Arch Virol. 2018;163:2531–2538. doi: 10.1007/s00705-018-3870-2. [DOI] [PubMed] [Google Scholar]
- 6.Tochetto C, Lima DA, Varela APM, Loiko MR, Paim WP, Scheffer CM, Roehe PM. Full-genome sequence of porcine circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound Emerg Dis. 2018;6(5):5–9. doi: 10.1111/tbed.12735. [DOI] [PubMed] [Google Scholar]
- 7.Li G, He W, Zhu H, Bi Y, Wang R, Xing G, Zhang C, Zhou J, Yuen KY, Gao GF, Su S. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. AdvSci (Weinh) 2018;5:1800275. doi: 10.1002/advs.201800275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saraiva GL, Vidigal PMP, Assao VS, Fajardo MLM, Loreto ANS, Fietto JLR, Bressan GC, Lobato ZIP, Almeida MR, Silva-Junior A. Retrospective detection and genetic characterization of porcine circovirus 3 (PCV-3) strains identified between 2006 and 2007 in Brazil. Viruses. 2019;11(3):201. doi: 10.3390/v11030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dal Santo AC, Cezario KC, Bennemann PE, Machado SA, Martins M. Full-genome sequences of porcine circovirus 3 (PCV3) and high prevalence in mummified fetuses from commercial farms in Brazil. Microb Pathog. 2020;141:104027. doi: 10.1016/j.micpath.2020.104027. [DOI] [PubMed] [Google Scholar]
- 10.Souza AE (2019) Detecção molecular de circovírus suíno (PCV-2), torque tenovirus suíno 1 e 2 (TTSUV-1 e TTSUV-2) e achados histopatológicos em vísceras de suínos sumetidos ao abate regular no estado do Espírito Santo. [Dissertação]. Pós-Graduação em Microbiologia e Parasitologia Aplicadas do Instituto Biomédico: Universidade Federal Fluminense
- 11.Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim D, Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/JCM.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, Qian P, Wu M, He Q. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. 2017;64(3):703–708. doi: 10.1111/tbed.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz ACM. (2019) Quantificação do circovirus suíno tipo 2 e detecção molecular do circovirus suíno tipo 3 em soro de suínos dos estados do Rio de Janeiro e Espírito Santo, Brasil. [Tese]. Pós-Graduação em Microbiologia e Parasitologia Aplicadas do Instituto Biomédico: Universidade Federal Fluminense
- 15.Hui RK, Zeng F, Chan CM, Yuen KY, Peiris JS, Leung FC. Reverse transcriptase PCR diagnostic assay for the coronavirus associated with severe acute respiratory syndrome. J Clin Microbiol. 2004;42(5):1994–1999. doi: 10.1128/JCM.42.5.1994-1999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond AJ, Rambaut A, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67(5):901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Librado PJR, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 19.Bandelt HJ, Forter P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 20.Fux R, Söckler C, Kathrin EL, Renken C, Krejci R, Sutter G, Ritzmann M, Eddicks M. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol J. 2018;15(25):1–9. doi: 10.1186/s12985-018-0929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mesquita RA, Anzai EK, Oliveira RN, Nunes FD. Avaliação de três métodos de extração de DNA de material parafinado para amplificação de DNA genômico pela técnica da PCR. Pesqui Odontol Bras. 2001;15(4):314–319. doi: 10.1590/S1517-74912001000400008. [DOI] [PubMed] [Google Scholar]
- 22.Ye X, Berg N, Fossum C, Wallgren P, Blomstrom A. Detection and genetic characterization of porcine circovirus 3 from pigs in Sweden. Virus Genes. 2018;54(4):466–469. doi: 10.1007/s11262-018-1553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Wei L, Lu Z, Mi S, Bao F, Guo H, Tu C, Zhu Y, Gong W. Retrospective study of porcine circovirus 3 infection in China. Transbound Emerg Dis. 2018;65:607–613. doi: 10.1111/tbed.12853. [DOI] [PubMed] [Google Scholar]
- 24.Fu X, Fang B, Ma J, Liu Y, Bu D, Zhou P, Wang H, Jia K, Zhang G. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg Dis. 2018;3(65):296–303. doi: 10.1111/tbed.12752. [DOI] [PubMed] [Google Scholar]
- 25.Yuzhakov AG, Raev SA, Alekseev KP, Grebennikova TV, Verkhovsky OA, Zaberezhny AD, Aliper TI. First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes. 2018;54:608–611. doi: 10.1007/s11262-018-1582-z. [DOI] [PubMed] [Google Scholar]
- 26.Klaumann F, Correa-Fiz F, Sibila M. Infection dynamics of porcine circovirus 3 (PCV3) in longitudinally sampled pigs from four Spanish farms. Vet Rec. 2019;184:619. doi: 10.1136/vr.105219. [DOI] [PubMed] [Google Scholar]
- 27.Franzo G, Grassi L, Tucciarone CM, Drigo M, Martini M, Pasotto D, Mondin A, Menandro ML. A wild circulation: high presence of porcine circovirus 3 in different mammalian wild hosts and ticks. Transbound Emerg Dis. 2019;66:1548.Disponívelem. doi: 10.1111/tbed.13180. [DOI] [PubMed] [Google Scholar]
- 28.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadejek T, Wozniak A, Mitek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. 2017;64(5):1350–1353. doi: 10.1111/tbed.12672. [DOI] [PubMed] [Google Scholar]
- 30.Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. 2017;64:1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen S, Sun W, Li Z, Zhuang X, Zhao G, Xie C, Zheng M, Jing J, Xiao P, Wang M, Han J, Ren J, Liu H, Lu H, Jin N. The detection of porcine circovirus 3 in Guangxi, China. Transbound Emerg Dis. 2018;65:27–31. doi: 10.1111/tbed.12754. [DOI] [PubMed] [Google Scholar]
- 32.Arruda B, Piñeyro P, Derscheid R, Hause B, Byers E, Dion K, Long D, Sievers C, Tangen T, Williams T, Schwartz K. PCV-3 associated disease in the United States swine herd. Emerg Microbes Infect. 2019;8(1):684–698. doi: 10.1080/22221751.2019.1613176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 19 kb).
(PDF 208 kb).