Abstract
The combination of plant extract and antibiotic represents a template for developing of antibiofilm drugs. This study investigated the synergistic effects of pomegranate/rosemary/antibiotic combinations against antibiotic resistance and biofilm formation of Pseudomonas aeruginosa. The results showed that 17 (85%) of total P. aeruginosa isolates were biofilm producers; however, 5 (25%) isolates were demonstrated as a strong biofilm producer. The highest MIC level (1024 μg/ml) of tested antibiotics against strong biofilm producer isolates was observed with piperacillin, however the MIC ranges of ceftazidime, gentamycin, imipenem, and levofloxacin against these isolates were reached to (256–1024 μg/ml), (32–1024 μg/ml), (8–1024 μg/ml), and (8–512 μg/ml), respectively. PS-1 was the representative isolate for strong biofilm formation and high antibiotic resistance. 16S rRNA gene analysis suggested that PS-1 (accession No. MN619678) was identified as a strain of P. aeruginosa POA1. Pomegranate and rosemary extracts were the most effective extracts in biofilm inhibition, which significantly inhibited 91.93 and 90.83% of PS-1 biofilm, respectively. Notably, the synergism between both plant extracts and antibiotics has significantly reduced the MICs of used antibiotics at the level lower than the susceptibility breakpoints. Pomegranate/rosemary/antibiotic combinations achieved the highest biofilm eradication, which ranging from 90.0 to 99.6%, followed by the eradication ranges of pomegranate/rosemary combination, rosemary, and pomegranate extracts, which reached to (76.5–85.4%), (53.1–73.7%), and (41.2–71.5%), respectively. The findings suggest that pomegranate/rosemary/antibiotic combinations may be an effective therapeutic agent for antibiotic resistance and biofilm formation of P. aeruginosa.
Keywords: Pseudomonas, Biofilm, Pomegranate, Rosemary, Antibiotic, Resistant, Inhibition, Eradication
Introduction
Pseudomonas aeruginosa is the most frequent opportunistic Gram-negative rods; it is capable of infecting variety of all tissues and becoming a major cause’s morbidity and mortality among hospital patients. It is the second causative agent of nosocomial pneumonia, the third common bacterial pathogen of urinary tract infection, the fourth common causes of surgical site infection, the fifth most frequently isolated pathogen from all sites, and the seventh regular pathogen isolated from the bloodstream [1] .
The chronic infections with P. aeruginosa are mainly due to form biofilm, which increases its resistance to conventional antibiotics by adding some mechanisms including: limited diffusion of antimicrobial agents, slow growth rate of biofilm cells in inner layers compared with outer layers, inactivation of antimicrobial agents by biofilm matrix, decreasing of biofilm cells permeability, resistance by using type IV secretion systems, multidrug efflux pumps expression, and the action of antibiotic-modifying enzymes [2–4]. In addition, the effectiveness of conventional antibiotics became limited due to their higher values of their MIC and MBC, which may results in vivo toxicity [5]. Hence, it is critically important to search new active compounds that can effectively inhibit and eradicate biofilm-related infections, as well as enhance the activity of the traditional antibiotics by decreasing their MIC and MBC values.
Medicinal plants are important in drug discovery as they often contain a vast number of bioactive compounds, which are less expensive, safer, and more readily available compared with synthetic compounds. The use of plant extracts or pure natural compounds in combination with conventional antibiotics may hold greater promise for inhibiting and eradicating microbial biofilms [6–8]. Recently, pomegranate and rosemary extracts have been studied in several systems of medicine for their pharmacological actions such as antitumor, antiviral, anti-inflammatory, antibacterial, and antifungal activities [9–11]. Therefore, the present work aims to investigate the activity of some plant extracts alone and in combination with antibiotic against antibiotic resistance and biofilm formation of Pseudomonas aeruginosa.
Materials and methods
P. aeruginosa isolates and growth media
The study was implemented on 20 P. aeruginosa isolates obtained from clinical lab of Kasr El-Aini, Hospital, Cairo, Egypt, during the period from July to September 2015. The identity of these isolates were confirmed by streaking on sterile cetrimide agar plates (Oxoid) and incubated overnight at 37 °C. The blue-green single colonies were picked up on sterile nutrient agar slants (Oxoid) for further confirmation by VITEK-automated microbiology system (Version: 07.01-Canada).
Tryptone soya broth (Oxoid) supplemented with sterile 1% glycerol (TSG) was used for the assaying of biofilm formation, inhibition, and eradication. Muller-Hinton agar (MHA, Oxoid) was used for the susceptibility testing of antibiotics and plant extracts against P. aeruginosa isolates. Muller-Hinton broth (MHB, Oxoid) was used for determining of the minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC), and fractional inhibitory concentration (FIC) of plant extracts and antibiotic powders. Semi-sold nutrient agar medium (Oxoid) supplemented with 5% glucose (SSM) was used for studying the inhibition activity of plant extracts against different bacterial motilities. This medium was solidified with 0.3, 0.5, and 1% agar bacteriology (Oxoid) according to the phenotype of motilities (swimming, swarming, and twitching, respectively).
Plant materials and antibiotics
The medicinal plants used in the current study were obtained from the local market, Haraz for spices and herbs, Cairo, Egypt. These plants including rosemary leaves (Rosmarinus officinalis), ginger roots (Zingiber officinale), cinnamon barks (Cinnamomum verum), thyme leaves (Thymus vulgaris), pomegranate peels (Punica granatum), clove flowers (Syzygium aromaticum), and peppermint leaves (Mentha piperita). The plant materials were individually powdered, mixed thoroughly, and stored at − 40 °C until use.
The antibiotic disks (Oxoid) used in this study were imipenem (IPM) 10 μg, ceftazidime (CDZ) 30 μg, cefepime (FEP) 30 μg, gentamycin (CN) 10 μg, norfloxacin (NOR) 10 μg, ofloxacin (OFX) 5 μg, ciprofloxacin (CIP) 5 μg, levofloxacin (LEV) 5 μg, amikacin (AK) 30 μg, gatifloxacin (GAT) 5 μg, nalidixic acid (NA) 30 μg, piperacillin (PRL) 100 μg, and tobramycin (TOB) 10 μg/disk. Antibiotic powders including PRL, CDZ, IPM, CN, and LEV were obtained from the United States pharmacopeia reference standards.
Biofilm formation assay
Screening of P. aeruginosa isolates for potential biofilm formation was studied by micro-dilution method [12] as follows: overnight culture of each P. aeruginosa isolate was separately diluted to 1.0 × 106 cfu/ml (equivalent to 0.5% McFarland standard) with TSG. Aliquots (200 μL) of the diluted cultures were dispensed into sterile wells of 96-well micro-titer plate. Wells containing 200 μL TSG were used as a negative control. The plates were incubated overnight at 37 °C. Next, the planktonic cells from each culture were decanted. The remaining biofilms were gently washed 3 times with 200 μL phosphate buffer saline solution pH 7.2 (PBS) and emptied by flicking the plate. After washing, each biofilm was stained with 200 μL crystal violet (0.5%, w/v) for 15 min at room temperature. Next, the content of each well was decanted and washed again with 200 μL PBS. After that, the wells were filled with 200 μL of 95% ethanol and incubated in shaker incubator (100 rpm) for 20 min at room temperature. Next, the established biofilms were measured at 570 nm against blank (95% ethanol) using a micro-titer plate reader (Shcheer SH9600-Shanghai) [13]. Based on ODs, the biofilm formation were classified into 3 categories as weak (OD570nm ≤ 0.5), intermediate (0.5 > OD570nm < 1.5), and strong (OD570nm ≥ 1.5) [14] .
Antibiotics susceptibility testing
The susceptibility of P. aeruginosa isolates to 13 different antibiotic disks (IPM, CDZ, FEP, CN, NOR, OFX, CIP, LEV, AK, GAT, NA, PRL, and TOB) was investigated by disk diffusion method [15]. Briefly, a sterile cotton wool swab was dipped into the bacterial suspension (adjusted to 1 × 106 cfu/ml) and spread evenly on the surface of sterile MHA plate and allowed to dry before placing the antibiotic disks. The plates were incubated for 24 h at 37 °C. Next, the inhibition zone diameters (mm) were measured around each disk and expressed as sensitive (S), intermediate (I), and resistant (R).
The MIC of PRL, CDZ, IPM, CN, and LEV against selected P. aeruginosa isolate was evaluated by micro-dilution method. Briefly, the antibiotics were separately dissolved with 0.5% dimethyl sulfoxide (DMSO) and sterilized through 0.22-μm syringe filter. Each well of 96-well micro-titer plate was dispensed with 100 μL of the tested bacterial suspension (2 × 106 cfu/ml in MHB) and 100 μL of each 2 fold serial dilutions of each tested antibiotic solution. Wells dispensed with 200 μL of and inoculated MHB containing 0.5% DMSO were considered as negative and positive controls, respectively. The MIC values of used antibiotics were interpreted as the lowest concentration of tested antibiotic that prevented visible growth after 24 h of incubation at 37 °C [16,17].
Plants extraction and analysis
The plant extract was prepared as follow: approximately 250 g of pulverized plant materials was individually suspended with 1250 ml ethanol (95%, v/v) and incubated for 3 days in the dark at room temperature. Next, each suspension was filtrated through a Whatman filter paper No.1 and concentrated to dryness under reduced pressure in a rotary evaporator (Heidolph, UK) at 40 °C. The dried extracts were separately stored in sterile Falcon tube at 4 °C until use [18]. Total polyphenol contents of the most effective plant extracts in biofilm inhibition was estimated by Folin–Ciocalteu method [19]. Phytochemical analysis of the selected extracts was measured by HPLC [20] at Department of Crops Technology, Food Technology Research Institute (FTRI)—Agricultural Research Center, Giza, Egypt.
Identification by 16S rRNA gene sequencing
The identity of representative isolate for strong biofilm formation and high antibiotic resistance was confirmed by molecular tools [21]. The basic local alignment search tool (BLAST) database [22] of National Center for Biotechnology Information (NCBI) was used to compare the sequence of 16S rDNA of the experimental isolate with known 16S rDNA sequences of bacteria. The obtained alignments were constructed using molecular evaluation genetic analysis (MEGA, version 5) program [23] .
Antibacterial activity, MIC, and MBC of plant extracts
The antibacterial activity of plant extracts (20%, w/v) against selected isolates was investigated by agar well diffusion method [24]. Each extract was dissolved with 0.5% DMSO and sterilized through 0.22-μm syringe filter. Aliquots (100 μL/well) of plant extract solution and control (0.5% DMSO) were loaded into MHA plates previously inoculated with tested Pseudomonas suspension (adjusted to 1 × 106 cfu/ml) and cut the well using 0.6-mm sterile cork borer. The plates were incubated overnight at 37 °C. Next, the antibacterial activity of each extract was evaluated by measuring the diameter (mm) of clear zone around each well.
The MIC and MBC of plant extracts against selected isolates were evaluated as previously described antibiotic MIC method. The MIC of plant extract was interpreted as the lowest concentration of the extract that prevented visible growth after 24 h of incubation at 37 °C. The MBC of plant extract was interpreted as the lowest concentration of the extract that killed 100% of bacterial inoculum after 24 h of incubation at 37 °C [16,17].
Biofilm inhibition assay
Biofilm inhibition activity of plant extracts against selected isolates was studied at different sub-MIC levels (0.5× MIC, 0.25× MIC and 0.125× MIC) by 2,3,5 triphenyltetrazolium chloride (TTC) method [25]. Each plant extract was dissolved with sterile TSG containing 0.5% DMSO and sterilized through 0.22-μm syringe filter. Each well of 96-well micro-titer plate was dispensed with 100 μL of plant extract solutions and 100 μL of TSB previously inoculated with 1 × 106 cfu/ml of the bacterial culture. Wells containing 200 μL of inoculating TSG (containing 0.5% DMSO) were considered as controls. Biofilm experiments were performed independently three times. The plates were incubated overnight at 37 °C. After incubation, the planktonic cells were decanted, and the remaining biofilm was gently washed 3 times with 200 μL PBS and emptied by flicking the plate. After washing, 150 μL of sterile TSG and 50 μL of 1% TTC were added to each well. The plates were incubated (protected from light) at 37 °C for 6 h. Next, biofilm inhibition activity of the treatment and control was estimated at 405 nm. Biofilm inhibition percentage was calculated using the formula: Biofilm inhibition % = [(OD405nm of control − OD405nm of the test)/(OD405nm of control)] × 100. The most effective plant extracts in biofilm inhibition were selected in the subsequent studies.
Motility inhibition assay
The motility inhibition assay of the selected plant extracts (1/4 MIC level) alone and in combination against swimming, swarming, and twitching motilities of the selected isolate was conducted as prior published method [26]. Briefly, the tested solutions were prepared as previously described for determining the MIC of antibiotics. The solutions were added to SSM before pouring. Swim and twitch plates were dried overnight before use, whereas swarm plates were dried 1.0 h before inoculation. After drying, the plates were inoculated with aliquots (5 μL) of bacterial suspension (1 × 106 cfu/ml). The inoculums were spotted at the center of swim SSM plate surfaces, while the inoculums of twitch and swarm plates were stabbed into the SSM plates. The inoculated swim, swarm, and twitch SSM plates containing 0.5% DMSO were considered as controls. Following the inoculation, the plates were incubated overnight at 37 °C. The diameters (mm) of the migration zones produced by the examined isolate on treated and control plates were compared. The motility experiments were performed independently three times.
Determination of FIC
The FIC of the selected plant extracts (1/4 × MIC) in combination with different antibiotics (concentration ranges, 2× MIC–1/32× MIC) against selected isolate was studied by checkerboard method 27. Briefly, the plant extracts and antibiotics solutions were prepared as previously described for antibiotic MIC method. Each well of 96-well micro-titer plate was dispensed with 100 μL of bacterial suspension in MHB (adjusted to 2 × 106 cfu/ml), 50 μL of selected plant extract solutions, and 50 μL of antibiotic solution. The plates were incubated overnight at 37 °C. The FIC index (∑FIC) is calculated as the sum of the MIC of each compound when used in combination divided by the MIC of the compound used alone. Synergism has traditionally been defined as an ∑FIC ≤ 0.5, indifferent when the ∑FIC is < 0.5 to < 2 and antagonistic when the ∑FIC is ≥ 2 [27].
Biofilm eradication assay
Biofilm eradication activity of the selected plant extracts (1/4× MIC) individually and in combination with antibiotics against selected bacterial isolate was studied at different MIC levels (concentration range, MIC to 1/32× MIC) of antibiotics using TTC method [25]. Briefly, the solutions of plant extracts and antibiotics were prepared as previously described for biofilm inhibition assay. The selected isolate was grown overnight in TSB at 37 °C, and then diluted (1:2, v/v) with sterile TSG. Each well of 96-well micro-titer plate was filled with 200 μL of the diluted culture (1 × 106 cfu/ml) and incubated overnight at 37 °C. After incubation, the planktonic cells were decanted, and the wells were filled again with 200 μL sterile TSG and incubated again overnight at 37 °C. Thereafter, planktonic cells were decanted, and the remaining biofilm in each well was gently washed 3 times with 200 μL PBS and emptied by flicking the plate. The performed biofilms were treated with 200 μL of tested solutions for 24 h at 37 °C. The biofilm treated with 0.5% DMSO is demonstrated as control. The eradication experiments were performed independently three times. After incubation, the tested solutions were decanted, and the remaining biofilm was washed 3 times with 200 μL PBS. Biofilm eradication was calculated using the formula: Biofilm eradication % = [(OD405 nm of control − OD405 nm of test)/(OD405 nm of control)] × 100. The most effective combinations in biofilm eradication were selected in the subsequent studies.
Scanning electron microscopy (SEM)
The eradication activity of the most effective combination against biofilm associated with the selected isolate was examined by SEM [28]. Briefly, formation and treatment of the examined biofilms were done as described in biofilm eradication with some modifications including each well of sterile 6-well micro-titer plate containing 3.5 ml of tested culture and sterile glass coverslips (3 × 3 mm), as well as the biofilms were treated with 3.5 ml of combination solution. Afterward, glass coverslips from treated and control (combination free) wells were gently washed 3 times with 3.5 ml PBS and initially fixed with 2.5% (v/v) glutaraldehyde at 4 °C for 2 h. The coverslips were rinsed with sterile PBS for 10 min and then dehydrated in a series ethanol solution (30, 50, 70, 80, 90% (v/v), and absolute). The dehydrated samples were coated with gold and then observed using SEM (SEM quanta FEG250-USA).
Statistical analysis
GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, USA) was used to perform the statistical analysis. Data were expressed as mean ± SD. Statistical differences between groups were performed using one-way and two-way ANOVA and Tukey’s post hoc test with significant level P < 0.05.
Results
Biofilm formation and antibiotic sensitivity testing
Screening of P. aeruginosa isolates for biofilm formation are summarized in Table 1. Out of the twenty isolates, 17 (85%) isolates were biofilm producers, 9 (45%) isolates were classified as weak biofilm producers, 5 (25%) isolates were strong biofilm producers, and 3 (15%) isolates were moderate biofilm producers. However, 3 (15%) isolates showed no ability to form biofilm.
Table 1.
Biofilm formation and antibiotic susceptibility pattern of Pseudomonas isolates.
| Isolates | Biofilm capacity | Antibiotics sensitivity testing | |||
|---|---|---|---|---|---|
| Resistance phenotype | Interpretative % | ||||
| R | I | S | |||
| PS-1 | ++++ | PRL, FEP, CDZ, IPM, AK, TOB, CN, OFX, GAT, NA, CIP, NOR, and LEV | 100 | 0.0 | 0.0 |
| PS-2 | +++ | PRL, FEP, CDZ, IPM, AK, TOB, OFX, GAT, NA, CIP, NOR, and LEV | 92.3 | 7.7 | 0.0 |
| PS-3 | ++ | FEP, CDZ, and IPM | 30.8 | 7.7 | 61.5 |
| PS-4 | +++ | PRL, FE,P and CDZ | 30.8 | 38.5 | 30.8 |
| PS-5 | +++ | FEP, CDZ, IPM, and NA | 30.8 | 38.5 | 30.8 |
| PS-6 | ++ | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-7 | + | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-8 | + | FEP, CDZ, IPM, and NA | 30.8 | 0.0 | 69.2 |
| PS-9 | + | FEP, CDZ, IPM, and NA | 38.5 | 15.4 | 46.2 |
| PS-11 | + | PRL, FEP, CDZ, IPM, and NA | 38.5 | 15.4 | 46.2 |
| PS-12 | + | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-13 | + | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-14 | – | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-15 | – | PRL, FEP, CDZ, IPM, AK, TOB, CN, OFX, GAT, NA, CIP, NOR, and LEV | 100 | 0.0 | 0.0 |
| PS-16 | – | PRL, FEP, CDZ, IPM, AK, TOB, CN, OFX, GAT, NA, CIP, NOR, and LEV | 100 | 0.0 | 0.0 |
| PS-17 | ++ | PRL, FEP, CDZ, IPM, AK, OFX, GAT, NA, CIP, NOR and LEV | 84.6 | 7.7 | 7.7 |
| PS-18 | + | FEP, CDZ, IPM, and NA | 30.8 | 7.7 | 61.5 |
| PS-19 | +++ | PRL, FEP, CDZ, IPM, AK, TOB, CN, OFX, GAT, NA, CIP, NOR, and LEV | 100 | 0.0 | 0.0 |
| PS-21 | + | PRL, FEP, CDZ, and IPM | 38.5 | 7.7 | 53.8 |
| PS-22 | + | FEP, CDZ, and IPM | 30.8 | 7.7 | 53.8 |
PRL, piperacillin; FEP, cefepime; CDZ, ceftazidime; IPM, imipenem; AK, amikacin; TOB, tobramycin; CN, gentamycin; OFX, ofloxacin; GAT, gatifloxacin; NA, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; LEV, levofloxacin; S, sensitive; I, intermediate; R, resistant; (−), non-biofilm producer; (+), weak biofilm producer; (++), moderate biofilm producer; (+++) and more, Strong biofilm producer isolates shown in bold letters
Antibiotic susceptibility of P. aeruginosa isolates showed that the highest antibiotic resistant isolates in the present study were PS-1, PS-2, PS-15, PS-16, and PS-19, which resistant to > 92.3% of tested antibiotics. In addition, the highest intermediate profile (> 30%) of the tested antibiotics were observed with PS-4, PS-5, and PS-8; however, PS-3, PS-6, PS-7, PS-9, PS-11, PS-12, and PS-13 were detected as the most sensitive isolates to antibiotics. These isolates were sensitive to > 30% of tested antibiotics (Table 1).
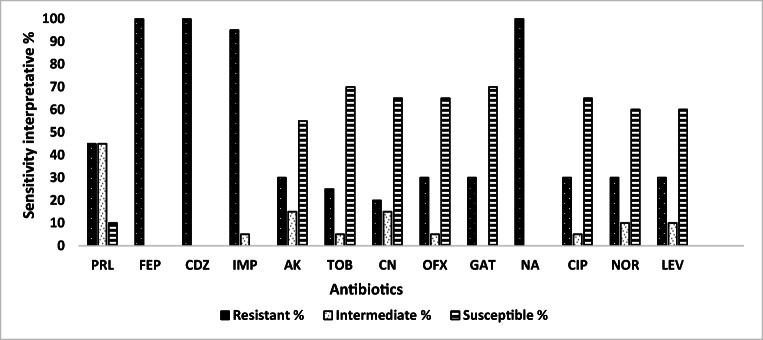
Overall tested antibiotics, the resistance of P. aeruginosa isolates to FEP, CDZ, NA, and IPM was the common (≥ 85%), followed by PRL (45%); however, the resistance of these isolates to AK, TOB, CN, OFX, GAT, CIP, NOR, and LEV were decreased to < 31%. The most effective antibiotics against P. aeruginosa isolates were GAT, TOB, CIP, CN, OFX, LEV, NOR, and AK, with susceptibility rates of 70, 70, 65, 65, 65, 60, 60, and 55%, respectively (Fig. 1).
Fig. 1.
Antibiotic sensitivity pattern overall tested Pseudomonas isolates. PRL, piperacillin; FEP, cefepime; CDZ, ceftazidime; IPM, imipenem; AK, amikacin; TOB, tobramycin; CN, gentamycin; OFX, ofloxacin; GAT, gatifloxacin; NA, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; LEV, levofloxacin
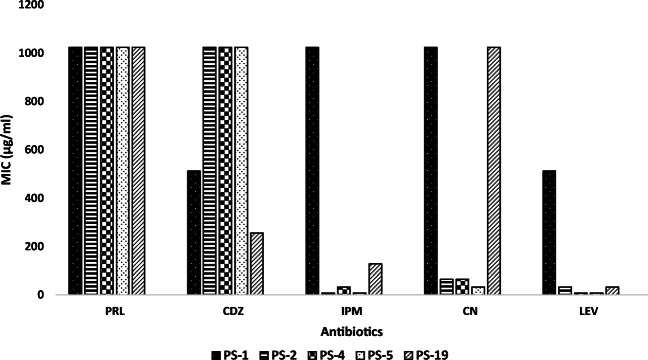
The MIC values of five different antibiotics (PRL, CDZ, IPM, CN, and LEV) against the strong biofilm producer isolates (PS-1, PS-2, PS-4, PS-5, and PS-19) are summarized in Fig. 2. The obtained results showed that the highest MIC level (1024 μg/ml) of tested antibiotics against strong biofilm producer isolates was observed with piperacillin, followed by the MIC ranges of CDZ, CN, IPM, and LEV, which reached to (256–1024 μg/ml), (32–1024 μg/ml), (8–1024 μg/ml), and (8–512 μg/ml), respectively. P. aeruginosa PS-1 was the most potent isolate in biofilm formation and antibiotic resistance in the present study; thus, it was selected as representative isolate for the subsequent studies.
Fig. 2.
The MIC of antibiotics against the strong biofilm producer isolates of P. aeruginosa. PRL, piperacillin; CDZ, ceftazidime; IPM, imipenem; CN, gentamycin; LEV, levofloxacin
Molecular characterization of PS-1
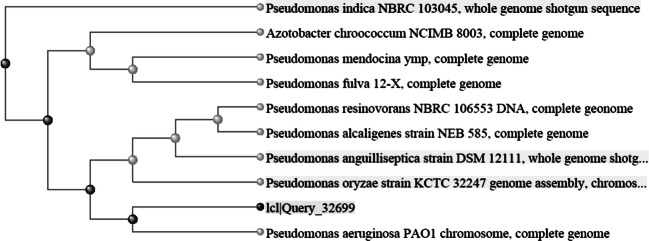
Phenotypic identity of P. aeruginosa PS-1 was confirmed by molecular characterization using 16S rRNA genes sequencing. BLAST analysis of the sequence data revealed that PS-1 (accession No. MN619678) belongs to family Pseudomonadaceae, displayed 98% sequence similarity with P. aeruginosa isolate POA1. Thus, PS-1 can be identified as an isolate of P. aeruginosa isolate POA1 (Fig. 3).
Fig. 3.
Phylogenetic tree of P. aeruginosa PS-1 (ICI/Query_32699)
Antibacterial activity, MIC, and MBC of ethanol plant extracts against PS-1
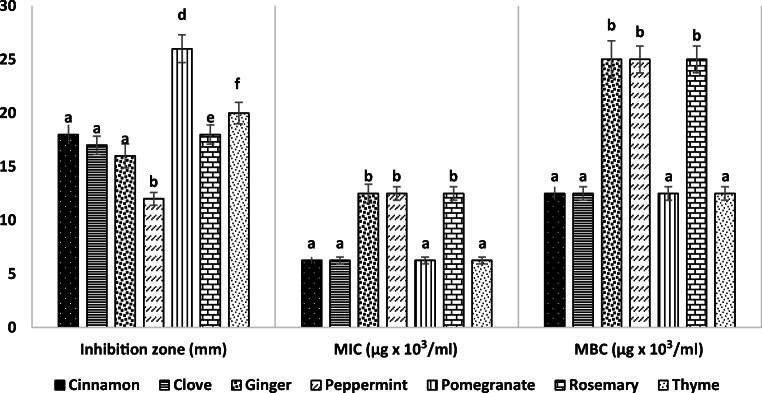
The antibacterial activity of seven plant extracts against PS-1 at a concentration 20% (w/v) are summarized in Fig. 4.The results indicated that the tested extracts exhibited variable degrees of inhibition zones (12–26 mm) against PS-1. Pomegranate extract had the highest inhibition zone (26 mm) against PS-1, followed by thyme (20 mm), cinnamon (18 mm), rosemary (18 mm), clove (17 mm), ginger (16 mm), and peppermint (12 mm) extracts. The MIC values of pomegranate, thyme, cinnamon, and clove extracts were reached to 6.25 mg/ml, whereas the MIC values of rosemary, peppermint, and ginger were significantly increased to 12.5 mg/ml, while the MBC versus MIC of each extract was equal 2 folds (Fig. 4).
Fig. 4.
Antibacterial activity, MIC, and MBC of curd extracts against PS-1. Result represents the mean ± SD of at least three independent experiments performed in triplicate. Different letters indicate significant differences between the test groups of each study (one-way ANOVA with Tukey’s post hoc test, P < 0.05)
Biofilm inhibition of plant extracts
Biofilm inhibition activities of seven plant extracts against PS-1 at different sub-MIC levels (0.5 x MIC, 0. 25 x MIC and 0.125 x MIC) are summarized in Table 2. All the tested extracts exhibited an inhibitory effect on biofilm formation of PS-1. In addition, biofilm inhibition activity of these extracts was significantly increased with increasing of sub-MIC level, except the difference in biofilm inhibition between 0.25× MIC and 0.125× MIC levels of thyme extract was not significant. Pomegranate and rosemary extracts at 0.5× MIC level were the most effective extracts in biofilm inhibition (≤ 91%), followed by peppermint (85.47%), ginger (71.56%), clove (67.41%), cinnamon (64%), and thyme (59.37%) extracts. There was no significantly difference in biofilm inhibition among pomegranate, rosemary, and peppermint extracts at 0.25× MIC level, which inhibited 71% of PS-1 biofilm; however, the biofilm inhibition of ginger, clove, cinnamon, and thyme extracts at this level was significantly decreased to 64.3, 61.0, 54.47, and 48.1%, respectively. Rosemary extract achieved the highest inhibition (72.33%) of PS-1 biofilm at 0.125× MIC level, followed by pomegranate (64.63%), ginger (51.7%), clove (50.87%), cinnamon (50.7%), peppermint (48.5%), and thyme (47.43%) extracts (Table 2). Previous data revealed that pomegranate and rosemary extracts were the most effective extracts in biofilm inhibition compared with other extracts. Thus, both extracts were selected for the remainder of the subsequent studies.
Table 2.
The inhibition activity of sub-MIC levels of plant extracts against biofilm formation of PS-1
| Sub-MIC Levels |
Plant extracts (MIC) | ||||||
|---|---|---|---|---|---|---|---|
| Cinnamon (6.25 mg/ml) |
Clove (6.25 mg/ml) |
Thyme (6.25 mg/ml) |
Rosemary (12.5 mg/ml) |
Peppermint (12.5 mg/ml) |
Pomegranate (6.25 mg/ml) |
Ginger (12.5 mg/ml) |
|
| Biofilm inhibition mean %1 ± SD | |||||||
| 0.5× MIC | 64.00 ± 0.12a | 67.41 ± 0.02d | 59.37 ± 0.02e | 90.83 ± 0.01g | 85.47 ± 0.01i | 91.93 ± 0.01g | 71.56 ± 0.01h |
| 0.25× MIC | 54.47 ± 0.57b | 61.00 ± 0.06e | 48.10 ± 0.02f | 72.33 ± 0.01h | 71.23 ± 0.57h | 72.30 ± 0.14h | 64.30 ± 0.01a |
| 0.125× MC | 50.70 ± 0.14c | 50.87 ± 0.07c | 47.43 ± 0.01f | 72.33 ± 0.02h | 48.50 ± 0.13f | 64.63 ± 0.06a | 51.70 ± 0.08c |
1: The inhibition % calculated from biofilm control (untreated with plant extract); SD, standard deviation. Statistical analysis was performed using two-way ANOVA and Tukey’s test (P < 0.05); different small letters represent the significant between means of the percentage of biofilm inhibition. The maximum biofilm inhibition activity shown in bold letters
Phytochemical analysis of pomegranate and rosemary extracts
The preliminary phytochemical screening of pomegranate and rosemary extracts showed that both extracts contained appreciable amount of tannins, saponins, quinones, terpenoids, steroids, flavonoids, phenols, and alkaloids; however, anthocyanin was almost negligible in both extracts. Coumarins and betacyanin were detected only in pomegranate, while glycosides detected only in rosemary extract. Additionally, the total phenolic contents of pomegranate was relatively high (255.41 mg/g) compared with rosemary (187.59 mg/g). It is noteworthy that the major polyphenol compounds identified in pomegranate were pyrogallol (4.4%), benzoic acid (1.4%), catechol (0.35%), gallic (0.21%), and ellagic (0.07%), while the rosmarinic acid (0.21%), benzoic acid (0.9%), and catechol (0.26%) were the major components in rosemary extract (Table 3).
Table 3.
Phytochemical analysis of 95% ethanol extracts of pomegranate and rosemary
| Phytochemical group |
Ethanol plant extracts | Phytochemical constituent |
Ethanol plant extracts | ||
|---|---|---|---|---|---|
| Pomegranate | Rosemary | Pomegranate | Rosemary | ||
| Identity | Yield % (W/W) | ||||
| Tannins | + | + | Pyrogallol | 4.44 | 1.09 |
| Saponins | + | + | Benzoic | 1.44 | 0.90 |
| Quinones | + | + | Chlorogenic | 0.53 | 0.63 |
| Terpenoids | + | + | Catechol | 0.35 | 0.26 |
| Steroids | + | + | Protocatchuic | 0.27 | 0.26 |
| Flavonoids | + | + | Vanillic | 0.22 | 0.24 |
| Phenols | + | + | Gallic | 0.21 | 0.22 |
| Alkaloids | + | + | P-OH-benzoic | 0.15 | 0.21 |
| Glycosides | – | + | Rosmarinic | 0.07 | 0.21 |
| Cardiac glycosides | + | + | Ellagic | 0.07 | 0.21 |
| Coumarins | + | – | Alpha-coumaric | 0.05 | 0.07 |
| Anthocyanin | – | – | Ferulic | 0.05 | 0.05 |
| Betacyanin | + | – | Caffeic | 0.04 | 0.05 |
| Salycilic | 0.04 | 0.05 | |||
| P-coumaric | 0.01 | 0.04 | |||
| Cinnamic | 0.01 | 0.03 | |||
| Iso-Ferulic | 0.01 | 0.02 | |||
| Total polyphenol contents [3] | 25.54 | 18.76 | |||
+, presence; −, absence
Inhibition of bacterial motilities
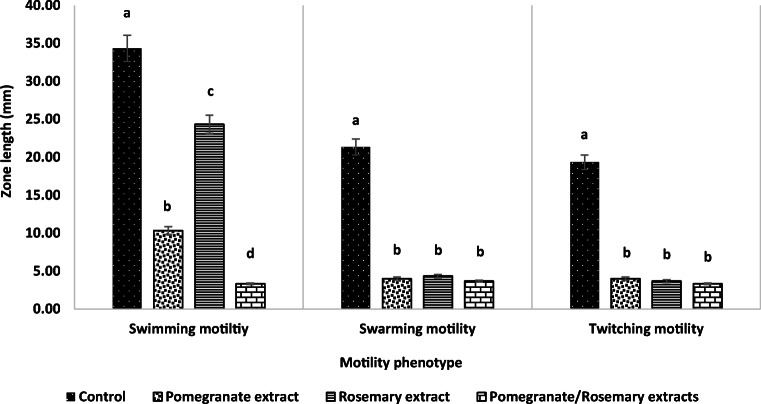
The inhibition activities of pomegranate and rosemary extracts alone and in combination against different phenotypes of PS-1 motilities at 0.25× MIC level are summarized in Fig. 5.The results showed that both extracts were significantly reduced the three phenotypes of motility without inhibiting of bacterial growth. Compared with control, pomegranate and rosemary extracts were significantly reduced > 95% of swarming and twitching motilities of PS-1. However, both extracts reduced the swimming motility by approximately 68.6 and 28.6%, respectively. Interestingly, the combination of pomegranate/rosemary extracts was significantly reduced > 95% of different PS-1 motilities. Consequently, this combination was selected for remainder of the subsequent studies.
Fig. 5.
Effect of pomegranate and rosemary PEP on P. aeruginosa PS-1 motilities. Result represents the mean ± SD of at least three independent experiments performed in triplicate. Different letters indicate significant differences between the test groups of each motility (one-way ANOVA with Tukey’s post hoc test, P < 0.05)
Synergistic potential of both plant extracts with antibiotics
The synergistic activity of plant extracts (pomegranate/rosemary) at 0.25× MIC level with PRL, CDZ, IPM, CN, or LEV was evaluated against the planktonic cells of PS-1. The results showed that all examined combinations were exhibited a synergistic effect against PS-1, with FIC index ≤ 0.5. In addition, the MIC values of these antibiotics were highly decreased from 1024, 1024, 1024, 512, and 512 μg/ml to 2, 4, 32, 2, and 2 μg/ml, respectively. Furthermore, the susceptibility of the examined isolate to these antibiotics was changed from resistant to sensitive phenotype (Table 4).
Table 4.
The FIC of combination (antibiotic/POM/ROS) against planktonic cells of P. aeruginosa PS-1
| Antibiotics | MIC (mg/ml) | FIC 1 of combinations (antibiotic/POM/ROS) | FIC index2 (∑FIC) |
Type of interaction3 | ||
|---|---|---|---|---|---|---|
| Antibiotic | POM/ROS | Combinations (antibiotic/POM/ROS) | ||||
| PRL | 1.024 | 3.13/6.25 | 0.032/1.56/3.125 | 0.030/0.5 | 0.531 | Additive |
| CDZ | 0.512 | 3.13/6.25 | 0.002/1.56/3.125 | 0.004/0.5 | 0.504 | Synergism |
| IPM | 1.024 | 3.13/6.25 | 0.004/1.56/3.125 | 0.004/0.5 | 0.504 | Synergism |
| CN | 1.024 | 3.13/6.25 | 0.002/1.56/3.125 | 0.002/0.5 | 0.502 | Synergism |
| LEV | 0.512 | 3.13/6.25 | 0.002/1.56/3.125 | 0.004/0.5 | 0.504 | Synergism |
FIC, fractional inhibitory concentration; 1, FIC = MIC combination/MIC alone; 2, FIC index (∑FIC) = FIC of plant mixture + FIC of antibiotic 3, synergism (FIC index < 0.5); additive (FIC index ranging from 0. 5 to1.0); indifferent (FIC index ranging from 1.0 to 2.0) and antagonism (FIC index > 2). POM: Pomegranate; ROS: Rosemary; PRL: Piperacillin; CDZ: Ceftazidime; IPM: Imipenem; CN: Gentamycin; LEV: Levofloxacin
Eradication of strong biofilms
The eradication activities of plant extracts (pomegranate and rosemary) alone and in combination with five different antibiotics at MIC level against the mature biofilms (48 h) of strong biofilm producer isolates were summarized in Table 5.The results showed that the combination of plant extracts with antibiotics achieved the highest biofilm eradications against various examined isolates, with eradication range from 90 to 99.6%, followed by plant extracts combination, rosemary, and pomegranate extracts, with eradication ranges (76.5–85.4%), (53.1–73.7%), and (41.2–71.5%), respectively. The most effective combination in biofilm eradication against various examined isolates was pomegranate/rosemary/CDZ, with eradication ranging from 97.3 to 99.6%, followed by pomegranate/rosemary/CN, pomegranate/rosemary/IPM, pomegranate/rosemary/LEV, and pomegranate/rosemary/PRL combinations, with biofilm eradication ranges (94.32–98.3%), (93.7–98.3%), (93.33–98.2%), and (90–95.5%), respectively.
Table 5.
The eradication activity of the synergistic combinations against biofilm associated with P. aeruginosa strains
| Tested strains | Plant extracts alone and combined with antibiotic at MIC level | |||||||
|---|---|---|---|---|---|---|---|---|
| POM | ROS | POM/ROS | POM/ROS/PRL | POM/ROS/CDZ | POM/ROS/IPM | POM/ROS/CN | POM/ROS/LEV | |
| Biofilm eradication mean %1 ± SD | ||||||||
| PS-1 | 41.20 ± 0.5a | 54.14 ± 0.3e | 82.35 ± 1.3f | 91.52 ± 0.6h | 99.60 ± 0.04 i | 93.33 ± 1.3h | 94.32 ± 0.60 h | 93.70 ± 1.3 h |
| PS-2 | 62.51 ± 0.8b | 55.26 ± 1.8e | 84.60 ± 1.5f | 93.80 ± 2.1h | 98.70 ± 1.70 i | 98.10 ± 2.1 i | 95.80 ± 2.10 h | 94.80 ± 1.4 h |
| PS-4 | 60.20 ± 1.1b | 70.70 ± 0.5c | 76.5 ± 1.0g | 95.50 ± 0.7h | 98.60 ± 1.70 i | 96.30 ± 1.3 h | 97.70 ± 0.70 i | 97.10 ± 1.8i |
| PS-5 | 71.5 ± 1.00c | 73.70 ± 1.4c | 84.10 ± 1.6f | 91.30 ± 1.7g | 97.30 ± 2.10 i | 95.10 ± 0.1 h | 98.30 ± 0.50 i | 97.40 ± 1.9 i |
| PS-19 | 48.40 ± 2.1d | 53.10 ± 0.2e | 85.40 ± 1.5f | 90.00 ± 1.3h | 98.30 ± 1.30 i | 98.20 ± 1.7i | 94.00 ± 0.40 h | 98.30 ± 1.1 i |
1, the eradication % calculated from biofilm control (untreated). POM, Pomegranate (1.6 mg/ml); ROS, Rosemary (3.1 mg/ml); PRL, piperacillin (0.032 mg/ml); IPM, imipenem (0.004 mg/ml); CDZ, ceftazidime (0.002 mg/ml); CN, gentamicin (0.002 mg/ml); LEV, levofloxacin (0.002 mg/ml); SD, standard division. Statistical analysis was performed using two-way ANOVA and Tukey’s test (P < 0.05); different small letters represent the significant between means of biofilm eradication %. The most effective combination shown in bold letters
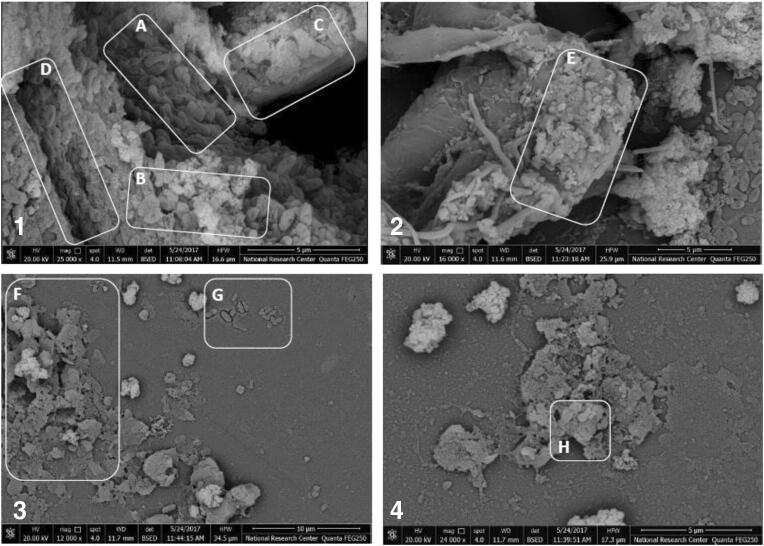
The SEM images of 48-h-old PS-1 biofilm treated with the most effective combination (pomegranate/rosemary/CDZ) compared with control (untreated) are depicted in Fig. 6. The SEM images of control samples revealed that PS-1 could produce a strong mature biofilm on the surface of glass slide cover within 48 h. A high density of compacted bacilli cells was observed in some areas of this biofilm, and most of these bacilli cells were embedded in extracellular polymeric matrix. Additionally, the obtained biofilm was comprised of water channels that function as a distribution system for oxygen and nutrients. However, the SEM images of biofilm after 24 h of treatment with the selected combination showed that most of biofilm structures were destroyed and removed from the surface of glass slide cover. The cells of this biofilm were detached from the biofilm structure, and most of them were eliminated.
Fig. 6.
SEM images of 48-h-old P. aeruginosa PS-1 biofilm before and after 24 h of treatment with MIC level of POM/ROS/CDZ combination. 1 and 2, images of untreated 48-h-old P. aeruginosa PS-1 biofilm; 3 and 4, images of 48-h-old P. aeruginosa PS-1 biofilm after 24 h of treatment with MIC level of POM/ROS/CDZ combination. a PS-1 micro-colonies; b extracellular polymeric substance; c bacterial cell of PS-1 capsulated by fibrous matrixes; d water channels; e Bacterial cell of PS-1 capsulated by fibrous matrixes; f destructed PS-1 biofilm after 24 h of combination treatment; g a few number of bacterial cells remained after 24 h of combination treatment; h damaged bacterial cell walls remained after 24 h of combination treatment
Discussion
The chronic infections with P. aeruginosa are mainly due to form biofilm, which increases its resistance to conventional antibiotics by adding some mechanisms. Thus, the effectiveness of conventional antibiotics became limited due to their higher values of their MIC and MBC, which may results in vivo toxicity [5,29].
The present study indicated the biofilm mass associated P. aeruginosa isolates were varied from weak to strong biofilm. The differences among these isolates in biofilm formation is probably due to the expression differences in genes responsible for exopolysaccharides, extracellular proteins, quorum-sensing molecules, flagella for swimming motilities, and pili for swarming and twitching motility [30,31]. Additionally, the capacity of biofilm formation is influenced by the clinical sources and the patient’s conditions [32] .
Antibiotic susceptibility testing in the current study revealed that the examined P. aeruginosa isolates were resistant to at least 30% of all tested antibiotics and elevated to 40% with PRL; however, 100% of these isolates were resistant to FEP, CDZ, IPM, and NA. The highest MIC value of antibiotics against strong biofilm producer isolates (PS-1, PS-2, PS-4, PS-5, and PS-19) was observed with PRL (1024 μg/ml) followed by CDZ, CN, IPM, and LEV, with MIC ranges (256–1024 μg/ml), (32–1024 μg/ml), (8–1024 μg/ml), and (8–512 μg/ml), respectively. The obtained results are consistent with previous studies, which noticed the high resistance of P. aeruginosa to conventional antibiotics, as a result of excessive antibiotic administration, which leads to the ineffectiveness of the empirical antibiotic therapy against this bacterium [33]. Pang et al. reported that P. aeruginosa are known to utilize their high levels of intrinsic and acquired resistance mechanisms to most conventional antibiotics [29]. In addition, adaptive antibiotic resistance of P. aeruginosa is a recently characterized mechanism, which includes biofilm-mediated resistance and formation of multidrug-tolerant persister cells, and is responsible for recalcitrance and relapse of infections.
The Present study demonstrated that there is no relationship between the capacity of biofilm formation and antibiotic resistance; our investigation revealed that PS-1 and PS-19 exhibited high antibiotic resistant associated with strong biofilm formation; however, PS-15 and PS-16 were high antibiotic resistant and non-biofilm producers; moreover, some other isolates in the present study were classified as moderate biofilm producers and resistant to one or more group of tested antibiotics. Thus, the resistance of bacteria to antibiotics is inappropriate marker for its efficiency to biofilm formation. Obtained results were supported by recent study [34], which revealed that there was no significant difference among strong, moderate, and weak biofilm producers in their resistance to penicillin, cefoxitin, and chloramphenicol. However, Qi et al. noticed that the correlation between the capacity of biofilm formation and the resistance of Acinetobacter baumannii isolates to antibiotics was negative, which mean that the biofilm forming isolates are less dependent on antibiotic resistance than no biofilm-forming isolates for survival [35]. Other studies demonstrated that biofilm resistance to antimicrobials is multifaceted, including reduced penetration of the agent into biofilms due to the presence of extracellular matrix, biofilm heterogeneity, and biofilm-specific phenotypes such as expression of efflux pump and persister cells [36,37].
The present study showed that pomegranate, thyme, cinnamon, rosemary, clove, and peppermint extracts exhibited different levels of antibacterial and antibiofilm activities against representative isolate (PS-1). Generally, the antibacterial and antibiofilm activities of these extracts were mainly due to presence of a large number of phytochemical compounds (tannins, saponins, flavonoids, phenols, etc.), which can affect multiple target sites against the bacterial cells [7,8,38,39]. The differences among these extracts in their antibacterial and antibiofilm activities are due to variation in their chemical constituents and volatile nature of their components [40] .
Data in the present study revealed that the most effective plant extracts in biofilm inhibition were pomegranate and rosemary extracts, which reduced > 95% the swarming and twitching motilities of PS-1, decreased swimming motility (68.6 and 28.6% of control, respectively), and reduced (91.93 and 90.83%, respectively) the biofilm formation of PS-1. The differences between both examined extracts in swimming motility might be due to their differences in the type and concentration of the active constituents, as well as the polyphenols contents (255.41 and 187.59 mg/g, respectively). The data of this study suggested that the biofilm inhibition activity of these extracts is due to their inhibition of swimming motility that led to a reduce of bacterial cells attaching to the surfaces, therefore decreasing of colony wetness and extra polymeric layer as well as quorum-sensing signals. These factors are necessary for swarming motility and development of bacterial biofilm [41,42]. Additionally, biofilm formation could be decreased by suppression of twitching motility, which is necessary for bacterial cells attaching and biofilm extending to new surface, as well as monolayer assembling of P. aeruginosa cells into micro-colonies [43]. Furthermore, suppression of swarming motility led to reduce of biofilm capacity because this motility is essential for biofilm development and maturation [44].
Biofilm inhibition activity of pomegranate and rosemary extracts might be explained by the presence polyphenol compounds, such as pyrogallol, catechol, gallic, ellagic, rosmarinic acid, and benzoic acid. Obtained results are consistent with previous studies, which revealed that the antagonistic activity of polyphenols against P. aeruginosa biofilms were due to disable quorum-sensing system, suppression of bacterial cell adherence and motilities, as well as inhibiting of polymeric matrix synthesis [39,45–47]. Defoirdt et al. found that pyrogallol inhibited quorum sensing through generation of H2O2 that somehow interfere with the expression of bioluminescence in Vibrio sp. [48]. Rudrappa et al. reported that biofilm inhibition of Bacillus subtilis was due to the physiological response by B. subtilis to the presence of catechol, which resulted in the down regulation of transcription of the yqxM-sipW-tasA and epsA-O operons, both of which were required for biofilm formation by B. subtilis [49]. Plyuta et al. found that the swarming motility of P. aeruginosa PAO1 was significantly decreased by 12–30% in the presence of 400–800 μg/mL of 4-hydroxybenzoic acid and by 20–50% at the same concentration of gallic acid; however, twitching motility zones were decreased by 10–15% in the presence of each phytochemical at the same concentration [50]. Ellagic acid exhibited high quorum-sensing inhibition in Chromobacterium violaceum, E. coli MT102, and Pseudomonas putida at concentrations of 40, 36, and 30 μg/mL, respectively [51]. The antagonistic activity of rosmarinic acid against the biofilm formation of P. aeruginosa PAO1, GU447238, and GU447238 was due to its inhibitory effect on the activity of protease, elastase, and hemolysin produced by these isolates [52] .
Despite the broad spectrum of antimicrobial and antibiofilm activities of a vast number of plant extracts against various pathogenic microorganisms, there were few evidences about their effectiveness alone (without chemical drugs) in clinical treatments [53,54]. However, the use of plant extracts or pure natural compounds in combination with conventional antibiotics may hold greater promise for inhibiting and eradicating microbial biofilms [7,55].
In this study, the combinations of both plants’ extracts (pomegranate and rosemary) with PRL, CDZ, IPM, CN, or LEV exhibited synergy effects (FIC index ≤ 0.5) against P. aeruginosa PS-1. The MIC values of these antibiotics were decreased to 32, 256, 256, 512, and 256 fold in these combinations compared with each antibiotic alone. The synergistic interaction between both extracts and antibiotics with different kinds, regardless of their mechanisms of action, suggested that it is not only one compound that is responsible for the observed synergistic effect but that each of the identified compounds contributes to this effect resulting in a pleiotropic effects of the both extracts. The obtained results were in agreement with past literature, which noticed that the combinations of plant extracts with antibiotics belonging to different families show synergy against clinical isolates of Gram-positive and negative bacteria, significantly reducing the MIC of all antibiotics tested [56,57]. Therefore, these combinations can be used for expanding the antimicrobial spectrum, preventing the emergence of antibiotic resistant bacteria, and diminishing toxicity, since lower concentration of these antibiotics can be used. The synergism effect in these combinations against PS-1 is due to the presence of the polyphenol compounds such as pyrogallol, catechol, gallic, ellagic, rosmarinic acid, and benzoic acid, which are capable of interacting with the cytoplasmic membrane, cell wall, nucleic acids, and energy transport and altering or inhibiting their functions [58–60].
Phytochemical analysis in the present study revealed that pyrogallol, catechol, gallic, ellagic, rosmarinic acid, and benzoic acid were the most abundant phenolic compounds identified in pomegranate and rosemary extracts. Pyrogallol exhibited synergistic interaction with aminoglycoside and quinolones antibiotics against Gram-positive bacteria; however, these combinations exhibited indifferent interaction against Gram-negative bacteria, especially P. aeruginosa [61]. Catechol is reported to cytoplasmic membrane damage and causing direct disruption of the lipid bilayers and alteration of the barrier function, which leads to enhanced penetration of antibiotics and decreasing their MICs [62,63]. The synergistic effect of both extracts with antibiotics was probably due to the presence of gallic, benzoic, and rosmarinic acids, which decreases the bacterial constituents (proteins, nucleic acids, and inorganic ions such as potassium or phosphate) by increasing the permeability of the bacterial cytoplasmic membrane. Another interesting mechanism of gallic acid is inhibiting supercoiling activity of bacterial gyrase by binding to the ATP binding site of gyrase B [64,65]. Chusri et al. found that ellagic act as efflux pump inhibitors in Gram negative bacteria and increased the effectiveness of some antibiotics against multi-drug-resistant Acinetobacter baumannii [66].
Biofilm mass of strong biofilm producer isolates were significantly reduced after 24 h of treatment with pomegranate and rosemary extracts alone and in combination with antibiotics (PRL, CDZ, IPM, CN, or LEV) at MIC level. Biofilm eradication of rosemary extract was relatively high (range, 53.1 to 73.7%) compared with the eradication activity (range, 41.2–71.5%) of pomegranate extract. In addition, the combination of both extracts was significantly increased the biofilm eradication to the range from 76.5 to 85.4%. Moreover, the eradication activity of both plant extracts/antibiotic combinations was significantly high (range, 76.5–99.6%) compared with other treatments in the present study. The differences between rosemary and pomegranate extracts in biofilm eradication can be explained by the differences in their polyphenol contents, concentrations, and mode of actions. The significant increase of biofilm eradication of the plant extracts combination compared with each single plant extract that might probably be attributed to the synergistic interaction between both extracts. The synergistic interaction of both extracts/antibiotic combinations had been proven in the current study, and that could explain the significant increasing of biofilm eradication of these combinations compared with the plant extracts alone and in combination. Furthermore, the eradication activities of these combinations against biofilms were supported by the images of electron microscope after 24 h of mature biofilm treatment with MIC level of pomegranate/rosemary/CDZ combination. These images showed high density of compacted bacilli cells in some areas of control biofilm (untreated), and most of these bacilli cells were embedded in extracellular polymeric matrix; however, the treated biofilm was completely destroyed and removed from the surface of glass slide cover, and most of the bacterial cell were eliminated.
Conclusion
Current study proposed new promising combinations of pomegranate and rosemary extracts with antibiotics (PRL, CDZ, IPM, CN, or LEV). These combinations disrupt P. aeruginosa biofilms by blocking of different bacterial motilities, destroying of biofilm architecture, and increasing the efficacy of antibiotics by decreasing their MIC levels. These findings may the first of many steps needed to complete this work. Further studies are required to assess the in vivo benefit of these combinations in treatment of P. aeruginosa biofilm.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Olivares E, Badel-Berchoux S, Provot C, Prévost G, Bernardi T, Jehl F. Clinical impact of antibiotics for the treatment of Pseudomonas aeruginosa biofilm infections. Front Microbiol. 2020;10:2894. doi: 10.3389/fmicb.2019.02894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyawali R, Ibrahim SA. Natural products as antimicrobial agents. Food Control. 2014;46:412–429. doi: 10.1016/J.FOODCONT.2014.05.047. [DOI] [Google Scholar]
- 3.Karuppiah P, Mustaffa M. Antibacterial and antioxidant activities of Musa sp. leaf extracts against multidrug resistant clinical pathogens causing nosocomial infection. Asian Pac J Trop Biomed. 2013;3(9):737–742. doi: 10.1016/S2221-1691(13)60148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lizana JA. Use of plant extracts to block bacterial biofilm formation. In: High School Students for Agricultural Science Research, Proceedings of the 3rd Congress PIIISA. (Olías, Raquel; Belver, Andrés; Sahrawy, Mariam; Serrato, Antonio Jesús; Cárdenas, Katiuska E.; Sandalio, Luisa M.; Rodríguez Serrano, María; Corpas, Francisco J.; Palma Martínez, José Manuel; Castro López AJ, ed.). CSIC - Estación Experimental del Zaidín (EEZ); 2013. http://digital.csic.es/handle/10261/99998. Accessed December 22, 2017
- 5.Roy R, Tiwari M, Donelli G, Tiwari V. (2018) Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action., 9:522, 554 10.1080/21505594.2017.1313372 [DOI] [PMC free article] [PubMed]
- 6.Cheesman MJ, Ilanko A, Blonk B, Cock IE. Developing new antimicrobial therapies: are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn Rev. 2017;11(22):57–72. doi: 10.4103/phrev.phrev_21_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kali A, Bhuvaneshwar D, Charles PMV, Seetha KS. Antibacterial synergy of curcumin with antibiotics against biofilm producing clinical bacterial isolates. J basic Clin Pharm. 2016;7(3):93–96. doi: 10.4103/0976-0105.183265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H, Deng Y, Wang H, Liu W, Zhuang X, Chu W. Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci Rep. 2015;5(1):16158. doi: 10.1038/srep16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassiri-Jahromi S. Punica granatum (pomegranate) activity in health promotion and cancer prevention. Oncol Rev. 2018;12(1):1–7. doi: 10.4081/oncol.2018.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mastrogiovanni F, Mukhopadhya A, Lacetera N et al (2019) Anti-inflammatory effects of pomegranate peel extracts on in vitro human intestinal caco-2 cells and ex vivo porcine colonic tissue explants. Nutrients 11(3). 10.3390/nu11030548 [DOI] [PMC free article] [PubMed]
- 11.Pérez-Sánchez A, Barrajón-Catalán E, Ruiz-Torres V et al (2019) Rosemary (Rosmarinus officinalis) extract causes ROS-induced necrotic cell death and inhibits tumor growth in vivo. Sci Rep. 9(1). 10.1038/s41598-018-37173-7 [DOI] [PMC free article] [PubMed]
- 12.O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. [6] Genetic approaches to study of biofilms Methods Enzymol 1999;310:91–109. doi:10.1016/S0076-6879(99)10008-9 [DOI] [PubMed]
- 13.Stiefel P, Rosenberg U, Schneider J, Mauerhofer S, Maniura-Weber K, Ren Q. Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Appl Microbiol Biotechnol. 2016;100(9):4135–4145. doi: 10.1007/s00253-016-7396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alcaráz LE, Satorres SE, Lucero RM, Puig De Centorbi ON. Species identification, slime production and oxacillin susceptibility in coagulase-negative staphylococci isolated from nosocomial specimens. Brazilian J Microbiol. 2003;34:45–51. doi: 10.1590/S1517-83822003000100010. [DOI] [Google Scholar]
- 15.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement Clinical and Laboratory Standards Institute. 2011;31(15). http://vchmedical.ajums.ac.ir/_vchmedical/documents/CLSI 2011.pdf. Accessed December 22, 2017
- 16.CLSI. Methods for determining bactericidal activity of antimicrobial agents; approved guideline M26-A. CLSI. 1999;19(18):7. https://clsi.org/media/1462/m26a_sample.pdf. Accessed December 22, 2017
- 17.CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved Standard-Ninth Edition. CLSI document M07-A9. 2012;32(2):92
- 18.Kawsud P, Puripattanavong J, Teanpaisan R. Screening for anticandidal and antibiofilm activity of some herbs in Thailand. Trop J Pharm Res. 2014;13(9):1495. doi: 10.4314/tjpr.v13i9.16. [DOI] [Google Scholar]
- 19.Ivanova V, Stefova M, Chinnici F (2010) Determination of the polyphenol contents in Macedonian grapes and wines by standardized spectrophotometric methods. J Serb Chem Soc 7585285366324(6345). 10.2298/JSC1001045I
- 20.Goupy P, Hugues M, Boivin P, Amiot MJ. Antioxidant composition and activity of barley (Hordeum vulgare) and malt extracts and of isolated phenolic compounds. J Sci Food Agric. 1999;79(12):1625–1634. doi: 10.1002/(SICI)1097-0010(199909)79:12<1625::AID-JSFA411>3.0.CO;2-8. [DOI] [Google Scholar]
- 21.Abd El-Salam AE, Abd-El-Haleem D, Youssef AS, Zaki S, Abu-Elreesh G, El-Assar SA. Isolation, characterization, optimization, immobilization and batch fermentation of bioflocculant produced by Bacillus aryabhattai strain PSK1. J Genet Eng Biotechnol. 2017;15:335–344. doi: 10.1016/j.jgeb.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manandhar S, Luitel S, Dahal RK (2019) In vitro antimicrobial activity of some medicinal plants against human pathogenic bacteria 2019. doi:10.1155/2019/1895340, 1, 5 [DOI] [PMC free article] [PubMed]
- 25.Sabaeifard P, Abdi-Ali A, Soudi MR, Dinarvand R. Optimization of tetrazolium salt assay for Pseudomonas aeruginosa biofilm using microtiter plate method. J Microbiol Methods. 2014;105:134–140. doi: 10.1016/j.mimet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 26.O’May C, Tufenkji N. The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl Environ Microbiol. 2011;77(9):3061–3067. doi: 10.1128/AEM.02677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White RL, Burgess DS, Manduru M, Bosso JA. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob Agents Chemother. 1996;40(8):1914–1918. doi: 10.1128/AAC.40.8.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceylan O, Uğur A, Saraç N, Ozcan F, Baygar T (2014) The in vitro antibiofilm activity of Rosmarinus officinalis L essential oil against multiple antibiotic resistant Pseudomonas sp and Staphylococcus sp 12(3&4):82–86
- 29.Pang Z, Raudonis R, Glick BR, Lin T-J, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Li M-Y, Zhang J, Lu P, Xu J-L, Li S-P. Evaluation of biological characteristics of bacteria contributing to biofilm formation. Pedosphere. 2009;19(5):554–561. doi: 10.1016/S1002-0160(09)60149-1. [DOI] [Google Scholar]
- 31.Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by <em>Staphylococcus aureus</em> isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. 2018;11:25–32. doi: 10.2147/IJGM.S153268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez LRR, Costa MCN, Freitas ALP, Barth AL. Evaluation of biofilm production by Pseudomonas aeruginosa isolates recovered from cystic fibrosis and non-cystic fibrosis patients. Brazilian J Microbiol. 2011;42:476–479. doi: 10.1590/S1517-83822011000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yayan J, Ghebremedhin B, Rasche K. Antibiotic Resistance of Pseudomonas aeruginosa in pneumonia at a Single University Hospital Center in Germany over a 10-year period. Webber MA, ed. PLoS One. 2015;10(10):e0139836. doi: 10.1371/journal.pone.0139836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Xu D, Shi L, Cai R, Li C, Yan H. Association between agr type, virulence factors, Biofilm formation and antibiotic resistance of Staphylococcus aureus isolates From Pork Production. Front Microbiol. 2018;9:1876. doi: 10.3389/fmicb.2018.01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi L, Li H, Zhang C, Liang B, Li J, Wang L, du X, Liu X, Qiu S, Song H. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet (London, England) 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 37.Akinbobola AB, Sherry L, Mckay WG, Ramage G, Williams C. Tolerance of Pseudomonas aeruginosa in in-vitro biofilms to high-level peracetic acid disinfection. J Hosp Infect. 2017;97(2):162–168. doi: 10.1016/j.jhin.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 38.Husain FM, Ahmad I, Khan MS, Ahmad E, Tahseen Q, Khan MS, Alshabib NA. Sub-MICs of Mentha piperita essential oil and menthol inhibits AHL mediated quorum sensing and biofilm of gram-negative bacteria. Front Microbiol. 2015;6:420. doi: 10.3389/fmicb.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero CM, Vivacqua CG, Abdulhamid MB, Baigori MD, Slanis AC, Allori MCG, Tereschuk ML. Biofilm inhibition activity of traditional medicinal plants from northwestern Argentina against native pathogen and environmental microorganisms. Rev Soc Bras Med Trop. 2016;49(6):703–712. doi: 10.1590/0037-8682-0452-2016. [DOI] [PubMed] [Google Scholar]
- 40.Mostafa AA, Al-Askar AA, Almaary KS, Dawoud TM, Sholkamy EN, Bakri MM. Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi J Biol Sci. 2018;25(2):361–366. doi: 10.1016/j.sjbs.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2(7):a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110(28):11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa : a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015;2015:1–17. doi: 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gopu V, Kothandapani S, Shetty PH. Quorum quenching activity of Syzygium cumini (L.) Skeels and its anthocyanin malvidin against Klebsiella pneumoniae. Microb Pathog. 2015;79:61–69. doi: 10.1016/j.micpath.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Liu Q, Meng X, Li Y, Zhao C-N, Tang G-Y, Li H-B. Antibacterial and antifungal activities of spices. Int J Mol Sci. 2017;18(6):1283. doi: 10.3390/ijms18061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva LN, Zimmer KR, Macedo AJ, Trentin DS. Plant natural products targeting bacterial virulence factors. Chem Rev. 2016;116(16):9162–9236. doi: 10.1021/acs.chemrev.6b00184. [DOI] [PubMed] [Google Scholar]
- 48.Defoirdt T, Pande GSJ, Baruah K, Bossier P. The apparent quorum-sensing inhibitory activity of pyrogallol is a side effect of peroxide production. Antimicrob Agents Chemother. 2013;57(6):2870–2873. doi: 10.1128/AAC.00401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudrappa T, Quinn WJ, Stanley-Wall NR, Bais HP. A degradation product of the salicylic acid pathway triggers oxidative stress resulting in down-regulation of Bacillus subtilis biofilm formation on Arabidopsis thaliana roots. Planta. 2007;226(2):283–297. doi: 10.1007/s00425-007-0480-8. [DOI] [PubMed] [Google Scholar]
- 50.Plyuta V, Zaitseva J, Lobakova E, Zagoskina N, Kuznetsov A, Khmel I. Effect of plant phenolic compounds on biofilm formation by Pseudomonas aeruginosa. APMIS. 2013;121(11):1073–1081. doi: 10.1111/apm.12083. [DOI] [PubMed] [Google Scholar]
- 51.Ta C, Freundorfer M, Mah T-F, Otárola-Rojas M, Garcia M, Sanchez-Vindas P, Poveda L, Maschek J, Baker B, Adonizio A, Downum K, Durst T, Arnason J. Inhibition of bacterial quorum sensing and biofilm formation by extracts of neotropical rainforest plants. Planta Med. 2014;80(04):343–350. doi: 10.1055/s-0033-1360337. [DOI] [PubMed] [Google Scholar]
- 52.Annapoorani A, Umamageswaran V, Parameswari R, Pandian SK, Ravi AV. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J Comput Aided Mol Des. 2012;26(9):1067–1077. doi: 10.1007/s10822-012-9599-1. [DOI] [PubMed] [Google Scholar]
- 53.Nasri H, Bahmani M, Shahinfard N, Moradi Nafchi A, Saberianpour S, Rafieian KM. Medicinal plants for the treatment of acne vulgaris: a review of recent evidences. Jundishapur J Microbiol. 2015;8(11):e25580. doi: 10.5812/jjm.25580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pramila DM, Xavier R, Marimuthu K, et al. Phytochemical analysis and antimicrobial potential of methanolic leaf extract of peppermint (Mentha piperita: Lamiaceae) J Med Plants Res. 2012;6(3):331–335. doi: 10.5897/JMPR11.1232. [DOI] [Google Scholar]
- 55.Cheesman M, Ilanko A, Blonk B, Cock I. Developing new antimicrobial therapies: are synergistic combinations of plant extracts/compounds with conventional antibiotics the solution? Pharmacogn Rev. 2017;11(22):57–72. doi: 10.4103/phrev.phrev_21_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luana Kamila A. Braga. Potentiation of in vitro antibiotic activity by Ocimum gratissimum L. African J Pharm Pharmacol. 2011;5(19). doi:10.5897/AJPP11.414
- 57.Stefanović OD, Stanković MS, Čomić LR. In vitro antibacterial efficacy of Clinopodium vulgare L. extracts and their synergistic interaction with antibiotics. 2011. https://www.semanticscholar.org/paper/In-vitro-antibacterial-efficacy-of-Clinopodium-L.-Stefanović-Stanković/604bae6a4a7f305d3cd647fdd3fbc743c426641a.
- 58.Miklasińska-Majdanik M, Kępa M, Wojtyczka R, Idzik D, Wąsik T. Phenolic compounds diminish antibiotic resistance of Staphylococcus aureus clinical strains. Int J Environ Res Public Health. 2018;15(10):2321. doi: 10.3390/ijerph15102321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie Y, Yang W, Tang F, Chen X, Ren L. Antibacterial activities of flavonoids: structure-activity relationship and mechanism. Curr Med Chem. 2015;22(1):132–149. doi: 10.2174/0929867321666140916113443. [DOI] [PubMed] [Google Scholar]
- 60.Sanhueza L, Melo R, Montero R, Maisey K, Mendoza L, Wilkens M. Synergistic interactions between phenolic compounds identified in grape pomace extract with antibiotics of different classes against Staphylococcus aureus and Escherichia coli. Agbor G, ed. PLoS One. 2017;12(2):e0172273. doi: 10.1371/journal.pone.0172273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lima VN, Oliveira-Tintino CDM, Santos ES, Morais LP, Tintino SR, Freitas TS, Geraldo YS, Pereira RLS, Cruz RP, Menezes IRA, Coutinho HDM. Antimicrobial and enhancement of the antibiotic activity by phenolic compounds: gallic acid, caffeic acid and pyrogallol. Microb Pathog. 2016;99:56–61. doi: 10.1016/j.micpath.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Amin MU, Khurram M, Khattak B, Khan J. Antibiotic additive and synergistic action of rutin, morin and quercetin against methicillin resistant Staphylococcus aureus. BMC Complement Altern Med. 2015;15(1):59. doi: 10.1186/s12906-015-0580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fazly Bazzaz BS, Sarabandi S, Khameneh B, Hosseinzadeh H. Effect of catechins, green tea extract and methylxanthines in combination with gentamicin against Staphylococcus aureus and Pseudomonas aeruginosa: - combination therapy against resistant bacteria. Aust J Pharm. 2016;19(4):312–318. doi: 10.3831/KPI.2016.19.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yi Z-B, Yu Y, Liang Y-Z, Bao Z. Evaluation of the antimicrobial mode of berberine by LC/ESI-MS combined with principal component analysis. J Pharm Biomed Anal. 2007;44(1):301–304. doi: 10.1016/j.jpba.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 65.Jayaraman P, Sakharkar MK, Lim CS, Tang TH, Sakharkar KR. Activity and interactions of antibiotic and phytochemical combinations against Pseudomonas aeruginosa in vitro. Int J Biol Sci. 2010;6(6):556–568. doi: 10.7150/ijbs.6.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chusri S, Villanueva I, Voravuthikunchai SP, Davies J. Enhancing antibiotic activity: a strategy to control Acinetobacter infections. J Antimicrob Chemother. 2009;64(6):1203–1211. doi: 10.1093/jac/dkp381. [DOI] [PubMed] [Google Scholar]