Abstract
Purpose
Five Lactobacillus strains isolated from sucuk (Turkish dry-fermented sausage) were studied for their genetic and technological properties.
Methods
For genotypic identification, strains 16S rRNA gene sequences were used. To determine the antimicrobial activity of strains, seven foodborne pathogens were tested. Strains technological properties were characterized.
Results
These strains were identified as Lactobacillus plantarum by 16S rRNA gene sequence analysis and the phylogenetic tree obtained by neighbor-joining method allowed grouping of these strains into three subgroups. L. plantarum strains showed antagonistic activities against Staphylococcus aureus, Listeria monocytogenes, Bacillus cereus, and Micrococcus luteus strains. PCR assay, using specific primers, showed the presence of bacteriocin (plantaricin) encoding genes in all L. plantarum strains tested. Antimicrobial metabolite production of these strains started at log phase and reached the maximum level at the end of the stationary phase. Regarding their technological properties, better growth was observed at 25 °C compared with 15 °C and 45 °C. The isolates which grown well within the pH scale pH 4.5–6.5 range additionally showed a decent growth at 6.5% salt concentration. It has been found that strains do not exhibit lipolytic and proteolytic activities nor have lysine, ornithine, and arginine decarboxylase activity. On the other hand, one strain showed weak nitrate reductase activity, and four strains produced acetoin from glucose. In addition, all strains were DL-lactic acid producers. Consequently, L. plantarum strains isolated exhibited some biochemical properties required for a starter culture in sucuk and similar products.
Conclusions
All identified strains may be a protective culture in the production of fermented meat products. In particular, L. plantarum S51 was distinguished from other isolates due to the inability to form acetoin from glucose. Further work will be needed to characterize L. plantarum strains as starter culture.
Keywords: Lactobacillus plantarum, Fermented sausage, Characterization, Antimicrobial activity
Introduction
Sucuk (Turkish dry-fermented sausage) is a very popular meat product in Turkey. It is a dry-fermented sausage variety produced traditionally and industrially. Traditional production is usually carried out in small-scale enterprises without the use of starter cultures, and sausages are ripened under natural conditions. In industrial production, starter cultures are used for controlled fermentation, and air conditioning units that can automatically control temperature, air current, and relative humidity for fermentation and drying processes.
Lactic acid bacteria (LAB) are the main bacteria group for sausage fermentation that improve the safety and sensorial properties of fermented sausages. The major role of this group is to provide rapid acidification by lactic acid production during sausage fermentation. They also produce other important products such as acetic acid, ethanol, volatile compounds, and antimicrobial metabolites. Some metabolic end products, like bacteriocins and bacteriocin-like inhibitory substance (BLIS), are responsible for antagonistic effects of LAB [1]. As the rate and extent of acid formation of LAB are effective, the antagonistic activity of the strains to be used against foodborne pathogens is also important for ensuring product safety. In terms of standardized production, the use of starter cultures is also of great importance. In many countries, strains isolated and identified from traditional products have been used in starter cultivation and are still being explored. Traditionally produced fermented products are preferred for their sensory properties [2]. However, it is not always possible to achieve the same level of quality and safety in traditional products [3]. The use of autochthonous lactic acid bacteria as a starter culture permits to produce sausages with similar sensory properties to the traditional production [4]. Autochthonous starter cultures isolated from traditional fermented products are well adapted to fermentation conditions [5, 6]. Many LAB strains were isolated and identified from sucuk [7–13]. Lactobacillus, which is the largest group in the lactic acid bacteria, is quite common in traditional fermented products and industrial starter preparations [14, 15]. Lactobacillus plantarum and Lactobacillus sakei are two important species isolated from sucuk [7–13]. L. brevis, L. curvatus, L. pentosus, and L. fermentum are the other species isolated from sausage [7, 8, 11, 13, 16]. In fermented meat products, L. plantarum stand out in lactobacilli because of its reliability, ability to use different sugars, easy adaptation to various media, and probiotic properties. L. plantarum is isolated from fermented sausages at different rates depending on fermentation conditions. In similar dry-fermented Greek sausages, Papamanoli et al. [17] found that L. plantarum strains represent 9.4% of LAB isolates, while Drosinos et al. [18, 19] reported that this species reach 37.2 to 45.6% of LAB isolates in these meat products. Considering the case of sucuk, Özdemir [9] and Kaban and Kaya [11] reported that L. plantarum strains constitute 2.8 to 45.7% of LAB isolates. To the best of our knowledge, no studies appeared to test the technological characteristics of sucuk originated LAB at the level tested in this study. In addition, the genetic determinants behind the antimicrobial activity of the autochthonous sucuk strains were not investigated previously, and the antimicrobial activity of these strains might be important for the food safety.
The aim of the present work was to characterize and genotypically identify sucuk originated LAB isolates and determine their technological characteristics including antimicrobial activity against food-borne pathogens in order to test their potential as a starter culture in fermented meat products.
Materials and methods
Isolates and indicator microorganisms
In this study, among the 150 LAB strains phenotypically identified previously by Kaban [20], 5 isolates were selected considering the antagonistic activities. For antagonistic activity tests, microorganisms that given in Table 1 were used.
Table 1.
Indicator microorganisms used in the study
| Indicator microorganisms used in study | Source |
|---|---|
| Listeria monocytogenes ATCC 3970 | ATCC |
| Listeria monocytogenes ATCC 7644 | ATCC |
| Staphylococcus aureus ATCC 29213 | ATCC |
| Staphylococcus aureus ATCC 25923 | ATCC |
| Staphylococcus aureus ATCC 25922 | ATCC |
| Bacillus cereus ATCC 11778 | ATCC |
| Micrococcus luteus NCIMB 8166 | NCIMB |
ATCC, American Type Culture Collection; NCIMB, National Collection of Industrial Food and Marine Bacteria
LAB strains were grown in de Man, Rogosa, and Sharpe broth (Merck, Darmstadt, Germany) at 30 °C for 24 h. Indicator microorganisms were grown in Tryptic Soy Broth (TSB; Oxoid, New Hampshire, UK) at 37 °C for 24 h. All microorganisms were stored at − 80 °C in stock solutions (40% glycerol concentration).
Determination of the antimicrobial activity of isolates
The antagonistic activities of the isolates against indicator strains listed in Table 1 were determined according to Schillinger and Lücke’s [21] well diffusion method. For this methodology, the culture supernatant was first prepared. Strains were inoculated in MRS liquid medium and incubated at 30 °C for 24 h. Centrifugation was then performed at 25 °C, 5700g for 10 min, and the supernatant was recovered. The supernatant was passed through a cellulose acetate filter having a pore diameter of 0.45 μm and used for well diffusion test. For the test, 7 ml of semisolid (tryptone soy agar) (Oxoid CM0131) medium containing 100 μL of an overnight culture of indicator microorganism was poured onto TSA plates. After solidification of the semisolid agar, wells of 5 mm were realized on the plates. Then 50 μL of the cell-free supernatant were transferred to the wells and incubated at 37 °C for 24 h. At the end of the incubation, the clear zones larger than 0.5 mm were evaluated as positive. Lactobacillus sakei Lb 706 (Federal Meat Research Institute, Kulmbach, Germany) strain was used as a positive control. Three microliters of proteinase K (10 mg / ml) were dropped into well containing cell-free supernatant. The absence of inhibition zone indicates the protein nature of the antimicrobial compounds [22].
Identification of LAB isolates
Genomic DNA of the isolates was obtained according to Barış [23]. The genotypic identification of the isolates was done by selecting 16S rRNA coding region sequence from genomic DNA.
For each sample to be subjected to PCR, 7 μl of 10× PCR buffer (Sigma), 1.4 μl of dNTP (deoxynucleotide triphosphates: dATP, dGTP, dCTP, dTTP, 10 mM), 0.7 μl of 50 μM of primer 27F forward (5′-AGA GTT TGA TCC TGG CTC AG-3 ′), 0.7 μl of 50 μM primer 1492R (reverse 5′-GGT TAC CTT GTT ACG ACT T-3′), 2.8 μl dimethyl sulfoxide, 4.2 μl MgCl2, 0.7 μl 5 units/μlTaq DNA polymerase, and 51 μl sterile distilled water were mixed to prepare a reaction mixture of 68.5 μl. Finally, 1.5 μl of template DNA was added to the final volume of 70 μl. Reaction mixtures were subjected to 2 min of initial denaturation at 95 °C and 36 cycle denaturation at 94 °C for 1 min, followed by 1 min at 53 °C and 2 min elongation at 72 °C and finalized with the elongation at 72 °C for 5 min. The electrophoresis of PCR products was performed in gel prepared with 0.7% agarose. Electrophoresis was carried out at 100 V for 45 min. After the run, the gel was visualized with a gel documentation system and analyzed with DNR BioImaging Systems Software. PCR products were sequenced by Macrogen Company (the Netherlands). The 16S rRNA gene sequences for all of the LAB strains were arranged in BioEdit version 7.2.5 and blasted on NCBI database using BLAST program (http://blast.ncbi.nlm.nih.gov). The 16S rRNA gene sequences for LAB species are regulated in MEGAX 10.1.7. Phylogenetic tree was obtained by using neighbor join (NJ) method with 1000 bootstrap replicates [24]. Phylogenetic tree analyses were performed with MEGAX [25].
Detection of bacteriocin encoding genes
Taking into account the bacteriocins that may be produced by the LAB identified, primers have been selected for genetic detection for bacteriocin production. All isolates were PCR tested for the presence of 13 plantaricin encoding genes with the primer sets given in Table 2. PCR mixes were prepared as described above with primer sets and reactions performed at different annealing temperatures suitable for each primer sets. PCR products were analyzed by electrophoresis on 1% (w/v) agarose gels at 100 V for 40 min. In the positive strains, appropriate amplicon sizes were detected.
Table 2.
The oligonucleotides used for determination of the presence of the bacteriocin encoding gene (Omar et al. [26])
| Plantaricin coding genes | Primer | Amplicon size (bp) | Annealing temp. (°C) |
|---|---|---|---|
| plnA |
F: GTA CAG TAC TAA TGG GAG R: CTT ACG CCA ATC TAT ACG |
450 | 53 |
| plnB |
F: TTC AGA GCA AGC CTA AAT GAC R: GCC ACT GTA ACA CCA TGA C |
165 | 51.5 |
| plnC |
F: AGC AGA TGA AAT TCG GCA G R: ATA ATC CAA CGG TGC AAT CC |
108 | 49.5 |
| plnD |
F: TGA GGA CAA ACA GAC TGG AC R: GCA TCG GAA AAA TTG CGG ATA C |
414 | 53 |
| plnEF |
F: GGC ATA GTT AAA ATT CCC CCC R: CAG GTT GCC GCA AAA AAA G |
428 | 53.2 |
| plnI |
F: CTC GAC GGT GAA ATT AGG TGT AAG R: CGT TTA TCC TAT CCT CTA AGC ATT GG |
450 | 52.5 |
| plnj |
F: TAA CGA CGG ATT GCT CTG 51475 R: AAT CAA GGA ATT ATC ACA TTA GTC |
475 | 51 |
| plnk |
F: CTG TAA GCA TTG CTA ACC AAT C R: ACT GCT GAC GCT GAA AAG |
246 | 52.9 |
| plnG |
F: TGC GGT TAT CAG TAT GTC AAA G R: CCT CGA AAC AAT TTC CCC C |
453 | 52.8 |
| plnN |
F: ATT GCC GGG TTA GGT ATC G R: CCT AAA CCA TGC CAT GCA C |
146 | 51.9 |
| plnNC8 |
F: GGTCTGCGTATAAGCATCGC R:AAATTGAACATATGGGTGCTTTAAATTCC |
207 | 60 |
| plnS |
F:GCCTTACCAGCGTAATGCCC R:CTGGTGATGCAATCGTTAGTTT |
320 | 60 |
| plnW |
F:TCACACGAAATATTCCA R:GGCAAGCGTAAGAAATAAATGAG |
165 | 55 |
The growth curve and bacteriocin-like metabolite production during growth
MRS broth was inoculated with 1% of the 24 h culture of LAB isolate and then absorbance values (Aquamate Thermo electron corporation, England) at 600 nm were measured in 2 h intervals at 30 °C for 24 h of incubation. Growth curves were formed with obtained optical density (OD) values. For determining the relation between growth and antimicrobial metabolite production, antimicrobial effects against L. monocytogenes ATCC 7644 of the samples taken at different times during the 24 h incubation period were examined. Antimicrobial activity of the cell-free supernatant was determined according to Biswas et al. [27]. Activity was calculated as AU/ml (Arbitrary unit/ml) (AU/ml = Dilution Factor * 1 ml/50 μl). Antimicrobial activities of samples were compared with growth curve analysis [28].
Biochemical and technological characterization of isolates
Isolates were characterized by the following biochemical tests: growth at pH 4.5, 5, 5.5, 6, or 6.5 in MRS broth, tolerance to NaCl by growth in NaCl containing MRS broth at concentrations of 6.5 and 10%; growth at different temperatures at 4 °C for 7 days, 15 °C for 3 days, 25 °C for 2 days, and 45 °C for 3 days in MRS broth [29]; production of acetoin from glucose, lypolytic activity in trybutyrin agar, gelatinase activity in gelatinase agar, and calcium hydrolyze activity in caseinate agar were determined according to Harrigan [29]. To determine whether isolates form acetoin from glucose, 5 ml methyl-red Voges-Proskauer (Merck, Darmstadt, Germany) broth was used. MR-VP broth was inoculated with 24-h cell culture and left for 48 h incubation at 30 °C. At the end of the incubation, 1 ml was taken from the tubes and transferred to a sterile tube and 0.2 ml of 40% KOH solution and 0.6 ml of alpha-naphthol were added. After 15 min, the presence of colored ring changing from pink to bright red on the liquid surface was evaluated as a positive result. Trybutyrin agar (Merck, Darmstadt, Germany) was used to determine lipolytic activity of isolates. Twenty-four-hour culture was transferred to wells drilled in agar plates. After the incubation at 30 °C for 6 days, clear zones were evaluated as positive results. Gelatin agar (Merck, Darmstadt, Germany) and calcium caseinate agar (Merck, Darmstadt, Germany) were used to determine proteolytic activity. The medium was inoculated with 24-h culture, and the presence of clear zone was evaluated as positive after 48-h incubation at 30 °C. For nitrate reductase activity, 5 ml nitrate broth (Sigma-Aldrich 72548) medium were inoculated with isolates, and after 24-h incubation at 30 °C, red color conversion was observed in the presence of N, N-dimethyl-1-naphtylamine as positive result [30]. Deamination of arginine, ornithine, and lysine were determined as follows:
Arginine, ornithine, and lysine amino acids were added to the Miller broth at a rate of 1%. The prepared media was filled into tubes of 3 ml each, and liquid paraffin was added to the tubes to ensure anaerobiosis [29]. Lactic acid configuration and quantities were detected enzymatically with D-lactic acid/L-lactic acid kit (Cat. No. 11112821035, Boehringer Mannheim/R-Biopharma, R-Biopharma GmbH, Darmstadt, Germany).
Results and discussion
In total, 150 LAB strains previously isolated from sucuk [20] were tested for antagonistic activity (Data not shown). According to antagonistic activity test, 5 strains (S50, S51, S72, S74, and S85) were selected for further technological and genotypic characterization. Comparison of 16S rRNA regions showed that S50, S51, S72, S74, and S85 are L. plantarum strains with 99% similarities. The accession numbers of LAB strains are given in Table 3.
Table 3.
Accession numbers of lactic acid bacteria isolates
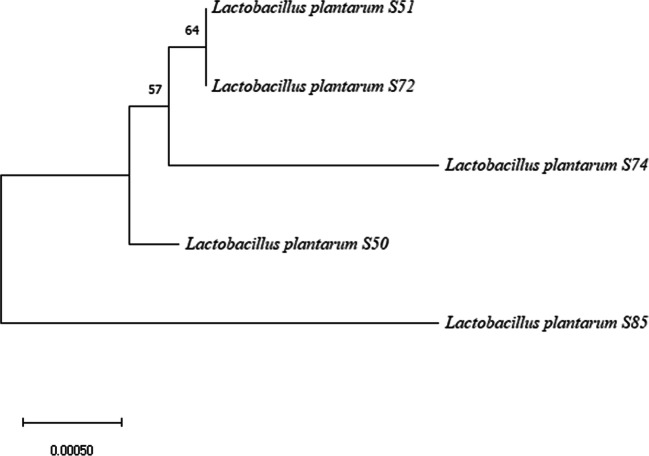
Figure 1 represents the phylogenetic relationship between MEGA alignments of 16S rRNA genes of isolates. In phylogenetic analysis, 3 subgroups were formed among L. plantarum strains. The first group consisted of S51, S72 and S74, second group S50 and third group S85, respectively.
Fig. 1.
A phylogenetic tree was constructed with the MEGA version 10.1.7 program using 16S rRNA gene sequences. Phylogenetic tree was inferred by Neighbor-Joining method [24]. Phylogenetic distances were calculated based on 1313 nt of 16S rRNA gene. The optimal tree with the sum of branch length = 0.00489278 is shown. The robustness of the NJ tree was tested by bootstrapping with 1000 replicates of data, and percentages are shown next to the branches (only values above 50% are reported) [31]. The evolutionary distances were computed using the p-distance method [32]
In fermented sausages such as sucuk, salami, and Rohwurst, LAB are important microorganisms for the inhibition of food pathogens such as S. aureus and L. monocytogenes [21, 33, 34]. As seen in Table 4, all of the strains showed antagonistic activity in the well diffusion test against seven indicator bacteria. Similar to our results, in previous studies, L. plantarum strains isolated from fermented sausages showed antimicrobial effects on several foodborne pathogens [17, 35–38]. Importantly using proteinase K during antimicrobial tests resulted in the loss of inhibition zones showing that the antimicrobial substances produced by the strains are of peptidic nature. Because of the protein-based antimicrobial activity, the presence of genes responsible for bacteriocin production in genomic DNA was investigated. Five L. plantarum strains were PCR screened for the 13 plantaricin genes (Table 5). Bacteriocins that produced from L. plantarum are known as plantaricins. Generally, many numbers of genetic determinants for plantaricin production are located in gene clusters. Among the studied 13 plantaricin genes, plnNC8, plnS, and plnW are structural genes, while others are plantaricin operons [26]. As a result of PCR products electrophoresis, amplicons were obtained with seven primer sets for plnB, plnC, plnD, plnI, plnJ, plnK, and plnN genes. All these amplicons showed expected lengths given in Table 2. The obtained amplifications indicate the presence of genes responsible for bacteriocin production [39]. These results were suggesting that protein-based antimicrobial activity of strains is due to plantaricin.
Table 4.
Antimicrobial activity of L. plantarum strains against selected foodborne pathogens
| Indicator microorganisms | S50 | S51 | S72 | S74 | S85 |
|---|---|---|---|---|---|
| Inhibition zone (mm) | |||||
| S. aureus ATCC29213 | + | + | + | + | + |
| S. aureus ATCC25923 | + | + | ++ | + | +++ |
| S. aureus ATCC25922 | ++ | ++ | ++ | ++ | ++ |
| L. monocytogenes ATCC 3970 | + | + | ++ | + | ++ |
| L. monocytogenes ATCC 7644 | +++ | ++ | +++ | +++ | ++ |
| Bacillus cereus ATCC 11778 | + | + | ++ | + | + |
| Micrococcus luteus NCIMB 8166 | + | + | +++ | + | + |
+, inhibition zone < 2 mm; ++, inhibition zone 3–4 mm; +++, inhibition zone >4 mm
Table 5.
Detection of the presence of genes responsible for plantaricin production
| Isolates | plnA | plnB | plnC | plnD | plnEF | plnI | plnJ | plnK | plnG | plnN | plnNC8 | plnS | plnW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S50 | – | + | + | + | – | + | + | + | – | + | – | – | – |
| S51 | – | + | + | + | – | + | + | + | – | + | – | – | – |
| S72 | – | + | + | + | – | + | + | + | – | + | – | – | – |
| S74 | – | + | + | + | – | + | + | + | – | + | – | – | – |
| S85 | – | + | + | + | – | + | + | + | – | + | – | – | – |
Results reveal the PCR detection of the related gene; + and − represent the presence and absence of the corresponding gene
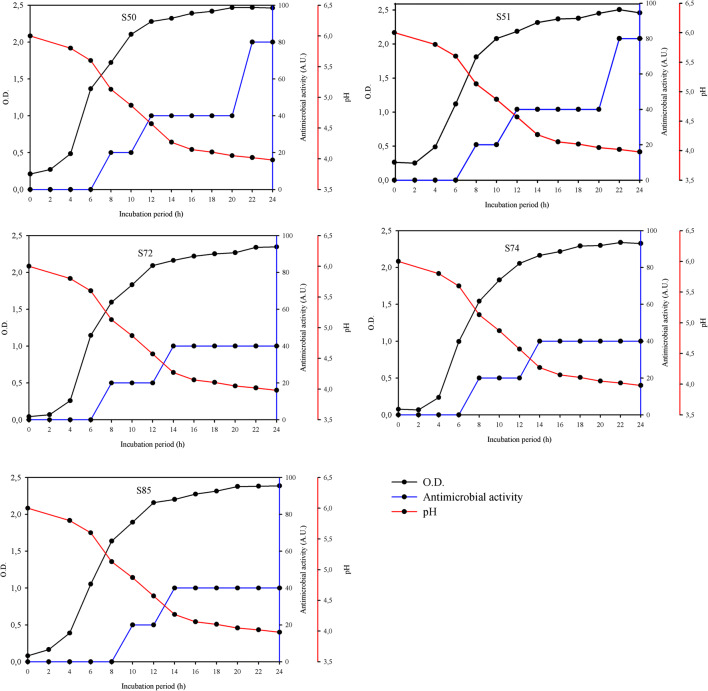
The growth curves for L. plantarum S50, S51, S72, S74, and S85 reveal that the first 4 h of lag phase for all strains were evident (Fig. 2). Antimicrobial activity during 24-h incubation was detected against L. monocytogenes ATCC 7644. Antimicrobial activity was started at the log phase for all strains and reached the maximum antimicrobial level at the stationary phase. Increasing cell numbers and critical nutrient level in the medium were thought to increase antimicrobial metabolite production. Similar results were given by Diep et al. [40]. Differences between strains were observed in terms of bacteriocin activity. The antimicrobial activities of L. plantarum S50 and S51 were found to be higher than those of S72, S74, and S85 strains. Antimicrobial activity showed similar changes in growth rate. We can say that the bacteriocin-like antimicrobial substance has the primary metabolite kinetic property like most of the plantaricins. Similar results were described in the previous study [41]. L. plantarum strains showed similar decreases in pH value after 24 h of incubation. The antimicrobial effect for all strains was started at pH 4.5–5.0, and the maximum antimicrobial activity was observed at pH 3.95–4.50. Similar results have been reported for the other bacteriocin producer L. plantarum strains [42–44].
Fig. 2.
Antimicrobial activity and pH of strains throughout the growth
The characteristics of L. plantarum strains were studied, and results are presented in Table 6; L. plantarum S50, S51, S72, S74, and S85 have optimum growth at 25 °C. Strains that grow at a temperature of 15 °C showed a poor growth at 45 °C. Schillinger and Lücke [45] reported that L. plantarum strains isolated from meat and meat products can grow at 45 °C. In accordance with sausage production conditions, L. plantarum is the usually dominant strain [11, 12]; while strains showed the best growth at pH 6 and 6.5, they showed medium level growth at low pH values (pH 4.5 and 5) and good growth at pH 5.5. These results suggested that the pH decreases to around 5 during fermentation, affecting the LAB growth otherwise. All of the strains grew at 6.5% salt concentration, whereas no growth was detected at 10% salt concentration. NaCl is an important additive for fermented sausages. It is used in 2–3% concentration for sausage batter [46]. The NaCl added to fermented sausages reduces the initial aw value and encourages the growth of Micrococcaceae family and LAB, which are technologically important groups [47, 48]. In fermented meat products, especially in formulations using nitrate, nitrate reductase activity of microorganisms is necessary for the development and stability of red color. This activity is provided especially by the members of Micrococcaceae family such as Staphylococcus xylosus, S. carnosus, and Kocuria varians strains in these products. These catalase-positive strains are often found as mixed cultures with LAB [49]. On the other side, there are also lactic acid bacteria which show nitrate reductase activities. Paik and Lee [50] reported that L. plantarum strains isolated from kimchi show weak nitrate reductase activity. In this study, a weak nitrate reductase activity was also observed in two L. plantarum strains (S72, S85). According to phylogenetic tree (Fig. 1), S51, S72 and S74 were quite close in subgroups. Despite the closeness of strains, nitrate reductase activities of S72 and S85 indicated phenotypical differences of strains.
Table 6.
Technological and biochemical properties of L. plantarum strains
| Technological and biochemical properties | S50 | S51 | S72 | S74 | S85 |
|---|---|---|---|---|---|
| 4 °C | – | – | – | * | – |
| 15 °C | ++ | ++ | ++ | + | ++ |
| 25 °C | +++ | +++ | +++ | +++ | +++ |
| 45 °C | + | + | + | * | + |
| pH 4.5 | + | + | + | + | + |
| pH 5.0 | + | + | + | + | + |
| pH 5.5 | ++ | ++ | ++ | ++ | ++ |
| pH 6.0 | +++ | +++ | +++ | +++ | +++ |
| pH 6.5 | +++ | +++ | +++ | +++ | +++ |
| 6.5% NaCl | ++ | ++ | ++ | ++ | ++ |
| 10% NaCl | – | – | – | – | – |
| Nitrate reductase | – | – | + | – | + |
| Lysine decarboxylase | – | – | – | – | – |
| Ornithine decarboxylase | – | – | – | – | – |
| Arginine decarboxylase | – | – | – | – | – |
| Casein hydrolysis | – | – | – | – | – |
| Gelatin hydrolysis | – | – | – | – | – |
| Tributyrin hydrolysis | – | – | – | – | – |
| Acetoin production from glucose | + | – | + | + | + |
| D-Lactic Acid (g/L) | 10. 54 | 6.47 | 5.70 | 6.73 | 8.97 |
| L-Lactic Acid (g/L) | 5.98 | 8.27 | 6.01 | 3.46 | 4.52 |
− not detected, * weak, + medium, ++ good, +++ very good
Biogenic amines are undesirable compounds in food. They may have toxic effect on human. Generally, biogenic amines are formed as a result of decarboxylation of amino acids by microbial enzymes. Lactobacilli can form different biogenic amines by decarboxylase activity depending on the species [50]. As a matter of fact, it has been determined that some strains of Lactobacilli produce biogenic amines [51, 52]. As can be seen in Table 6, all of the strains were negative for lysine, ornithine, and arginine decarboxylase activity. No strain hydrolyzed tributyrin, gelatin, and casein. These results showed that the tested strains did not show proteolytic and lipolytic activity. It has also been reported that L. plantarum strains do not have lipolytic activity [17, 53–56] and proteolytic activities [54].
L. plantarum, Pediococcus pentosaceus, and P. acidilactici are LAB used as starter cultures that can form acetoin. L. sakei is rare, while L. curvatus is very rare producer for this compound [47]. In the present study, except for L. plantarum S51, the others were positive in the Voges-Proskauer test. The formation of acetic acid or acetoin in very high amounts in fermented meat products is not desirable [48].
The stereochemistry of lactic acid produced by LAB is an important feature. Lactic acid isomer produced during fermentation varies according to the species. The ratio of D-lactic acid and L-lactic acid varies in the racemic mixture they produce based on lactate dehydrogenase (LDH) activities of LAB. The racemic mixture is not always uniformly formed. Since D (−)-lactic acid cannot be hydrolyzed by lactate dehydrogenase in humans, L (+)-lactic acid producing strains are preferred as starter cultures [56]. We determined that all L. plantarum strains tested produced DL-lactic acid. In two strains, L (+)-lactic acid production was more than 50% (L. plantarum S51, S72). L. plantarum strains differed from each other with their technological characteristics.
Conclusion
In conclusion, some technological properties of L. plantarum strains isolated from traditionally fermented sausage were determined. All strains showed good antagonistic activity against Listeria monocytogenes. The amplification of genes responsible for the production of plantaricin (plnB, plnC, plnD, plnI, plnJ, plnK, and plnN) by PCR analysis supported this result. According to our findings, these strains may be potential protective cultures. When considered in terms of biochemical and technological properties, L. plantarum strains exhibited some of the properties required for a starter culture in fermented sausage and similar products. The L. plantarum strain S51 did not produce acetoin from glucose, which is a major drawback for this strain. Montel et al. [57] showed that acetoin producer starter cultures can cause undesirable dairy-like product odor in dry sausages. In terms of the importance of acetoin level on product flavor, it is important to investigate the effect of aroma formation in model systems for other strains in detail. Further work will characterize the role of the identified L. plantarum strains as starter cultures in fermented meat products as well as their probiotic and functional properties will be investigated.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dalié DKD, Deschamps M, Richard F. Lactic acid bacteria – potential for control of mould growth and mycotoxins: a review. Food Control. 2010;21(4):370–380. doi: 10.1016/j.foodcont.2009.07.011. [DOI] [Google Scholar]
- 2.Ojha KS, Kerry JP, Duffy G, Beresford T, Tiwari BK. Technological advances for enhancing quality and safety of fermented meat products. Trends Food Sci Technol. 2015;44(1):105–116. doi: 10.1016/j.tifs.2015.03.010. [DOI] [Google Scholar]
- 3.Leroy F, Scholliers P, Amilien V. Elements of innovation and tradition in meat fermentation: conflicts and synergies. Int J Food Microbiol. 2015;212:2–8. doi: 10.1016/j.ijfoodmicro.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Palavecino Prpich NZ, Garro OA, Romero M, Judis MA, Cayré ME, Castro MP. Evaluation of an autochthonous starter culture on the production of a traditional dry fermented sausage from Chaco (Argentina) at a small-scale facility. Meat Sci. 2016;115:41–44. doi: 10.1016/j.meatsci.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Aquilanti L, Garofalo C, Osimani A, Clementi F. Ecology of lactic acid bacteria and coagulase negative cocci in fermented dry sausages manufactured in Italy and other Mediterranean countries: an overview. Int Food Res J. 2016;23(2):429–445. [Google Scholar]
- 6.Nediani M, García L, Saavedra L, Martínez S, López Alzogaray S, Fadda S. Adding value to goat meat: biochemical and technological characterization of autochthonous lactic acid Bacteria to achieve high quality fermented sausages. Microorganisms. 2017;5(2):26. doi: 10.3390/microorganisms5020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gürakan GC, Bozoglu TF, Weiss N. Identification of Lactobacillus strains from Turkish-style dry fermented sausages. LWT-Food Sci Technol. 1995;28(1):139–144. doi: 10.1016/S0023-6438(95)80026-3. [DOI] [Google Scholar]
- 8.Özdemir H. Yuksek sicaklik derecesinde olgunlastirilan Turk fermente sucuklarinda laktobasillerin seyir, izolasyon ve identifikasyonu. Gida. 1996;21(6):465–470. [Google Scholar]
- 9.Özdemir H. Türk fermente sucugunun florasındaki dominant laktobasil türlerinin sucugun organoleptik nitelikleri ile ilişkisi. Ankara Üniversitesi Veteriner Fakültesi Dergisi. 1998;46:189–198. [Google Scholar]
- 10.Çon AH, Gokalp HY. Production of bacteriocin-like metabolites by lactic acid cultures isolated from sucuk samples. Meat Sci. 2000;55:89–96. doi: 10.1016/S0309-1740(99)00129-1. [DOI] [PubMed] [Google Scholar]
- 11.Kaban G, Kaya M. Identification of lactic acid bacteria and gram-positive catalase-positive cocci isolated from naturally fermented sausage (sucuk) J Food Sci. 2008;73(8):385–388. doi: 10.1111/j.1750.3841.2008.00906.x. [DOI] [PubMed] [Google Scholar]
- 12.Adiguzel G, Atasever M. Phenotypic and genotypic characterization of lactic acid bacteria isolated from Turkish dry fermented sausage. Rom Biotechnol Lett. 2009;14(1):4130.4138. [Google Scholar]
- 13.Kesmen Z, Yetiman E, Gulluce Kacmaz N, Sagdic O, Cetin B, Adigüzel A, Şahin F, Yetim H. Combination of culture-dependent and culture independent molecular methods for the determination of lactic microbiota in sucuk. Int J Food Microbiol. 2012;153(3):428–435. doi: 10.1016/j.ijfoodmicro.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 14.De Vries MC, Vaughan EE, Kleerebezem M, de Vos WM. Lactobacillus plantarum survival, functional and potential probiotic properties in the human intestinal tract. Int Dairy J. 2006;16(9):1018.1028. doi: 10.1016/j.idairyj.2005.09.003. [DOI] [Google Scholar]
- 15.Giraffa G, Chanishvili N, Widyastuti Y. Importance of lactobacilli in food and feed biotechnology. Res Microbiol. 2010;161(6):480–487. doi: 10.1016/j.resmic.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Dinçer B, Özdemir H, Mutluer B, Yaglı Ö, Erol İ, Akgün S. Türk fermente sucuğuna özgü starter kültür bakterilerinin izolasyon, identifikasyon ve üretimleri. Ankara Üniv Vet Fak Derg. 1995;42:285–293. [Google Scholar]
- 17.Papamanoli E, Tzanetakis N, Litopoulou, Tzanetaki E, Kotzekidou P. Characterization of lactic acid bacteria isolated from a Greek dry-fermented sausage in respect of their technological and probiotic properties. Meat Sci. 2003;65(2):859–867. doi: 10.1016/S0309-1740(02)00292-9. [DOI] [PubMed] [Google Scholar]
- 18.Drosinos EH, Mataragas M, Xiraphi N, Moschonas G, Gaitis F, Metaxopoulos J. Characterization of the microbial flora from a traditional Greek fermented sausage. Meat Sci. 2005;69(2):307–317. doi: 10.1016/j.meatsci.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Drosinos EH, Paramithiotis S, Kolovos G, Tsikouras I, Metaxopoulos I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in southern Greece. Food Microbiol. 2007;24(3):260–270. doi: 10.1016/j.fm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Kaban G (2007) Geleneksel olarak üretilen sucuklardan laktik asit bakterileri ile katalaz pozitif kokların izolasyonu identifikasyonu, üretimde kullanılabilme imkânları ve uçucu bileşikler üzerine etkileri. Doktora Tezi, Atatürk Üniversitesi. Fen Bilimleri Enstitüsü, Erzurum
- 21.Schillinger U, Lücke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55(8):1901–1906. doi: 10.1128/AEM.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altuntas EG, Cosansu S, Ayhan K. Some growth parameters and antimicrobial activity of a bacteriocin-producing strain Pediococcus acidilactici 13. Int J Food Microbiol. 2010;141(1–2):28–31. doi: 10.1016/j.ijfoodmicro.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 23.Barış Ö (2009) Erzurum İlindeki Mağaralarda Damlataşı Oluşumunda Etkili Bakterilerin İzolasyonu Karakterizasyonu Ve Tanısı. Doktora Tezi, Atatürk Üniversitesi, Fen Bilimleri Enstitüsü, Biyoloji Anabilim Dalı, Erzurum
- 24.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omar NB, Abriouel H, Keleke S, Valenzuela AS, Martínez-Cañamero M, López RL, Ortega E, Gálvez A. Bacteriocin-producing Lactobacillus strains isolated from poto poto, a Congolese fermented maize product, and genetic fingerprinting of their plantaricin operons. Int J Food Microbiol. 2008;127(1–2):18–25. doi: 10.1016/j.ijfoodmicro.2008.05.037. [DOI] [PubMed] [Google Scholar]
- 27.Biswas SR, Ray P, Johnson MC, Ray B. Influence of growth conditions on the production of a bacteriocin, pediocin AcH, by Pediococcus acidilactici H. Appl Environ Microbiol. 1991;57(4):1265–1267. doi: 10.1128/AEM.57.4.1265-1267.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Reenen CA, Dicks LMT, Chikindas ML. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol. 1998;84(6):1131–1137. doi: 10.1046/j.1365-2672.1998.00451.x. [DOI] [PubMed] [Google Scholar]
- 29.Harrigan WF (1998) Laboratory methods in food microbiology. Academic Press. California 92101.4495, USA, 100
- 30.Kloos WE, Schleifer KH. Simplified scheme for routine ıdentification of human Staphylococcus species. J Clin Microbiol. 1975;1(1):82–88. doi: 10.1128/JCM.1.1.82-88.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford: Oxford university press; 2000. [Google Scholar]
- 33.Kaban G, Kaya M. Effect of starter culture on growth of Staphylococcus aureus in sucuk. Food Control. 2006;17(10):797–801. doi: 10.1016/j.foodcont.2005.05.003. [DOI] [Google Scholar]
- 34.Lewus CB, Kaiser A, Montville TJ. Inhibition of food-borne bacterial pathogens by bacteriocins from lactic acid bacteria isolated from meat. Appl Environ Microbiol. 1991;57(6):1683–1688. doi: 10.1128/AEM.57.6.1683-1688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todorov SD, Stojanovski S, Iliev I, Moncheva P, Nero LA, Ivanova IV. Technology and safety assessment for lactic acid bacteria isolated from traditional Bulgarian fermented meat product “lukanka.”. Braz J Microbiol. 2017;48(3):576–586. doi: 10.1016/j.bjm.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbosa M, Todorov SD, Ivanova I, Chobert JM, Haertlé T, de Melo Franco BDG. Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiol. 2015;46:254–262. doi: 10.1016/j.fm.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Sawitzki MC, Fiorentini ÂM, Bertol TM, Sant'Anna ES. Lactobacillus plantarum strains isolated from naturally fermented sausages and their technological properties for application as starter cultures. Food Sci Technol. 2009;29(2):340–345. doi: 10.1590/S0101-20612009000200016. [DOI] [Google Scholar]
- 38.Toksoy A, Beyatli Y, Aslim B. Sucuk ve sosislerden izole edilen Lactobacillus plantarum suşlarının bazı metabolik ve antimikrobiyal aktivitelerinin incelenmesi. Turk J Vet Anim Sci. 1999;23(6):533–540. [Google Scholar]
- 39.Remiger A, Ehrmann M, Vogel RF. Identification of Bacteriocin encoding genes in lactobacilli by polymerase chain reaction (PCR) Syst Appl Microbiol. 1996;19(1):28–34. doi: 10.1016/S0723.2020(96)80005.1. [DOI] [Google Scholar]
- 40.Diep DB, Håvarstein LS, Nes IF. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178(15):4472–4483. doi: 10.1128/JB.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.François ZN, Marie KP, Azemfack T, Noëlle H (2013) Antimicrobial activity of a bacteriocin produced by Lactobacillus plantarum 29V and strain’ s viability in palm kernel oil, International Journal of Food Sciences and Nutrition. 2(3):102–108. 10.11648/j.ijnfs.20130203.12
- 42.Barbosa MS, Todorov SD, Ivanova IV, Belguesmia Y, Choiset Y, Rabesona H, Chobert JM, Haertle T, Franco BDGM. Characterization of a two-peptide plantaricin produced by Lactobacillus plantarum MBSa4 isolated from Brazilian salami. Food Control. 2016;60:103–112. doi: 10.1016/j.foodcont.2015.07.029. [DOI] [Google Scholar]
- 43.Todorov SD, Gotcheva B, Dousset X, Onno B, Ivanova I. Influence of growth medium on bacteriocin production in Lactobacillus plantarum ST31. Biotechnol Biotechnol Equip. 2014;14(1):50–55. doi: 10.1080/13102818.2000.10819062. [DOI] [Google Scholar]
- 44.Todorov SD. Bacteriocin production by Lactobacillus plantarum AMA.K isolated from Amasi, a Zimbabwean fermented milk product and study of the adsorption of bacteriocin AMA.K to Listeria sp. Braz J Microbiol. 2008;39(1):178–187. doi: 10.1590/S1517-83822008000100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schillinger U, Lücke FK. Identification of lactobacilli from meat and meat products. Food Microbiol. 1987;4(3):199–208. doi: 10.1016/0740-0020(87)90002-5. [DOI] [Google Scholar]
- 46.Toldrá F, Rico E, Flores J. Activities of pork muscle proteases in model cured meat systems. Biochimie. 1992;74(3):291–296. doi: 10.1016/0300-9084(92)90128-2. [DOI] [PubMed] [Google Scholar]
- 47.Lücke FK, Hechelmann H. Starter cultures for dry sausages and raw ham composition and effect. Fleischwirtschaft. 1987;67(3):307.314. [Google Scholar]
- 48.Kaya M, Kaban G. Fermente Et Ürünleri. In: Aran N, editor. Gıda Biyoteknolojisi İstanbul. Nobel Yayın: Türkiye; 2010. pp. 157–190. [Google Scholar]
- 49.Kaban G, Kaya M, Lücke FK (2012) Meat starter cultures. Encycl Biotechnol Agric Food, 1–4
- 50.Paik HD, Lee JY. Investigation of reduction and tolerance capability of lactic acid bacteria isolated from kimchi against nitrate and nitrite in fermented sausage condition. Meat Sci. 2014;97(4):609–614. doi: 10.1016/j.meatsci.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 51.Bover-cid S, Hugas M, Izquierdo-pulido M, Vidal-carou MC. Amino acid-decarboxylase activity of bacteria isolated from fermented pork sausages. Int J Food Microbiol. 2001;66(3):185–189. doi: 10.1016/S0168-1605(00)00526-2. [DOI] [PubMed] [Google Scholar]
- 52.Moracanin SV, Stefanovic S, Radicevic T, Borovic B, Djukic D. Production of biogenic amines by lactic acid Bacteria isolated from Uzicka sausages. Proce Food Sci. 2015;5:308–311. doi: 10.1016/j.profoo.2015.09.068. [DOI] [Google Scholar]
- 53.Essid I, Medini M, Hassouna M. Technological and safety properties of Lactobacillus plantarum strains isolated from a Tunisian traditional salted meat. Meat Sci. 2009;81(1):203–208. doi: 10.1016/j.meatsci.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 54.Speranza B, Racioppo A, Beneduce L, Bevilacqua A, Sinigaglia M, Corbo MR. Autochthonous lactic acid bacteria with probiotic aptitudes as starter cultures for fish based products. Food Microbiol. 2017;65:244–253. doi: 10.1016/j.fm.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Zeng X, Xia W, Wang J, Jiang Q, Xu Y, Qiu Y, Wang H. Technological properties of Lactobacillus plantarum strains isolated from Chinese traditional low salt fermented whole fish. Food Control. 2014;40:351–358. doi: 10.1016/j.foodcont.2013.11.048. [DOI] [Google Scholar]
- 56.Holzapfel WH, Haberer P, Snel J, Schillinger U. Overview of gut flora and probiotics. Int J Food Microbiol. 1998;41:85–101. doi: 10.1016/S0168.1605(98)00044.0. [DOI] [PubMed] [Google Scholar]
- 57.Montel MC, Reitz J, Talon R, Berdagué JL, Rousset-Akrim S. Biochemical activities of Micrococcaceae and their effects on the aromatic profiles and odours of a dry sausage model. Food Microbiol. 1996;13(6):489–499. doi: 10.1006/fmic.1996.0056. [DOI] [Google Scholar]