Abstract
Carbapenem-resistant Enterobacterales (CREs) have been recognized as an important threat to global health. CRE cause the majority of the difficult-to-treat infections in health-care settings and are associated with high mortality. Klebsiella pneumoniae carbapenemase (KPC)–producing CREs, in particular Klebsiella pneumoniae, are globally disseminated and responsible for a large number of outbreaks. Development of rapid methods for KPC detection can provide great clinical and epidemiological benefits to prevent KPC dissemination. The aim of this study was to standardize and validate a LC-MS/MS method to detect KPC. This method was also tested against a broad variety of species, including CRE with other carbapenemase genes and the recently reported mcr-1. For validation, 111 isolates with reduced susceptibility to carbapenems were selected (49 KPC-positive and 62 KPC-negative). The presence of four tryptic peptides related to the KPC enzyme was evaluated, and the identification of at least two of them classified the isolate as “KPC-positive.” The LTLGSALAAPQR and LALEGLGVNGQ peptides were both detected in 47 of 49 isolates with the blaKPC gene. The other two peptides, GFLAAAVLAR and APIVLAVYTR, were detected in 46 and 19 isolates with the blaKPC gene, respectively. The method correctly classified 47 of 49 KPC-positive and all KPC-negative isolates yielding 96.07% of sensitivity and 100% of specificity. In conclusion, our results demonstrate that the KPC peptide markers were robustly detected by the method which presented high sensitivity and full specificity and therefore can be used as a reliable method to identify this resistance mechanism.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00222-y) contains supplementary material, which is available to authorized users.
Keywords: Enterobacterales, Carbapenemases, KPC, LC-MS/MS, Mass spectrometry
Introduction
Carbapenem-resistant Enterobacterales (CREs) are a serious public health threat responsible for the majority of the drug-resistant infections and associated with high morbidity and mortality [1–3]. CREs carrying Klebsiella pneumoniae carbapenemase (KPC) gene, in particular Klebsiella pneumoniae (KP), are widely disseminated and responsible for a large number of outbreaks around the world [4]. One critical point on tackling CREs worldwide is the development of rapid detection methods in order to allow the implementation of infection control measures to prevent CRE dissemination [5].
Recently, a variety of mass spectrometry (MS) methods for carbapenemase detection have been developed. Most MS methods aimed to detect the enzymatic activity [6–16] while other focused on the detection of a protein genetically linked to certain plasmids that carry the blaKPC gene [17, 18]. A limitation of these methods is that they use indirect and nonspecific approaches. An interesting direct approach that uses MS to detect tryptic peptides of carbapenemases has been proposed [19], and several variations of this method have been tested [20]. The main drawback of these approaches is the turnaround time needed to perform the whole analysis. A method based on liquid chromatography-electrospray-tandem mass spectrometry (LC-ESI-MS/MS) and multiple reaction monitoring (MRM) which detected tryptic peptides of PBP2a and PBP2c in methicillin-resistant S. aureus in 60–80 min was described [21] and indicates that such methods can provide faster results.
In a similar way, Wang et al. have validated a rapid LC-MS/MS method for the direct detection of 4 specific tryptic peptides of the KPC protein in clinical isolates with an isolate-to-result time of less than 90 min and reported a sensibility and sensitivity of 100% [22]. Our aim was to validate a simplified version of the method proposed by Wang and to evaluate its performance against a variety of species, including CRE with other carbapenemase genes.
Materials and methods
Bacterial isolates
A total of 119 bacterial isolates were selected from an epidemiological study evaluating Enterobacterales with reduced susceptibility to carbapenems in several hospitals in the southernmost state of Brazil [23]. Real-time PCR was used to determine the presence of blaNDM-1, blaKPC, blaVIM-type, blaGES-type, blaOXA-48-like, blaIMP-type, and mcr-1 [24]. All isolates were identified at specie level by MALDI-TOF MS (Bruker AutoFlex Speed mass spectrometer, Bruker Daltonics, Billerica, MA) with MALDI Biotyper (v3.1 Bruker Daltonics, Inc.) using the same extract which was used to LC-MS/MS analysis.
Standardization
Eight well-characterized K. pneumoniae clinical isolates (4 KPC-positive and 4 KPC-negative) were used for the initial experiments. All bacterial isolates were grown on Mueller-Hinton agar (MHA) plates for 18–24 h at 35 ± 1 °C and lysed with formic acid (FA) and acetonitrile (ACN) as described previously [20] with minor modifications. Briefly, for each isolate, a 1 μL calibrated loop of fresh bacterial cells was suspended in 1 mL 70% ethanol, vortexed for 10 s, and centrifuged at 16,200×g for 2 min. Supernatant was removed, and the residual ethanol was evaporated at room temperature for 5 min. The pellet was re-suspended in 50 μL of 70% FA and vortexed for 10 s, followed by addition of 50 μL of ACN. The resulting solution was vortexed for 10 s and centrifuged at 16,200×g for 2 min. A volume of 100 μL of the supernatant was stored at − 20 °C for the assay. The KPC specific peptide markers were determined previously, as described by Wang et al., 2017. For experimental method standardization, these 8 isolates were run in technical triplicates to set retention time (RT) and check its reproducibility (data not shown).
Tryptic protein digestion
Fifty microliters of FA/ACN lysate was evaporated in an incubator at 55 °C. The proteins were re-suspended in 100 μL of 50 mM of NH4HCO3 and vortexed for 15 min. Protein digestion was carried out in an incubator for 30 min at 55 °C with addition of 1 μg of Trypsin (Sigma-Aldrich®). Digested extracts were vortexed for 15 s and centrifuged for 4 min at 16,200×g. Ninety microliters of the supernatant was transferred to a vial with fixed insert and submitted to LC-MS/MS analysis.
Validation
A total of 111 Enterobacterales (including blaKPC, blaNDM, blaGES, blaVIM, blaIMP, and mcr-1 positive) isolates were used for the validation of the method. These isolates were processed as described above.
External standard (positive control)
Two hundred microliters of FA/ACN lysate of KPC-positive K. pneumoniae isolate previously characterized during the standardization was mixed with 200 μL FA/ACN lysate of a KPC-negative K. pneumoniae isolate to create a “half-concentration” solution to approximate a middle point of the expected signal. Protein extract was chosen as opposed to purified or labeled peptides to replicate as closely as possible the complex protein matrix of the test samples. A volume of 20 μL of this positive control was treated identically to the blinded validation isolated extracts and was run as the first and the last sample of the testing day to confirm assay performance and for the purpose of MS/MS spectral comparisons of KPC peptide markers with unknowns in the validation set [22].
Targeted LC-MS/MS
The method was run on a liquid chromatograph Agilent 1260 Infinity coupled to an AB Sciex 5500 QTRAP-system. The chromatographic separation was carried out on a Luna C18 column (5 μm, 150mm × 2.0 mm; Phenomenex, Torrance/CA). The mobile phases were water and acetonitrile, both containing 0.1% FA. The flow rate was 300 μL min−1 and a 4-min equilibrate time was applied. The gradient started with 10% of B increasing to 35% in 5 min, 90% in 10 min, kept for 3 min, decreasing to 10% in 14 min, and maintained until 15 min. The column temperature was maintained at 40 °C and sample vials kept at 40 °C. The injection volume was 2 μL. Mass spectrometer resolution in multiple reaction monitoring (MRM) was unitary, and dwell time applied was 300 ms for all transitions. The retention times for the selected four high abundance core KPC peptides were determined with 4 known KPC isolates by targeted LC-MS/MS. The collision energy of the chosen precursor was optimized to produce the strongest signals. Targeted LC-MS/MS was run without labeled peptides. For automatic peak integration a retention time window of 30 s was applied. A 40-min column washing/regeneration was run after 12 sample-run to clean the column. Multiquant™ software (v.2.1.1, AB Sciex) was used for data treatment.
Blinded evaluation set
We perform a validation set with 111 carbapenem-resistant isolates (49 KPC-positives and 62 KPC-negative) randomly injected into the LC-MS/MS. A positive control was tested at the beginning and at the end of the run and after each set of 25 protein extractions. One operator blinded to the isolates was designated to interpret resulting spectra. The operator submitted the final list of results to a second operator whom matched this determination with previous results. To score an isolate in this blinded set, the operator followed the instructions described previously [22].
Investigation of false-negative results
To evaluate isolates that presented false-negative results in LC-MS/MS method, we performed different strategies as follows: (1) the injection volume was increased from 2 to 10 μL to verify sensitivity problems. We also increased the injection volume from the positive control extracts; (2) carbapenemase production was evaluated by Blue-Carba carbapenemase test [25], and the presence of other possible carbapenemases was evaluated by next generation sequencing (NGS) (MiSeq, Illumina) as well as the genetic environment of blaKPC gene [26–28]; (3) the LC-MS/MS method was performed after bacterial incubation on MHA supplemented with 2 μg/mL of meropenem.
Results
Instrument performance and blinded evaluation set
The characterization of the clinical isolates and the performance of the method are described in the Table 1.
Table 1.
Characterization of 111 clinical isolates and performance of KPC detection on LC-MS/MS
| Bacterial species | Mechanism (HRMa) | No. of isolates | % of isolates classified as KPC producers (No. of positive/total No. of isolates) |
|---|---|---|---|
| Citrobacter freundii | blaNDMb | 1 | 0 (0/1) |
| blaKPCc | 2 | 100 (2/2) | |
| Enterobacter asburiae | blaKPCc | 1 | 100 (1/1) |
| Enterobacter cloacae complex | blaKPCc | 3 | 100 (3/3) |
| blaKPCc + blaNDMb | 2 | 100 (2/2) | |
| blaNDMb | 5 | 0 (0/5) | |
| blaOXAd-48like | 3 | 0 (0/3) | |
| blaNDMb + blaOXAd-48like | 5 | 0 (0/5) | |
| blaVIMe | 2 | 0 (0/2) | |
| Negative | 4 | 0 (0/4) | |
| Enterobacter hormaechei | blaNDMb | 1 | 0 (0/1) |
| Escherichia coli | blaKPCc | 3 | 66.6 (2/3) |
| blaKPCc + mcr-1f | 2 | 100 (2/2) | |
| blaOXAd-48like | 1 | 0 (0/1) | |
| Klebsiella aerogenes | Negative | 2 | 0 (0/2) |
| Klebsiella oxytoca | blaKPCc | 1 | 100 (1/1) |
| blaNDMb | 1 | 0 (0/1) | |
| Negative | 4 | 0 (0/4) | |
| Klebsiella pneumoniae | blaGESg | 1 | 0 (0/1) |
| blaIMPh | 2 | 0 (0/2) | |
| blaKPCc | 26 | 100 (26/26) | |
| blaKPCc + mcr-1f | 1 | 0 (0/1) | |
| blaKPCc + blaNDMb | 4 | 100 (4/4) | |
| blaKPCc + blaOXAd-48like | 2 | 100 (2/2) | |
| blaNDMb | 1 | 0 (0/1) | |
| blaOXAd-48like | 1 | 0 (0/1) | |
| blaVIMe | 1 | 0 (0/1) | |
| Negative | 15 | 0 (0/15) | |
| Kluyvera intermedia | blaGESg | 1 | 0 (0/1) |
| Morganella morganii | blaOXAd-48like | 2 | 0 (0/2) |
| Negative | 1 | 0 (0/1) | |
| Providencia rettgeri | blaKPCc + blaGESg | 1 | 100 (1/1) |
| blaNDMb | 1 | 0 (0/1) | |
| Serratia marcescens | blaKPCc + blaNDMb | 1 | 100 (1/1) |
| blaGESg | 5 | 0 (0/5) | |
| Negative | 2 | 0 (0/2) |
aHigh-resolution melting real-time PCR; bNew Delhi metallo-β-lactamase; cKlebsiella pneumoniae carbapenemase; doxacillinase; eVerona integron-encoded metallo-β-lactamase; fmobile colistin resistance–1; gGuiana-extended spectrum; himipenemase metallo-β-lactamase
No significant variations were observed on retention time and intensity during standardization. Since method’s standardization was performed by a single expert operator and did not intend to verify method’s performance, we did not include an internal standard for intensity ratio calculation in this phase.
The average intensity of the positive control was 1.21 × 104 for APIVLAVYTR, 2.22 × 104 for LTLGSALAAPQR, 3.13 × 104 for GFLAAAVLAR, and 1.27 × 104 for LALEGLGVNGQ. The intensity ratios for the peptide markers of the positive isolates are described in the Online Resource 1. If the intensity ratio for a given peptide was < 0.1, it was scored negative. If the ratio was > 0.1, it was scored positive.
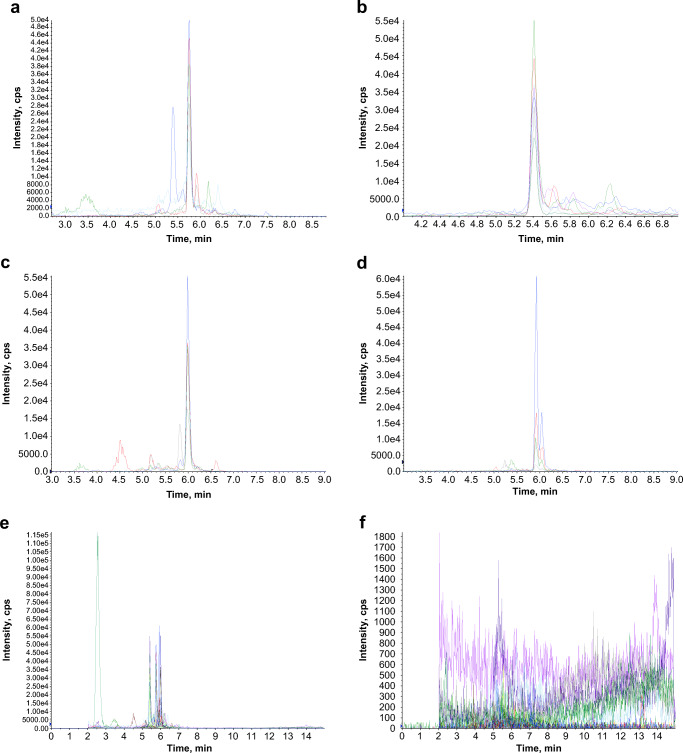
The presence of four tryptic peptides related to the KPC enzyme was evaluated, and the identification of at least two of them classified the isolate as “KPC-positive.” The LTLGSALAAPQR and LALEGLGVNGQ peptides were both detected in 47 of 49 isolates with the blaKPC gene. The other peptides, GFLAAAVLAR and APIVLAVYTR, were detected in 46 and 19 isolates with the blaKPC gene, respectively. No peptide was detected on negative isolates. The blinded operator correctly classified 47 of 49 KPC-positive isolates and all KPC-negative isolates yielding an overall performance of 96.07% of sensitivity and 100% of specificity to the method. None of the four tryptic peptides were detected in the isolates with other carbapenemase genes (blaNDM, blaGES, blaVIM, and blaIMP) or mcr-1. The representative total chromatograms of a KPC-positive and a KPC-negative results and a representative chromatogram of each one of the four KPC peptide markers to illustrate the RT are demonstrated in the Fig. 1.
Fig. 1.
The LC-MS/MS chromatograms of the four KPC peptide markers and the total chromatogram of a positive and a negative isolate. a APIVLAVYTR, b LTLGSALAAPQR, c GFLAAAVLAR, d LALEGLGVNGQ, e a total chromatogram of a positive isolate, and f a total chromatogram of a negative isolate
Despite that GFLAAAVLAR and APIVLAVYTR appear to have no contribution to method’s sensitivity, the evaluation of these peptides is still important: first—for confirmation purposes and—second—peptides’ individual sensitivity appears to be influenced by instrument factors, so the best peptide may vary from one analytical system to another.
Only one Escherichia coli and 1 Klebsiella pneumoniae harboring blaKPC presented negative results in LC-MS/MS. In the additional investigation, we observed a positive correlation between the injection volume and the average intensity of the positive controls (r = 0.83 for APIVLAVYTR and r > 0.99 for the other three peptides); however, none of the four tryptic peptides was detected in these blaKPC isolates after increased injection volume. Noteworthy, the Blue-Carba test presented positive result for these isolates and the results of NGS confirmed the presence of blaKPC-2 with 100% of homology and no other carbapenemase genes were found [26].
A detailed analysis was performed in the polymorphic region of Tn4401 to evaluate the KPC promoters. We observed the presence of 3 previously reported promoters (P1, P2, and P3). Both isolates have only one copy of blaKPC, and no deletions in the upstream genetic environment were found.
Finally, after incubation in the presence of meropenem, one false-negative isolate (K. pneumoniae) presented positive result for all peptides evaluated and the other isolate (E. coli) presented positive results to LTLGSALAAPQR, GFLAAAVLAR, and LALEGLGVNGQ in LC-MS/MS.
Discussion
We aimed to evaluate a LC-MS/MS method to direct detection of KPC protein in clinical isolates based on a previously published study [22]. Method validation and performance evaluation were carried out by submitting a broad variety of Enterobacterales harboring the most prevalent carbapenemase genes. Each isolate was tested in a randomly interleaved run and analyzed by one blinded expert operator. The analyses were performed according to previously established parameters [22], and the method proved to be very satisfactory presenting high sensitivity (96.07%) and an absolute specificity. The presence of other carbapenemases did not interfere in the method performance, corroborating previous findings about the robustness of the method.
We have made minor adaptations to the digestion protocol, resulting in a simple, low cost, and rapid procedure. Moreover, it could be performed on a FA/ACN protein extract identical to that used on MALDI-TOF MS routine.
As previously mentioned [22], targeted LC-MS/MS provided a rapid approach to peptide assay without labeled peptides and presented no false-positive results. After the overnight incubation period, the lysis and digestion took around 60 min and the LC-MS/MS run took around 18 min for each isolate, with an isolate-to-result time of less than 90 min.
Differently from the previous study [22], we choose to include LALEGLGVNGQ in the method. Despite the peptide chemical modifications and the presence of an interfering peak, we considered that this peptide could be used to discriminate KPC positive from negative isolates. Without this peptide, one KPC-positive isolate would have been misidentified as negative, reducing methods sensitivity.
Carryover effect may lead to false positive results in samples following strong positive signals. This effect can be influenced by instrument factors (e.g., needle, injector, injection volume, valves, and tubes), frequency and method of column cleaning and peptide’s intrinsic chemistry. The methods sensitivity allowed a volume of injection of 2 μL, which mitigates the carryover, besides offering longest chromatographic column lifetime. Retention time shifts are another source of variability in the peak selection [22]. In our study, since we had no carryover effect, the use of positive controls was more important due to RT shifts then to rule out carryover effect.
According to NGS, both false-negative isolates presented one copy of blaKPC, the 3 previously reported promoters (P1, P2, and P3) and no deletion in the upstream genetic environment of this gene. These results correlate with Tn4401b isoform and are associated with lower levels of KPC production and carbapenem resistance [28, 29].
Since the false-negatives KPC type (KPC-2) have been included in the genoproteomic phase of method’s development [22] and we did not find in the genome analysis another carbapenemase gene that justify the Blue-Carba positivity, we inferred that the presence of meropenem in Blue-Carba test could be inducing KPC expression. In a previous study, Jousset et al. suggest that there is no genetic argument for an inducible expression of blaKPC-2. This study was performed with transcriptomic analysis of an E. coli TOP10 transformant exposed to suprainhibitory levels of carbapenem. Carbapenem exposure did not influence the transcription of blaKPC while it had a major impact on chromosomal genes and adaptive responses. However, the blaKPC-2 gene was already highly expressed even before antibiotic exposure. Besides that, the genetic background has probably an impact on the expression of plasmid genes [30]. Indeed, Roth et al. have demonstrated that there are differences in blaKPC expression between clinical isolates and transformants [31]. Another study from the same group has demonstrated an increase in KPC expression when exposed to sub, MIC and suprainhibitory concentrations of antibiotic, even non-β-lactam drugs. Their findings still suggest that strain and genera have a great influence in KPC expression [32]. Therapeutic failure with carbapenems against KPC producers with low MICs has been reported, corroborating the idea of induced expression [33]. Few studies with important limitations were involved in evaluating KPC production. Thus, mechanisms of blaKPC expression are far away to be fully comprehended.
In conclusion, in this study we have submitted a previously reported rapid LC-MS/MS method for the direct detection of tryptic KPC peptides to a broader variety of bacterial species and different resistance mechanisms. The peptides were robustly identified, and the method presented high sensitivity and full specificity and therefore can be used as a reliable method to identify this resistance mechanism among Enterobacterales.
Electronic supplementary material
(XLSX 35 kb)
(PDF 138 kb)
Funding information
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Ministry of Science, Technology, Inovation and Comunications, Brasília, Brazil; FIPE/HCPA (Research and Events Support Fund at Hospital de Clínicas de Porto Alegre); MAPA (Ministry of Agriculture, Livestock and Food Supply of Brazil). A.L.B. and A.F.M are research fellows from the CNPq, Ministry of Science, Technology, Inovation and Comunications, Brazil. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Centers for Disease Control (2013) Antibiotic resistance threats in the United States. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf Acessed 30 August 2019
- 2.Jacob JT, Klein E, Laxminarayan R, Beldavs Z, Lynfield R, Kallen AJ, Ricks P, Edwards J, Srinivasan A, Fridkin S, Rasheed JK, Lonsway D, Bulens S, Herrera R, McDonald C, Patel J, Bell M, Cardo D. Vital signs: carbapenem-resistant Enterobacteriaceae. Morb Mortal Wkly Rep. 2013;62(9):165–170. [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (2014) Antimicrobial resistance: global report on surveillance. http://wwwwhoint/drugresistance/documents/surveillancereport/en/ Acessed 30 August 2019
- 4.Campos AC, Albiero J, Ecker AB, Kuroda CM, Meirelles LE, Polato A, Tognim MC, Wingeter MA, Teixeira JJ. Outbreak of Klebsiella pneumoniae carbapenemase–producing K. pneumoniae: a systematic review. Am J Infect Control. 2016;44(11):1374–1380. doi: 10.1016/j.ajic.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 5.White House National Action Plan for Combating Antibiotic-Resistant Bacteria (2015) https://www.whitehouse.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf. Acessed 30 August 2019
- 6.Hrabák J, Studentová V, Walková R, Zemlicková H, Jakubu V, Chudácková E, Gniadkowski M, Pfeifer Y, Perry JD, Wilkinson K, Bergerová T. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peaper DR, Kulkarni MV, Tichy AN, Jarvis M, Murray TS, Hodsdon ME. Rapid detection of carbapenemase activity through monitoring ertapenem hydrolysis in Enterobacteriaceae with LC-MS/MS. Bioanalysis. 2013;5:147–157. doi: 10.4155/bio.12.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni MV, Zurita AN, Pyka JS, Murray TS, Hodsdon ME, Peaper DR. Use of imipenem to detect KPC, NDM, OXA, IMP, and VIM carbapenemase activity from gram-negative rods in 75 minutes using liquid chromatography-tandem mass spectrometry. J Clin Microbiol. 2014;52:2500–2505. doi: 10.1128/JCM.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sauget M, Cabrolier N, Manzoni M, Bertrand X, Hocquet D. Rapid, sensitive and specific detection of OXA-48-like producing Enterobacteriaceae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Microbiol Methods. 2014;105:88–91. doi: 10.1016/j.mimet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Lasserre C, De Saint ML, Cuzon G, Bogaerts P, Lamar E, Glupczynski Y, Naas T, Tandé D. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J Clin Microbiol. 2015;53:2163–2171. doi: 10.1128/JCM.03467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foschi C, Franza V, Conti M, Tamburini MV, Roncarati G, Cordovana M, Smirnova V, Patrono D, Mancini R, Landini MP, Ambretti S. Use of liquid chromatography-tandem mass spectrometry (LC-MS/MS) to detect carbapenemase production in Enterobacteriaceae by a rapid meropenem degradation assay. New Microbiol. 2015;38:571–576. [PubMed] [Google Scholar]
- 12.Mirande C, Canard I, Buffet Croix Blanche S, Charrier JP, van Belkum A, Welker M, Chatellier S. Rapid detection of carbapenemase activity: benefits and weaknesses of MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2015;34:2225–2234. doi: 10.1007/s10096-015-2473-z. [DOI] [PubMed] [Google Scholar]
- 13.Ghebremedhin B, Halstenbach A, Smiljanic M, Kaase M, Ahmad-Nejad P. MALDI-TOF MS based carbapenemase detection from culture isolates and from positive blood culture vials. Ann Clin Microbiol Antimicrob. 2016;15:5. doi: 10.1186/s12941-016-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber CA, Sidjabat HE, Zowawi HM, Kvaskoff D, Reed S, McNamara JF, McCarthy KL, Harris P, Toh B, Wailan AM, Paterson DL. Detection of carbapenemase activity in Enterobacteriaceae using LC-MS/MS in comparison with the neo-rapid CARB kit using direct visual assessment and colorimetry. J Microbiol Methods. 2016;131:68–72. doi: 10.1016/j.mimet.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Monteferrante CG, Sultan S, Ten Kate MT, Dekker LJ, Sparbier K, Peer M, Kostzrewa M, Luider TM, Goessens WH, Burgers PC. Evaluation of different pretreatment protocols to detect accurately clinical carbapenemase-producing Enterobacteriaceae by MALDI-TOF. J Antimicrob Chemother. 2016;71:2856–2867. doi: 10.1093/jac/dkw208. [DOI] [PubMed] [Google Scholar]
- 16.Ramos AC, Carvalhaes CG, Cordeiro-Moura JR, Rockstroh AC, Machado AMO, Gales AC. Influence of culture media on detection of carbapenem hydrolysis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2016;54:1896–1898. doi: 10.1128/JCM.00749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2014;52:2804–2812. doi: 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youn JH, Drake SK, Weingarten RA, Frank KM, Dekker JP, Lau AF. Clinical performance of a matrix-assisted laser desorption ionization-time of flight mass spectrometry method for detection of certain blaKPC-containing plasmids. J Clin Microbiol. 2016;54:35–42. doi: 10.1128/JCM.01643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charretier Y, Charrier JP, Franceschi C, Zambardi G, Cecchini T, Degout-Charmette E (2012) Method of detecting at least one mechanism of resistance to carbapenems by mass spectrometry. Patent US 20150031063
- 20.Charretier Y, Schrenzel J. Mass spectrometry methods for predicting antibiotic resistance. Proteomics Clin Appl. 2016;10:964–981. doi: 10.1002/prca.201600041. [DOI] [PubMed] [Google Scholar]
- 21.Charretier Y, Dauwalder O, Franceschi C, Degout-Charmette E, Zambardi G, Cecchini T, Bardet C, Lacoux X, Dufour P, Veron L, Rostaing H, Lanet V, Fortin T, Beaulieu C, Perrot N, Dechaume D, Pons S, Girard V, Salvador A, Durand G, Mallard F, Theretz A, Broyer P, Chatellier S, Gervasi G, Van Nuenen M, Roitsch CA, Van Belkum A, Lemoine J, Vandenesch F, Charrier JP. Rapid bacterial identification, resistance, virulence and type profiling using selected reaction monitoring mass spectrometry. Sci Rep. 2015;5:13944. doi: 10.1038/srep13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Drake SK, Youn JH, Rosenberg AZ, Chen Y, Gucek M, Suffredini AF, Dekker JP. Peptide markers for rapid detection of KPC carbapenemase by LC-MS/MS. Sci Rep. 2017;7(1):2531. doi: 10.1038/s41598-017-02749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magagnin CM, Rozales FP, Antochevis L, Nunes LS, Martins AS, Barth AL, Sampaio JM, Zavascki AP. Dissemination of blaOXA-370 gene among several Enterobacteriaceae species in Brazil. Eur J Clin Microbiol Infect Dis. 2017;36:1907. doi: 10.1007/s10096-017-3012-x. [DOI] [PubMed] [Google Scholar]
- 24.Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67(4):906–909. doi: 10.1093/jac/dkr563. [DOI] [PubMed] [Google Scholar]
- 25.Pires J, Novais A, Peixe L. Blue-carba, an easy biochemical test for detection of diverse carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013;51:4281–4283. doi: 10.1128/JCM.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geneious Prime 2019.1.3. (https://www.geneious.com)
- 28.Naas T, Cuzon G, Truong H, Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother. 2012;56(9):4753–4759. doi: 10.1128/AAC.00334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitchel B, Rasheed JK, Endimiani A, Hujer AM, Anderson KF, Bonomo RA, Patel JB. Genetic factors associated with elevated carbapenem resistance in KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54(10):4201–4207. doi: 10.1128/AAC.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jousset AB, Rosinski-Chupin I, Takissian J, Glaser P, Bonnin RA, Naas T (2018) Transcriptional landscape of a blaKPC-2 plasmid and response to Imipenem exposure in Escherichia coli TOP10. Front Microbiol 9(2929) [DOI] [PMC free article] [PubMed]
- 31.Roth AL, Kurpiel PM, Lister PD, Hanson ND. blaKPC RNA expression correlates with two transcriptional start sites but not always with gene copy number in four genera of gram-negative pathogens. Antimicrob Agents Chemother. 2011;55(8):3936–3938. doi: 10.1128/AAC.01509-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth AL, Lister PD, Hanson ND. Effect of drug treatment options on the mobility and expression of blaKPC. J Antimicrob Chemother. 2013;68:2779–2785. doi: 10.1093/jac/dkt280. [DOI] [PubMed] [Google Scholar]
- 33.Daikos GL, Markogiannakis A. Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect. 2011;17:1135–1141. doi: 10.1111/j.1469-0691.2011.03553.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 35 kb)
(PDF 138 kb)