Abstract
Long non-coding RNA (LncRNA) LINC00525 has been shown to be upregulated in several human cancers and deduced to possess caner regulatory role. The regulation of molecular mechanics of human glioma by lncRNA-LINC00525 through microRNA sponging in glioma is elusive. The lncRNA-LINC00525 showed significant (P < 0.05) upregulation in glioma cancer cells. The upregulation of lncRNA-LINC00525 was upto 6.6-fold in glioma cells relative to the normal cells. Knockdown of lncRNA-LINC00525 significantly declined the proliferation of the glioma cancer cells. Additionally, the colony formation was inhibited by around 60% in glioma cells. The wound healing and transwell assays revealed significant (P < 0.05) inhibition of migration and invasion potential under lncRNA-LINC00525 knockdown. The western blotting study of biomarkers of epithelial to mesenchymal transition (EMT) revealed that lncRNA-LINC00525 gene silencing reduced the expression of mesenchymal molecular markers but increased the protein levels of epithelial markers. miR-338-3p was predicted to be interacting with lncRNA-LINC00525 in glioma and was shown to mediated the regulatory role of lncRNA-LINC00525. Taken together, the results of present study are supportive of the prognostic applicability of lncRNA-LINC00525 against human glioma together with its therapeutic potential against the said malignancy.
Keywords: Glioma, Long non-coding RNA, microRNA, Proliferation, Migration, Invasion, Glioblastoma
Introduction
The human gliomas are considered as the most prevalent types of human central nervous system and are broadly classified into the three broad pathological categories, which include oligodendrogliomas, astrocytomas and ependymomas (Goodenberger and Jenkins 2012). Approximately, five new cases of glioma per 1 lakh of human population are diagnosed annually. Out of the total cases diagnosed, more than 50% cases are found to be manifesting the grade IV astrostomas referred as glioblastomas (Schwartzbaum et al. 2006). The glioblastoma is highly aggressive and patients affected with this disorder have 5 year survival rate of not more than year and a half (Bush et al. 2017). Moreover, it is seen to more commonly affect the male population with 1.5 times higher prevalence rates (Nizamutdinov et al. 2018). The glioblastoma is currently being treated through maximal safe surgical resection combined with the controlled application of radio- and chemotherapeutic treatments (Bianco et al. 2017). Despite such clinical treatment measures, the over-all survival rate of glioblastoma has not enhanced much. The inefficiency of the treatment strategy is attributed to difficulty of disease prognosis at earlier stages (Bush et al. 2017). The researchers are thus in continuous exploration of the effective prognostic and treatment tactics for obtaining better clinical outcomes against the malignant type of human gliomas. The research approaches seen with promising results towards this direction include the discovery of specific diagnostic and therapeutic targets to be utilized for the management of human glioma. The non-coding RNAs consisting of long non-coding RNAs (lncRNAs) and microRNAs (miRs) are being extensively studied for their cancer regulatory effects in humans (Slaby et al. 2017). The research findings have clearly indicated that lncRNAs and miRs regulate the molecular mechanics of growth and proliferation of cancer including the human gliomas and there is growing support for these RNA bio-molecules to serve as the prognostic and therapeutic agents against the glioma (Peng et al. 2018; Shi et al. 2017). The dysregulation of lncRNA-LINC00525 and miR-338-3p has been implicated to be associated with several human cancers (Wang et al. 2017; Zhang et al. 2017). However, the regulatory interactional interplay of lncRNA-LINC00525 and miR-338-3p has not been worked out in glioma. We, in the present study, found that the glioma cancer cell lines exhibited significantly higher transcript levels of lncRNA-LINC00525 and the experimental silencing of lncRNA-LINC00525 not only reduced the growth of the cancer cells but also rendered them to migrate and invade at significantly lower rates, in vitro. Interestingly, it was deduced that lncRNA-LINC00525 positively regulates the epithelial–mesenchymal transition (EMT) of glioma cells and its knockdown markedly enhanced the expression of epithelial molecular marker proteins but declined the protein levels of mesenchymal biomarkers. The lncRNA-LINC00525 was found to be sponging the expression of miR-338-5p as the latter was confirmed to interact with lncRNA-LINC00525 in sequence directed manner. The anti-proliferative effects of lncRNA-LINC00525 knockdown were shown to be operated through transcript level enhancement of tumor suppressor miRNA-338-3p. The study thus deduced that lncRNA-LINC00525 might emerge as crucial prognostic marker against human glioma and showed the therapeutic value of lncRNA-LINC00525 targeting through miR-338-3p.
Materials and methods
Growth and maintenance of cell lines and their transfection
The American Type Collection Center (ATCC) provided the glioma cancer cell lines (U87, MO59K, U118, Hs683 and LN-18) as well as the normal astrocyte cell line. Dulbecco's Modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, Missouri, USA) supplemented with high concentration glucose was used for culturing of all the cell lines using 37 °C temperature and 5% CO2 concentration conditions which were maintained using humidified growth incubator.
Transfection
The transfection of glioma cell lines was performed with the help of Lipofectamine 2000 (Thermo Fisher Scientific) reagent as per the manufacturer’s protocol. Various constructs used for transfection of cancer cells were ordered from Guangzhou RiboBio Co. Ltd (Guangzhou, China).
Synthesis of complementary DNA (cDNA) and expression analysis
Prior to the real time polymerase chain reaction (RT-PCR) study, the extraction of total RNA from the normal astrocyte and cancer cell lines and its subsequent reverse transcription to cDNA was performed with the help of TRIzol reagent (Thermo Fisher Scientific) and HiScript II qRT SuperMix kit (Vazyme, China), respectively. The cDNA was used as template and SYBR Green PCR master mix (Thermo Fisher Scientific) was used for performing the RT-PCR reaction. The PCR was run on the QuantStudio 5.0 PCR system (Applied Biosystems). The 2−ddCt method based calculations were performed to examine the relative gene expression levels. For the normalization of the RT-PCR expression analysis, the human GADPH gene was used an internal control.
Cell viability assay
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (sigma-Aldrich) assay was used to determine the viability of transfected glioma cells. Briefly, 2 × 104 cells were added per well of a 96-well plate and cultured in DMEM medium supplemented with high glucose concentration at 37 °C for 0 h, 12 h, 24 h, 36 h, 48 h, 72 h or 96 h. The MTT solution at final concentration of 0.5% was added to each well of the plate and 4 h incubation was given at 37 °C. The dissolution of formazan crystals was made by the addition of dimethyl sulfoxide (DMSO). Finally, the absorbance values were measured at 570 nm using ELx-800 Universal Microplate Reader. At least three replicas were kept for each experimental sample.
Colony formation assay
The clonogenic colony forming assay was determined to assess the relative colony formation by the transfected glioma cells, in vitro. Here, about 2000 transfected cells were added to each well of the 6-well plate and cell culturing was performed for a period of 2 weeks at 37 °C in humidified incubator. After, 2 weeks the colonies were visualized by light microscope after their washing with PBS, fixation by ethanol and staining with 0.1% crystal violet solution. The relative percentage of the colonies formed was calculated using five random microscopic fields per sample.
Wound-heal assay
The migration of the transfected glioma cells was determined through the wound-heal assay. Briefly, the transfected cells were cultured in 6-well plates at 2 × 105 cells per well in DMEM at 37 °C until a regular cell surface was obtained. The surface was then carefully scratched to carve a linear wound with the tip of a 10 μl micropipette which was photographed under the light microscope. The 37 °C incubation was continued for 24 h following which the wound was again visualized using microscope and its width was compared with the initial wound width. Three replicates were used for carrying out the experiment.
Transwell chamber assays
The transwell assay was used for the estimation of invasion of the transfected glioma cancer cells through the transwell membrane coated with Matrigel. The transfected cells were inoculated at cellular density of 1 × 104–1 × 105 cells per well in the upper chamber of transwell while the lower chamber was filled with DMEM medium only. The plate was incubated at 37 °C for 48 h. The cells which invaded the lower chamber were fixed with 70% ethanol, crystal violet (0.1%) staining was performed. The cells were then examined under light microscope. The cell counting was done and the relative percentage of the cell invasion was estimated using multiple random microscopic fields. The experiments were performed with three replicates.
In silico analysis
To specifically identify the microRNA which targets lncRNA-LINC00525 in glioma, the in silico analysis was performed using StarBase v2.0 (https://starbase.sysu.edu.cn/starbase2/index.php) online database. The online bioinformatics was further used to identify the specific lncRNA-LINC00525 sequence used for microRNA binding.
Western blotting
Total cellular proteins from the transfected cells were extracted with the help of RIPA lysis and extraction buffer (Thermo Fisher Scientific) according to the manufacture’s protocol. The protein quantification was done using Bio-Rad protein assay kit. The separation of the protein extracts was performed using 10% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). Afterwards, the proteins were electrophoretically transferred to the nitrocellulose membranes which were exposed to specific antibodies for 12 h at 4 °C in dark. This was followed by the incubation of the membranes to peroxidase-linked secondary antibodies 2 h at room temperature. The protein bands were visualized using the enhanced chemiluminescence. Human β-actin protein was recognized as the internal control in the protein blotting studies.
Statistical analysis
The experiments were performed in triplicate and expressed as mean ± SD. Student’s t-test (for comparison between two samples) and one way analysis of variance followed by Tukey’s test (for comparison between more than two samples) were used for statistical analysis using GraphPad Prism software (version 7; GraphPad Software, Inc., La Jolla, CA, USA. P < 0.05 was considered to indicate a statistically significant difference.
Results
Glioma cells exhibit transcript upregulation of lncRNA-LINC00525
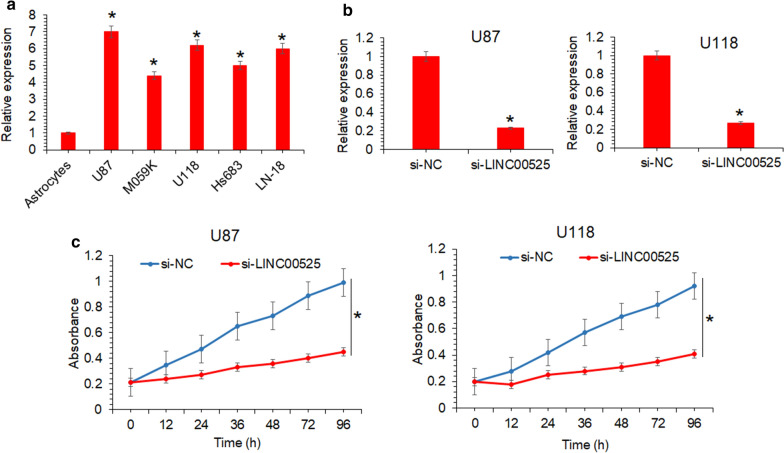
To know about the expression of lncRNA-LINC00525 in human glioma, the RT-PCR was performed for analyzing its transcript abundance in glioma cell lines (U87, MO59K, U118, Hs683 and LN-18) in comparison to the normal astrocyte cell line. It was seen that all the cancer cell lines exhibited significantly (P < 0.05) higher lncRNA-LINC00525 expression than the normal astrocytes (Fig. 1a). The upregulation of lncRNA-LINC00525 was upto 6.6 folds in glioma cells. Highest expression levels were noted from the U87 and U187 cell lines and thus these were taken for the further characterization of role lncRNA-LINC00525 in glioma. The results are thus indicative of dysregulation of lncRNA-LINC00525 in glioma suggesting its probable molecular regulatory role in the malignancy.
Fig. 1.
The silencing of glioma upregulated lncRNA-LINC00525 led to significant decline of glioma cancer cell growth. a Expression analysis of lncRNA-LINC00525 from glioma cancer cell lines (U87, MO59K, U118, Hs683 and LN-18) in comparison to normal astrocytes, b RT-PCR expression study of lncRNA-LINC00525 in U87 and U118 cancer cells transfected with si-NC or si-LINC00525, c the estimation of proliferation of U87 and U118 cancer cells transfected with si-NC or si-LINC00525 by MTT assay. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05)
LncRNA-LINC00525 knockdown inhibited the glioma cell growth
The U87 and U187cell lines were transfected with si-LINC00525 construct to perform the experimental gene silencing of lncRNA-LINC00525 in these cell lines. The RT-PCR expression study of lncRNA-LINC00525 showed that the si-LINC00525 transfected cells exhibited significant transcriptional repression (5.3-fold) of lncRNA-LINC00525 in comparison to the respective normal control cell lines transfected with si-NC (Fig. 1b). The proliferation of cancer cells under lncRNA-LINC00525 knockdown was shown to be significantly (P < 0.05) lower in comparison to the control cells, as deduced by MTT assay (Fig. 1c).
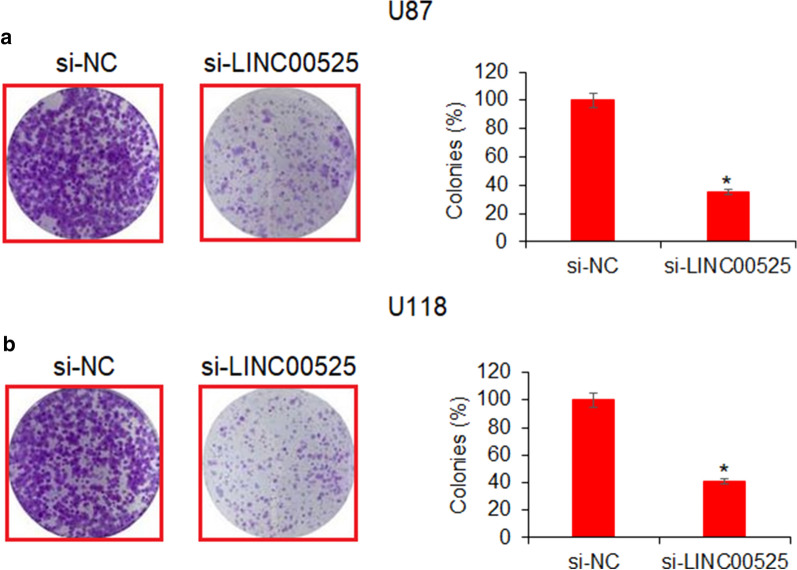
LncRNA-LINC00525 knockdown inhibited the colony formation of glioma cells
The effects of lncRNA-LINC00525 was assessed by colony formation assay. The results showed that U118 and U87 glioma cell colonies decreased significantly upon RNA-LINC00525 silencing (Fig. 2a, b). The colonies were inhibited by almost 60% in both U118 and U87 glioma cells. These results suggest that lncRNA-LINC00525 regulates the growth of glioma cancer cells and its intentional silencing might serve a therapeutic role against the human glioma.
Fig. 2.
The transcriptional knockdown of lncRNA-LINC00525 reduced the colony formation by glioma cells, markedly. a Relative colony formation by U87 cancer cells transfected with si-NC or si-LINC00525, b relative colony formation by U118 cancer cells transfected with si-NC or si-LINC00525. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05)
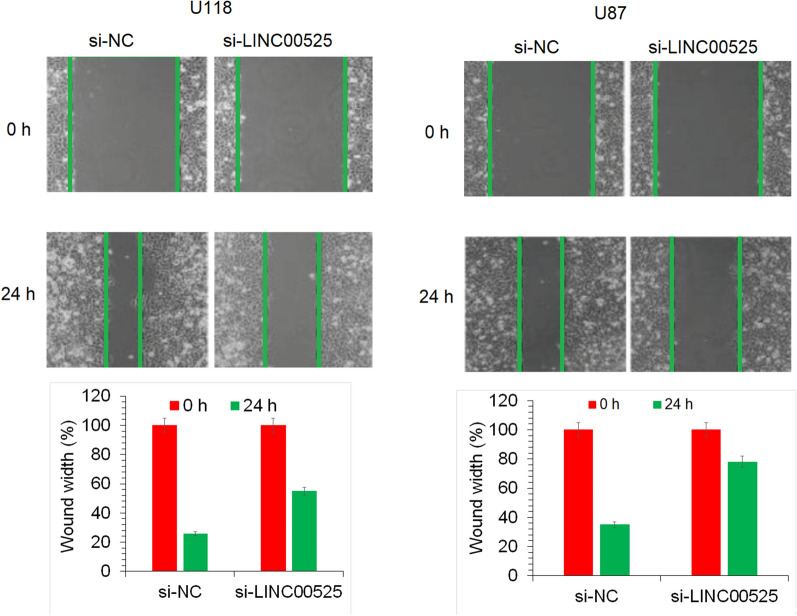
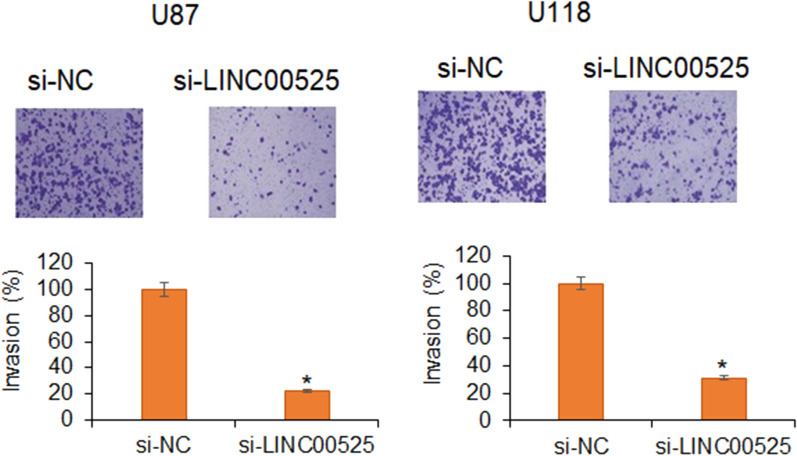
LncRNA-LINC00525 silencing suppressed glioma cell migration and invasion
The metastasis of cancer cells is one of the crucial aspects of aggressiveness of a particular human cancer (Steeg 2016). Thus, scientists are continuously looking for the measures to contain the motility of cancer cells. The assessment of the migration of glioma cells showed that the cancer cells showed significantly lower migration under lncRNA-LINC00525 knockdown (Fig. 3). The invasion of the glioma cells was also seen to significantly fall under lncRNA-LINC00525 knockdown (Fig. 4). Together the results show that lncRNA-LINC00525 might be positively regulating the metastasis of glioma cells and thus its molecular targeting may assist is restricting the glioma spread.
Fig. 3.
lncRNA-LINC00525 knockdown restricts the glioma cell migration. Analysis of migration U87 and U118 cancer cells transfected with si-NC or si-LINC00525 by wound-heal assay. The experiments were performed in triplicate and expressed as mean ± SD (P < 0.05)
Fig. 4.
lncRNA-LINC00525 knockdown restricts the glioma cell invasion. Analysis of invasion U87 and U118 cancer cells transfected with si-NC or si-LINC00525 by transwell assay. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05)
The regulation of epithelial to mesenchymal transition of glioma cells by lncRNA-LINC00525
The study of the protein expressions of the molecular markers of epithelial–mesenchymal transition (EMT) showed that the knockdown of lncRNA-LIN00525 led to the enhancement in the protein levels of the epithelial molecular markers (E-cadherin and α-catenin) while it significantly reduced the expression of the mesenchymal protein markers (fibronectin and vimentin) (Fig. 5). Similar trends were observed for U87 and U118 cell lines. The results that show that lncRNA-LINC00525 might be regulating the EMT of glioma cells further revealing its therapeutic against the human glioma.
Fig. 5.
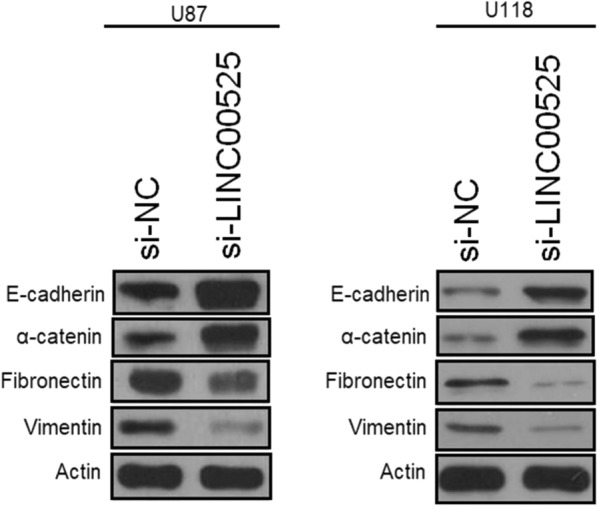
Knockdown of lncRNA-LINC00525 enhances the protein expression of epithelial markers while it declines the expression of mesenchymal marker proteins. Western blotting of EMT marker proteins from U87 and U118 cancer cells transfected with si-NC or si-LINC00525. The experiments were performed in triplicate
LncRNA-LINC00525 exercised glioma regulatory role through miR-338-5p
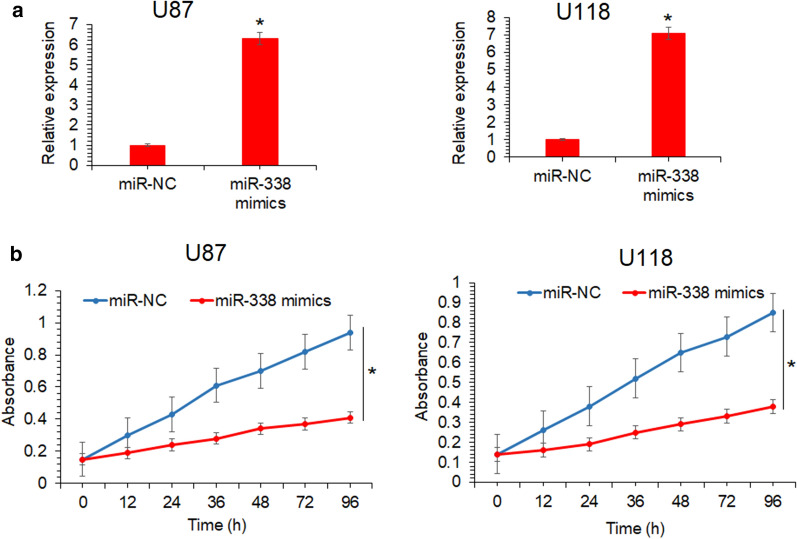
To specifically predict the microRNA targeting lncRNA-LINC00525 in glioma, in silico analysis was used. The bioinformatics showed that miR-338-3p is interacting with lncRNA-LINC00525 in a sequence specific manner in glioma (Fig. 6a). The prediction was supported by the RT-PCR study of miR-338-3p in glioma cell lines. The cancer cell lines exhibited significantly lower miR-338-3p expression corresponding to higher lncRNA-LINC00525 levels in comparison to the normal astrocytes (Fig. 6b). miR-338-3p was over-expressed in U87 and U118 cell lines (Fig. 7a). The miR-338-3p over-expression further reduced the proliferation of glioma cancer cells, in vitro (Fig. 7b). The results thus suggest that lncRNA-LINC00525 might be regulating the mechanics of growth and proliferation of glioma cancer cells via its sponging of miR-338-3p.
Fig. 6.
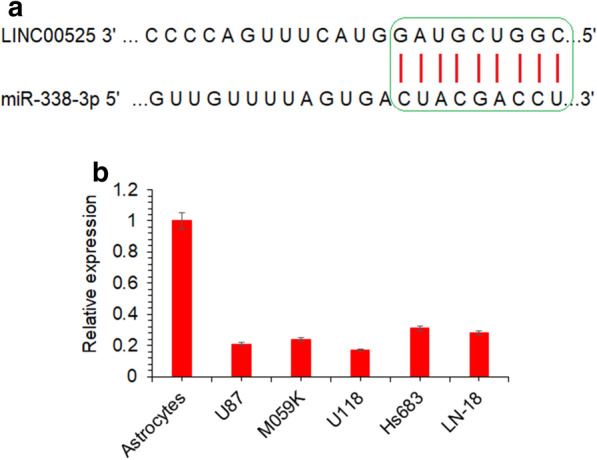
lncRNA-LINC00625 is targeted by miR-338-3p in human glioma. a In silico analysis for the target prediction of microRNA specifically targeting lncRNA-LINC00525, b expression analysis of miR-338-3p from glioma cancer cell lines (U87, MO59K, U118, Hs683 and LN-18) in comparison to normal astrocytes. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05)
Fig. 7.
The regulatory effects of lncRNA-LINC00525 are exerted though miR-338-3p in human glioma. a Expression analysis of miR-338-3p from U87 and U118 cancer cells transfected with miR-NC or miR-338 mimics, b expression analysis of lncRNA-LINC00525 from U87 and U118 cancer cells transfected with miR-NC or miR-338 mimics, c the estimation of proliferation of U87 and U118 cancer cells transfected with miR-NC or miR-338 mimics by MTT assay. The experiments were performed in triplicate and expressed as mean ± SD (*P < 0.05)
Discussion
Gliomas are the most aggressive types of human cancers mainly affecting central nervous system. The gliomas constitute 50% of all the primary malignant tumors of central nervous system and exhibit high mortality and incidence (Vallée et al. 2017). The grade four glioma patients have an average survival period of just 16 months (Yu et al. 2017). The human glioma is currently been treated though the application of careful surgical resection combined with radiotherapy/radiotherapy and chemotherapy, particularly the administration of temozolomide (Bianco et al. 2017). However, the over-all survival rate of glioma is still very low and the glioma patients hardly survive for a year and half (Bush et al. 2017). Thus, the currently attempted clinical are not much admirable and it is therefore needed to look for the alternative therapeutic procedures which may prove more effective and to lessen the burden of this malignancy. Originally described as the noise of eukaryotic genome, the long-coding RNAs (lncRNAs) have emerged as the extensively studied bio-molecules for their prognostic role and therapeutic value against the human cancer (Lin and Yang 2018). The human cancers have been shown to express aberrant transcript levels of lncRNAs. Numbers of lncRNAs display either significant higher or lower expression in human gliomas and have been suggested to serve in the disease prognosis (Rao et al. 2017; Lin et al. 2019; Wang et al. 2019). In the present study, the expression of lncRNA-LINC00525 was significantly higher in the glioma cancer cells than the normal astrocyte cell line and this observation fits well with the already established findings making it suitable to serve as the prognostic biomarker in diagnosing human glioma (Peng and Zheng 2019). The silencing of lncRNA-LINC00525 declined the growth of glioma cells and glioma cells proliferated at remarkably lower rates. The basis of such anti-proliferative effects of lncRNA-LINC00525 was shown to be its regulatory control on the cell apoptosis (Zheng and Peng 2019). Interestingly, the results of the present research study indicated that lncRNA-LINC00525 has pro-metastatic regulatory role in human glioma. The lncRNA-LINC00525 knockdown in glioma cells negatively affected their migration and invasion potential. The reason behind such anti-motility effect might be decline of cell viability by the transcriptional silencing of lncRNA-LINC00525 together with its microRNA sponging action in glioma. The suppression of migration and invasion by lncRNA-LINC00525 has been also reported for the non-small lung cancer cells (Yang et al. 2020). Moreover, the key finding of the present study was the elucidation of regulation of epithelial–mesenchymal transition (EMT) of glioma cells by lncRNA-LINC00525. EMT is responsible for the tumorogenesis of human cancer and known to fundamentally mediate the cancer cell metastasis (Kiesslich et al. 2013). During EMT, the cells lose their adhesion with the basement membrane and undergo several morphological transformations with the alterations of expression of proteins which are used as the bio-markers for this biological cellular transition (Moreno-Bueno et al. 2009). The silencing of lncRNA-LINC00525 increased the expression of epithelial markers (E-cadherin and α-catenin) while it was seen to decrease the expression of mesenchymal marker proteins (vimentin and fibronectin) (Sethi et al. 2011). At the molecular level, LINC00525 was shown to mediate its regulatory effects through the sponging of miR-338-3p which has been also confirmed by the previous studies in non-small lung cancer (Yang et al. 2020). Summing up, the study signifies the prognostic application and therapeutic potential of lncRNA-LINC00525 against the human glioma and emphasized on the importance of molecular regulatory axis, miR-338-3p/lncRNA-LINC for its therapeutic targeting against human glioma.
Taken together, the study showed glioma cells expressed significantly higher transcripts of lncRNA-00525. This transcriptional dysregulation together with the findings that lncRNA-LINC00525 through sponging of miR-338-3p regulates the growth and metastasis of human glioma cancer cells and is exerting a regulatory control over their EMT revealed that lncRNA-00525 might serve as potential prognostic marker against the human glioma and the miR-338-3p/lncRNA-LINC0025 molecular axis might be employed as a vital therapeutic target against human glioma in future.
Acknowledgements
All the author of this manuscript is thankful to The First Affiliated Hospital of Nanchang University, Nanchang, Jiangxi Province, 330006, China, to conduct the presented protocol.
Authors’ contributions
YW, MW and YL designed the protocol of the study. YW, MW, FL and YL performed the experimental work and collect the data for presented study. MW and YL involve in the statistical analysis. MW and YL supervised the work and drafted the manuscript, although all author contributes for the preparation of manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1186/s13568-022-01498-4
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yilv Wan and Feng Liang contributed equally to this work
Change history
12/21/2022
This article has been retracted. Please see the Retraction Notice for more detail: 10.1186/s13568-022-01498-4
Contributor Information
Minjun Wei, Email: wmj0111@163.com.
Ying Liu, Email: liuyingoomumu@163.com.
References
- Bianco J, Bastiancich C, Jankovski A, Des Rieux A, Préat V, Danhier F. On glioblastoma and the search for a cure: where do we stand? Cell Mol Life Sci. 2017;74(13):2451–2466. doi: 10.1007/s00018-017-2483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush NA, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–4. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205(12):613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Kiesslich T, Pichler M, Neureiter D. Epigenetic control of epithelial–mesenchymal-transition in human cancer. Mol Clin Oncol. 2013;1(1):3–11. doi: 10.3892/mco.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Hou PF, Meng S, Chen F, Jiang T, Li ML, Shi ML, Liu JJ, Zheng JN, Bai J. Emerging roles of p53 related lncRNAs in cancer progression: a systematic review. Int J Biol Sci. 2019;15(6):1287. doi: 10.7150/ijbs.33218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, Santos V, Palacios J, Portillo F, Cano A. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc. 2009;4(11):1591. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- Nizamutdinov D, Stock EM, Dandashi JA, Vasquez EA, Mao Y, Dayawansa S, Zhang J, Wu E, Fonkem E, Huang JH. Prognostication of survival outcomes in patients diagnosed with glioblastoma. World Neurosurg. 2018;109:67–74. doi: 10.1016/j.wneu.2017.09.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Zheng Q. Long non-coding RNA Linc00525 as a prognostic factor to regulate proliferation and apoptosis in colorectal cancer. J Clin Oncol. 2019;37(15):15085. doi: 10.1200/JCO.2019.37.15_suppl.e15085. [DOI] [Google Scholar]
- Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AK, Rajkumar T, Mani S. Perspectives of long non-coding RNAs in cancer. Mol Biol Rep. 2017;44(2):203–218. doi: 10.1007/s11033-017-4103-6. [DOI] [PubMed] [Google Scholar]
- Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503. doi: 10.1038/ncpneuro0289. [DOI] [PubMed] [Google Scholar]
- Sethi S, Macoska J, Chen W, Sarkar FH. Molecular signature of epithelial–mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am J Transl Res. 2011;3(1):90. [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dong B, Cao J, Mao Y, Guan W, Peng Y, Wang S. Long non-coding RNA in glioma: signaling pathways. Oncotarget. 2017;8(16):27582. doi: 10.18632/oncotarget.15175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. doi: 10.1042/BCJ20170079. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée A, Lecarpentier Y, Guillevin R, Vallée JN. Thermodynamics in gliomas: interactions between the canonical WNT/beta-catenin pathway and PPAR gamma. Front Physiol. 2017;2017(8):3. doi: 10.3389/fphys.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Huang Z, Gao R, Zeng Z, Yang W, Sun Y, Wei W, Wu Z, Yu L, Li Q, Zhang S. Expression of long noncoding RNA urothelial cancer associated 1 promotes cisplatin resistance in cervical cancer. Cancer Biother Radiopharm. 2017;32(3):101–110. doi: 10.1089/cbr.2016.2156. [DOI] [PubMed] [Google Scholar]
- Wang S, Li J, Yang X. Long non-coding RNA LINC00525 promotes the stemness and chemoresistance of colorectal cancer by targeting miR-507/ELK3 axis. Int J Stem Cells. 2019;12(2):347. doi: 10.15283/ijsc19041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lin X, Zhang P, Liu Y, Liu Z, Qian B, Liu X, Shao G. Long non-coding RNA LINC00525 promotes the non-small cell lung cancer progression by targeting miR-338-3p/IRS2 axis. Biomed Pharmacother. 2020;124:109858. doi: 10.1016/j.biopha.2020.109858. [DOI] [PubMed] [Google Scholar]
- Yu Y, Fu X, Ran Q, Yang K, Wen Y, Li H, Wang F. Globularifolin exerts anticancer effects on glioma U87 cells through inhibition of Akt/mTOR and MEK/ERK signaling pathways in vitro and inhibits tumor growth in vivo. Biochimie. 2017;142:144–151. doi: 10.1016/j.biochi.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Zhang G, Zheng H, Zhang G, Cheng R, Lu C, Guo Y, Zhao G. MicroRNA-338-3p suppresses cell proliferation and induces apoptosis of non-small-cell lung cancer by targeting sphingosine kinase 2. Cancer Cell Int. 2017;17(1):46. doi: 10.1186/s12935-017-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Peng P. P-028 Long non-coding RNA Linc00525 is an unfavorable prognostic factor and regulates proliferation and apoptosis in colorectal cancer. Ann Oncol. 2019;30:155–157. doi: 10.1093/annonc/mdz155.027. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.