Abstract
Portulaca oleracea is a C4 herb capable of performing CAM under drought stress. It is distributed worldwide and is either considered a polymorphic species or a complex of subspecies, due to its numerous morphological variations. We evaluated CAM plasticity within P. oleracea genotypes since the complexity surrounding this species may be reflected in intraspecific variations in photosynthetic behavior. Eleven subspecies of P. oleracea from distant geographical locations and one cultivar were morphologically and physiologically characterized. C4 and CAM photosynthesis were monitored in plants exposed to well-watered, droughted and rewatered treatments, and data obtained were compared among individual genotypes. All subspecies expressed CAM in a fully-reversible manner. Transcript abundance of C4–CAM signature genes was shown to be a useful indicator of the C4–CAM–C4 switches in all genotypes. C4-related genes were down-regulated and subsequently fully expressed upon drought and rewatering, respectively. CAM-marker genes followed the opposite pattern. A gradient of morphological traits and drought-induced nighttime malate accumulation was observed across genotypes. Therefore, different combinations of CAM expression levels, plant sizes and shapes are available within the P. oleracea complex, which can be a valuable tool in the context of C4/CAM photosynthesis research.
Subject terms: Plant physiology, Abiotic, Drought, C4 photosynthesis
Introduction
C4 photosynthesis and the crassulacean acid metabolism (CAM) are carbon (C) concentrating mechanisms (CCMs), similar in their biochemical pathways, as both use phosphoenolpyruvate carboxylase (PPC) to perform the primary fixation of CO2 into 4-C acids1,2. These acids are subsequently decarboxylated, regenerating CO2 in the vicinity of ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) to minimize O2 binding. The oxygenase activity leads to 3-phosphoglycerate (3PGA) and 2-phosphoglycolate (2PG) formation, the latter molecule being toxic, and requiring processing and elimination via photorespiration3. Despite the shared similarities between C4 and CAM, each CCM is usually associated with a specific set of anatomical characteristics and regulatory mechanisms, rendering uncommon the co-occurrence of both syndromes within a single plant4.
Overall, C4 acts as a spatial specialization involving the transfer of CO2 acceptor molecules between a mesophyll and a bundle sheath cell (MC and BSC, respectively)5. In contrast, CAM works as a temporal specialization, where acid formation and mobilization occur in a single MC but at different times of the day6. Also, CAM has been shown to vary in the degree of diel acid fluctuation and gas exchange patterns, with the contribution of the nocturnal CO2 primary fixation to total C assimilation varying across species7. These features characterize different types of CAM, ranging from strong CAM, where virtually all CO2 assimilation derives from CAM activity8 (Nobel 1988), to weak CAM (e.g., CAM cycling), in which atmospheric CO2 uptake takes place exclusively during the day, and the refixation of respiratory CO2 leads to a small nighttime accumulation of organic acids9.
At the weak-CAM end of the spectrum, Portulaca species are C3–C4 intermediates and C4 plants capable of performing CAM under drought. Facultative CAM was shown to occur in species belonging to the six phylogenetic clades of Portulaca, some of which performing CAM-cycling when water is deprived10–16. In addition to this complex scenario, different decarboxylating systems occur in Portulaca, with NADP-malic enzyme (ME) and NAD-ME representatives such as P. grandiflora and P. oleracea, respectively17,18.
Portulaca oleracea is a promising candidate for a C4/CAM model species due to its fast growth, efficient seed production, and accumulating literature on CCM-related biochemical and gene expression data19–26. The uncommon C4-to-CAM transition in leaves of P. oleracea is associated with the transcriptional induction of specific genes. They include a CAM-specific PPC isoform (PPC-1E1c), an aluminum-activated malate transporter (ALMT-12E.1) and a dicarboxylate carrier (DIC-1.1)23,27, and their relative transcript abundances have been suggested as a valuable tool to assess CAM induction in this species27. ALMT proteins have a role in nocturnal malate uptake into the vacuole28,29, while DIC transporters mediate C-skeleton transport across mitochondria30. Besides, several C4-related transcripts were identified to be exclusively expressed in leaves under well-watered conditions in P. oleracea, including a C4-related PPC isoform (PPC-1E1a’)23, a NAD-ME (NADME-2E.1) and an aspartate aminotransferase (ASPAT-1E1)27.
Commonly known as purslane, P. oleracea can germinate over a wide temperature range (10–40 °C)31, thriving in various light intensities, photoperiods, soil types and moisture conditions32–34, even being considered a noxious weed for agriculture35. It also presents a cosmopolitan distribution, occurring in most tropical and subtropical regions36. Variations in chromosome number37, vegetative and reproductive morphology38–42 are already described between accessions of this species. Because of such high phenotypical plasticity, P. oleracea is sometimes referred to as a polymorphic species33,37, or even subdivided into different subspecies43,44 or microspecies45–47 that form a taxonomic aggregate or complex. The most up-to-date phylogeny detailing Portulaca infra-familiar relationships still refers to the traditional P. oleracea subspecies system to highlight that this clade is paraphyletic and clusters with other species, e.g. P. molokiniensis44,48.
However, comparative information on the physiological performance, particularly in the context of CAM plasticity and CCM-related transcriptional reprogramming, is missing for the P. oleracea complex. Understanding CAM plasticity across P. oleracea genotypes may be a valuable source of information for future biotechnological applications seeking to explore C4/CAM compatibility using this species as a model26,49. Therefore, we hypothesized that there might be different degrees of CAM expression among members of the P. oleracea complex, particularly when comparing genotypes with distinctive morphological traits (e.g., leaf succulence and size) and originally from contrasting environmental conditions.
To this end, we assembled a collection of twelve P. oleracea genotypes, composed of eleven subspecies from different geographic regions and one cultivar, to compare physiological variation in CAM response. Subspecies were characterized based on climatic conditions of their place of origin or morphological traits using a clustering approach by principal component and hierarchical clustering analyses (PCA and HCA, respectively). Nighttime malate accumulation (Δmalate) and transcript abundance of CAM- and C4-signature genes revealed that weakly expressed, facultative CAM is a shared trait among all genotypes analyzed. We also validated the previously published recommendation of specific C4 and CAM genes27 by monitoring their relative transcript abundance and detecting similar expression patterns across various genotypes. Finally, our findings indicate that different combinations of drought-induced CAM expression intensities, plant sizes and shapes are available within the P. oleracea complex, an array that offers various possibilities for future C4/CAM photosynthesis research.
Results
Characterizing P. oleracea genotypes using clustering approaches
Previously identified P. oleracea subspecies and one cultivar genotype (Table 1) were grown side-by-side under greenhouse conditions for three generations before the start of the experiments. Seed attributes were analyzed under scanning electron microscopy (SEM) to confirm subspecies identification and the purity of lots (Supplementary Figure S1). A commercial cultivar was included as a reference genotype for physiological analyses since it was previously used in physio-transcriptomic studies27. Additional Brazilian wild accessions were identified via SEM analysis either as subsp. granulatostellulata or subsp. nitida (Supplementary Figure S1), and were not included in the physiological experiments, as these subspecies were already represented in our collection.
Table 1.
Identification and geographical origin of Portulaca oleracea genotypes.
| Genotype identification | Taxonomic identificationa | Geographical origin | Refs. |
|---|---|---|---|
| trituberculata | Portulaca trituberculata Danin, Domina & Raimondo | Greece, 38° 13′37′′ N, 25° 59′57′′ E | [1] |
| sicula | Portulaca sicula Danin, Domina & Raimondo | Italy, 43° 27′53′′ N, 11° 52′39′′ E | [1] |
| oleracea |
Portulaca oleracea subsp. oleracea/Portulaca trituberculata [1] |
Chile, 36° 49′ S, 73°03′ W | [1] |
| rausii | Portulaca rausii Danin | Greece, 36° 50′56′′ N, 27° 04′31′′ E | [1] |
| zaffranii | Portulaca zaffranii Danin | South Africa, 23° 04′04,1′′ E, 34° 02′36,5′′ S | [1] |
| nitida | Portulaca nitida (Danin & H.G.Baker) Ricceri & Arrigoni | Israel, 31° 47′11′′ N, 34° 42′33′′ E | [2] |
| edulis | Portulaca edulis Danin et Bagella [2] | Greece, 35° 05′08′′ N, 33° 16′46′′ E | [1] |
| papillatostellulata | Portulaca papillatostellulata (Danin & H.G.Baker) Danin | Austria, 48° 17′43′′ N, 16° 52′18′′ E | [1] |
| sativa |
Portulaca oleracea subsp. sativa (Haw.) Čelak |
Austria, 48° 11′33′′ N, 16° 23′04′′ E | [1] |
| tuberculata | Portulaca tuberculata León | Peru, 12° 35′39′′ S, 69° 11′38′′ W | [1] |
| granulatostellulata |
Portulaca granulatostellulata (Poelln.) Ricceri & Arrigoni |
Israel, 32° 23′60′′ N, 34° 52′58′′ E | [2] |
| cultivar | commercial cultivar | Agristarb | [3] |
aTaxonomic identification according to the registry at The Plant List (2013), except for P. edulis since it is not listed. [1] Walter et al., 2015. [2] Danin et al., 2012. [3] Ferrari et al. 2020.
bSeeds bought from Agristar do Brasil Ltda.—São Paulo, Brazil.
The climatic conditions of the place of origin and morphological attributes were compared for the eleven subspecies, and the cultivar was included in the morphological analysis only. Data obtained was analyzed via principal component analysis (PCA) for cluster identification, and then further evaluated using hierarchical clustering analysis (HCA).
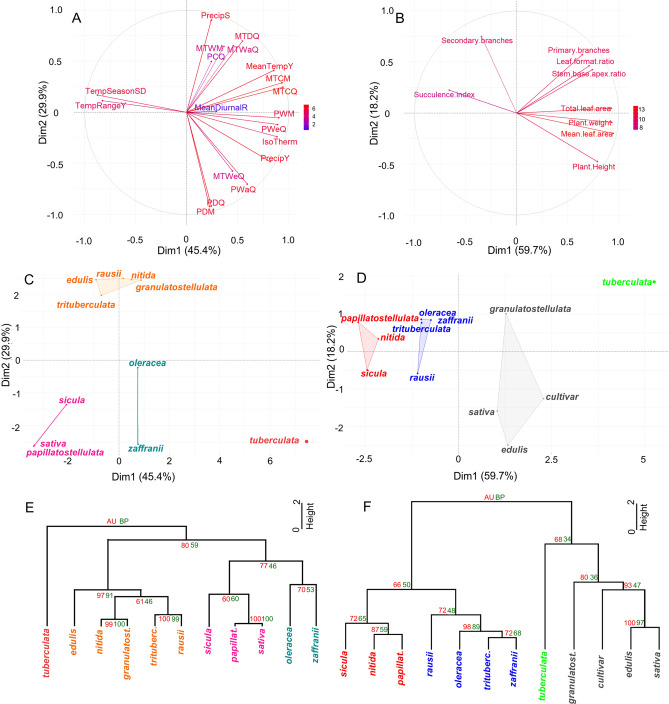
First, 19 climatic variables were retrieved from the World Clim database50 using the geographic coordinates from the sampling place of each subspecies (Supplementary Table S1, Fig. 1A,C,E). Variables presented >|0.64| correlations to the first 4 PCs, but we focused on PC 1 and 2 since 16 out of the 19 variables were significantly correlated to these PCs. One genotype, tuberculata, was isolated from the other subspecies in PC1 by scoring high in winter temperature parameters. On the other hand, precipitation variables reflecting on seasonality contributed to separating the remaining subspecies along PC2. The following groups were formed: 1—consisting of tuberculata alone, 2—formed by edulis, trituberculata, rausii, nitida and granulatostellulata; 3—formed by sicula, sativa and papillatostellulata; 4—including oleracea and zaffranii (Fig. 1C,E).
Figure 1.
Characterization of the Portulaca oleracea complex based on climatic and morphometric variables via principal component analysis (PCA) and hierarchical clustering analysis (HCA). (A,B) Variable contribution to each PC. The first two PCs explain 75% and 77% of the variance in A and B, respectively. The color scale indicates the relative variable contribution to each PC. (C,D) Groups of subspecies formed—the first two PCs harbor the most significant correlations to the variables. (E,F) HCA groups subspecies into clusters and values of approximately unbiased (AU) and bootstrap (BP) are presented in red and green, respectively (see Materials and methods section for details). Data for these analyses are presented in Tables S1 and S2. In (A,C,E), the following climatic variables were analyzed: latitude, longitutude, annual mean temperature (MeanTempY), mean diurnal range (MeanDiurnalR), isothermality (IsoTherm), temperature seasonality standard deviation (TempSeasonSD), max. temperature of warmest month (MTWM), min. temperature of coldest month (MTCM), temperature annual range (TempRangeY), mean temperature of wettest quarter (MTWeQ), mean temperature of driest quarter (MTDQ), mean temperature of warmest quarter (MTWaQ), mean temperature of coldest quarter (MTCQ), annual precipitation (PrecipY), precipitation of wettest month (PWM), precipitation of driest month (PDM), precipitation seasonality (PrecipS), precipitation of wettest quarter (PWeQ), precipitation of driest quarter (PDQ), precipitation of warmest quarter (PWaQ), precipitation of coldest quarter (PCQ).
Second, nine morphometric parameters were measured in the 12 genotypes, using at least 25 well-watered, two-month-old individuals that were grown side-by-side under controlled conditions (Supplementary Table S2, Fig. 1B,D,F). These data were subsequently analyzed by PCA and HCA. The first three principal components explained a total of 90.16% of the variance, but only PCs 1 and 2 (77.83% of the variance) were kept since PC3 showed low correlation coefficients to the variables (<|0.52|). While the number of primary and secondary branches was positively correlated to PC2, succulence was negatively correlated to PC1, and the remaining characteristics were positively correlated to PC1 (Fig. 1B). Considering the contribution of each variable to the PCs, leaf size and stem branching were important factors when separating the four clusters. The groups were composed of: A—containing exclusively tuberculata; B—papillatostellulata, nitida and sicula; C—oleracea, zaffranii, trituberculata and rausii; and D—sativa, edulis, and granulatostellulata and the cultivar (Fig. 1D,F). We tested if morphological correlated with each other (Supplementary Table S3), and positive correlations (r > 0.61, p-value < 0.05) revealed that large plants have large leaves and harbor more primary branches. Succulence was negatively correlated (r < − 0.59, p-value < 0.05) with plant robustness parameters and leaf area. Overall, taller plants harbored thinner and larger leaves, whereas smaller plants were more branched with smaller, thicker leaves (Supplementary Table S3).
When comparing the clusters formed using either the climate or morphological approaches, tuberculata was placed in an isolated cluster, while the following pairs clustered together using both approaches: sicula + papillatostellulata; trituberculata + rausii; and oleracea + zaffranii.
Drought represses C4 and promotes CAM pathways across genotypes
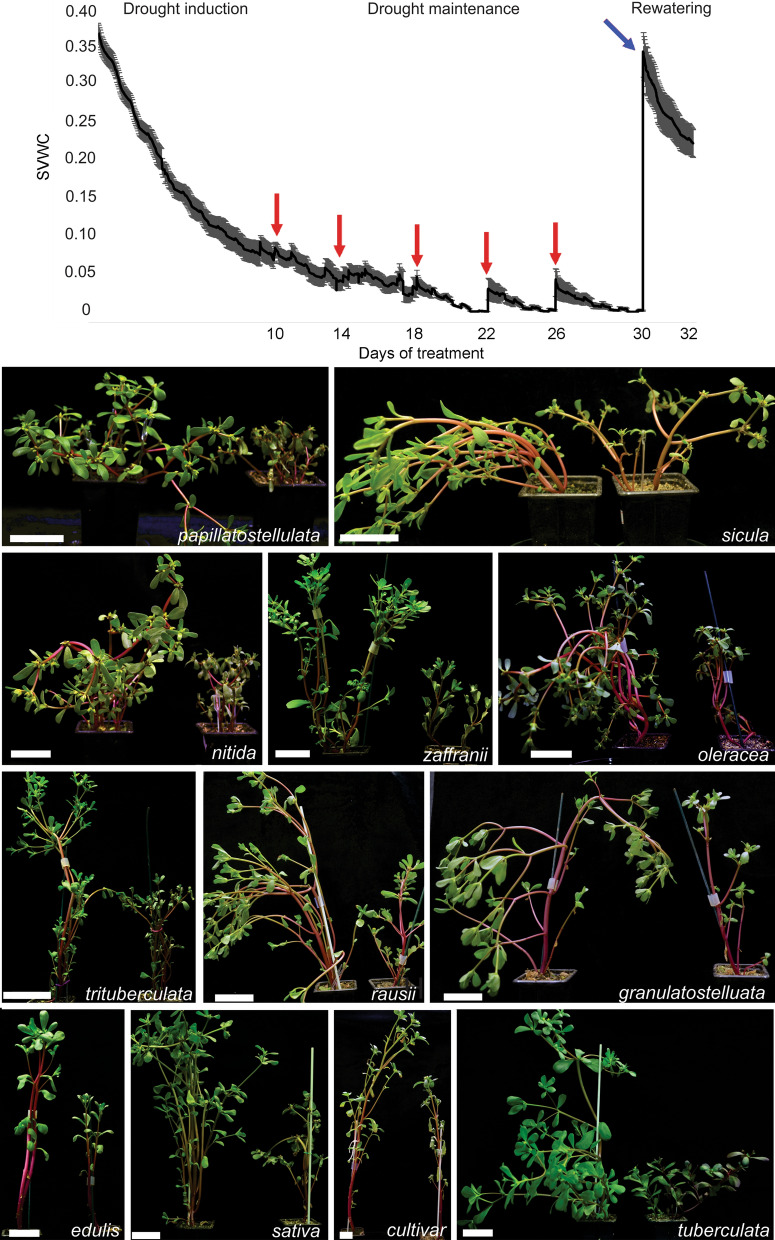
After characterizing the morpho-climatic traits of our collection, the impacts of water availability on CAM photosynthesis across genotypes was investigated by comparing CAM-related traits. To this goal, one-month-old plants were kept side-by-side under well-watered or droughted conditions for 30 days, or exposed to drought for 30 days followed by two days of complete rewatering (Fig. 2A). To prevent plants from dying, the drought treatment consisted of withholding water for ten consecutive days, followed by a 20 day period in which a small water volume (10 ml) was added to the pots whenever the soil water content reached values close to zero (usually every four days). The drought treatment promoted a marked reduction in plant size in all genotypes compared to the well-watered counterparts (Fig. 2B).
Figure 2.
Drought treatment impacts on soil volumetric water content (SVWC) and overall plant morphology. (A) Changes in SVWC during drought and rewatering treatments in P. oleracea, with red and blue arrows indicating partial (10 ml per pot) and full watering events, respectively (see Methods for details). Data are means ± SE for monitored genotypes. (B) Representative images of 2-month-old plants kept under well-watered (left) and droughted (right) conditions.
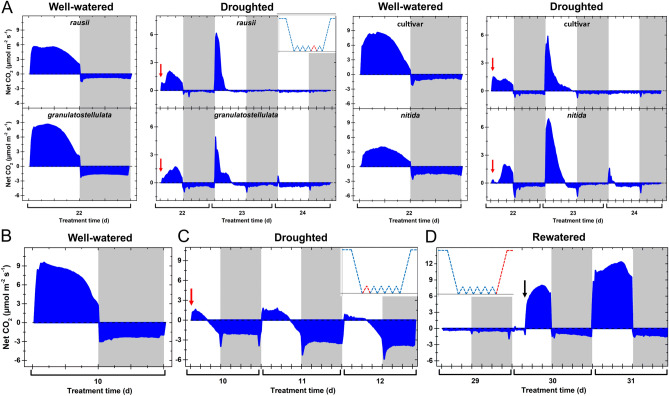
We monitored continuous net CO2 exchange in rausii, granulatostellulata, nitida and the cultivar, which are representatives of each of the four morphological clusters identified in this work (Fig. 3). A multi-chamber IRGA system was used to monitor the entire shoot of 2-weeks-old plants, revealing a similar behavior for all genotypes monitored (Fig. 3). Well-watered plants displayed CO2 assimilation throughout the entire light period. In contrast, CO2 uptake was limited to a brief burst in the early morning at the start of drought treatment, whereas daytime CO2 uptake was undetectable at the end of the drought period. Daytime CO2 uptake was recovered within hours after full rewatering of the plants, indicating the activation of C4 photosynthesis when the water supply returned. Nocturnal net CO2 uptake was not observed across the genotypes analyzed.
Figure 3.
Similar drought-triggered changes in diel gas exchange are shared by Portulaca oleracea subspecies. (A) Net CO2 exchange of shoots of representative subspecies after 16 days of drought. (B-D) Net CO2 exchange by shoots of granulatostellulata in well-watered conditions (B), after the initial 10-day-period of water withholding (C), and after 20 days of drought and into rewatering (D). In (A–D), two-week-old individuals were used due to the size restriction of the cuvette. Also, data were normalized against the leaf area. Shaded areas indicate the dark period, red arrows indicate partial watering event (5 ml), and the blue arrow indicates full rewatering. Inserts schematically illustrate soil water content following water deprivation (see Methods for details), and the time points corresponding to the gas exchange measurements are highlighted in red.
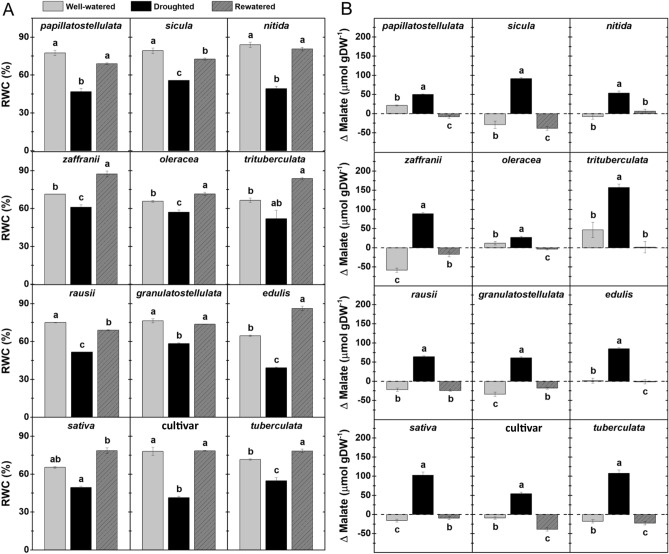
Leaf relative water content (RWC) values were significantly lower under drought compared to well-watered conditions for all genotypes (Fig. 4A). Rewatering recovered leaf RWC to values similar, or almost similar (e.g., sicula and rausii), to those detected in well-watered plants. To investigate whether all genotypes were able to switch to CAM photosynthesis in response to drought, leaf malate levels were determined at dawn and dusk, and nocturnal malate accumulation (Δmalate) was calculated for each of them (Fig. 4B, Supplementary Table S4). Under well-watered conditions, papillatostellulata, oleracea, trituberculata and edulis, the latter at very low levels, displayed accumulation of malate overnight (positive Δmalate), whereas malate levels in the remaining genotypes decreased overnight (negative Δmalate) (Fig. 4B). All genotypes presented positive Δmalate under drought treatment, with the lowest and highest Δmalate values (26.7 and 157.2 µmol malate per g dry weight, respectively) detected in oleracea and trituberculata, respectively. Nocturnal malate accumulation was consistently reduced following rewatering in all subspecies (Fig. 4B).
Figure 4.
Impacts of water availability P. oleracea on relative water content (RWC) and nocturnal malate accumulation. (A) Leaf RWC of well-watered, droughted and rewatered plants. (B) Nocturnal malate accumulation (Δ malate) in well-watered, droughted and rewatered plants. Data are means ± SE of at least three biological replicates, and different letters indicate statistically significant differences (p < 0.05) among the treatments for each subspecies. In (B), standard error = √((standard errorwell-watered)2 + (standard errordroughted)2.
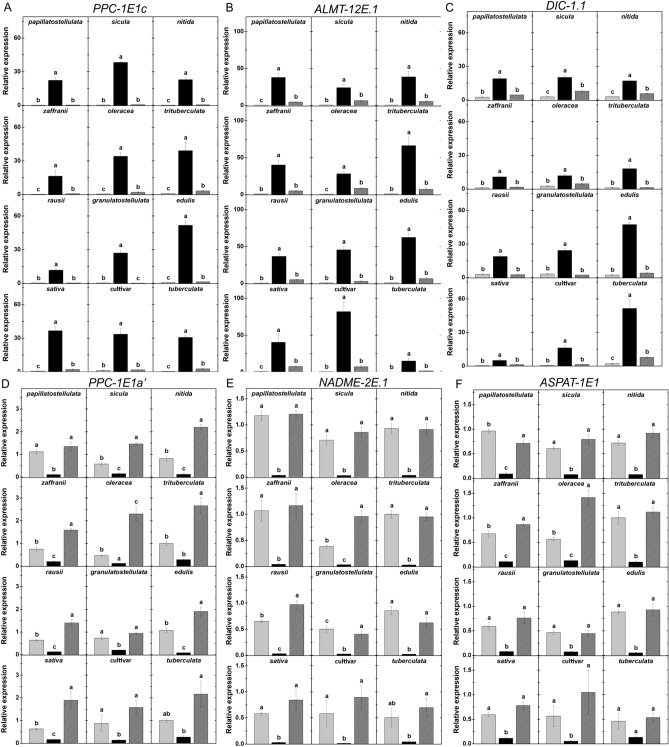
We then monitored the transcriptional patterns for CCM-related genes across the studied genotypes. Transcriptional profiling of CAM-related genes under drought, when compared to well-watered conditions, showed increments of up to 165, 123 and 25 times in mRNA levels of PPC-1E1c, ALMT-12E.1 and DIC-1.1, respectively (Fig. 5A–C, Supplementary Table S4). Rewatering reduced relative transcript levels for all three CAM-marker genes significantly in most cases, reaching values as low as those detected in well-watered plants.
Figure 5.
Impacts of water availability on transcript abundance of C4- and CAM-marker genes. (A-C) Relative abundance of CAM-specific transcripts: PPC1E1c (A), ALMT-12E.1 (B), DIC-1.1 (C). (D–F) Relative abundance of CAM-specific transcripts: PPC1E1a’ (D), NADME-2E.1 (E), ASPAT-1E1 (F). Mean relative expression was normalized against well-watered trituberculata samples, and mRNA levels were determined in samples harvested at dawn (A–C) or dusk (D–F). Data are means ± SE of at least three biological replicates, and different letters indicate statistically significant differences (p < 0.05) among the treatments for each subspecies.
For C4-related genes such as PPC-1E1a’, NADME-2E.1 and ASPAT-1E1, mRNA levels were between 3 and 48 times lower under drought than in well-watered plants across genotypes (Fig. 5D–F, Table S4). Rewatering resulted in PPC-1E1a’ mRNA levels relatively higher than those detected in well-watered plants, except for papillatostellulata, granulatostellulata and the cultivar, which exhibited similar abundance in both rewatered and well-watered plants (Fig. 5F). Upon rewatering, relative transcript abundance of NADME-2E.1 and ASPAT-1E1 returned to levels as high as those detected in well-watered individuals (Fig. 5D–F, Supplementary Table S4).
Lastly, we assessed drought resilience across the genotypes by performing Chl a fluorescence imaging in source leaves of plants exposed to long-term, continuous water withholding, i.e., 20 and 35 days (Supplementary Figure S2–4). In all genotypes, PSII operating efficiency (Fq’/Fm’) was similar comparing well-watered and droughted plants, and only trituberculata, rausii and the cultivar exhibited slightly reduced Fq’/Fm’ after prolonged droughted (Supplementary Figure S2). Non-photochemical quenching (NPQ) significantly increased after drought only in tuberculata (Supplementary Figure S3). The most prominent reduction in maximum quantum efficiency of PSII photochemistry (Fv/Fm) was detected after 34 days of drought stress in tuberculata, papillatostellulata and the cultivar (Supplementary Figure S4).
Comparing CCM-related physio-molecular traits across the genotypes
After characterizing C4–CAM photosynthesis for P. oleracea genotypes individually using malate quantification and transcript abundance of key C4–CAM-related genes, we compared the genotypes at each water availability condition separately (Supplementary Tables S4, S5). Nighttime malate accumulation was significantly different (p-value < 0.05) across genotypes at all three water availability conditions. However, such a pattern was not statistically supported in terms of CAM-related transcript accumulation. Although tuberculata clustered apart from the other genotypes at the initial morpho-climate characterization, its Δmalate levels and transcript profiles did not stand out compared to the remaining genotypes, except for the lower accumulation of ALMT-12E.1 mRNA levels under drought. Also, genotype pairs that initially clustered together (sicula + papillatostellulata; trituberculata + rausii; and oleracea + zaffranii) did not show similar trends for any of the CCM-related parameters.
Surprisingly, no significant correlations were observed between Δmalate and CAM-related transcript abundance (including PPC-1E1c mRNA levels) under drought. However, a positive correlation (r > 0.76, p-value < 0.05) was observed between (1) Fv/Fm and Fq’/Fm’, (2) PPC-1E1a' mRNA levels and either RWC and Δmalate, and (3) between NADME-2E.1 and ASPAT-1E.1 mRNA levels (Supplementary Table S6). Also, ALMT-12E.1 mRNA abundance was negatively correlated to ASPAT-1E.1 transcript levels (r = − 0.74, p-value < 0.05, Supplementary Table S6).
Discussion
CAM has long been described as a highly plastic adaptive syndrome, operating in different modes and magnitudes depending on the lineage51. CAM-related literature includes a vast array of studies on interspecific variability in the contribution of nocturnal CO2 uptake to net daily carbon gain (i.e., weak vs. strong CAM), the diel pattern of gas exchange, the occurrence of CAM throughout the plant life cycle or as environmental conditions change (i.e., constitutive versus facultative CAM), and molecular evolution of CAM-specific gene lineages1,7,52,53. Among CAM physiotypes, facultative CAM species, as P. oleracea, are recognized as particularly convenient systems for understanding the discrete changes in genetic architecture and gene expression associated with the CAM pathway54,55.
Different studies have already addressed the pronounced evolutionary changes forming a gradient ranging from C3 to obligatory CAM species56–58, but C4/CAM-performing species comprehend a new and yet little explored scenario16,23,58,59. In addition, studies linking CAM intensity, environmental conditions and plant morphoanatomical variations have shown that different trends may occur in different plant lineages60. For example, at the plant family level, some Orchidaceae show a correlation between decrescent CAM intensity and increasing altitude61, whereas Eulophiinae terrestrial orchids evolved higher CAM expression during the transition to drier habitats62. Also, tropical Oncidiinae epiphytes that express weak CAM possess thinner leaves, while strong CAM orchids have thicker leaves63. However, when comparing morphologically similar C3–CAM cycling Talinum species, differences in nocturnal acidity were more inconspicuous, although correlated with low humidity coefficients (r = − 0.55) from each species place of sampling64. Although comparing leaf thickness and cell size from C3 and obligate CAM Yucca species showed a positive correlation to nocturnal gas exchange and higher leaf acidification65, the comparison of 24 genotypes of C3–CAM Yucca gloriosa showed no correlation between leaf anatomy and CAM intensity66. This highlights that among intermediate phenotypes, the evolutionary trends may be more challenging to identify. Therefore, each plant lineage may show specific trends for the group, not necessarily matching the typical CAM traits as traditionally described67.
In this context, the P. oleracea complex represents a valuable system both for exploring the intraspecific variability of CAM and for providing additional biochemical and genetic information about the rare co-occurrence of C4 and CAM pathways. Traditionally, P. oleracea has been considered an aggregate of subspecies or microspecies43,46,47, also sometimes referred to as different species68–70. To our knowledge, taxonomic reports list 19 subspecies/microspecies distinguished according to their seed size and coat ornaments46. On the other hand, P. oleracea is sometimes considered a polymorphic species, and due to its cosmopolitan distribution and high adaptability, it is somewhat expected to present high variability in morphological traits among populations, even forming a continuum33,37,38,71. Such plasticity might also affect seed attributes, making seed morphology and size alone inconclusive to differentiate subspecies, especially considering that hybrid subspecies have already been reported in mixed populations33,37. A comprehensive phylogeny including as many accessions of P. oleracea as possible will be needed to solve the species paraphyletic scenario, but its cosmopolitan distribution may prove to be a challenge to this goal. In the present study, we selected genotypes sampled from independent populations and identified as different subspecies, and the clustering methods applied here confirmed previously described trends38, where weedy (small, prostrate and branched) phenotypes were clustered separately from more robust and erect phenotypes, e.g. commercial cultivars.
Regarding the intraspecific metabolic plasticity in P. oleracea, CAM was found to be expressed in a completely reversible way in all genotypes analyzed in the present study. Our findings indicate that, although there are significant intraspecific differences in drought-induced Δmalate, these are not directly correlated with the transcript abundances of CAM-specific genes (PPC-1E1c, ALMT-12E.1 and DIC-1.1). Also, there was no correlation between the transcript levels of these CAM-related genes that would suggest a causal relation. Therefore, the widespread occurrence of low-level CAM expression across the P. oleracea genotypes seems to be achieved without a strict balance between the expression levels of key CAM-related genes and the intensity of nighttime acidification, which further reflects the complexity behind the CAM syndrome7,55.
Over the last years, molecular and bioinformatics tools have been progressively applied to characterize the large gradient of expression found in CAM plants72–77. Thus, understanding the molecular processes behind CAM photosynthesis has gained further interest as a source of information for bioengineering endeavors seeking to improve crop resistance to extreme drought conditions49,78,79. However, a comparative study across different genotypes using CCM transcript abundances of key genes in C4–CAM species was missing, despite its potential to provide relevant information for future attempts of engineering CAM into C426,49. Here we show that, at least at the transcriptional level, major components of C4 and CAM photosynthesis were regulated in opposite directions by water availability across all P. oleracea genotypes analyzed. All three CAM-marker genes (PPC-1E1c, ALMT-12E.1 and DIC-1.127), previously identified exclusively in one genotype (here referred to as cultivar), were expressed at significantly high levels under drought in all genotypes analyzed. This indicates that, in overall terms, the transcriptional control of key components of CAM machinery by water availability is conserved within the P. oleracea complex. Similarly, the C4-related genes PPC-1E1a’, NADME-2E.1 and ASPAT-1E1 were also clearly down- and up-regulated across subspecies by drought and rewatering, respectively. Still, individual puzzle pieces belonging to different CCMs, such as the significant negative correlation between ALMT-12E.1 (CAM) and ASPAT-1E.1 (C4) mRNA levels under drought, indicates that there is still much room for investigating the intricate connection and concomitant modulation between C4 down- and CAM up-regulation.
Weak, inducible CAM has been reported for different Portulaca species, with the drought-promoted increase in nocturnal acidification either associated with a small nocturnal CO2 uptake (e.g. P. cyclophylla and P. cryptopela) or as a result of CAM-cycling (e.g. P. digyna)15,16. The diel CO2 uptake patterns observed in the present study support the occurrence of CAM-cycling for all four P. oleracea analyzed. Therefore, it is safe to assume that the malate formed overnight in droughted P. oleracea genotypes is derived from recycling nocturnal respiratory CO254 . The drought-induced Δmalate values reported here for P. oleracea are comparable to previous works detecting malate in the same species21, and close to the range of acidity detected for other droughted Portulaca species (e.g. P. digyna and P. cyclophylla showed 45.8 and 8.5 µmol H+ gFW−1, respectively14). Overall, when considering a combination of morpho-groups and Δmalate values under drought stress, a small array of combinations was formed, with robust (sativa—102.6 µmol gDW−1 and granulatostellulata—60.94 µmol gDW−1) and weedier (sicula—91.15 µmol gDW−1 and papillatostellulata—50.13 µmol gDW−1) phenotypes with representatives showing contrasting Δmalate values.
Facultative CAM may provide adaptive advantages other than carbon gain, even for weak cyclers80. In this context, the drought-resilient phenotypes of P. oleracea reported here, confirmed by the maintenance of photosystem operation throughout prolonged drought (Supplementary Figure S2–S4), and its full and rapid recovery of C4 upon rewatering (Fig. 3D), may be supported by the persistence of daytime CO2 assimilation behind closed stomata from the decarboxylation of malate accumulated overnight80,81. Also, without undermining the contribution of other traits (e.g., abundant seed production, resistance to abiotic stresses)82, the growth rates offered by C4 photosynthesis when water is available, combined with drought resilience facilitated by CAM expression under drought, can contribute to the weediness of P. oleracea. Other morpho-physiological traits as differences in water-capture strategies, cuticle thickness, epicuticular wax, stomatal density, stomatal responsiveness and root architecture, which remains to be determined for the subspecies, may be behind the remarkable drought resilience observed across the P. oleracea complex. Moreover, the high antioxidant capacity typically found in P. oleracea leaves24,25, may also be particularly important to maintain photosystem operation and avoid oxidative damage during severe drought spans.
In conclusion, drought was shown to simultaneously downregulate C4 and promote CAM in all P. oleracea genotypes. The mode of CAM expression (i.e., weak, facultative CAM, CAM-cycling), and the C4/CAM-marker gene expression profiles were conserved across the genotypes, further emphasizing the occurrence of inducible CAM as a common trait shared by all members of the Portulaca genus. As facultative C4/CAM photosynthesis is found across P. oleracea complex, future studies on the tissue organization and molecular regulatory requirements for the occurrence of concomitant C4/CAM photosynthesis could be performed in genotypes with significant morphological differences. Such studies may provide critical insights for future attempts aiming at incorporating C4/CAM into crop species. Moreover, from the perspective of photosynthetic plasticity, virtually any of the 12 genotypes of P. oleracea analyzed here could serve as model systems for further studies on C4–CAM transition. Therefore, other aspects, including plant morphology, genome size, ploidy level, and amenability to genetic transformation, could be taken into consideration before the selection of a particular P. oleracea genotype to become a genetic model for C4–CAM research.
Materials and methods
Studied Portulaca oleracea subspecies
To investigate the potential differences in CAM plasticity within the P. oleracea complex, a collection of twelve purslane accessions from different geographical locations was assembled (Table 1). Although seed size is not trustworthy for subspecies identification, seed ornamentation is so far considered the best-described structural attribute for subdividing of the P. oleracea complex46, allowing cross-referencing with previous studies in this species. Therefore, at least 15 seeds from each genotype were critical point-dried (Balzers CPD 030), coated with gold (Balzers SCD 050 Sputter Coater), and examined in a scanning electron microscope (Sigma VP—Zeiss, Oberkochen, Germany) to confirm the purity of the lots. The status for each subspecies was checked at The Plant List83 (Table 1). The genotype here referred to as “cultivar” was previously used in transcriptome studies27 and was included in this study as a reference for physiological and molecular data. Seeds from each accession were grown under well-watered conditions for at least three generations before the experiments.
Morphometric analysis
Morphometric measurements were performed in at least 25 individuals of each subspecies grown for 8–10 weeks under well-watered conditions. The following parameters were selected based on existing literature38 and analyzed: plant height (cm), fresh plant weight (g), number of primary and secondary branches, diameter of stem base and apex (cm, measured at approximately 1 cm from plant edges and used to calculate stem base/apex ratio), total and mean leaf area (cm2), leaf length and width (used to calculate leaf format ratio), and succulence (saturated water content84).
Plant material and growth conditions for drought experiments
Plants were grown in 300-mL pots containing a 1:1 mixture of commercial substrate (Plantmax HT, Eucatex, São Paulo, Brazil) and expanded vermiculite supplemented with 1 g L−1 NPK 10:10:10, 4 g L−1 of dolomite limestone (MgCO3 + CaCO3) and 2 g L−1 thermophosphate (Yoorin Master, Brazil). Plants were kept in a growth chamber at approximately 600 µmol m2 s−1 incident to the top of the chamber, 12 h photoperiod, air temperature of around 27 °C day/22 °C night and air humidity of approximately 60% day/80% night. All plants were watered daily to field capacity until the start of the treatments.
For assessing CAM plasticity, one-month-old plants were separated into three experimental groups subjected to different watering regimes: (1) well-watered, (2) droughted, and (3) rewatered. Well-watered plants were continuously irrigated to field capacity throughout the experiment. Water was withheld from droughted and rewatered groups for 10 days, and subsequently, 10 mL water was added per pot whenever soil humidity reached values close to zero, usually every 4 days, for 20 consecutive days. Plants of the rewatered group were irrigated to field capacity for the next 4 days. From the start of the drought treatment, soil volumetric water content (SVWC) was continuously monitored using Decagon soil moisture meter EC-5 (Fig. 2). At the end of each treatment, samples were harvested 1 h after the onset of illumination (dawn samples) and 1 h before the end of the light period (dusk samples). For all analyses, four biological replicates, each replicate composed of all fully-expanded and non-senescent leaves of at least three plants, were harvested at each sampling time. Samples were frozen, powdered and stored at − 80 °C until use. Plants from all genotypes were grown side-by-side, and downstream analyses were performed with samples for all genotypes at the same time to avoid introducing unnecessary variation.
Relative water content (RWC)
Fresh weight (FW) was determined in 10 leaf discs (~ 0.8 cm diameter) immediately after harvesting. Subsequently, the leaf discs were fully-hydrated by incubation in deionized water for 24 h, followed by measuring the turgid weight (TW). Finally, the samples were dried to a constant weight at 65 °C and allowed to cool down before determining the dry weight (DW). RWC was calculated using the formula [(FW − DW)/(TW − FW) × 100]85.
Organic acids quantification
For organic acid profiling, approximately 200 mg FW of frozen leaf samples were extracted in 1 ml of 80% (v/v) ethanol for 15 min at 80 °C, and the supernatants were recovered by centrifugation (5,000 g, 15 min). Pellets were re-extracted three times, and all supernatants were combined and reduced to dryness under vacuum. Aliquots of 1 mL of the supernatant were dried under vacuum and resuspended in 300 µL ultrapure water. Chromatography was carried out on an Agilent Technologies series 1,200 coupled with a diode array detector (DAD) on a reverse-phase column (SupelcoGel C610H-6% Cross Linked HPLC Column 300 mm × 7.8 mm, 9 µm) and with a guard column (SupelcoGel H Guard Column 50 mm × 4.6 mm, 9 µm) at 30 °C, using 0.1% (v/v) phosphoric acid as mobile phase running isocratically at 0.5 mL min−1. Eluted compounds were detected at 210 nm and quantified through external calibration. Endogenous metabolite concentrations were obtained by comparing the peak areas of the chromatograms against commercial standards.
Continuous gas exchange
Gas exchange was monitored using a 12-channel, custom-built IRGA system (PP Systems), as described previously86. The twelve cuvettes were housed in a growth chamber at approximately 600 μmol m−2 s−1 incident to the top of the chamber, 12 h photoperiod, air temperature of 28 °C day/18 °C night and air humidity of 60% day/80% night. Gas exchange was monitored using an infra-red gas exchange system based on a CIRAS-DC analyzer (PP Systems, USA), and calculated using SC-DC software (PP Systems, USA). The contribution of soil respiration and soil moisture to the environment in the cuvette was minimized by wrapping multiple-layers of parafilm around the rim of the pot, with a little hole for the stem. CO2 exchange rates were based on leaf area, measured using ImageJ 1.50i (NIH, Bethesda, MD, USA).
Chlorophyll a fluorescence imaging
The maximum quantum efficiency of PSII photochemistry (Fv/Fm), PSII operating efficiency (Fq’/Fm’) and non-photochemical quenching (NPQ) were determined through a non-modulated imaging fluorometer (CF Imager, Technologica, UK), as described by Baker (2008). All measurements were taken between 2 and 5 h after the start of the light period, and values of minimal (Fo) and maximal (Fm) fluorescence were obtained from dark-adapted leaves for 30 min before receiving a saturating light pulse (~ 6,000 µmol photons m-2 s-1 for 1 s). Measurements were performed in at least two leaves from three different plants for each treatment.
RNA isolation and quantitative RT-PCR (qPCR) analysis
Total RNA was extracted from approx. 80 mg of frozen leaves using the ReliaPrep RNA Tissue Miniprep System (Promega) for fibrous tissues, with careful homogenization steps. RNA samples were quantified using a microvolume spectrophotometer (NanoDrop ND-1000, Thermo Scientific, USA). Purity was assessed by keeping ratios in between the following intervals: 1.8 < A260/280 < 2.3; 1.6 < A260/230 < 2.3. The extracted RNA was treated with DNase (DNase I Amplification Grade, Thermo Fisher Scientific) for 10 min at room temperature. Complementary DNA (cDNA) synthesis was synthesized using SuperScript IV Reverse Transcriptase kit (Thermo Fisher Scientific) and qPCR reactions were performed in a StepOnePlus Real-Time PCR System (Applied Biosystems), using 10 μl mix reaction composed of 5 μl Power SYBR green 2X (Thermo Fisher Scientific), 2 μl cDNA sample and 300 nM of forward and 300 nM of reverse primers. The amplification program consisted of 10 min initial step at 95 °C, followed by 40 cycles with 15 s 95 °C, 30 s 60 °C and 30 s 72 °C. In all cases, the melting curve was analyzed to detect unspecific amplification and primer dimerization. The relative transcript abundance was calculated by applying the 2−ΔΔCT method (Livak and Schmittgen 2001). All primer sequences used are listed in Supplementary Table S7.
Climate space data
Using the coordinates from their places of sampling (Table 1), climate information (19 variables) for all 11 subspecies was retrieved from the WorldClim database (https://www.worldclimorg/bioclim)50 at a spatial scale of 10 min of a degree, using the raster package87 in R 3.6.188.
Statistical analysis
All statistical analyses were performed using R (version 3.6.188) via RStudio (version 1.2.1335). The data were checked for normality using the Shapiro–Wilk test, and for homogeneity of variances using the Levene test. When appropriate, two means were compared using: two-sample t-test (normal, homoscedastic), Welch’s t-test (normal, heteroscedastic), Mann–Whitney test (non-normal, homoscedastic) or transformed by square root or log and fitted into any of the other descriptions (non-normal, heteroscedastic). For comparison between three or more means, we used: ANOVA and post-hoc Tukey test (normal, homoscedastic), Welch ANOVA + Games-Howell post-hoc test (normal, heteroscedastic); Kruskal–Wallis + Mann–Whitney U-paired post-hoc test without adjusted p-value (non-normal, homoscedastic) or transformed by square root or log and fitted into any of the other descriptions (non-normal, heteroscedastic).
Morphometric and climate data were normalized using z-scale and analyzed by principal component analysis (PCA) using the pre-installed R function prcomp() (visualized using factoextra package89). Hierarchical clustering analysis was done using the pvclust package (version 2.0-090) with Euclidean distance and the Ward’s method criterion. For each dendrogram, AU (Approximately Unbiased) and BP (Bootstrap Probability) values were computed using 1,000 bootstraps—see pvclust manual (Available at: https://cran.r-project.org/web/packages/pvclust/pvclust.pdf). The AU value is an unbiase p-value, more accurate than BP.
Supplementary information
Acknowledgements
We kindly thank Dr. Johannes Walter (Natural History Museum Vienna, Austria), Dr. Avinoam Danin (The Hebrew University of Jerusalem, Israel), Dr. Jose Geraldo de Aquino Assis and MSc. Thaíla Vieira A. Santos (Universidade Federal da Bahia, Brazil) for donating seeds of P. oleracea subspecies. This work was supported by the São Paulo Research Foundation, Brasil (FAPESP – Grant No. 2016/04755-4), by the Royal Society, UK (Newton Advanced Fellowship – Grant No. NA140007), and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES – Finance Code 001).
Author contributions
L.F. and J.H. conceived the project and supervised the experiments; R.C.F. conducted most of the experiments; B.C.C., E.C.F., T.S. and S.F.B conducted part of the experiments; V.D.G. performed the statistical analysis; R.C.F., L.F. and J.H. wrote the article with contributions from other authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-71012-y.
References
- 1.Winter K, Smith JAC. An introduction to crassulacean acid metabolism. Biochemical principles and ecological diversity. In: Winter K, Smith JAC, editors. Crassulacean Acid Metabolism Biochemistry, Ecophysiology and Evolution. Berlin: Springer; 1996. pp. 1–10. [Google Scholar]
- 2.Kanai R, Edwards GE, Introduction I. The Biochemistry of Photosynthesis C4 Plant Biology. Singapore: Woodhead Publishing Limited; 1999. [Google Scholar]
- 3.Bauwe H, Hagemann M, Fernie AR. Photorespiration: players, partners and origin. Trends Plant Sci. 2010;15:330–336. doi: 10.1016/j.tplants.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Sage RF. Are crassulacean acid metabolism and C4 photosynthesis incompatible? Funct. Plant Biol. 2002;29:775–785. doi: 10.1071/PP01217. [DOI] [PubMed] [Google Scholar]
- 5.Hatch MD. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Bioehim. Biophys. Acta. 1987;895:81–106. [Google Scholar]
- 6.Osmond C. Crassulacean acid metabolism: a curiosity in context. Annu. Rev. Plant Physiol. 1978;29:379–414. [Google Scholar]
- 7.Winter K. Ecophysiology of constitutive and facultative CAM photosynthesis. J. Exp. Bot. 2019;70:6495–6508. doi: 10.1093/jxb/erz002. [DOI] [PubMed] [Google Scholar]
- 8.Nobel PS. Gas exchange. In: Nobel PS, editor. Environmental Biology of Agaves and Cacti. Cambridge: University Press; 1988. pp. 43–66. [Google Scholar]
- 9.Irwin P. T. Crassulacean acid metabolism. Curr. Biol. 1985;36:595–622. [Google Scholar]
- 10.Koch K, Kennedy RA. Characteristics of crassulacean acid metabolism in the succulent C4 dicot Portulaca oleracea L. Plant Physiol. 1980;65:193–197. doi: 10.1104/pp.65.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch KE, Kennedy RA. Crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L. Under natural environmental conditions. Plant Physiol. 1982;69:757–761. doi: 10.1104/pp.69.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ku SB, Shieh YJ, Reger BJ, Black CC. Photosynthetic characteristics of Portulaca grandiflora, a succulent C4 dicot. Plant Physiol. 1981;68:1073–1080. doi: 10.1104/pp.68.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guralnick LJ, Jackson MD. The occurrence and phylogenetics of crassulacean acid metabolism in the Portulacaceae. Int. J. Plant Sci. 2001;162:257–262. [Google Scholar]
- 14.Holtum JAM, Hancock LP, Edwards EJ, Winter K. Optional use of CAM photosynthesis in two C4 species, Portulaca cyclophylla and Portulaca digyna. J. Plant Physiol. 2017;214:91–96. doi: 10.1016/j.jplph.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Winter K, Holtum JAM. Facultative crassulacean acid metabolism (CAM) in four small C3 and C4 leaf-succulents. Aust. J. Bot. 2017;65:103–108. [Google Scholar]
- 16.Winter K, Sage RF, Edwards EJ, Virgo A, Holtum JAM. Facultative crassulacean acid metabolism in a C3–C4 intermediate. J. Exp. Bot. 2019;70:6571–6579. doi: 10.1093/jxb/erz085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G. Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3–C4 intermediate in genus Portulaca L. (Portulacaceae) J. Exp. Bot. 2010;61:3647–3662. doi: 10.1093/jxb/erq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G. Unique photosynthetic phenotypes in Portulaca (Portulacaceae): C3–C4 intermediates and NAD-ME C4 species with Pilosoid-type Kranz anatomy. J. Exp. Bot. 2017;68:225–239. doi: 10.1093/jxb/erw393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazen AMA. Changes in levels of phosphoenolpyruvate carboxylase with induction of crassulacean acid metabolism (CAM)-like behavior in the C4 plant Portulaca oleracea. Physiol. Plant. 1996;98:111–116. [Google Scholar]
- 20.Mazen AMA. Changes in properties of phosphoenolpyruvate carboxylase with induction of crassulacean acid metabolism (CAM) in the C4 plant Portulaca oleracea. Photosynthetica. 2000;38:385–391. [Google Scholar]
- 21.Lara MV, Disante K, Podestá FE, Andreo CS, Drincovich M. Induction of a crassulacean acid like metabolism in the C4 succulent plant, Portulaca oleracea L.: physiological and morphological changes are accompanied by specific modifications in phosphoenolpyruvate carboxylase. Photosynth. Res. 2003;77:241–254. doi: 10.1023/A:1025834120499. [DOI] [PubMed] [Google Scholar]
- 22.Lara MV, Drincovich MF, Andreo CS. Induction of a crassulacean acid-like metabolism in the C4 succulent plant, Portulaca oleracea L.: study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell Physiol. 2004;45:618–626. doi: 10.1093/pcp/pch073. [DOI] [PubMed] [Google Scholar]
- 23.Christin PA, et al. Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. J. Exp. Bot. 2014;65:3609–3621. doi: 10.1093/jxb/eru087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D’Andrea RM, Andreo CS, Lara MV. Deciphering the mechanisms involved in Portulaca oleracea (C4) response to drought: metabolic changes including crassulacean acid-like metabolism induction and reversal upon rewatering. Physiol. Plant. 2014;152:414–430. doi: 10.1111/ppl.12194. [DOI] [PubMed] [Google Scholar]
- 25.Jin R, et al. Physiological changes of purslane (Portulaca oleracea L.) after progressive drought stress and rehydration. Sci. Hortic. (Amsterdam) 2015;194:215–221. [Google Scholar]
- 26.Ferrari RC, Freschi L. C4/CAM as a means to improve plant sustainable productivity under abiotic-stressed conditions: regulatory mechanisms and biotechnological implications. In: Khan MIR, Reddy PS, Ferrante A, Khan NA, editors. Plant Signaling Molecules. Amsterdam: Elsevier; 2019. pp. 517–532. [Google Scholar]
- 27.Ferrari RC, et al. C4 and crassulacean acid metabolism within a single leaf: deciphering key components behind a rare photosynthetic adaptation. New Phytol. 2020;225:1699–1714. doi: 10.1111/nph.16265. [DOI] [PubMed] [Google Scholar]
- 28.Kovermann P, et al. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007;52:1169–1180. doi: 10.1111/j.1365-313X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- 29.Borland AM, Griffiths H, Hartwell J, Smith JAC. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 2009;60:2879–2896. doi: 10.1093/jxb/erp118. [DOI] [PubMed] [Google Scholar]
- 30.Palmieri L, et al. Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem. J. 2008;410:621–629. doi: 10.1042/BJ20070867. [DOI] [PubMed] [Google Scholar]
- 31.Singh KP. Thermoresponse in Portulaca. Curr. Sci. 1968;17:506–507. [Google Scholar]
- 32.Singh KP. Effect of different photoperiods on growth and flowering in Portulaca oleracea L. Curr. Sci. 1972;41:573–574. [Google Scholar]
- 33.Matthews JF, Ketron DW, Zane SF. The biology and taxonomy of the Portulaca oleracea L. (Portulacaceae) complex in North America. Rhodora. 1993;95:166–183. [Google Scholar]
- 34.Rahdari P, Hoseini SM. Effect of different levels of drought stress (PEG 6000 concentrations) on seed germination and inorganic elements content in purslane (Portulaca oleraceae L.) leaves. J. Stress Physiol. Biochem. 2012;8:51–61. [Google Scholar]
- 35.Singh J, Singh K. Contibution to the ecology of ten noxious weeds. J. Indian Bot. Soc. 1967;46:440–451. [Google Scholar]
- 36.Hernandes-Lopes J, Oliveira-Neto MA, Melo-de-Pinna GFA. Different ways to build succulent leaves in Portulacineae (Caryophyllales) Int. J. Plant Sci. 2016;177:198–208. [Google Scholar]
- 37.Walter J, Vekslyarska T, Dobeš C. Flow cytometric, chromosomal and morphometric analyses challenge current taxonomic concepts in the Portulaca oleracea complex (Portulacaeae, Caryophyllales) Bot. J. Linn. Soc. 2015;179:144–156. [Google Scholar]
- 38.Gorske SF, Rhodes AM, Hopen HJ. A numerical taxonomic study of Portulaca oleracea. Weed Sci. 1979;27:96–102. [Google Scholar]
- 39.Danin A, Bagella S. A new cultivar microspecies of the Portulaca oleracea aggregate from the E Mediterranean. Willdenowia. 2012;42:63–65. [Google Scholar]
- 40.Egea-Gilabert C, Ruiz-Hernández MV, Parra MÁ, Fernández JA. Characterization of purslane (Portulaca oleracea L.) accessions: suitability as ready-to-eat product. Sci. Hortic. (Amsterdam) 2014;172:73–81. [Google Scholar]
- 41.El-Bakatoushi R. Intra-specific genetic differentiation shaping three Portulaca oleracea L. micro-species. Pak. J. Bot. 2015;47:2309–2320. [Google Scholar]
- 42.Salah KBH, Chemli R. Variabilité phénotypique de quelques populations de Pourpier (Portulaca oleracea L.) en Tunisie. Acta Bot. Gall. 2004;151:111–119. [Google Scholar]
- 43.Danin A, Baker I, Baker HG. Cytogeography and taxonomy of the Portulaca oleracea L. polyploid complex. Isr. J. Bot. 1978;27:177–211. [Google Scholar]
- 44.Ocampo G, Columbus JT. Molecular phylogenetics, historical biogeography, and chromosome number evolution of Portulaca (Portulacaceae) Mol. Phylogenet. Evol. 2012;63:97–112. doi: 10.1016/j.ympev.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 45.Danin A, Domina G, Raimondo FM. Microspecies of the Portulaca oleracea aggregate found on major Mediterranean islands (Sicily, Cyprus, Crete, Rhodes) Flora Mediterr. 2008;18:89–107. [Google Scholar]
- 46.Danin, A. & Raus, T. A key to 19 microspecies of the Portulaca oleracea aggregate. In Caryophyllales: New Insights into Phylogeny, Systematics, and Morphological Evolution of the Order, Proceedings of the Symposium Held on 24th–27th (ed. Timonin, A.K.), 70–83 (Lomonosov State University, 2012).
- 47.Danin A, Buldrini F, Mazzanti MB, Bosi G. The history of the Portulaca oleracea aggregate in the Emilia-Romagna Po Plain (Italy) from the Roman age to the present. Plant Biosyst. 2014;148:622–634. [Google Scholar]
- 48.Ocampo G, et al. Evolution of leaf anatomy and photosynthetic pathways in Portulacaceae. Am. J. Bot. 2013;100:2388–2402. doi: 10.3732/ajb.1300094. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, et al. A roadmap for research on crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol. 2015;207:491–504. doi: 10.1111/nph.13393. [DOI] [PubMed] [Google Scholar]
- 50.Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. [Google Scholar]
- 51.Dodd AN, Borland AM, Haslam RP, Griffiths H, Maxwell K. Crassulacean acid metabolism: plastic, fantastic. J. Exp. Bot. 2002;53:569–580. doi: 10.1093/jexbot/53.369.569. [DOI] [PubMed] [Google Scholar]
- 52.Lüttge U. Photosynthetic flexibility and ecophysiological plasticity: questions and lessons from Clusia, the only CAM tree, in the neotropics. New Phytol. 2006;171:7–25. doi: 10.1111/j.1469-8137.2006.01755.x. [DOI] [PubMed] [Google Scholar]
- 53.Heyduk K, Moreno-Villena JJ, Gilman IS, Christin P, Edwards EJ. The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. 2019;20:485–493. doi: 10.1038/s41576-019-0107-5. [DOI] [PubMed] [Google Scholar]
- 54.Winter K, Holtum JAM. Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 2014;65:3425–3441. doi: 10.1093/jxb/eru063. [DOI] [PubMed] [Google Scholar]
- 55.Winter K, Holtum JAM, Smith JAC. Crassulacean acid metabolism: a continuous or discrete trait? New Phytol. 2015;208:73–78. doi: 10.1111/nph.13446. [DOI] [PubMed] [Google Scholar]
- 56.Winter K, Garcia M, Holtum JAM. On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchoë, and Opuntia. J. Exp. Bot. 2008;59:1829–1840. doi: 10.1093/jxb/ern080. [DOI] [PubMed] [Google Scholar]
- 57.Silvera K, et al. Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 2010;37:995–1010. [Google Scholar]
- 58.Hancock L, Edwards EJ. Phylogeny and the inference of evolutionary trajectories. J. Exp. Bot. 2014;65:3491–3498. doi: 10.1093/jxb/eru118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards EJ. Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytol. 2019;223:1742–1755. doi: 10.1111/nph.15851. [DOI] [PubMed] [Google Scholar]
- 60.Edwards EJ, Ogburn MR. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. Int. J. Plant Sci. 2012;173:724–733. [Google Scholar]
- 61.Silvera K, Santiago LS, Cushman JC, Winter K. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 2009;149:1838–1847. doi: 10.1104/pp.108.132555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bone RE, Smith JAC, Arrigo N, Buerki S. A macro-ecological perspective on crassulacean acid metabolism (CAM) photosynthesis evolution in Afro-Madagascan drylands: Eulophiinae orchids as a case study. New Phytol. 2015;208:469–481. doi: 10.1111/nph.13572. [DOI] [PubMed] [Google Scholar]
- 63.Silvera K, Winter K, Rodriguez BL, Albion RL, Cushman JC. Multiple isoforms of phosphoenolpyruvate carboxylase in the Orchidaceae (subtribe Oncidiinae): implications for the evolution of crassulacean acid metabolism. J. Exp. Bot. 2014;65:3623–3636. doi: 10.1093/jxb/eru234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris FS, Martin CE. Plasticity in the degree of CAM-cycling and its relationship to drought stress in five species of Talinum (Portulacaceae) Oecologia. 1991;86:575–584. doi: 10.1007/BF00318325. [DOI] [PubMed] [Google Scholar]
- 65.Heyduk K, Burrell N, Lalani F, Leebens-Mack J. Gas exchange and leaf anatomy of a C3-CAM hybrid, Yucca gloriosa (Asparagaceae) J. Exp. Bot. 2016;67:1369–1379. doi: 10.1093/jxb/erv536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karolina, H., Ray, J. N. & Leebens-Mack, J. Leaf anatomy is not correlated to CAM function in a C3+CAM hybrid species, Yucca gloriosa. bioRxiv.10.1101/726737 (2019). [DOI] [PMC free article] [PubMed]
- 67.Herrera A. Are thick leaves, large mesophyll cells and small intercellular air spaces requisites for CAM? Ann. Bot. 2020;125:859–868. doi: 10.1093/aob/mcaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ricceri C, Arrigoni PV. L’aggregato Di Portulaca Oleracea L. (Portulacaceae) in Italia. Parlatorea. 2000;4:91–97. [Google Scholar]
- 69.Domina G, Raimondo FM. A new species in the Portulaca oleracea aggregate (Portulacaceae) from the Island of Soqotra (Yemen) Webbia. 2009;64:9–12. [Google Scholar]
- 70.Danin A, Reyes Betancort J. The status of Portulaca oleracea in Tenerife, the Canary Islands. Lagascalia. 2006;26:71–81. [Google Scholar]
- 71.El-Bakatoushi R, Alframawy AM, Samer M, El-Sadek L, Botros W. Evolution of the Portulaca oleracea L. aggregate in Egypt on molecular and phenotypic levels revealed by morphology, inter-simple sequence repeat (ISSR) and 18S rDNA gene sequence markers. Flora Morphol. Distrib. Funct. Ecol. Plants. 2013;208:464–477. [Google Scholar]
- 72.Brilhaus D, Bräutigam A, Mettler-Altmann T, Winter K, Weber APM. Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare. Plant Physiol. 2016;170:102–122. doi: 10.1104/pp.15.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abraham PE, et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat. Plants. 2016;2:1–10. doi: 10.1038/nplants.2016.178. [DOI] [PubMed] [Google Scholar]
- 74.Yang X, et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017;8:1–15. doi: 10.1038/s41467-017-01491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maleckova E, Brilhaus D, Wrobel TJ, Weber APM. Transcript and metabolite changes during the early phase of ABA-mediated induction of CAM in Talinum triangulare. J. Exp. Bot. 2019;70:6581–6596. doi: 10.1093/jxb/erz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wai CM, et al. Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. Plant J. 2017;92:19–30. doi: 10.1111/tpj.13630. [DOI] [PubMed] [Google Scholar]
- 77.Boxall SF, et al. Kalanchoë PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell. 2020;32:1136–1160. doi: 10.1105/tpc.19.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borland AM, et al. Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 2014;19:327–338. doi: 10.1016/j.tplants.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hartwell J, Dever LV, Boxall SF. Emerging model systems for functional genomics analysis of crassulacean acid metabolism. Curr. Opin. Plant Biol. 2016;31:100–108. doi: 10.1016/j.pbi.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 80.Herrera A. Crassulacean acid metabolism and fitness under water deficit stress: If not for carbon gain, what is facultative CAM good for? Ann. Bot. 2009;103:645–653. doi: 10.1093/aob/mcn145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams WW, Osmond CB. Internal CO2 supply during photosynthesis of sun and shade grown cam plants in relation to photoinhibition. Plant Physiol. 1988;86:117–123. doi: 10.1104/pp.86.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zimmerman CA. Growth characteristics of weediness in Portulaca Oleracea L. Ecology. 1976;57:964–974. [Google Scholar]
- 83.The Plant List. The plant list—a working list of all plant species. Version 1.1, Accessed 11 Aug 2020. https://www.theplantlist.org/ (2013).
- 84.Ogburn RM, Edwards EJ. Quantifying succulence: a rapid, physiologically meaningful metric of plant water storage. Plant Cell Environ. 2012;35:1533–1542. doi: 10.1111/j.1365-3040.2012.02503.x. [DOI] [PubMed] [Google Scholar]
- 85.Barss HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Bot. 1962;15:413–428. [Google Scholar]
- 86.Dever LV, Boxall SF, Kneřová J, Hartwell J. Transgenic perturbation of the decarboxylation phase of crassulacean acid metabolism alters physiology and metabolism but has only a small effect on growth. Plant Physiol. 2015;167:44–59. doi: 10.1104/pp.114.251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hijmans, R. J. & van Etten, J. raster: Geographic analysis and modeling with raster data. R package version 1.4. (2012).
- 88.R Core Team. R: a language and environment for statistical computing, Accessed 11 Aug 2020. https://www.R-project.org/ (2019).
- 89.Kassambara, A. & Mundt, F. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.3. (2017).
- 90.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.