Abstract
Background
The resurgence of interest in the comparative developmental study of chelicerates has led to important insights, such as the discovery of a genome duplication shared by spiders and scorpions, inferred to have occurred in the most recent common ancestor of Arachnopulmonata (a clade comprising the five arachnid orders that bear book lungs). Nonetheless, several arachnid groups remain understudied in the context of development and genomics, such as the order Amblypygi (whip spiders). The phylogenetic position of Amblypygi in Arachnopulmonata posits them as an interesting group to test the incidence of the proposed genome duplication in the common ancestor of Arachnopulmonata, as well as the degree of retention of duplicates over 450 Myr. Moreover, whip spiders have their first pair of walking legs elongated and modified into sensory appendages (a convergence with the antennae of mandibulates), but the genetic patterning of these antenniform legs has never been investigated.
Results
We established genomic resources and protocols for cultivation of embryos and gene expression assays by in situ hybridization to study the development of the whip spider Phrynus marginemaculatus. Using embryonic transcriptomes from three species of Amblypygi, we show that the ancestral whip spider exhibited duplications of all ten Hox genes. We deploy these resources to show that paralogs of the leg gap genes dachshund and homothorax retain arachnopulmonate-specific expression patterns in P. marginemaculatus. We characterize the expression of leg gap genes Distal-less, dachshund-1/2 and homothorax-1/2 in the embryonic antenniform leg and other appendages, and provide evidence that allometry, and by extension the antenniform leg fate, is specified early in embryogenesis.
Conclusion
This study is the first step in establishing P. marginemaculatus as a chelicerate model for modern evolutionary developmental study, and provides the first resources sampling whip spiders for comparative genomics. Our results suggest that Amblypygi share a genome duplication with spiders and scorpions, and set up a framework to study the genetic specification of antenniform legs. Future efforts to study whip spider development must emphasize the development of tools for functional experiments in P. marginemaculatus.
Keywords: Arachnida, Arachnopulmonata, Gene duplication, Paralogs, Leg gap genes, Sensory biology
Background
In the past 25 years, comparative developmental study of chelicerates has resurged with the integration of molecular biology and genomics, spearheaded by developmental genetic investigations of the spiders Cupiennius salei and Parasteatoda tepidariorum [1–6]. Such works have investigated several important aspects of chelicerate development, such as the molecular mechanism of dorso-ventral axis patterning [3, 7–9], segmentation of the prosoma [1, 10–13] and opisthosoma [2, 13], and specification of prosomal versus opisthosomal fate [14, 15]. A few non-spider chelicerate models have also emerged in recent years and provided new perspectives on chelicerate development, such as the horseshoe crab Limulus polyphemus (Xiphosura) [16, 17], the mites Archegozetes longisetosus [18–21] and Tetranychus urticae (Acariformes) [22–24], the tick Rhipicephalus microplus (Parasitiformes) [25], the Arizona bark scorpion Centruroides sculpturatus (Scorpiones) [26] and the harvestmen Phalangium opilio (Opiliones) [27–30]. Studies in A. longisetosus and C. sculpturatus have suggested that changes in Hox gene number and spatial expression are responsible for such phenomena as the reduced segmentation of mites and the supernumerary posterior appendage identities of scorpions [18, 26].
In addition to the insights into chelicerate development, one of the major recent outcomes of increasing availability of genomic resources in the group was the discovery of a shared whole (or partial) genome duplication in spiders and scorpions. This genome duplication is inferred to trace back to the most recent common ancestor of the recently proposed clade Arachnopulmonata, which includes spiders, scorpions, and three other arachnid orders with book lungs [6, 26, 31–33]. This duplication event is also inferred to be independent from the multiple rounds of whole genome duplication undergone by horseshoe crabs (Xiphosura) [34, 35]. The duplication in spiders and scorpions encompasses several important developmental genes, such as those in the homeobox family [6, 32]. Excitingly, there is increasing evidence of divergent expression and function of paralogs, such as in the case of Hox genes, leg gap genes and retinal determination gene network (RDGN) homologs [6, 26, 36–40].
Nevertheless, knowledge on the development of several arachnid orders still remains scarce or entirely unexplored in the contexts of embryological study and genomic architecture. One particularly interesting arachnid order whose genomic evolution and developmental biology remains largely unexplored is Amblypygi. Commonly known as whip spiders, Amblypygi comprise approximately 220 described species of nocturnal predators that are distributed primarily in the tropics and subtropics worldwide [41, 42]. Amblypygi is part of the clade Pedipalpi (together with the orders Thelyphonida and Schizomida), which in turn is sister group to spiders (Araneae) [31, 43–45]. Considering the phylogenetic position of Amblypygi within Arachnopulmonata, a better understanding of Amblypygi genomics is anticipated to facilitate exploration of the evolutionary outcomes of the genome duplication in Arachnopulmonata, and better characterizing the extent to which arachnopulmonate orders retained and specialized the ensuing paralogous genes. Unfortunately, the embryology of the order Amblypygi is known only from the works of Pereyaslawzewa [46], a brief mention in Strubell [47] (“Phrynidae”), Gough [48], and the comprehensive study of Weygoldt [49] in Phrynus marginemaculatus (formerly Tarantula marginemaculata).
Beyond the interest in their genomic architecture, whip spiders possess a fascinating biology and natural history. Whip spiders are emerging as model systems in behavioral ecology, learning, and neurophysiology in arachnids [50, 51]. Their pedipalps, the second pair of appendages, are robust raptorial devices used for striking prey, including mostly arthropods, and even small vertebrates [41, 50, 52]. However, their most conspicuous characteristic and namesake is their “whip”, a modified antenniform first walking leg that is not used for locomotion, but instead as a sensory appendage. The antenniform legs are extremely elongated relative to the other leg pairs, and the tibia and tarsus are pseudo-segmented into hundreds of articles that harbor an array of chemo-, thermo-, hygro- and mechanoreceptive sensilla [53–55]. The peripheral circuitry of the antenniform legs is complex, exhibiting peripheral synapses and giant neurons which are possibly involved in fast sensory responses [55–57]. The antenniform legs also have been shown to be important for foraging, mating, and intrasexual contests [58, 59]. Notably, concentration of sensory structures, elongation and pseudo-segmentation of these legs constitute a striking convergence with the antennae of mandibulate arthropods (Pancrustacea and Myriapoda). Evidence from Hox gene expression and functional experiments support the view that the antenna of mandibulates is positionally homologous to the chelicera of chelicerates (deutocerebral appendage) [21, 30]. Despite their different positions along the antero-posterior axis of the body, the serial homology of both antennae (mandibulates) and antenniform legs (whip spiders) to walking legs constitutes a potentially useful comparison to address whether a striking morphological convergence is associated with convergence in mechanisms of genetic patterning.
Much of our knowledge about the genetic basis for appendage fate specification in arthropods was discovered in the fruit fly Drosophila melanogaster and involves regulation by leg gap genes and Hox genes. Broadly overlapping domains of leg gap genes homothorax (hth), dachshund (dac) and Distal-less (Dll) in the antenna promote antennal fate through activation of downstream antennal determinants such as spineless [60–63] (reviewed by [64]); leg identity in thoracic appendages is mediated by mutually antagonistic hth (proximal), dac (median) and Dll (distal) domains, and repression of hth by the Hox gene Antennapedia (Antp) [62, 65, 66]. Ectopic expression of hth on the leg discs results in leg-to-antenna transformation, whereas ectopic Antp in the antennal disc results in antenna-to-leg transformation [62, 65, 66]. In chelicerates, similar to D. melanogaster, the chelicera (antennal positional homolog) has broadly overlapping domains of Dll and hth; legs and pedipalps lack a distal hth, and a proximal Dll expression domain, indicating a conservation of expression of positional homologs [28, 67–69]. In line with this evidence, knockdown of hth in the harvestmen Phalangium opilio results in chelicera-to-leg transformation (and also partial pedipalp-to-leg transformation) [30]. In contrast to D. melanogaster and other insects, arachnid Antp is not expressed in the leg-bearing segments, and knockdown of this gene in the spider Parasteatoda tepidariorum has revealed a role in repressing limbs in the opisthosoma [14]. Expression of dac in a medial domain of chelicerate pedipalps and legs suggests a conserved role in patterning the medial leg territory; accordingly, knockdown of dac in the harvestmen P. opilio and the spider P. tepidariorum results in defects in medial podomeres, namely, the femur, patella, and tibia [27, 38]. Additionally, loss of a dac domain of the chelicera of spiders in comparison to harvestman has been implicated in the transition from three- to two-segmented chelicerae [27, 28].
Towards revitalizing embryological studies in Amblypygi, we generated herein protocols and comprehensive genomic resources for developmental genetic study of the whip spider P. marginemaculatus (Fig. 1a, b), resuming the seminal studies begun by Peter Weygoldt with this promising model species. Leveraging a comprehensive developmental transcriptome we established for this species, we first investigated the presence and retention of systemic gene duplications in Amblypygi predicted from their phylogenetic position in Arachnopulmonata; to mitigate biases stemming from survey of a data point from a single exemplar, we included as well the embryonic transcriptomes we recently generated for two species of the distantly related family Charinidae (Charinus ioanniticus and Charinus israelensis) [40, 41, 70]. As a first step towards elucidating the patterning of antenniform legs, we investigated a possible role of the leg gap genes homothorax, dachshund and Distal-less in specifying antenniform leg fate, by characterizing their expression pattern in developing embryonic stages of P. marginemaculatus using newly established procedures for colorimetric whole mount in situ hybridization.
Fig. 1.
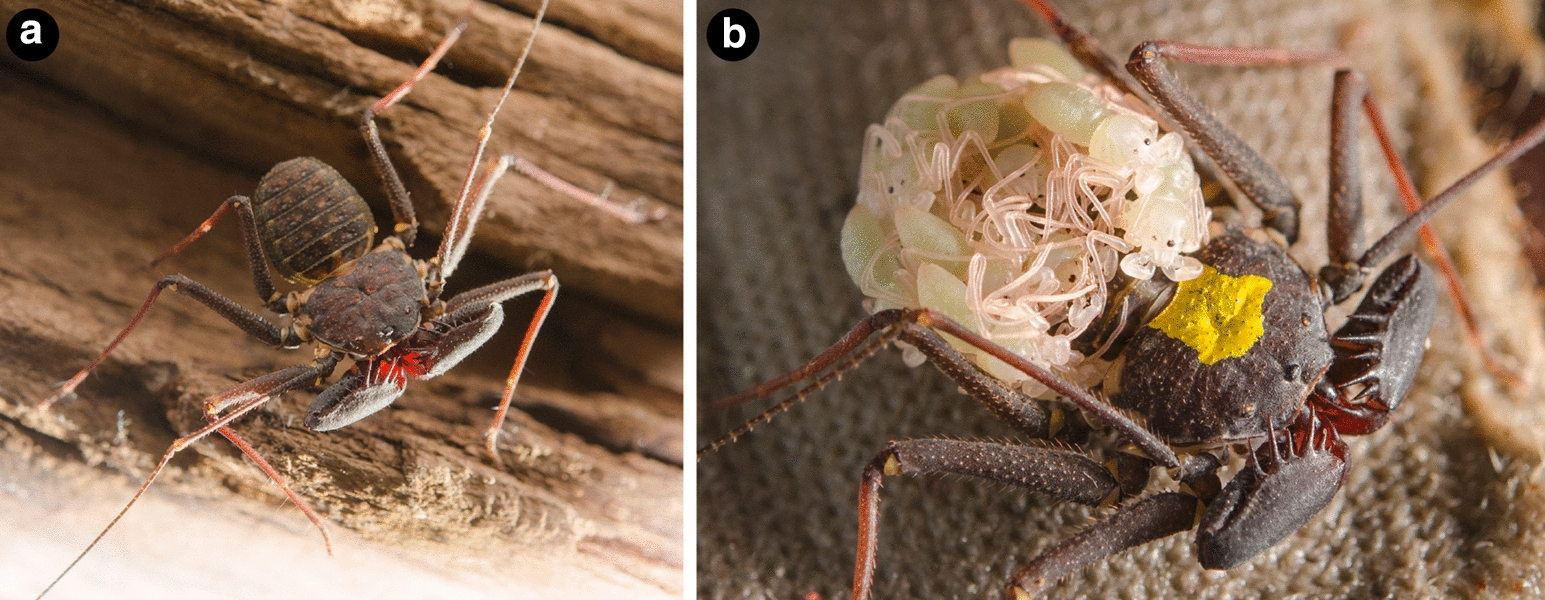
Phrynus marginemaculatus as a model to study Amblypygi development. a Adult male P. marginemaculatus, dorsal view. b Adult female P. marginemaculatus, carrying praenymphae on the opisthosoma. The yellow ink is used to mark the females in the colony
Results
Overview of embryogenesis in Amblypygi
A characterization of the postembryonic development of P. marginemaculatus is available in the works of Weygoldt [71, 72]. The following summary is after Weygoldt [49], complemented with observations from the present study. We note that the timing of embryonic stages is approximate, as the temperature in a previous work [49] was kept in a range of 24–27 °C. Details on the culture and collection of embryos are described in “Methods”.
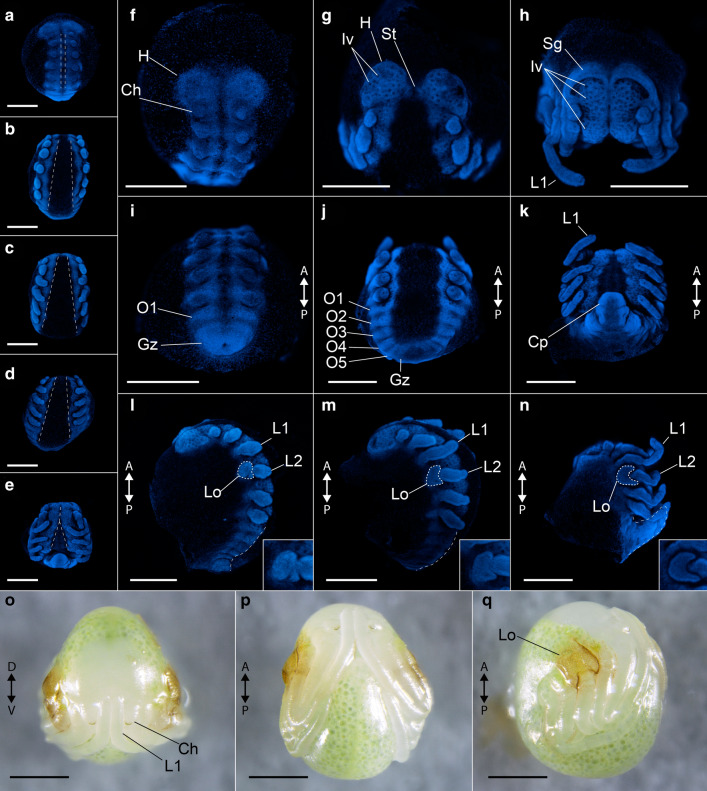
In our colony, a sample of 18 clutches from different females had an average of 19 eggs (minimum: 8 eggs; maximum: 39 eggs). Each egg is ovoid and measures 1.2–1.4 mm at the major axis. The yolk granules are light green, and fixation turns them light yellow. Upon secreting the brood sac and eggs into the ventral side of the opisthosoma, the brood sac is transparent, but gradually hardens and becomes dark brown in the course of two days. Around six days after egg laying (6 dAEL), a blastoderm is formed. At 7–8 dAEL, the yolk is more condensed, a perivitelline space is formed, and cells move into the yolk presumably in the region of the future blastopore. At 8–9 dAEL, a blastopore is recognizable on the dorsal surface of the embryo, and a germ disc is formed. Towards 10 dAEL, the germ band starts to take form, the blastopore region is displaced posteriorly, and the first signs of segmentation of the prosoma are visible. A presumable cumulus, a group of migrating cells with dorso-ventral organizing properties, was reported by Weygoldt [49] at this stage, but was not investigated by us. At 11–12 dAEL, the nascent prosomal segments elongate anteriorly and by 13–14 dAEL the segmented germ band occupies the ventral hemisphere of the egg. The limb buds of prosomal appendages begin to appear (Fig. 2a). The process of reversion begins, with a sagittal split of the germ band (Fig. 2a). From this point, until 19 dAEL, the ventral sulcus progressively enlarges with the dorsal displacement of both halves, and then begins to ventrally fuse again starting anteriorly (Fig. 2a–e). The head lobes on 15 dAEL have a circular outline and uniform ectoderm (Fig. 2f). From 15–19 dAEL, ectodermal cells invaginate along the ventral midline and head lobes (neural precursor cells), which can be seen as evenly spaced dark dots (Fig. 2f–h). Concomitantly, the anterior rim of the head lobes forms semi-lunar grooves (i.e., folds over itself) (Fig. 2f–h). Opisthosomal segments 1–12 sequentially appear from the posterior growth zone (Fig. 2i–k). At 19 dAEL, the yolk moves into the caudal papilla (growth zone) and the opisthosoma flexes ventrally (Fig. 2k, n). The first pair of legs (future antenniform legs) is already longer than the other appendages in an early limb bud stage (15 dAEL), and more clearly in subsequent stages (Fig. 2l–n). A protuberance on the proximal end of the second walking leg pair (L2), the nascent lateral organ (osmoregulatory embryonic organ), is discernible at 16 dAEL and later stages, and becomes reniform (Fig. 2l–n). From 20–21 dAEL, the embryo begins to secrete a cuticle, which is darker on the fully formed lateral organ. The head lobes have completely folded onto themselves, and the distal segment of the chelicera is sclerotized. The embryo then ruptures the egg shell, and is now termed deutembryo (Fig. 2o–q). Deutembryos remain in the brood sac for further 70 days without discernible external changes. The primordia of the eyes are formed towards the end of deutembryo development (70–106 dAEL), and eye spots of the median and lateral eyes become visible. From 90–106 dAEL the deutembryos molt into praenymphae, leave the brood sac and climb onto the adult female’s back (Fig. 1b). For a histological description of gastrulation, of the mesoderm, brain and nervous system, digestive system and gonad formation, we refer the reader to the exquisite original descriptions of Weygoldt [49].
Fig. 2.
Overview of the embryonic development of Phrynus marginemaculatus. Maximum intensity projections of Z-stacks of embryos imaged with nuclear staining (Hoechst) fluorescence. a–e Five embryonic stages, in ventral view, spanning the elongation of appendages, and showing germ band inversion and development of the ventral midline. f–h Head development, frontal view. f Undifferentiated head lobes. g Head lobe with neuron precursor cell invaginations. h Head lobes with anterior rim folding (semi-lunar grooves). i–k Opisthosomal development, posterior view. i Embryo with one opisthosomal segment. j Embryo with five opisthosomal segments. k Embryo with ventrally curved opisthosoma and most opisthosomal segments formed (number unclear). l, n Lateral organ formation and opisthosomal curving, lateral view. l Lateral organ as an undifferentiated bud proximal in leg II. m Lateral organ with a discreet “C” shape, and opisthosomal segments curved up. n Lateral organ in its final “kidney” shape, and opisthosoma curved ventrally. Insets in l–n show the lateral organ (Lo), which is outlined with dashed lines in the respective main panel. Dashed lines on the posterior regions of the embryos in l–n mark the curvature of the opisthosoma. o–q Deutembryo, in frontal, ventral and lateral view, respectively. Ch chelicera, Gz growth zone, H head lobe, Iv neural precursor cell invagination, St stomodeum. Scale bars: 500 µm
Phrynus marginemaculatus embryonic transcriptome
In order to provide a comprehensive resource for facilitating developmental genetic study and comparative genomics in Amblypygi, we assembled an embryonic transcriptome spanning different embryonic stages. The assembly of P. marginemaculatus transcriptome resulted in 544,372 transcripts composed of 277,432,101 bp and N50 of 440 bp. BUSCO scores, which reflect the presence and number of copies of widely conserved single-copy genes (BUSCO-Arthropoda; n = 1066), indicated 94.2% completeness with 5.9% of genes duplicated. From all transcripts (including isoforms), we predicted 37,540 peptide sequences with the Trinotate pipeline with a top blastp hit in Swiss-Prot database. A total of 801 of these peptides (2.1%) had a top hit to non-metazoan proteins, which we presumed to derive from microbial contaminants. The total number of predicted proteins is higher in comparison with the predicted protein set from the genome of the spider Parasteatoda tepidariorum (27,900) and the scorpion Centruroides sculpturatus (30,456) [6], which is attributable to the typical fragmentation of genes in such transcriptomic assemblies.
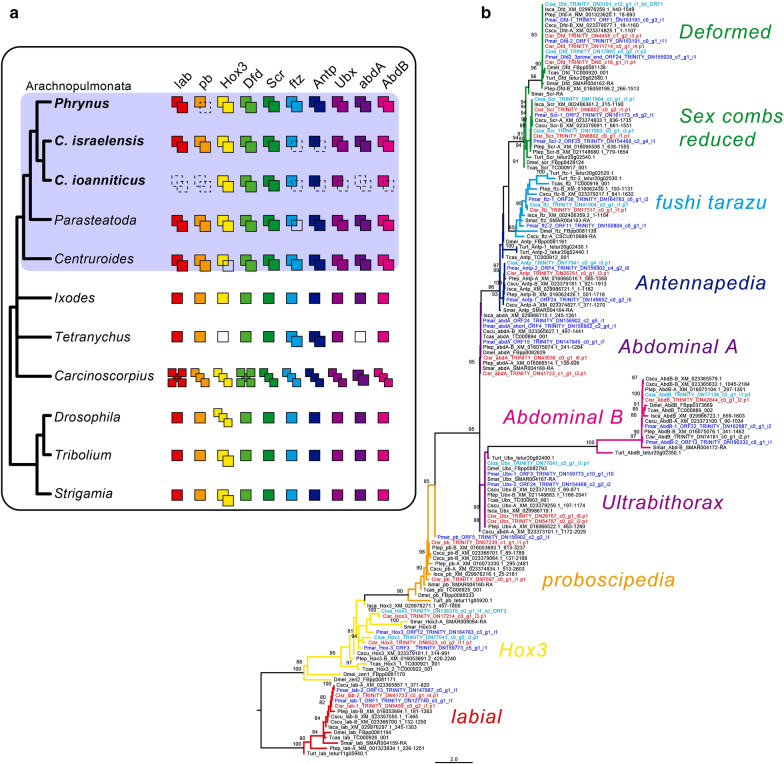
To investigate the possibility of systemic gene duplication in Amblypygi and further test the transcriptome completeness, we searched for all ten Hox genes, emphasizing discovery of paralogs known from the genomes of the spider P. tepidariorum (except for fushi tarazu) and the scorpion C. sculpturatus (except for Hox3) [6, 26, 32]. One of the Hox3 copies of P. tepidariorum has a highly divergent homeodomain sequence, constituting an unusual long branch in our phylogenetic analysis (Additional file 1: Fig. S1). The same is true for the Hox3 paralog bicoid of Drosophila melanogaster (Additional file 1: Fig. S1). We therefore removed both paralogs from our main analysis. We found orthologs of all ten Hox genes in P. marginemaculatus, namely labial, proboscipedia, Hox3, Deformed, Sex combs reduced, fushi tarazu, Antennapedia, Ultrabithorax, abdominal-A and Abdominal-B (Fig. 3a, b). We found evidence that all Hox genes, except proboscipedia exhibited two copies, and the predicted protein sequences of these copies overlapped with clear amino acid differences in P. marginemaculatus transcriptome (Fig. 3b). A Pmar-Dfd and Pmar-abdA were assembled in a third gene, but their mostly non-overlapping sequences on the alignment and nearly identical nucleotide or amino acid sequence to their respective paralog 2 indicate that they are fragmented assemblies of the same copy. We additionally analyzed two embryonic transcriptomes of Charinus whip spiders recently generated by us [40]. For C. israelensis, we discovered orthologs of all ten Hox genes, with two genes for all cases except ftz and Antp (Fig. 3a, b). A third gene was annotated as Cisr-Dfd, but we interpret it as an assembly error, given its largely non-overlapping sequence with respect to the other Cisr-Dfd sequences. For C. ioanniticus, we discovered one gene each for ftz, Antp, Ubx, AbdB, and two genes each for Hox3, Dfd, and Scr. We did not detect orthologs of lab, pb and AbdB in C. ioanniticus transcriptome (Fig. 3a, b).
Fig. 3.
Hox gene duplications in Amblypygi. a Schematic representation of the number of copies of each of the ten Hox genes in the selected arthropod terminals. Dotted squares in the amblypygids Phrynus marginemaculatus, Charinus israelensis, and C. ioanniticus indicate absence from the transcriptome. Empty squares in Parasteatoda, Centruroides, and Tetranychus indicate absence from the genome. Number of Hox genes in Carcinoscorpius rotundicauda after [35] (but see [102] for evidence of additional copies). b Tree topology inferred from maximum likelihood analysis of a conserved region (71 amino acid characters) using the same terminals of the schematics (ln L = − 3442.810). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80% shown). Species: Phrynus marginemaculatus (Pmar); Charinus israelensis (Cisr); C. ioanniticus (Cioa); Parasteatoda tepidariorum (Ptep); Centruroides sculpturatus (Cscu); Ixodes scapularis (Isca); Tetranychus urticae (Ttur); Strigamia maritima (Smar); Drosophila melanogaster (Dmel); Tribolium castaneum (Tcas). lab: labial; pb: proboscipedia/maxillopedia; Hox3: Hox3/zerknullt/z2; Dfd: Deformed; Scr: Sex combs reduced; ftz: fushi tarazu; Antp: Antennapedia/prothorax-less; Ubx: Ultrabithorax; abdA: abdominal-A; AbdB: Abdominal-B
These absences almost certainly reflect incompleteness of transcriptomic assemblies, and cannot be equated with true losses. We therefore consider the union of the Hox gene counts across the three species as the reconstruction of the whip spider ancestral Hox complement. Our results suggest that the ancestral Hox complement in Amblypygi consists of 20 genes (two copies of every Hox gene), a trait they share with spiders and scorpions. While a genome is not available for whip spiders, the phylogenetic placement of this order, as well as the pattern of Hox duplication, strongly suggests that whip spiders share the condition of two Hox clusters observed in spider and scorpion genomes.
Gene expression analyses in Phrynus marginemaculatus
All three methods we trialled for fixing embryos of P. marginemaculatus yielded embryos which are suitable for in situ hybridization (ISH). However, fixation of intact embryos in formaldehyde/heptane phase sometimes results in the vitelline membrane adhering to the embryo. Upon dissection of the vitelline membrane for ISH, parts of the embryo are commonly lost. Piercing a hole in the embryo for fixation in formaldehyde achieves a similar result, and is therefore not recommended. Dissecting the vitelline membrane under PBS and immediately fixing the embryos in formaldehyde yielded the best results. We did not detect any difference in the signal-to-background ratio of colorimetric ISH between embryos fixed for 2 h or 24 h. To test gene expression assays and investigate the genetic patterning of antenniform legs, we selected the leg gap genes Distal-less, dachshund and homothorax, which constitute the subset of PD axis patterning genes associated with functional (RNAi) datasets in other arachnid model species [27, 30, 73].
Distal-less identification and expression
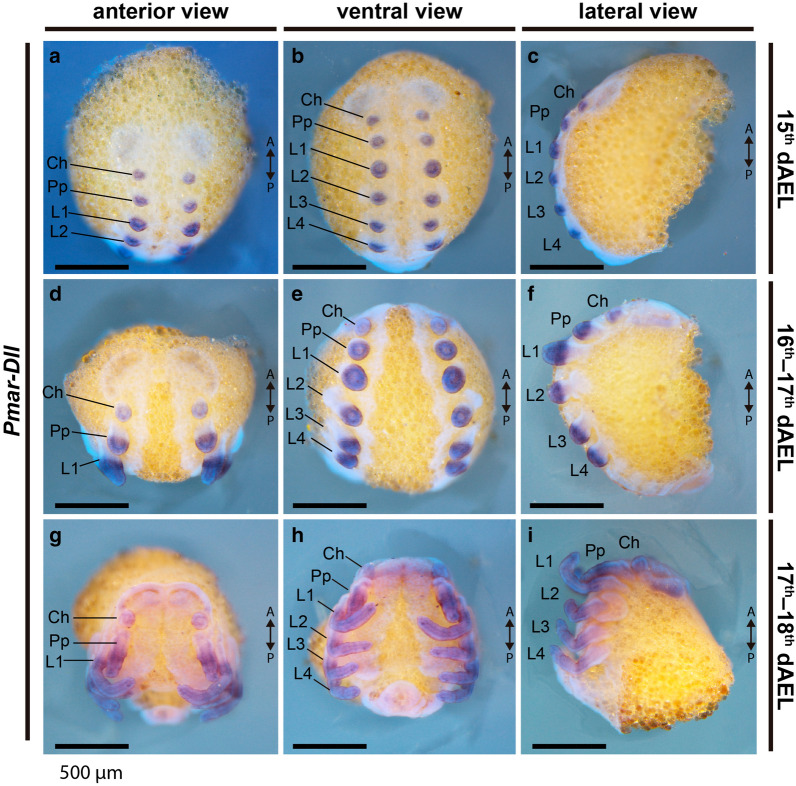
Similar to other arachnopulmonates and arthropods in general, a single Distal-less gene, Pmar-Dll, was found by similarity searches and its orthology to other metazoan Distal-less genes was confirmed with a phylogenetic analysis (Additional file 2: Fig. S2). Pmar-Dll is strongly expressed in all developing appendages, from early limb bud stage (15 dAEL) and is localized to the distal portion of the chelicera, pedipalp and legs throughout leg elongation (Figs. 4a–i; 5a–l). In 17–18 dAEL embryos, the expression is heterogeneous and forms bands of stronger expression on the forming podomeres of the pedipalps and legs (Fig. 5h–l). We did not detect expression on the ventral midline (contra [16, 28]), or in the opisthosoma, and observed only diffuse expression on the head lobes of the stages surveyed. Expression on L1 of 16–17 dAEL embryos is subtly stronger on the proximal part of the expression domain (Fig. 5c–c′), but we did not detect clear differences in younger or older embryonic stages. The proximal and distal boundaries of strong Distal-less expression do not differ between L1 and the other legs, spanning two-thirds of the legs from the tip of the tarsus and excluding the most proximal third (Fig. 5c, i).
Fig. 4.
Pmar-Distal-less (Pmar-Dll) colorimetric in situ hybridization. Whole mount bright field images (Z-stack maximum intensity projection) overlaid with nuclear staining (Hoechst). Ch chelicera, Pp pedipalp, L1–4 leg 1–4; A/P anterior/posterior, dAEL days after egg laying. Scale bars: 500 µm
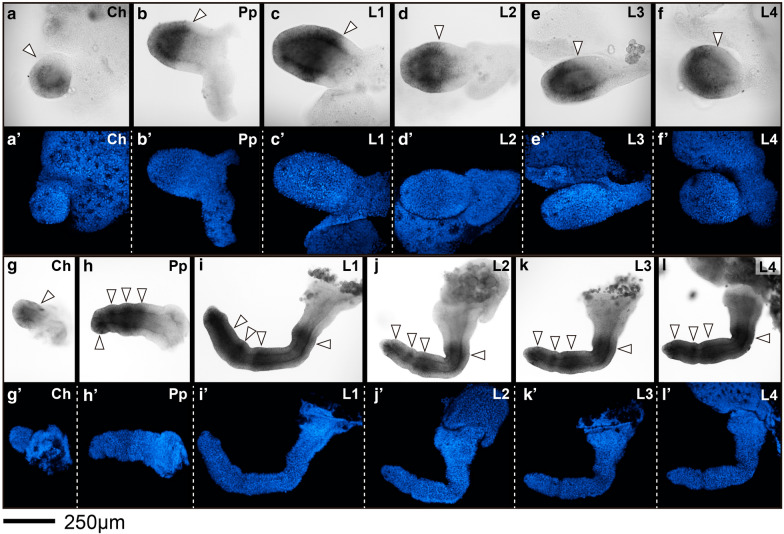
Fig. 5.
Pmar-Distal-less (Pmar-Dll) colorimetric in situ hybridization, appendage dissections in lateral view. Distal is to the left. a–l Bright field. a′–l′ Nuclear staining (Hoechst), confocal Z-stack maximum projection. a–f 16–17 dAEL embryo. g–l 17–18 dAEL embryo. j, j′ have been vertically reflected to adjust orientation. Distal is to the left. Arrowheads in a–f indicate proximal boundary of expression. Arrowheads in g–l indicate stronger domains of expression. Ch chelicera, Pp pedipalp, L1–4 leg 1–4. Scale bar: 250 µm
In contrast to other arachnid embryos (mygalomorph spiders [69]; araneomorph spiders [67, 73]; Opiliones [28]; and mites [20, 74]), Dll expression was not detected as a strong domain in the endite of pedipalp, which is very inconspicuous at these embryonic stages. We were not able to assess Dll expression in the pedipalpal endite in deutembryo stages due to deposition of cuticle.
dachshund identification and expression
dachshund is typically present as a single copy in Arthropoda, but is found independently duplicated in Arachnopulmonata and Xiphosura [38, 75]. We found two genes with high similarity to Dmel-dac and confirmed their identity as dachshund paralogs with a phylogenetic analysis (Additional file 3: Fig. S3). Pmar-dac1 and Pmar-dac2 are each nested in a clade with paralogs 1 and 2 of other arachnopulmonates, which appears as independent from horseshoe crab duplications (Additional file 3: Fig. S3).
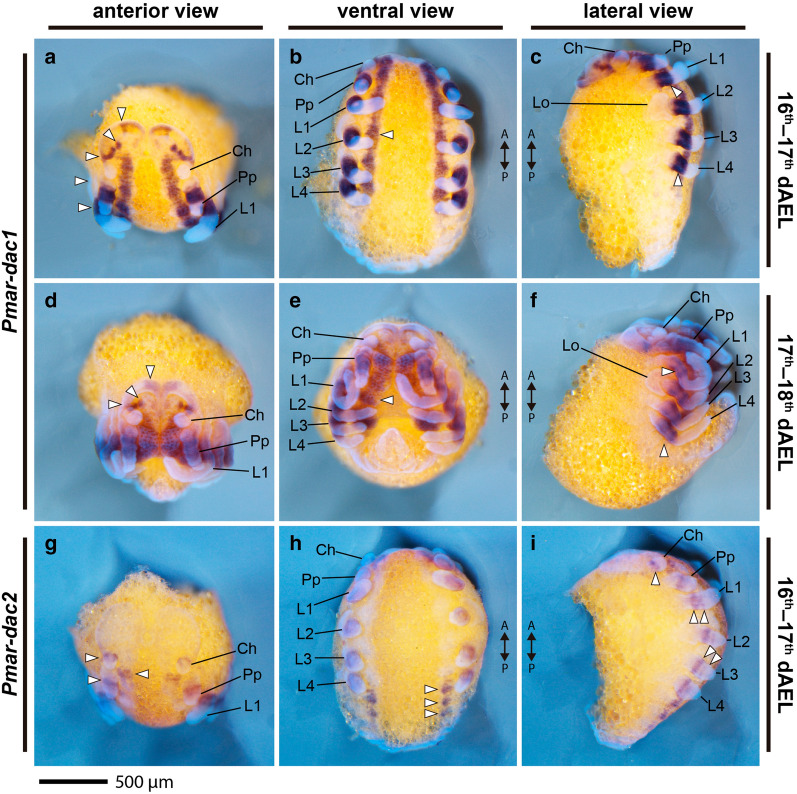
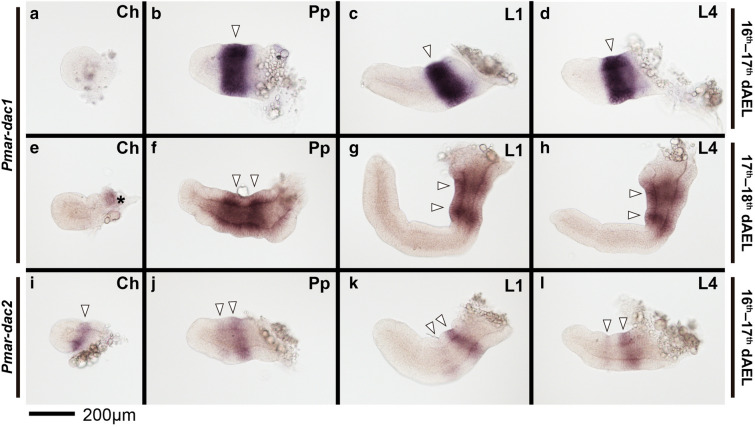
Pmar-dac1 is strongly expressed as broad medial band in the pedipalp and the four pairs of legs (Fig. 6a–f). This sharp medial band excludes the most proximal and the distal domains of these appendages (Fig. 7). No staining was detected on the chelicera (Figs. 6a–f, 7a, e). Additionally, expression occurs in three domains in the head lobes and strongly on the ventral midline of the prosoma. The continuous expression on the ventral midline stops before the opisthosoma (Fig. 6a–f).
Fig. 6.
Pmar-dachshund 1 (Pmar-dac1) (a–f) and Pmar-dachshund 2 (Pmar-dac2) (g–i) colorimetric in situ hybridization. Whole mount bright field images (Z-stack automontage) overlaid with nuclear staining (Hoechst). Ch chelicera, Pp pedipalp, L1–4 leg 1–4, Lo lateral organ, A/P anterior/posterior; arrowhead: expression domain; dAEL days after egg laying. Scale bars: 500 µm
Fig. 7.
Pmar-dachshund 1 (Pmar-dac1) (a–h) and Pmar-dachshund 2 (Pmar-dac2) (i–l) colorimetric in situ hybridization, appendage dissections in lateral view. Distal is to the left. White arrowhead indicates discreet domains of expression. Asterisk in e marks expression from the lateral margin of the headlobe. Ch: chelicera; Pp: pedipalp; L1, 4: leg 1, 4. dAEL: days after egg laying. Scale bar: 200 µm
Pmar-dac2 is expressed as ring at the proximal region of chelicera, pedipalp and legs (Fig. 6g–i). On 16–17 dAEL embryos, a second thinner ring of expression is also present on the pedipalps and legs (Figs. 6g–i, 7i–l). Expression domains also occur on the neuromere of L1 and pedipalp, being larger on the pedipalpal neuromere (Fig. 6g). Three spot domains also occur on O1, O2 and O3 segments (Fig. 6h–i).
homothorax identification and expression
Similar to the case with arachnopulmonate dachshund, homothorax is also found duplicated in arachnopulmonates [32, 39]. We annotated two genes in P. marginemaculatus transcriptome and the phylogenetic analysis shows that each copy is nested in two clades with the arachnopulmonate paralogs (Additional file 4: Fig. S4).
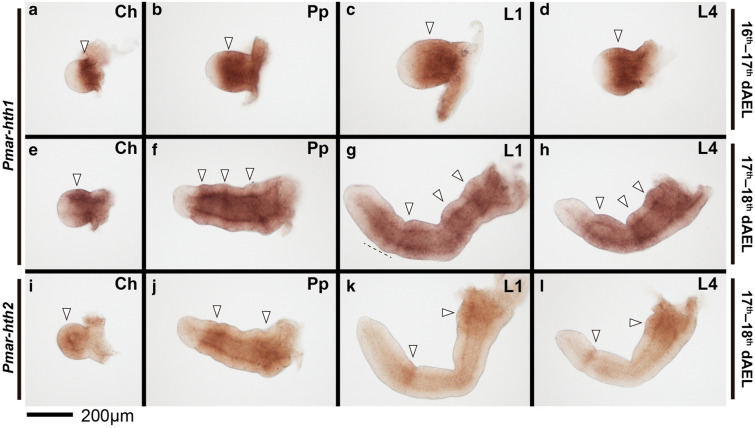
Pmar-hth1 is conspicuously expressed on the developing appendages throughout the stages studied (15–18 dAEL; Figs. 8, 9). On 16–17 dAEL embryos, Pmar-hth1 is expressed more strongly on the proximal parts of the chelicera, pedipalp, and legs, and is absent in a small distal domain (Fig. 9a–d). On 17–18 dAEL, expression in all elongated appendages is similar, and spans most of the appendage length except for a small distal territory (Fig. 9e–h). Additionally, three stronger stripe domains occur distal to the coxa of pedipalp and legs. Expression in L1 and other legs is similar in the studied stages, save for a slightly expanded distal domain on the proximal tarsus of L1 (Fig. 9g, h). Expression on the ventral ectoderm and head lobes is weak and homogenous in early stages, and becomes stronger in 17–18 dAEL (not shown)
Fig. 8.
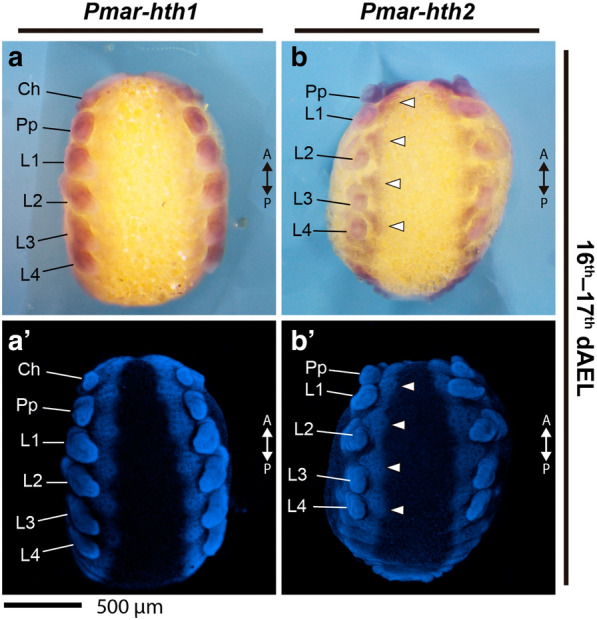
Pmar-homothorax 1 (Pmar-hth1) (a–a′) and Pmar-homothorax 2 (Pmar-hth2) (b–b′) colorimetric in situ hybridization, ventral view. a, b Whole mount bright field images (Z-stack automontage). a′, b′ Nuclear staining (Hoechst) (Z-stack automontage). Ch chelicera, Pp pedipalp, L1–4 leg 1–4, A/P anterior/posterior; arrowhead: expression domain; dAEL days after egg laying. Scale bar: 500 µm
Fig. 9.
Pmar-homothorax 1 (Pmar-hth1) (a–h) and Pmar-homothorax 2 (Pmar-hth2) (i–l) colorimetric in situ hybridization, appendage dissections in lateral view. Distal is to the left. White arrowhead indicates discreet domains of expression. Dotted line in “G” marks expanded distal expression domain. Ch: chelicera; Pp: pedipalp; L1, 4: leg 1, 4. dAEL: days after egg laying. Scale bar: 200 µm
Pmar-hth2 is expressed in the developing appendages, but is reduced in intensity and different in pattern in comparison to Pmar-hth1 (Fig. 8b–b′). Pmar-hth2 spans the whole chelicera (Fig. 9i). Expression on the pedipalp spans most of the appendage, excluding only a distal territory (Fig. 9j). In addition, there is a proximal and a sub-terminal strong domain (Fig. 9j). Expression on L1 and other legs is identical in all stages studies. Pmar-hth2 expression on the legs is consistently weaker in comparison to the pedipalp, but likewise possess a proximal and a distal sub-terminal domain (Fig. 9k, l). Expression on the ventral ectoderm, opisthosoma and head lobes is ubiquitous (Fig. 8b).
Discussion
Genomic resources for whip spiders corroborate widespread retention of duplicates across Arachnopulmonata
Changes in Hox gene expression, regulation and targets are linked to changes in morphology and Hox gene duplications have also been implicated in morphological innovations [76–79]. Duplication in the Hox cluster is common in the vertebrates, which may have up to eight Hox clusters (teleost fishes) due to successive rounds of genome duplications [76]. On the other hand, genome and Hox cluster duplications are considered rare in Arthropoda [80]. Complete Hox cluster duplication in the phylum has only been reported in horseshoe crabs (Xiphosura) [34, 35, 81, 82], and in spiders and scorpions (presumed ancestral in Arachnopulmonata). The phylogenetic placement of Amblypygi makes this group an opportune system to test the downstream prediction that P. marginemaculatus retains and expresses Hox paralogs shared by spiders and scorpions. In accordance with this hypothesis, we discovered that the whip spider P. marginemaculatus presents homologs of all ten Hox genes, and all excepting proboscipedia appear to be duplicated, as supported by our alignment and phylogenetic analysis. Our survey in the two Charinus transcriptomes confirmed the duplication of several Hox genes, and additionally revealed that two copies of proboscipedia (absent from P. marginemaculatus transcriptome) are present at least in C. israelensis. Our results suggest that Amblypygi retain a complete duplicated Hox complement with 20 genes. Similarly, a recent transcriptomic survey in Charinus acosta and Euphrynichus bacillifer reported duplications of nearly all Hox genes (excepting proboscipedia), as well as Wnt and Frizzled genes [83]. Taken together with the recent discovery of duplicated RDGN genes in Charinus, our results suggest that Amblypygi retain ohnologs of numerous developmental patterning genes, as a function of an ancient, shared whole genome duplication.
A second line of evidence of a shared genome duplication event comes from conserved expression patterns of leg gap gene paralogs. It has recently been shown that duplicates of leg patterning genes extradenticle, optomotor blind, dachshund (dac) and homothorax (hth) present one copy with an expression domain resembling a stereotypical panarthropod pattern, and a second copy with a divergent or novel expression pattern, with these two sets of expression patterns shared by a spider and a scorpion [75]. For instance, dac1 of surveyed arachnopulmonates retains the plesiomorphic expression pattern of a broad median band in the appendages, the condition observed in the single-copy dac of all non-arachnopulmonate arachnids surveyed to date [20, 28], whereas dac2 is expressed as a proximal and a separate medial domain at the patella-tibia boundary [38, 67, 69, 75]. Similarly, hth1 retains a plesiomorphic expression pattern on the proximal-median appendage excluding the tarsus, whereas hth2 presents a strong short proximal domain and stripes of expression at podomere boundaries [39, 67, 69, 75]. In the whip spider P. marginemaculatus, we indeed recovered two copies of dachshund and homothorax, with paralogs nested in two clades of arachnopulmonate copies with high nodal support (Additional file 3: Fig. S3; Additional file 4: Fig. S4). Moreover, Pmar-dac and Pmar-hth have paralog-specific expression patterns shared with spiders and a scorpion: Pmar-dac1/2 and Pmar-hth1/2 conserve the plesiomorphic expression pattern of arachnids, while copy 2 retains the arachnopulmonate-specific pattern. These data, together with the evidence from Hox gene duplications and the topologies of the dac and hth gene trees, provide compelling support for genome duplication as a synapomorphy of Arachnopulmonata.
A dachshund2 domain in the two-segmented clasp-knife chelicera of P. marginemaculatus
Expression of Pmar-Dll, Pmar-dac1/2 and Pmar-hth1/2 on the appendages is largely similar to spiders and scorpions (see above) [38, 39, 67–69, 75]. A notable difference in the chelicera of the whip spider is the retention of proximal expression of one of the dachshund paralogs. In arachnids with three-segmented chelicerae, such as the daddy-long-legs Phalangium opilio, dac is expressed in the proximal segment of the appendage [28]. The same is true for the three-segmented chelicera of scorpions, in which both dac1 and dac2 are expressed in the proximal segment [75]. In the two-segmented chelicera of spiders, dac1 is not expressed in the chelicera save for a non-ectodermal faint staining in late embryonic stages [38, 67–69, 84]. dac2 of spiders is also largely not expressed in the chelicera, save for a small ventral domain at the base of the chelicera of Parasteatoda tepidariorum and Pholcus phalangioides [38]. It has been proposed that the loss of a proximal dac domain is associated with the loss of the proximal segment in the chelicera of spider and other arachnids. This hypothesis is supported by functional data in the daddy-long-leg P. opilio: knockdown of the single-copy dac resulted in the loss of the proximal segment of the chelicera (as well as deletions of medial podomeres in pedipalps and legs) [27]. Similar to spiders, the two-segmented chelicera of the whip spider P. marginemaculatus does not express Pmar-dac1. Nonetheless, a clear complete ring of proximal expression was detected for Pmar-dac2.
In contrast to the classic leg gap phenotype incurred by knockdown of the ancestral dac copy in the harvestman, knockdown of dac2 in the spider P. tepidariorum resulted in the loss of the patella-tibia segment boundary in the pedipalp and walking legs; effects of this knockdown on the chelicera were not reported by the authors, nor is the function of dac1 known in P. tepidariorum [38]. Given the interpretation that dac2 has a derived role in patterning a segment boundary in the spider, the incidence of the Pmar-dac2 stripe in the middle of the whip spider chelicera (and the absence of Pmar-dac1 in the chelicera) is tentatively hypothesized here to constitute evidence of a segment boundary patterning activity that separates the basal and distal cheliceral segment of Amblypygi (rather than evidence of a proximal segment identity in whip spiders that is homologous to the proximal cheliceral segment of groups like harvestmen and scorpions). Exploring this hypothesis in future will require the establishment of RNA interference tools in P. marginemaculatus.
Antenniform leg distal allometry is specified early in development
To investigate a possible role of changes in leg gap gene expression in the patterning of the antenniform leg pair (L1) of whip spiders, we specifically searched for changes in expression domain between embryonic antenniform leg (L1) and the posterior legs (L2–4). The expression patterns of Pmar-Dll, Pmar-dac1/2 and Pmar-hth1/2 are mostly identical between the antenniform leg L1 and the posterior legs. We found only a slightly expanded Pmar-hth1 domain in the distal leg (compare Fig. 9g, h). It is unclear if this difference bears any functional significance in the differential fate of L1. Similar distal expression dynamics appear to distinguish the identity of the pedipalp and walking legs of the harvestman, as knockdown of the single copy of hth in P. opilio results in homeotic transformation of pedipalps to legs [30].
Besides the difference in sensory equipment, the antenniform leg pair of adults differ from the posterior walking legs in being much longer, and presenting numerous articles (pseudo-segments) on the tibia, metatarsus and especially the tarsus, whereas the walking legs have pseudo-segments only in the tarsus [41, 53, 55]. Antenniform legs may be more than 2.5 times longer than the walking legs, and this allometry in adults is mostly accounted for by the length added by the distal leg segments, which may attain more than 100 articles (tibiomeres + tarsomeres) [53]. Understanding the timing of the ontogenetic divergence of L1 relative to the walking legs is crucial for circumscribing target stages for investigation of the genetic patterning of this phenomenon.
Current evidence indicates that this allometry on the antenniform legs takes place both embryonically and postembryonically. The praenympha, the stage that molts from the deutembryo and climbs to the mother’s back, has the full number of tibiomeres, but not of tarsomeres. The final number of tarsomeres is attained in the next molt to the protonymph [53, 85], and from this moment onwards the subsequent growth is isometric [53, 85]. In embryos of P. marginemaculatus, Weygoldt [49] observed that the primordia of L1 are the first to appear in the germ band and are already longer than the remaining prosomal appendages, an observation paralleling the allometric growth of the L2 limb bud of the harvestman P. opilio (wherein L2 is the longest leg and also bears the most tarsomeres) [29]. It is presently unclear at what point in development the distal podomeres of antenniform legs attain their allometry with respect to the proximal podomeres in the whip spider. In other words, are embryonic legs simply growing longer overall with respect to walking legs, or are they longer due to distal growth? The expression pattern dachshund in P. marginemaculatus early embryos reveals a further interesting aspect of this allometry. The expression of dac (including paralogs) in the legs of arachnids surveyed to date occurs on podomeres proximal to the tibia, so that the tibia–metatarsus–tarsus territory may be identified as the a dac-free domain [20, 27, 38, 67, 69, 75, 84]. The distal boundary of Pmar-dac1 and Pmar-dac2 in stages long before podomere formation reveals that the distal domain (Pmar-dac1-free cells) of L1 is already longer with respect to the proximal leg and the other legs (compare Fig. 7c, d). These data support the inference that the distal allometry, and by extension the antenniform leg fate, are already specified from the onset of limb bud development.
Conclusion
The study of the embryology of Amblypygi has faced 45 years of hiatus since the seminal work describing the development of Phrynus marginemaculatus [49]. Here, we take the first steps in establishing P. marginemaculatus as a chelicerate model for modern evolutionary developmental study and for comparative genomics. We provide evidence for a genome duplication in Amblypygi that is shared with the remaining arachnopulmonate orders; show that the distal allometry of antenniform legs is specified early in appendage embryogenesis; and observe that expression patterns of the leg gap genes Distal-less, dachshund and homothorax do not substantiate their role in fate specification of the sensory legs of whip spiders. Future efforts to uncover the genetic underpinnings of this fascinating convergence with the mandibulate antenna should prioritize discovery of leg fate specification mechanisms in early whip spider development. Despite the establishment of a few promising arachnid models for studying chelicerate embryology, a pervasive challenge has been the establishment of toolkits for testing gene function [3, 22, 27, 73]. Therefore, future efforts to study whip spider development must emphasize the development of tools for functional experiments in P. marginemaculatus.
Methods
Establishment of amblypygid colony in the laboratory
Adult individuals of Phrynus marginemaculatus were purchased from a commercial supplier that collects animals in the Florida Keys (USA). The maintenance of a colony of P. marginemaculatus (Fig. 1a, b) in the laboratory followed guidelines provided in the works of Peter Weygoldt [49, 71, 72] and notes on behavioral studies of this species [86–88]. While most species of Amblypygi have been reported as aggressive and better kept in separate containers [41], individuals of P. marginemaculatus can be kept in a stable laboratory colony in large community containers [86]. A maximum of 20 animals may be allowed to interact and mate freely in plastic boxes (67.3 × 41.9 × 32.4 cm) with a 2 cm layer of damp coconut fiber saturated with distilled water. The substrate is continuously dampened with approximately 500 mL of dH2O on the substrate every week. All animals are kept in an acclimatized room at 26–28 °C in a controlled reversed 16L:8D light cycle to simulate summer months in Florida. To prevent flies from reproducing in this humid environment, the lid of the container is provided with a 10 × 20 cm fine mesh cloth, which also allows for air circulation. Each container is furnished with pieces of tree bark, egg carton, and retreats made from polystyrene foam or florist’s foam, to provide enough space for animals to hide and suspend themselves during molting and egg-laying. After about 2 weeks of setting up a terrarium, the substrate hosted a stable colony of collembolans (possibly brought with the tree bark). The co-occurrence of collembolans does not appear to be disadvantageous in any way. Collembolans are invariably observed feasting on carcasses and likely contribute to preventing mite infestations (see also [41]). Animals are fed at least twice a week with nymphs of Acheta domestica ad libitum. Carcasses are removed as soon as detected to prevent the spread of mites. Freshly hatched praenymphae are kept with the female, and aggregate on the back of the brooding female (Fig. 1b). The next instar, the protonymph, disperse from the female upon molting. Protonymphs are transferred to community terraria as described above. Protonymphs and subsequent instars are fed pinheads of A. domestica and Gryllus bimaculatus ad libitum three times per week until adulthood. The first adult individuals from our laboratory-bred animals were detected 8 months after hatching, which accords to what has been described by Weygoldt [49].
Terraria with adults are inspected daily for freshly molten individuals or ovigerous females, which are immediately separated from the main colony to avoid aggressive interactions. Ovigerous females carry the eggs outside the body in a brood sac attached to the ventral side of the opisthosoma [41]. Eggs are allowed to develop with the female until the desired stage for developmental work. Approximate stages may be inferred from days after egg laying (dAEL) at 26 °C following the staging in [49]. For obtaining the eggs, females are anesthetized with CO2 and immobilized in customized plastic petri dishes (35 × 11 mm) with a central whole and foam (Fig. 10a, b). With a blunt forceps pair, the pleura of the opisthosoma embracing the eggs may be gently displaced from the brood sac, and the brood sac removed as whole (Fig. 10c). The female may be returned to the colony unharmed. The brood sac easily dissolves when immersed in a phosphate buffer solution (PBS), and eggs may be cleaned with forceps under PBS. We obtained high mortality in keeping isolated eggs developing in petri dishes with humid (wet filter paper) or dry in 26 °C incubators, as mold frequently develops. However, we found out that embryos will survive and develop immersed in PBS supplemented with kanamycin (1:5000) at 26 °C incubator and even hatch into deutembryos (~ 20 dAEL).
Fig. 10.
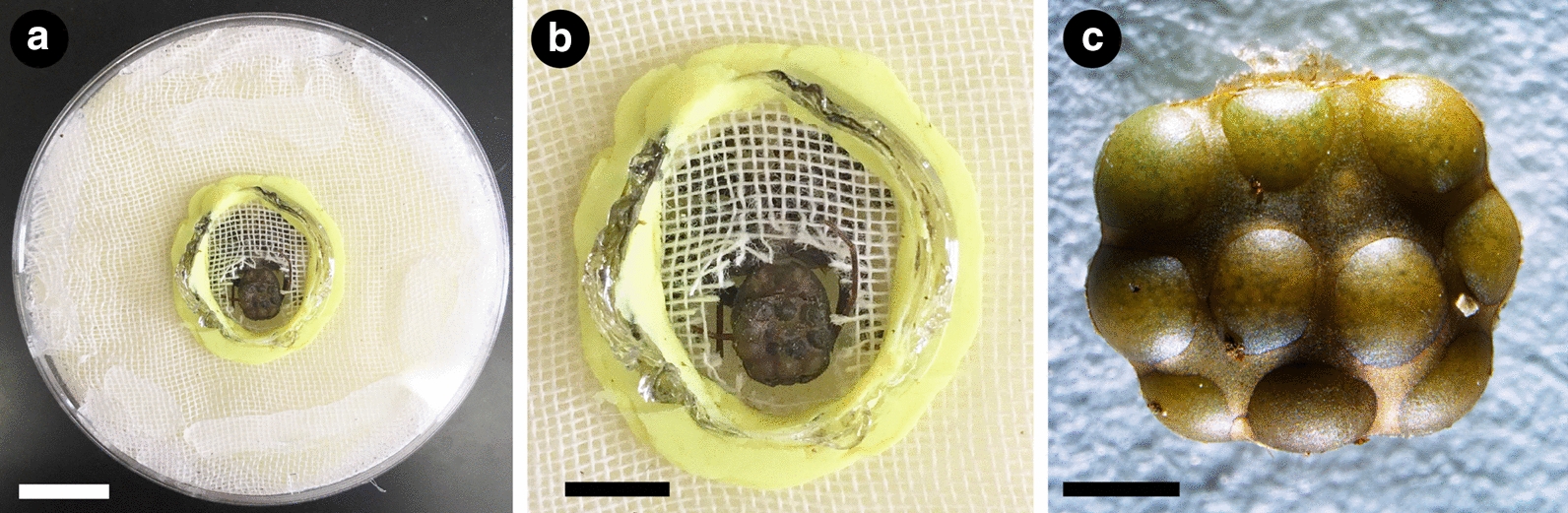
Tool for removing the brood sac from whip spiders. a Dissecting dish made of plastic petri dish and foam. b Detail of an immobilized female ready to have the brood sac dissected. c Intact brood sac dissected from the female. Scale bars: a 1 cm (approximate); b 5 mm (approximate); c 1 mm
Phrynus marginemaculatus transcriptome assembly
We collected embryos of different embryonic stages as described above, and conducted total RNA extractions of each stage separately: hatched deutembryo (female #5, n = 3; undetermined age, before eyespots), unhatched deutembryo (female #2 n = 3; approx. 21 dAEL), and leg elongation stage (female #3, n = 2; approx. 16–17 dAEL), late limb bud stage (female 6, n = 1; 16 dAEL) and early limb bud stage (female 6, n = 1; 14 dAEL) (voucher images are available upon request). Embryos were homogenized in Trizol TRIreagent (Invitrogen) and stored at − 80 °C until extraction. The combined total RNA of the five extractions was submitted to cDNA library preparation and sequencing in an Illumina High-Seq 2500 platform using paired-end short read sequencing strategy (2 × 100 bp) at the Center for Gene Expression, Biotechnology Center of University of Wisconsin-Madison (USA). The quality of the raw reads was assessed with FastQC (Babraham Bioinformatics). De novo assembly of compiled paired-end reads was conducted with the software Trinity v. 2.6.6 [89], using Trimmomatic v. 0.36 [90] to remove low quality reads and adaptors. Completeness of the transcriptome was assessed with the Trinity script ‘TrinityStats.pl’, and BUSCO v. 3 [91] on the longest isoforms using the database ‘Arthropoda’ and analysis of Hox genes. Whole transcriptome annotation was conducted following the Trinotate pipeline, by predicting proteins with TransDecoder v. 5.2.0 [92] and searching best hits with blastx and blastp against Swiss-Prot and Pfam databases [93].
Gene identification
Hox genes (labial, proboscipedia, Hox3, Deformed, Sex combs reduced, fushi tarazu, Antennapedia, Ultrabithorax, abdominal-A and Abdominal-B), Distal-less, dachshund, and homothorax orthologs were identified from the embryonic transcriptome of Phrynus marginemaculatus using BLAST searches (tblastn) with peptide queries of the homologs of Drosophila melanogaster and Parasteatoda tepidariorum. The best hits were reciprocally blasted to NCBI database and further assessed with a phylogenetic analysis.
For the phylogenetic analysis of Hox genes, we selected arthropod terminals with available genomes and well annotated references: the fruit fly Drosophila melanogaster (FlyBase), the beetle Tribolium castaneum (GCA_000002335.3) (Insecta), the centipede Strigamia maritima (GCA_000239455.1) (Myriapoda), the tick Ixodes scapularis (GCA_002892825.2), the mite Tetranychus urticae (GCA_000239435.1), the spider Parastetoda tepidarioum (GCA_000365465.2) and the scorpion Centruroides sculpturatus (GCA_000671375.2) (Chelicerata) [6, 94–97] (Additional file 5: Table S1). For the analysis of Distal-less, dachshund, and homothorax we selected terminals used in Nolan et al. [75] and included the peptide sequence for P. marginemaculatus candidates (Additional file 5: Table S1). Peptides were predicted with ORF Finder (NCBI), TransDecoder v. 5.5.0 [92], or obtained as CDS when possible from publicly available NCBI database. Peptide sequences were aligned using Clustal Omega [98] in the software SeaView v. 4 [99]. Gene trees were inferred under maximum likelihood using IQTREE v. 1.6.8 [100] with automatic model selection (-m TEST) and 1000 ultrafast bootstrap resampling (-bb 1000). Alignments for the Hox gene analysis are available in Additional files 6, 7. Genes referred throughout text refer to the longest isoform assembled by Trinity in a contig, and annotated by the procedure detailed above. Paralogs were discerned from gene fragments using as criteria the presence of largely overlapping sequences with amino acid differences.
Fixation and in situ hybridization
Collecting freshly laid eggs and keeping them in controlled temperature with the female or incubator (26 °C) allows to select desirable stages for fixation. We fixed embryos for in situ hybridization with three methods: (1) intact eggs in a phase of 4% formaldehyde/PBS and heptane overnight, after an established spider fixation protocol [3]; (2) piercing a hole in the vitelline membrane, and immersing eggs in 4% formaldehyde/PBS overnight; (3) dissecting the embryo out of the vitelline membrane and fixing in 4% formaldehyde/PBS for periods varying from 2 to 24 h. All three procedures were conducted in a shaking platform at room temperature (~ 22 °C). Following the fixation period, embryos were rinsed several times in 1× PBS/0.1% Tween-20 (Sigma Aldrich) (PBS-T), and gradually dehydrated into pure methanol. Embryos were then stored in 20 °C freezer until next procedures.
cDNA was synthesized with SuperScriptIII kit (Thermo Fisher) following the manufacturer’s instructions, using the same total RNA used to generate the embryonic transcriptome. Riboprobe templates for the genes Pmar-dac1, Pmar-dac2, Pmar-hth1, and Pmar hth-2 in situ hybridizations were generated as follows: genes were PCR amplified from cDNA using gene-specific primers designed with Primer3 v. 4.1.0 [101] and complemented with T7 linker sequences (5′-ggccgcgg-3′; 5′-cccggggc-3′), and a second PCR using universal T7 primer to generate T7 polymerase templates specific for sense (control) and anti-sense (signal) probes (Additional file 8: Table S2). Sense and antisense riboprobes for Pmar-Dll were generated with T7 and T3 polymerases using a plasmid template. For cloning of Pmar-Dll, gene-specific primers were used to amplify an 848 bp fragment (Additional file 8: Table S2), which was incorporated to vectors and E. coli using TOPO TA cloning kit (Thermo Fisher) following the manufacturer’s instructions, and Sanger sequenced to confirm their identities.
For colorimetric in situ hybridization, we adapted a protocol for the spider Parasteatoda tepidariorum after Akiyama-Oda and Oda [3]. The staining reactions lasted between 2 and 8 h at room temperature using nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-3′-indolyphosphate (BCIP) staining reactions (Sigma Aldrich). Embryos were then rinsed in PBS-T (0.05% Tween-20), counter-stained with 10 µg/mL Hoechst 33342 (Sigma-Aldrich, St. Louis, MO, USA), post-fixed in 4% formaldehyde/PBS-T and stored at 4 °C. Images were taken in a petri dish with agar under PBS-T, using a Nikon SMZ25 fluorescence stereomicroscope mounted with a DS-Fi2 digital color camera (Nikon Elements software). Appendage mounts were imaged in 70% glycerol/PBS-T in a Zeiss LCM 710 confocal microscope, or with an Olympus DP70 color camera mounted on an Olympus BX60 epifluorescence compound microscope.
Supplementary information
Additional file 1: Fig. S1. Tree topology inferred from maximum likelihood analysis of a conserved region (75 amino acid characters) (ln L = − 4780.167) using the same terminals of Fig. 3 and including Parasteatoda tepidariorum Hox3-A paralog and Drosophila melanogaster bicoid (Hox3) (red arrow). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). For abbreviations, see Additional file 5: Table S1.
Additional file 2: Fig. S2. Tree topology of Distal-less (Dll) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 15,272.780). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). Accession numbers are available in Additional file 5: Table S1. The terminal for the whip spider Phrynus marginemaculatus Dll ortholog is in boldface.
Additional file 3: Fig. S3. Tree topology of dachshund (dac) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 21,699.907). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). The terminals for the whip spider Phrynus marginemaculatus dac orthologs are in boldface. The original protein alignment of all terminals (except P. marginemaculatus) is from Nolan et al. [75].
Additional file 4: Fig. S4. Tree topology of homothorax (hth) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 10,459.647). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). The terminals for the whip spider Phrynus marginemaculatus hth orthologs are in boldface. The original protein alignment of all terminals (except P. marginemaculatus) is from Nolan et al. [75].
Additional file 5: Table S1. List of species, accession numbers and abbreviations of terminals used in the phylogenetic analysis of Hox genes, Distal-less, dachshund and homothorax homologs.
Additional file 6. Protein alignment used in the phylogenetic analysis depicted in Fig. 3.
Additional file 7. Protein alignment used in the phylogenetic analysis depicted in Additional file 1: Figure S1.
Additional file 8: Table S2. List of primers for Pmar-Dll, Pmar-dac1/2, Pmar hth1/2, and cloned fragments for Pmar-Dll.
Acknowledgements
This work is dedicated to Peter Weygoldt, whose pioneer work in the development of Phrynus marginemaculatus and contributions to the study of Amblypygi biology continue to inspire the authors of this paper. Comments from two anonymous reviewers improved an early draft of this manuscript. Sequencing was performed at the University of Wisconsin-Madison BioTechnology Center. Microscopy was performed at the Newcomb Imaging Center, Department of Botany, University of Wisconsin-Madison.
Authors’ contributions
Both authors designed the study, analyzed data, developed protocols and contributed to writing. GG conducted gene searches, orthology assessment, phylogenetic analysis, in situ hybridization, assembled figures and drafted the manuscript. Both authors read and approved the final manuscript.
Funding
GG was supported by a Wisconsin Alumni Research Foundation Fall Research Competition award. Materials for the study were supported by an Oscar Franke student competition award to GG from the International Society of Arachnology. This material is based on work supported by the National Science Foundation under Grant IOS-1552610 to PPS.
Availability of data and materials
The data generated and analyzed in this study is included in the main article, additional information and available upon request. Raw reads are deposited in NCBI Sequence Read Archive under accession SRR12232018.
Ethics approval and consent to participate
Studies with arachnids do not require ethics approval or consent to participate.
Consent for publication
Not applicable.
Competing interests
The authors declare having no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13227-020-00163-w.
References
- 1.Damen WGM, Hausdorf M, Seyfarth E-A, Tautz D. A conserved mode of head segmentation in arthropods revealed by the expression pattern of Hox genes in a spider. PNAS. 1998;95:10665–10670. doi: 10.1073/pnas.95.18.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stollewerk A, Schoppmeier M, Damen WGM. Involvement of Notch and Delta genes in spider segmentation. Nature. 2003;423:863–865. doi: 10.1038/nature01682. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama-Oda Y, Oda H. Early patterning of the spider embryo: a cluster of mesenchymal cells at the cumulus produces Dpp signals received by germ disc epithelial cells. Development. 2003;130:1735–1747. doi: 10.1242/dev.00390. [DOI] [PubMed] [Google Scholar]
- 4.Akiyama-Oda Y, Oda H. Cell migration that orients the dorsoventral axis is coordinated with anteroposterior patterning mediated by Hedgehog signaling in the early spider embryo. Development. 2010;137:1263–1273. doi: 10.1242/dev.045625. [DOI] [PubMed] [Google Scholar]
- 5.Schwager EE, Schoppmeier M, Pechmann M, Damen WG. Duplicated Hox genes in the spider Cupiennius salei. Front Zool. 2007;4:10–11. doi: 10.1186/1742-9994-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwager EE, Sharma PP, Clarke T, Leite DJ, Wierschin T, Pechmann M, et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017;15:62. doi: 10.1186/s12915-017-0399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akiyama-Oda Y, Oda H. Axis specification in the spider embryo: dpp is required for radial-to-axial symmetry transformation and sog for ventral patterning. Development. 2006;133:2347–2357. doi: 10.1242/dev.02400. [DOI] [PubMed] [Google Scholar]
- 8.Oda H, Akiyama-Oda Y. Differing strategies for forming the arthropod body plan: lessons from Dpp, Sog and Delta in the fly Drosophila and spider Achaearanea. Dev Growth Differ. 2008;50:203–214. doi: 10.1111/j.1440-169X.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- 9.Pechmann M, Benton MA, Kenny NJ, Posnien N, Roth S. A novel role for Ets4 in axis specification and cell migration in the spider Parasteatoda tepidariorum. eLife. 2017;6:e27590. doi: 10.7554/eLife.27590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwager EE, Pechmann M, Feitosa NM, McGregor AP, Damen WGM. hunchback functions as a segmentation gene in the spider Achaearanea tepidariorum. Curr Biol. 2009;19:1333–1340. doi: 10.1016/j.cub.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 11.Pechmann M, Khadjeh S, Turetzek N, McGregor AP, Damen WGM, Prpic N-M. Novel function of Distal-less as a gap gene during spider segmentation. PLoS Genet. 2011;7:e1002342. doi: 10.1371/journal.pgen.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setton EVW, Sharma PP. Cooption of an appendage-patterning gene cassette in the head segmentation of arachnids. PNAS. 2018;128:201720193-10. doi: 10.1073/pnas.1720193115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paese CLB, Schoenauer A, Leite DJ, Russell S, McGregor AP. A SoxB gene acts as an anterior gap gene and regulates posterior segment addition in a spider. eLife. 2018;7:1735. doi: 10.7554/eLife.37567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khadjeh S, Turetzek N, Pechmann M, Schwager EE, Wimmer EA, Damen WGM, et al. Divergent role of the Hox gene Antennapedia in spiders is responsible for the convergent evolution of abdominal limb repression. PNAS. 2012;109:4921–4926. doi: 10.1073/pnas.1116421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pechmann M, Schwager EE, Turetzek N, Prpic N-M. Regressive evolution of the arthropod tritocerebral segment linked to functional divergence of the Hox gene labial. Proc R Soc B Biol Sci. 2015;282:20151162-6. doi: 10.1098/rspb.2015.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mittmann B, Scholtz G. Distal-less expression in embryos of Limulus polyphemus (Chelicerata, Xiphosura) and Lepisma saccharina (Insecta, Zygentoma) suggests a role in the development of mechanoreceptors, chemoreceptors, and the CNS. Dev Genes Evol. 2001;211:232–243. doi: 10.1007/s004270100150. [DOI] [PubMed] [Google Scholar]
- 17.Blackburn DC, Conley KW, Plachetzki DC, Kempler K, Battelle B-A, Brown NL. Isolation and expression of Pax6 and atonal homologues in the American horseshoe crab, Limulus polyphemus. Dev Dyn. 2008;237:2209–2219. doi: 10.1002/dvdy.21634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnett AA, Thomas RH. Posterior Hox gene reduction in an arthropod: Ultrabithorax and Abdominal-B are expressed in a single segment in the mite Archegozetes longisetosus. EvoDevo. 2013;4:23. doi: 10.1186/2041-9139-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnett AA, Thomas RH. The delineation of the fourth walking leg segment is temporally linked to posterior segmentation in the mite Archegozetes longisetosus (Acari: Oribatida, Trhypochthoniidae) Evol Dev. 2012;14:383–392. doi: 10.1111/j.1525-142X.2012.00556.x. [DOI] [PubMed] [Google Scholar]
- 20.Barnett AA, Thomas RH. The expression of limb gap genes in the mite Archegozetes longisetosus reveals differential patterning mechanisms in chelicerates. Evol Dev. 2013;15:280–292. doi: 10.1111/ede.12038. [DOI] [PubMed] [Google Scholar]
- 21.Telford MJ, Thomas RH. Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. PNAS. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khila A, Grbić M. Gene silencing in the spider mite Tetranychus urticae: dsRNA and siRNA parental silencing of the Distal-less gene. Dev Genes Evol. 2007;217:241–251. doi: 10.1007/s00427-007-0132-9. [DOI] [PubMed] [Google Scholar]
- 23.Grbić M, Khila A, Lee K-Z, Bjelica A, Grbić V, Whistlecraft J, et al. Mity model: Tetranychus urticae, a candidate for chelicerate model organism. BioEssays. 2007;29:489–496. doi: 10.1002/bies.20564. [DOI] [PubMed] [Google Scholar]
- 24.Dearden PK, Donly C, Grbić M. Expression of pair-rule gene homologues in a chelicerate: early patterning of the two-spotted spider mite Tetranychus urticae. Development. 2002;129:5461–5472. doi: 10.1242/dev.00099. [DOI] [PubMed] [Google Scholar]
- 25.Santos VT, Ribeiro L, Fraga A, de Barros CM, Campos E, Moraes J, et al. The embryogenesis of the Tick Rhipicephalus (Boophilus) microplus: the establishment of a new chelicerate model system. Genesis. 2013;51:803–818. doi: 10.1002/dvg.22717. [DOI] [PubMed] [Google Scholar]
- 26.Sharma PP, Schwager EE, Extavour CG, Wheeler WC. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc Biol Sci. 2014;281:20140661. doi: 10.1098/rspb.2014.0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma PP, Schwager EE, Giribet G, Jockusch EL, Extavour CG. Distal-less and dachshund pattern both plesiomorphic and apomorphic structures in chelicerates: RNA interference in the harvestman Phalangium opilio (Opiliones) Evol Dev. 2013;15:228–242. doi: 10.1111/ede.12029. [DOI] [PubMed] [Google Scholar]
- 28.Sharma PP, Schwager EE, Extavour CG, Giribet G. Evolution of the chelicera: a dachshund domain is retained in the deutocerebral appendage of Opiliones (Arthropoda, Chelicerata) Evol Dev. 2012;14:522–533. doi: 10.1111/ede.12005. [DOI] [PubMed] [Google Scholar]
- 29.Sharma PP, Schwager EE, Extavour CG, Giribet G. Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol Dev. 2012;14:450–463. doi: 10.1111/j.1525-142X.2012.00565.x. [DOI] [PubMed] [Google Scholar]
- 30.Sharma PP, Tarazona OA, Lopez DH, Schwager EE, Cohn MJ, Wheeler WC, et al. A conserved genetic mechanism specifies deutocerebral appendage identity in insects and arachnids. Proc R Soc B Biol Sci. 2015;282:20150698. doi: 10.1098/rspb.2015.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, et al. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol. 2014;31:2963–2984. doi: 10.1093/molbev/msu235. [DOI] [PubMed] [Google Scholar]
- 32.Leite DJ, Baudouin-Gonzalez L, Iwasaki-Yokozawa S, Lozano-Fernandez J, Turetzek N, Akiyama-Oda Y, et al. Homeobox gene duplication and divergence in arachnids. Mol Biol Evol. 2018;35:2240–2253. doi: 10.1093/molbev/msy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma PP, Santiago MA, González-Santillán E, Monod L, Wheeler WC. Evidence of duplicated Hox genes in the most recent common ancestor of extant scorpions. Evol Dev. 2015;17:347–355. doi: 10.1111/ede.12166. [DOI] [PubMed] [Google Scholar]
- 34.Kenny NJ, Chan KW, Nong W, Qu Z, Maeso I, Yip HY, et al. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity. 2015;116:190–199. doi: 10.1038/hdy.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shingate P, Ravi V, Prasad A, Tay B-H, Garg KM, Chattopadhyay B, et al. Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution. Nat Commun. 2020;11:2322. doi: 10.1038/s41467-020-16180-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schomburg C, Turetzek N, Prpic N-M. Candidate gene screen for potential interaction partners and regulatory targets of the Hox gene labial in the spider Parasteatoda tepidariorum. Dev Genes Evol. 2020;230:105–120. doi: 10.1007/s00427-020-00656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samadi L, Schmid A, Eriksson BJ. Differential expression of retinal determination genes in the principal and secondary eyes of Cupiennius salei Keyserling (1877) EvoDevo. 2015;6:16. doi: 10.1186/s13227-015-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turetzek N, Pechmann M, Schomburg C, Schneider J, Prpic N-M. Neofunctionalization of a duplicate dachshund gene underlies the evolution of a novel leg segment in arachnids. Mol Biol Evol. 2015;33:109–121. doi: 10.1093/molbev/msv200. [DOI] [PubMed] [Google Scholar]
- 39.Turetzek N, Khadjeh S, Schomburg C, Prpic N-M. Rapid diversification of homothorax expression patterns after gene duplication in spiders. BMC Evol Biol. 2017;17:168. doi: 10.1186/s12862-017-1013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gainett G, Ballesteros JA, Kanzler CR, Zehms JT, Zern M, Aharon S, et al. How spiders make their eyes: Systemic paralogy and function of retinal determination network homologs in arachnids. bioRxiv. 2020 doi: 10.1101/2020.04.28.067199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weygoldt P. Whip Spiders (Chelicerata: Amblypygi). Their Biology, Morphology and Systematics. Stenstrup: Apollo Books; 2000. [Google Scholar]
- 42.de Miranda GS, Giupponi APL, Prendini L, Scharff N. Weygoldtia, a new genus of Charinidae Quintero, 1986 (Arachnida, Amblypygi) with a reappraisal of the genera in the family. Zool Anz J Comp Zool. 2018;273:23–32. [Google Scholar]
- 43.Giribet G. Current views on chelicerate phylogeny—a tribute to Peter Weygoldt. Zool Anz J Comp Zool. 2018;273:7–13. [Google Scholar]
- 44.Ballesteros JA, Sharma PP. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst Biol. 2019;33:896–917. doi: 10.1093/sysbio/syz011. [DOI] [PubMed] [Google Scholar]
- 45.Lozano-Fernandez J, Tanner AR, Giacomelli M, Carton R, Vinther J, Edgecombe GD, et al. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat Commun. 2019;10:2295. doi: 10.1038/s41467-019-10244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereyaslawzewa S. Développement embryonnaire des phrynes. Ann Sci Nat Zool. 1901;13:104–117. [Google Scholar]
- 47.Strubell A. Zur Entwicklungsgeschichte der Pedipalpen. Zool Anz. 1892;15:87–93. [Google Scholar]
- 48.Gough LH. The development of Admetus pumilio Koch; a contribution to the embryology of the Pedipalps. Q J Microsc Sci New Ser. 1902;45:595–630. [Google Scholar]
- 49.Weygoldt P. Untersuchungen zur Embryologie und Morphologie der Geißelspinne Tarantula marginemaculata C. L. Koch (Arachnida, Amblypygi, Tarantulidae) Zoomorphologie. 1975;82:137–199. [Google Scholar]
- 50.Chapin KJ, Hebets EA. The behavioral ecology of amblypygids. J Arachnol. 2016;44:1–14. [Google Scholar]
- 51.Wiegmann DD, Hebets EA, Gronenberg W, Graving JM, Bingman VP. Amblypygids: model organisms for the study of arthropod navigation mechanisms in complex environments? Front Behav Neurosci. 2016;10:47. doi: 10.3389/fnbeh.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seiter M, Lemell P, Gredler R, Wolff JO. Strike kinematics in the whip spider Charon sp. (Amblypygi: Charontidae) J Arachnol. 2019;47:260–267. [Google Scholar]
- 53.Igelmund P. Morphology, sense organs, and regeneration of the forelegs (whips) of the whip spider Heterophrynus elaphus (Arachnida, Amblypygi) J Morphol. 1987;193:75–89. doi: 10.1002/jmor.1051930108. [DOI] [PubMed] [Google Scholar]
- 54.Foelix RF, Chu-Wang IW, Beck L. Fine structure of tarsal sensory organs in the whip spider Admetus pumilio (Amblypygi, Arachnida) Tissue Cell. 1975;7:331–346. doi: 10.1016/0040-8166(75)90009-9. [DOI] [PubMed] [Google Scholar]
- 55.Santer RD, Hebets EA. The sensory and behavioural biology of whip spiders (Arachnida, Amblypygi). In: Spider physiology and behaviour. 1st ed. Elsevier Ltd; 2011. pp. 1–64.
- 56.Foelix R, Troyer D, Igelmund P. Peripheral synapses and giant neurons in whip spiders. Microsc Res Tech. 2002;58:272–282. doi: 10.1002/jemt.10136. [DOI] [PubMed] [Google Scholar]
- 57.Foelix RF. Occurrence of synapses in peripheral sensory nerves of arachnids. Nature. 1975;254:146–148. doi: 10.1038/254146a0. [DOI] [PubMed] [Google Scholar]
- 58.Santer RD, Hebets EA. Agonistic signals received by an arthropod filiform hair allude to the prevalence of near-field sound communication. Proc R Soc B Biol Sci. 2008;275:363–368. doi: 10.1098/rspb.2007.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santer RD, Hebets EA. Evidence for air movement signals in the agonistic behaviour of a nocturnal arachnid (Order Amblypygi) PLoS ONE. 2011;6:e22473-6. doi: 10.1371/journal.pone.0022473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong PDS, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–1974. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- 61.Dong PD, Chu J, Panganiban G. Proximodistal domain specification and interactions in developing Drosophila appendages. Development. 2001;128:2365–2372. doi: 10.1242/dev.128.12.2365. [DOI] [PubMed] [Google Scholar]
- 62.Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 63.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Setton EVW, March LE, Nolan ED, Jones TE, Cho H, Wheeler WC, et al. Expression and function of spineless orthologs correlate with distal deutocerebral appendage morphology across Arthropoda. Dev Biol. 2017;430:224–236. doi: 10.1016/j.ydbio.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 65.Struhl G. A homoeotic mutation transforming leg to antenna in Drosophila. Nature. 1981;292:635–638. doi: 10.1038/292635a0. [DOI] [PubMed] [Google Scholar]
- 66.Struhl G. Genes controlling segmental specification in the Drosophila thorax. PNAS. 1982;79:7380–7384. doi: 10.1073/pnas.79.23.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prpic N-M, Damen WM. Expression patterns of leg genes in the mouthparts of the spider Cupiennius salei (Chelicerata: Arachnida) Dev Genes Evol. 2004;214:1–7. doi: 10.1007/s00427-004-0393-5. [DOI] [PubMed] [Google Scholar]
- 68.Prpic N-M, Janssen R, Wigand B, Klingler M, Damen WGM. Gene expression in spider appendages reveals reversal of exd/hth spatial specificity, altered leg gap gene dynamics, and suggests divergent distal morphogen signaling. Dev Biol. 2003;264:119–140. doi: 10.1016/j.ydbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Pechmann M, Prpic N-M. Appendage patterning in the South American bird spider Acanthoscurria geniculata (Araneae: Mygalomorphae) Dev Genes Evol. 2009;219:189–198. doi: 10.1007/s00427-009-0279-7. [DOI] [PubMed] [Google Scholar]
- 70.Garwood RJ, Dunlop JA, Knecht BJ, Hegna TA. The phylogeny of fossil whip spiders. BMC Evol Biol. 2017;17:105. doi: 10.1186/s12862-017-0931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weygoldt P. Beobachtungen zur fortpflanzungsbiologie und zum verhalten der geißelspinne Tarantula marginemaculata C. L. Koch (Chelicerata, Amblypygi) Z Morph Tiere. 1969;64:338–360. [Google Scholar]
- 72.Weygoldt P. Lebenszyklus und postembryonale entwicklung der geibelspinne Tarantula marginemaculata C. L. Koch (Chelicerata, Amblypygi) im laboratorium. Z Morph Tiere. 1970;67:58–85. [Google Scholar]
- 73.Schoppmeier M, Damen WGM. Double-stranded RNA interference in the spider Cupiennius salei: the role of Distal-less is evolutionarily conserved in arthropod appendage formation. Dev Genes Evol. 2001;211:76–82. doi: 10.1007/s004270000121. [DOI] [PubMed] [Google Scholar]
- 74.Thomas RH, Telford MJ. Appendage development in embryos of the oribatid mite Archegozetes longisetosus (Acari, Oribatei, Trhypochthoniidae) Acta Zool. 1999;80:193–200. [Google Scholar]
- 75.Nolan ED, Santibáñez López CE, Sharma P. Developmental gene expression as a phylogenetic data class: support for the monophyly of Arachnopulmonata. Dev Genes Evol. 2020;230:137–153. doi: 10.1007/s00427-019-00644-6. [DOI] [PubMed] [Google Scholar]
- 76.Wagner GP, Amemiya C, Ruddle F. Hox cluster duplications and the opportunity for evolutionary novelties. PNAS. 2003;100:14603–14606. doi: 10.1073/pnas.2536656100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142x.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- 78.Ronshaugen M, McGinnis N, McGinnis W. Hox protein mutation and macroevolution of the insect body plan. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 79.Hughes CL, Kaufman TC. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the Hemipteran head. Development. 2002;127:3683–3694. doi: 10.1242/dev.127.17.3683. [DOI] [PubMed] [Google Scholar]
- 80.Pace RM, Grbić M, Nagy LM. Composition and genomic organization of arthropod Hox clusters. EvoDevo. 2016;7:11. doi: 10.1186/s13227-016-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nossa CW, Havlak P, Yue J-X, Lv J, Vincent KY, Brockmann HJ, et al. Joint assembly and genetic mapping of the Atlantic horseshoe crab genome reveals ancient whole genome duplication. GigaSci. 2014;3:708–721. doi: 10.1186/2047-217X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Y, Liang Y, Yan Q, Zhang L, Chen D, Ruan L, et al. The draft genome of horseshoe crab Tachypleus tridentatus reveals its evolutionary scenario and well-developed innate immunity. BMC Genomics. 2020;21:137. doi: 10.1186/s12864-020-6488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harper A, Baudouin-Gonzalez L, Schönauer A, Seiter M, Holzem M, Arif S, et al. Widespread retention of ohnologs in key developmental gene families following whole genome duplication in arachnopulmonates. bioRxiv. 2020 doi: 10.1101/2020.07.10.177725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abzhanov A, Kaufman TC. Homologs of Drosophila appendage genes in the patterning of arthropod limbs. Dev Biol. 2000;227:673–689. doi: 10.1006/dbio.2000.9904. [DOI] [PubMed] [Google Scholar]
- 85.Beck L, Foelix R, Gödeke E, Kaiser R. Morphology, larval development, and hair sensilla of the antenniform legs of the whip spider Heterophrynus longicornis Butler (Arach., Amblypygi) Zoomorphologie. 1977;88:259–276. [Google Scholar]
- 86.Rayor LS, Taylor LA. Social behavior in amblypygids, and a reassessment of arachnid social patterns. J Arachnol. 2006;34:399–421. [Google Scholar]
- 87.Santer RD, Hebets EA. Tactile learning by a whip spider, Phrynus marginemaculatus C.L. Koch (Arachnida, Amblypygi) J Comp Physiol A. 2009;195:393–399. doi: 10.1007/s00359-009-0417-8. [DOI] [PubMed] [Google Scholar]
- 88.Fowler-Finn KD, Hebets EA. An examination of agonistic interactions in the whip spider Phrynus marginemaculatus (Arachnida, Amblypygi) J Arachnol. 2006;34:62–76. [Google Scholar]
- 89.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, et al. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.The UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2018;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 2016;7:10507. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herndon N, Shelton J, Gerischer L, Ioannidis P, Ninova M, Dönitz J, et al. Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics. 2020;21:47. doi: 10.1186/s12864-019-6394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chipman AD, Ferrier DEK, Brena C, Qu J, Hughes DST, Schröder R, et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12:e1002005-24. doi: 10.1371/journal.pbio.1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:1–6. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gouy M, Guindon S, Gascuel O. SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 100.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 102.Nong W, Qu Z, Li Y, Barton-Owen T, Wong AYP, Yip HY, et al. Horseshoe crab genomes reveal the evolutionary fates of genes and microRNAs after three rounds (3R) of whole genome duplication. bioRxiv. 2020 doi: 10.1101/2020.04.16.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Tree topology inferred from maximum likelihood analysis of a conserved region (75 amino acid characters) (ln L = − 4780.167) using the same terminals of Fig. 3 and including Parasteatoda tepidariorum Hox3-A paralog and Drosophila melanogaster bicoid (Hox3) (red arrow). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). For abbreviations, see Additional file 5: Table S1.
Additional file 2: Fig. S2. Tree topology of Distal-less (Dll) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 15,272.780). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). Accession numbers are available in Additional file 5: Table S1. The terminal for the whip spider Phrynus marginemaculatus Dll ortholog is in boldface.
Additional file 3: Fig. S3. Tree topology of dachshund (dac) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 21,699.907). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). The terminals for the whip spider Phrynus marginemaculatus dac orthologs are in boldface. The original protein alignment of all terminals (except P. marginemaculatus) is from Nolan et al. [75].
Additional file 4: Fig. S4. Tree topology of homothorax (hth) inferred from maximum likelihood analysis of amino acid sequences (ln L = − 10,459.647). Numbers on the notes are ultrafast bootstrap resampling frequencies (only > 80 shown). The terminals for the whip spider Phrynus marginemaculatus hth orthologs are in boldface. The original protein alignment of all terminals (except P. marginemaculatus) is from Nolan et al. [75].
Additional file 5: Table S1. List of species, accession numbers and abbreviations of terminals used in the phylogenetic analysis of Hox genes, Distal-less, dachshund and homothorax homologs.
Additional file 6. Protein alignment used in the phylogenetic analysis depicted in Fig. 3.
Additional file 7. Protein alignment used in the phylogenetic analysis depicted in Additional file 1: Figure S1.
Additional file 8: Table S2. List of primers for Pmar-Dll, Pmar-dac1/2, Pmar hth1/2, and cloned fragments for Pmar-Dll.
Data Availability Statement
The data generated and analyzed in this study is included in the main article, additional information and available upon request. Raw reads are deposited in NCBI Sequence Read Archive under accession SRR12232018.