Significance
The PRL phosphatases are highly oncogenic when overexpressed. However, the mechanism by which they promote tumorigenesis is unknown. Here, we reveal PTEN as a putative PRL2 substrate and define a mechanism for PTEN degradation through PRL2-mediated PTEN dephosphorylation. These insights immediately place the PRL2 phosphatase into the PI3K/AKT pathway, one of the critical signaling networks altered in cancer. We further demonstrate in a preclinical model that removal of Prl2 in Pten+/− mice increases the level of PTEN and inhibits PTEN heterozygosity-induced tumorigenesis. Given the observed inverse correlation between PRL2 expression and PTEN level, targeting PRL2 serves as a potential PTEN restoration strategy to treat cancers caused by PTEN deficiency.
Keywords: PTEN, PRL2, protein tyrosine phosphatases, NEDD4, ubiquitination
Abstract
Tumor suppressor PTEN (phosphatase and tensin homologue deleted on chromosome 10) levels are frequently found reduced in human cancers, but how PTEN is down-regulated is not fully understood. In addition, although a compelling connection exists between PRL (phosphatase of regenerating liver) 2 and cancer, how this phosphatase induces oncogenesis has been an enigma. Here, we discovered that PRL2 ablation inhibits PTEN heterozygosity-induced tumorigenesis. PRL2 deficiency elevates PTEN and attenuates AKT signaling, leading to decreased proliferation and increased apoptosis in tumors. We also found that high PRL2 expression is correlated with low PTEN level with reduced overall patient survival. Mechanistically, we identified PTEN as a putative PRL2 substrate and demonstrated that PRL2 down-regulates PTEN by dephosphorylating PTEN at Y336, thereby augmenting NEDD4-mediated PTEN ubiquitination and proteasomal degradation. Given the strong cancer susceptibility to subtle reductions in PTEN, the ability of PRL2 to down-regulate PTEN provides a biochemical basis for its oncogenic propensity. The results also suggest that pharmacological targeting of PRL2 could provide a novel therapeutic strategy to restore PTEN, thereby obliterating PTEN deficiency-induced malignancies.
The PRL (phosphatase of regenerating liver) phosphatases (PRL1, 2, and 3) are overexpressed in a wide range of human cancers, where their expression is correlated with late-stage metastasis as well as poor clinical outcomes (1, 2). Although there is a compelling connection between PRLs and cancer, the mechanism by which they promote tumorigenesis is unknown. To elucidate the biological function of the PRLs, the effect of PRL deletion was analyzed at the organismic level (1). Mice lacking Prl2, the most ubiquitously and abundantly expressed PRL, display developmental abnormalities associated with placenta insufficiency, impaired spermatogenesis, and hematopoietic stem cell self-renewal, likely due to impaired AKT activity as a result of increased level of PTEN (phosphatase and tensin homologue deleted on chromosome 10) in affected tissues (3–5). However, the oncogenic role of PRL2 in the context of tumorigenesis has not been fully evaluated in vivo. Interestingly, AKT is among the most hyperactivated oncoproteins (6), whereas PTEN is the second most frequently inactivated tumor suppressor after p53 in human cancers (7, 8). The discovery that phosphatidylinositol 3,4,5-phosphate (PIP3) is a PTEN substrate places this phosphatase into one of the most commonly abrogated signaling pathways in cancer, the phosphoinositide 3-kinase (PI3K) pathway. Hence, a decrease in PTEN activity will trigger a rise in PIP3 concentration, leading to unopposed activation of the protooncogene AKT. Thus, our previous findings with Prl2−/− mice (3–5) suggest that PRL2 may function to amplify AKT signaling by reducing the level of PTEN that normally antagonizes PI3K activity.
Unlike classical tumor suppressors, PTEN is haploinsufficient and Pten heterozygous mice develop tumors spontaneously (9–14). Importantly, decreased PTEN or partial loss of PTEN function is prevalent in human cancers, and PTEN deficiency in the absence of genetic loss or mutation can increase cancer risk in a PTEN dose-dependent manner (7, 15–19). Given the strong cancer susceptibility to subtle variations in PTEN level, the ability of PRL2 to down-regulate PTEN offers a plausible mechanism for PRL2-mediated tumorigenesis. However, the detailed molecular mechanism underpinning PRL2-mediated PTEN down-regulation remains unclear due to the lack of credible substrates identified for PRL2.
To unequivocally define the role of PRL2 in oncogenesis and to elucidate the mechanism by which PRL2 down-regulates PTEN, we studied the effect of PRL2 ablation on PTEN heterozygosity-induced tumorigenesis. We discovered that deletion of Prl2 in Pten+/− mice potently impedes tumor progression and prolongs survival. We found that PRL2 deficiency increases the level of PTEN and reduces AKT signaling, leading to decreased cell proliferation and increased survival. Consistent with the mouse genetic data, we uncovered that high PRL2 expression is correlated with low PTEN level and decreased overall clinical outcome. Mechanistically, we identified PTEN as a putative PRL2 substrate and demonstrated that PRL2 down-regulates PTEN by dephosphorylating Y336 in PTEN, thereby stimulating NEDD4-mediated PTEN ubiquitination and proteasomal degradation. Collectively, the work establishes that PRL2 promotes tumorigenesis by lowering PTEN and unveils a mechanism for PTEN down-regulation. The results also provide support for targeting PRL2 as a therapeutic approach for PTEN restoration to treat cancers caused by PTEN deficiency.
Results
PRL2 Deficiency Increases PTEN Protein In Vivo.
To determine whether PRL2 promotes tumorigenesis by down-regulating PTEN, we investigated whether loss of PRL2 increases PTEN and reduces tumor formation in the context of PTEN heterozygous deficiency. Pten+/− mice display an increased incidence of cancers (10, 12, 13, 20) and serve as an excellent model for PTEN deficiency-induced human cancers. We generated wild-type (WT), Prl2−/−, Pten+/−, and Pten+/−;Prl2−/− mice on a C57BL/6 genetic background (Fig. 1A). As reported previously, we did not observe Pten−/− mice, as they die early during embryogenesis, indicating that PRL2 is upstream of PTEN and loss of PRL2 cannot rescue embryonic lethality resulting from homozygous PTEN deletion. Similarly, Prl2−/− mice were 20% smaller when compared with WT or Pten+/− littermates (4) (SI Appendix, Fig. S1A), and the body weight of Pten+/− mice was not different from that of the WT controls. Interestingly, however, the body weight of male Pten+/−;Prl2−/− mice was 10% greater than that of Prl2−/− male mice, although that of female mice was similar between Prl2−/− and Pten+/−;Prl2−/− (SI Appendix, Fig. S1A). These data suggest that PTEN heterozygous deletion can partially rescue the stunted growth caused by PRL2 deficiency, supporting that PTEN is downstream of PRL2.
Fig. 1.
PRL2 deficiency increases PTEN protein in vivo. (A) Representation of the crossing scheme used to produce Pten+/−;Prl2−/− mice. Four experimental mice, WT, Prl2−/−, Pten+/−, and Pten+/−;Prl2−/−, are marked as bold. Mouse 4 indicates experimental Pten+/−;Prl2−/− mice. KO, knockout. (B) Western blotting analysis of PTEN, PRL1, and PRL2 expression in 3-mo-old WT, Prl2−/−, Pten+/−, and Pten+/−;Prl2−/− mouse spleen tissue. Anti–β-actin and GAPDH served as the loading controls. (C) Quantification of PTEN levels corresponding to B, showing PTEN levels in mouse spleen tissue. Each bar represents the mean SD from three independent experiments. *P < 0.01, ***P < 0.0001. (D) Immunofluorescence analysis showing decreased PTEN protein levels in PRL2-overexpressing cells. Cells were stained with Myc-PTEN (green) and Flag-PRL2 (red). (D, Right) Overlay of PRL2, PTEN, and DAPI staining of the same field. (Scale bar, 10 μm.) (E) Relative fluorescence intensity values corresponding to D were detected by ImageJ software. Each bar represents the mean SD from three independent experiments. **P < 0.001.
Similar to previous findings (3–5), PTEN levels were 25% higher in Prl2-deficient mice than in the WT controls (Fig. 1 B and C and SI Appendix, Fig. S1 B and D). Importantly, we also observed a 35.7% increase of PTEN in Pten+/−;Prl2−/− mice in comparison with Pten+/− mice (Fig. 1 B and C). The levels of PTEN increase determined by Western blots are in agreement with immunohistochemistry measurements (SI Appendix, Fig. S1 C, E, and F). PRL2 deletion did not affect PTEN transcription, since Pten messenger RNA (mRNA) levels were similar in WT and Prl2−/− mice (SI Appendix, Fig. S1G). To further corroborate that PRL2 functions to suppress PTEN protein, we performed immunofluorescence analysis of cells stably expressing Myc-tagged PTEN. As expected, PRL2, but not its catalytically inactive mutant CS/DA (where the active-site nucleophilic C101 and the general acid/base D69 were mutated to S and A, respectively), is able to suppress the PTEN level inside the cell (Fig. 1 D and E and SI Appendix, Fig. S1 H and I). These results further demonstrate that deletion of PRL2 increases PTEN protein in vivo, and PRL2 functions to down-regulate PTEN in a phosphatase activity-dependent manner.
Loss of PRL2 Suppresses Tumor Development and Extends Life Span.
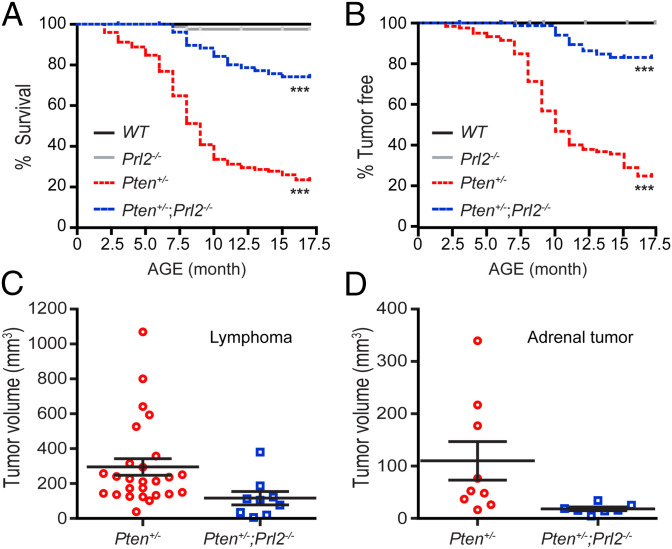
To determine the impact of PRL2 removal on PTEN heterozygosity-induced tumorigenesis, we monitored a cohort of WT (n = 87), Prl2−/− (n = 89), Pten+/− (n = 125), and Pten+/−;Prl2−/− (n = 85) mice for 17 mo for survival and tumor formation. In agreement with previous studies (10, 12, 13), ∼80% of Pten+/− mice developed tumors during the first 2 to 17 mo of age, and 80% of them died before 17 mo (Fig. 2 A and B and SI Appendix, Table S1). The mean survival for Pten+/− mice was 9 mo. In contrast, tumor onset was dramatically delayed in the Pten+/−;Prl2−/− cohort, with the first tumor appearing at 7 mo, and 80% of Pten+/−;Prl2−/− mice survived past the 17-mo observation window (Fig. 2 A and B). In addition, significantly fewer and smaller tumors were found in Pten+/−;Prl2−/− mice compared with the Pten+/− cohort (Fig. 2 C and D and SI Appendix, Fig. S2 and Table S1). Detailed characterization of tumors in Pten+/− and Pten+/−;Prl2−/− mice is provided in SI Appendix. Overall, Pten+/− mice harbored 71 tumors (78.9%) while Pten+/−;Prl2−/− mice only had 11 tumors (19.3%). None of the WT and Prl2−/− mice developed tumors (Fig. 2B) during the same period of time. These in vivo data show that PRL2 deficiency dramatically reduces the tumor-forming potential and considerably increases the tumor-free survival of Pten+/− mice, indicating a central role for PRL2 in PTEN deficiency-induced tumorigenesis.
Fig. 2.
Loss of PRL2 suppresses tumor development and extends the life span. (A) Comparative analysis of Kaplan–Meier curves showing survival rates for WT (n = 87), Prl2−/− (n = 89), Pten+/− (n = 125), and Pten+/−;Prl2−/− (n = 85) mice up to 17 mo of age. Black line, WT mice; gray line, Prl2−/− mice; red dashed line, Pten+/− mice; blue dashed line, Pten+/−;Prl2−/− mice. ***P < 0.0001 by log-rank test. (B) The percentage of tumor-free mice. PRL2 deficiency suppresses tumor formation in Pten+/−;Prl2−/− mice (n = 57) compared with Pten+/− mice (n = 90). ***P < 0.0001 by log-rank test. (C) Deletion of PRL2 suppresses lymphoid tumor growth (P < 0.04). (D) Deletion of PRL2 suppresses adrenal tumor growth. Adrenal tumor size was decreased in the Pten+/−;Prl2−/− group compared with Pten+/− mice (P < 0.05).
Deletion of PRL2 Decreases Proliferation and Increases Apoptosis in the Tumor.
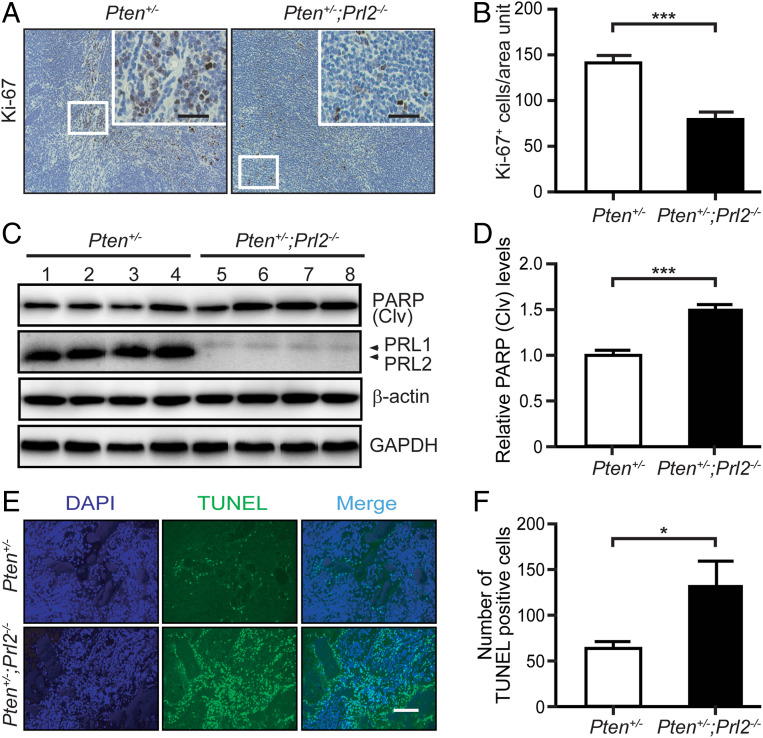
Given that loss of PRL2 attenuates tumor progression, we examined primary lymphomas, the most predominant cancer type in Pten+/− mice (13) (SI Appendix, Table S1), from the Pten+/− and Pten+/−;Prl2−/− cohorts in order to determine the mechanism by which PRL2 obliteration inhibits tumorigenesis. We first evaluated whether PRL2 deficiency has any effect on angiogenesis. We stained the lymphomas (and adrenal tumors) for the endothelial marker CD31 and observed no differences in microvessel density between tumors in Pten+/− and Pten+/−;Prl2−/− mice (SI Appendix, Fig. S3 A–D). By contrast, the number of Ki-67–positive cells in lymphomas was 44.5% lower in Pten+/−;Prl2−/− mice as compared with Pten+/− mice (Fig. 3 A and B). In addition, cleaved PARP expression and the number of TUNEL (terminal deoxynucleotidyl dUTP nick end labeling)-positive cells were increased by 50 and 107%, respectively, in lymphomas from Pten+/−;Prl2−/− mice compared with those from Pten+/− mice (Fig. 3 C–F). Taken together, these data indicate that PRL2-deficient tumor cells are less proliferative and more apoptotic than their PRL2-containing counterparts. The decreased proliferative and survival propensities exhibited by PRL2-deficient cells impede cellular growth and tumorigenesis.
Fig. 3.
Deletion of PRL2 decreases proliferation and increases apoptosis in the tumor. (A) Representative Ki-67 staining from lymphomas of Pten+/− and Pten+/−;Prl2−/− mice. (Scale bars, 50 μm.) (B) Quantification of Ki-67–positive cells per area unit. Reduction of Ki-67–positive cells in Pten+/−;Prl2−/− mouse tumor cells compared with Pten+/− mice. Error bars show SD from three independent experiments. ***P < 0.001. (C) Western blotting analysis showing increased cleaved PARP level in lymphoma samples of Pten+/−;Prl2−/− compared with Pten+/−. Anti–β-actin and GAPDH served as the loading controls. (D) Quantification of cleaved PARP protein levels in mouse lymphomas corresponding to C. Error bars represent SD from three independent experiments. ***P < 0.001. (E) Representative TUNEL staining from lymphomas of Pten+/− and Pten+/−;Prl2−/− mice. (Scale bar, 100 μm.) (F) Quantification of the apoptotic index corresponding to E. Error bars represent SD from three independent experiments. *P = 0.02.
PRL2 Deficiency Attenuates Tumorigenesis by Up-Regulating PTEN to Reduce AKT Activity.
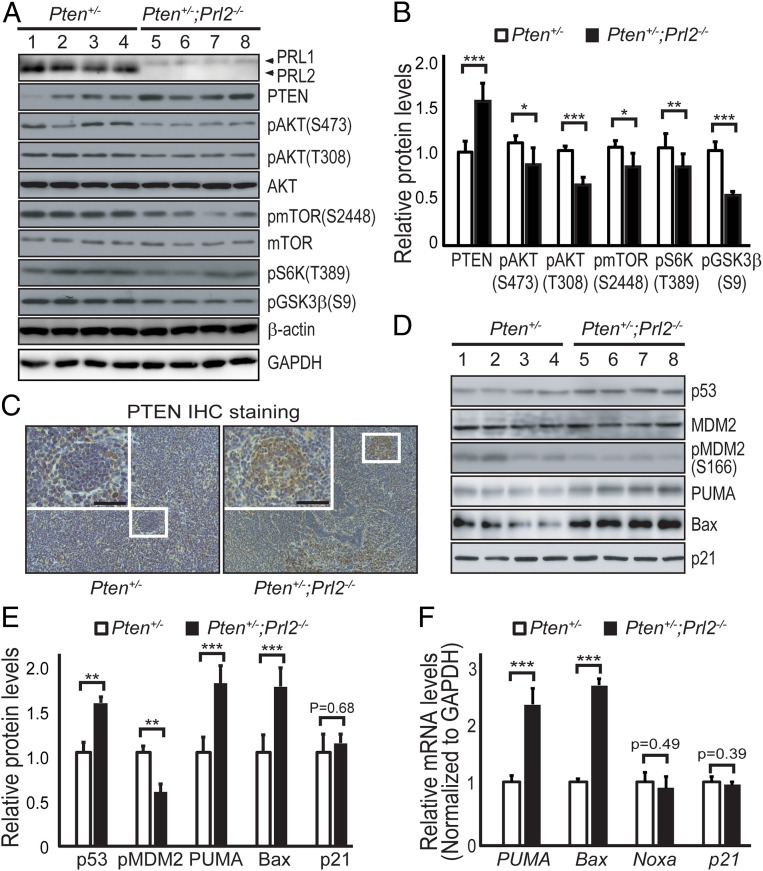
We posited that PRL2 promotes tumorigenesis by down-regulating PTEN and that deletion of PRL2 should restore PTEN, thereby obliterating PTEN deficiency-induced malignancies. Indeed, PTEN protein level was 50% higher in lymphomas from Pten+/−;Prl2−/− mice compared with those from Pten+/− mice (Fig. 4 A and B). Similar results were obtained from immunohistochemistry staining of the tumor tissues (Fig. 4C). Together, the data indicate that loss of PRL2 also increases PTEN in lymphomas. As observed previously (4), PRL2 deletion has no effect on either PI3K expression or activity (SI Appendix, Fig. S3 E and F). Theoretically, an increase in PTEN will decrease PIP3 concentration, which reduces AKT localization to the membrane, where it can be activated by phosphorylation (6). To ascertain whether an increase in PTEN leads to a hypoactive AKT kinase, we measured the activating phosphorylation status of AKT. We found that pAKT/T308 and pAKT/S473 were 36 and 15% lower in tumors from Pten+/−;Prl2−/− mice, respectively (Fig. 4 A and B). We did not detect any change in the levels of PH-domain leucine-rich repeat protein phosphatases (PHLPP1 and 2) (SI Appendix, Fig. S3G), which dephosphorylates AKT at S473 (21). Therefore, the lower AKT phosphorylation (and activity) in Pten+/−;Prl2−/− tumors is most likely due to up-regulation of PTEN as a result of PRL2 deletion.
Fig. 4.
PRL2 deficiency attenuates tumorigenesis by up-regulating PTEN to reduce ATK activity. (A) Western blotting analysis of mouse lymphoma samples showing that deletion of PRL2 increases PTEN and decreases AKT signaling as measured by pAKT(S473), pAKT(T308), p-mTOR(S2448), pS6K(T389), and pGSK3β(S9). Anti–β-actin and GAPDH served as the loading controls. (B) Quantification of protein levels in mouse lymphomas corresponding to A. Error bars show SD from three independent experiments. *P < 0.01, **P = 0.004, ***P < 0.001. (C) Deletion of PRL2 increases PTEN protein expression in the tumor. Immunohistochemistry (IHC) staining of mouse lymphomas from each genotype with an anti-PTEN antibody. (Scale bars, 50 μm.) (D) Deletion of PRL2 significantly increases p53-mediated apoptosis. Western blotting analysis of p53, MDM2, pMDM2(S166), PUMA, Bax, and p21 protein expression from Pten+/− and Pten+/−;Prl2−/− mouse lymphomas. (E) Quantification of protein levels in D. Error bars represent SD from three independent experiments. *P < 0.01, **P < 0.001, ***P < 0.0001. (F) qRT-PCR analysis showing deletion of PRL2 increases transcriptional levels of p53 downstream target genes. Primer sequences are listed in SI Appendix, Table S2. Error bars represent SD from three independent experiments. ***P < 0.0001.
To further substantiate that impaired tumorigenesis in Pten+/−;Prl2−/− mice is indeed due to increased PTEN expression and decreased AKT activity, we investigated several pathways that are known to be regulated by AKT (6). AKT promotes cell growth through activation of mammalian target of rapamycin complex 1 (mTORC1). To determine whether deletion of PRL2 reduces mTORC1 signaling, we analyzed the activation status of mTOR and its downstream target, the S6 kinase, in lymphomas from Pten+/− and Pten+/−;Prl2−/− mice. Consistent with our expectation, we found that p-mTOR/S2448 and pS6K/T389 were 19 and 20% lower in tumors from Pten+/−;Prl2−/− mice, respectively (Fig. 4 A and B). AKT also promotes cell proliferation and metabolism by exerting an inhibitory phosphorylation on Ser9 of GSK3β, thereby blocking the accessibility of GSK3β substrates. We observed a 47% decrease in pGSK3β/S9 in Pten+/−;Prl2−/− lymphomas compared with those from Pten+/− mice (Fig. 4 A and B). These data indicate that loss of PRL2 may decrease tumor cell proliferation and growth by inhibiting the mTORC1/S6K pathway and activating GSK3β, consistent with AKT inactivation in Prl2−/− cells.
AKT can also regulate cell survival through direct phosphorylation of BAD and FoxOs (6). No notable differences in pBAD/S136 and pFoxO1(T24)/pFoxO3(T32)/pFoxO4(T28) were observed between tumors from Pten+/− and Pten+/−;Prl2−/− mice (SI Appendix, Fig. S3G). We then explored whether the phosphorylation of MDM2, another substrate of AKT and the E3 ubiquitin ligase for the tumor suppressor p53, is altered. Phosphorylation of MDM2 on S166 by AKT triggers its translocation from the cytoplasm to the nucleus and/or increases its ubiquitin ligase activity, both of which promote p53 degradation. Therefore, we analyzed pMDM2/S166 and p53 levels as well as several p53 target genes from lymphoma samples derived from Pten+/− and Pten+/−;Prl2−/− mice. Deletion of Prl2 in Pten+/− mice decreased the pMDM2/S166 level by 42%, leading to a 53% increase in p53 protein (Fig. 4 D and E).
p53 activates the intrinsic cell-death pathway by directly up-regulating the proapoptotic Bcl-2 family member Bax as well as the BH3-only proapoptotic protein PUMA (p53 up-regulated modulator of apoptosis) (22). Consistent with the increase in p53, we found that the levels of PUMA and Bax proteins are 1.74- and 1.70-fold higher in tumors from Pten+/−;Prl2−/− mice than those from Pten+/− mice, respectively (Fig. 4 D and E). In line with these findings, the mRNA levels of PUMA and Bax are also increased by 2.2- and 2.5-fold, respectively, in lymphomas from Pten+/−;Prl2−/− mice (Fig. 4F). Interestingly, deletion of PRL2 has no effect on the level of Noxa (Fig. 4F), another BH3-only proapoptotic protein that is also p53-dependent. No change in cyclin-dependent kinase (CDK) inhibitor p21 protein (Fig. 4 D and E) or mRNA (Fig. 4F) was observed when PRL2 was deleted from Pten+/− tumors. We also found no change in cyclin D1, one of the major cyclins for CDKs (SI Appendix, Fig. S3G). These observations indicate that p53-dependent cell-cycle progression was not affected by PRL2 deletion. Taken together, these results suggest that the increased apoptosis in Pten+/−;Prl2−/− cells is likely brought about by p53-mediated selective expression of proapoptotic Bcl-2 family proteins Bax and PUMA, as a result of impaired AKT phosphorylation on MDM2.
We also analyzed if any other previously implicated PRL-mediated pathways are also perturbed when PRL2 is deleted in PTEN heterozygosity-induced tumors. Ectopic PRL expression in cultured cells activates several signaling pathways, including the Rho family of small GTPases, Src, ERK1/2, STAT3, and AKT (1). However, no appreciable changes were observed in the phosphorylation status of pSrc/Y527, pERK1/2, and pSTAT3/Y705 (SI Appendix, Fig. S3 G and H). We also did not find any change in Jak2 or variation in RhoA and Ras activity (SI Appendix, Fig. S3 G–I). PRLs have been reported to regulate intracellular [Mg2+] in cell culture through interaction with the CNNM family of proteins (2). However, no differences in CNNM1 and 3 expression or Mg2+ concentration were found between samples from Pten+/− and Pten+/−;Prl2−/− mice (SI Appendix, Fig. S4). Taken together, these results indicate that ablation of PRL2 attenuates tumorigenesis primarily through up-regulating PTEN and impeding AKT activity.
PRL2 Expression Is Inversely Correlated with PTEN Level in Human Cancers.
To assess the clinical relevance of PTEN regulation by PRL2, we investigated whether there is a correlation between PRL2 and PTEN in human cancers. From the cBioPortal database (www.cbioportal.org/), we discovered that PRL2 is amplified in many cancers (SI Appendix, Fig. S5A). PTEN levels are frequently down-regulated in human malignancies, even in the absence of genetic loss or mutation (19). Furthermore, partial loss of PTEN function or decreased PTEN expression is sufficient to promote tumor progression (7, 17). Consistent with the mouse genetic data, we found that PRL2 mRNA inversely correlated with PTEN protein with statistical significance in multiple human cancers (Fig. 5 B, E, and H and SI Appendix, Fig. S5 D and G). In line with the inverse correlation between PRL2 and PTEN, we also observed that PRL2 mRNA expression positively correlated with the activation of the PI3K/AKT pathway components pAkt/S473, p-mTOR/S2448, pS6K/T389, pS6/S235/S236, and pS6/S240/S244 in prostate adenocarcinoma (SI Appendix, Fig. S5B), consistent with our biochemical findings (Fig. 4 A and B). Furthermore, when these samples were divided into two groups based on PRL2 mRNA level (low and high), the mean PTEN protein level was significantly lower in the group with high PRL2 expression compared with the group with lower PRL2 expression (Fig. 5 A, D, and G and SI Appendix, Fig. S5 C and F), supporting an inverse correlation between PRL2 mRNA and PTEN protein in these cancer patients. More importantly, cancer patients with a high PRL2 mRNA level have a worse prognosis for survival than those with a lower PRL2 expression (Fig. 5 C, F, and I and SI Appendix, Fig. S5 E and H). In contrast, a high PRL2 level has little predictive value in assessing patient survival in cancer types where there is no clear correlation between PRL2 mRNA and PTEN levels (SI Appendix, Fig. S5 I and J). The results are in accordance with our findings that removal of PRL2 in Pten+/− mice increases the level of PTEN and inhibits PTEN heterozygosity-induced tumorigenesis and suggest that PRL2 overexpression could contribute to the reduced survival rate in multiple human cancers by down-regulating PTEN. To further corroborate the inverse correlation between PRL2 and PTEN, we examined the relationship between PRL2 amplification and PTEN deletion in The Cancer Genome Atlas (TCGA) database. We found 466 patients with PTEN deletion, but only 4 had PRL2 amplification. Moreover, we found 61 patients with PRL2 amplification, but 57 out of the 61 exhibited no PTEN deletion. These observations suggest that these two events, namely PRL2 amplification and PTEN deletion, are most likely mutually exclusive, which is in line with the model proposed.
Fig. 5.
PRL2 expression is inversely correlated with PTEN level in human cancers. (A) Boxplots of RPPA data showing the reverse correlation between the mRNA levels of PRL2 and protein levels of PTEN in kidney renal clear cell carcinoma patients (n = 476). Patient samples were divided into two groups (low PRL2 and high PRL2) based on the mean PRL2 mRNA level, and then the PTEN protein levels of the two groups were plotted. PTEN protein level was significantly lower in the high PRL2 mRNA group than the low PRL2 mRNA group by Mann–Whitney U tests. (B) The mRNA level of PRL2 and protein level of PTEN from KIRC were plotted and Spearman rank-correlation analyses were performed. Negative correlation coefficients suggest an inverse correlation between PRL2 mRNA and PTEN protein in the cancer samples. (C) Patients with high PRL2 mRNA levels have significantly reduced overall survival in KIRC by Kaplan–Meier survival analysis. HR, hazard ratio. (D) Reverse correlation of PRL2 mRNA and PTEN protein levels in liver hepatocellular carcinoma (n = 181). (E) The mRNA level of PRL2 and protein level of PTEN from LIHC were plotted and Spearman rank-correlation analyses were performed. (F) Patients with high PRL2 mRNA levels have significantly reduced overall survival in LIHC by Kaplan–Meier survival analysis. (G) Reverse correlation of PRL2 mRNA and PTEN protein levels in pancreatic adenocarcinoma (n = 116). (H) The mRNA level of PRL2 and protein level of PTEN from PAAD were plotted and Spearman rank-correlation analyses were performed. (I) Patients with high PRL2 mRNA levels have significantly reduced overall survival in PAAD by Kaplan–Meier survival analysis.
PRL2 Promotes NEDD4-Mediated PTEN Polyubiquitination and Degradation.
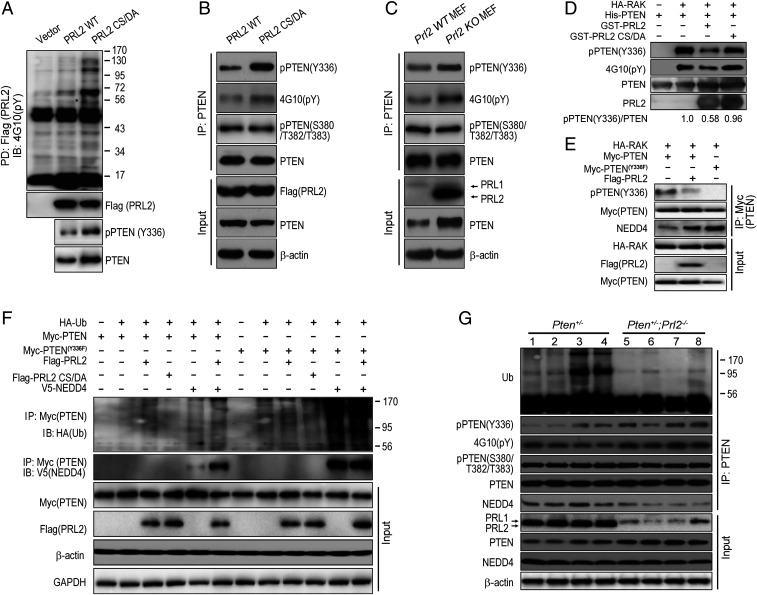
The results described above suggest that PRL2 promotes AKT signaling by down-regulating PTEN. To determine how PRL2 controls PTEN, we found that PRL2-overexpressing cells, but not the vector control or the catalytically inactive mutant PRL2 CS/DA cells, exhibit accelerated PTEN protein degradation (Fig. 6 A and B). Furthermore, treatment of cells with the proteasome inhibitor MG132 restored PTEN protein levels in PRL2-overexpressing cells but not in the PRL2 CS/DA and vector control cells (Fig. 6C). These results indicate that PRL2 reduces PTEN through proteasome-dependent degradation and that PRL2 phosphatase activity is essential for PRL2-mediated PTEN down-regulation.
Fig. 6.
PRL2 promotes NEDD4-mediated PTEN polyubiquitination and degradation. (A) PRL2 destabilizes PTEN through the proteasomal pathway. Wild-type PRL2 but not the catalytically inactive mutant promotes PTEN degradation in cells. HEK293 control vector cells, PRL2-overexpressing stable cells, and PRL2 mutant cells were treated with cycloheximide (CHX) at 100 μg/mL for the indicated times, and cell lysates were analyzed by Western blotting. Data represent three independent experiments. Anti–β-actin and GAPDH served as the loading controls. (B) Quantification of the results in A. PTEN and actin signals were measured using the ImageJ program. The ratio of PTEN/actin was determined for each sample and plotted as fold of time 0 for each cell line. Each bar represents the mean SD from three independent experiments. (C) Overexpression of PRL2 decreases PTEN protein level. PTEN levels (relative to β-actin) were measured using ImageJ. Data are representative of at least three independent experiments. Anti–β-actin and GAPDH served as the loading controls. (D) Ubiquitination of PTEN was accelerated in the presence of PRL2. HEK293 cells were cotransfected with plasmids encoding HA-tagged ubiquitin, Flag-PRL2, Flag-PRL2 CS/DA, and Myc-PTEN. Forty-eight hours after transfection, cells were harvested and lysed. Myc-tagged PTEN was pulled down with Myc antibody and subjected to immunoblotting against the HA tag to detect ubiquitinated PTEN. IB, immunoblotting; IP, immunoprecipitation. (E) PRL2-mediated PTEN ubiquitination is K48-linked polyubiquitination. HEK293 cells transfected with Flag-PRL2 and Myc-PTEN in the presence of HA-tagged WT-Ub or lysine mutants Ub-K48R or Ub-K63R. The expressed PTEN was pulled down by Myc antibody. HA-tagged ubiquitin was used to detect ubiquitinated PTEN. (F) PRL2 increases NEDD4-catalyzed PTEN ubiquitination. HEK293 cells were cotransfected with plasmids encoding for HA-tagged ubiquitin, Flag-tagged PRL2, mutant PRL2 CS/DA, Myc-tagged PTEN, and V5-tagged NEDD4, XIAP, and WWP2. The expressed PTEN was pulled down from cell lysates by Myc antibody. Subsequently, Western blotting against HA-tagged ubiquitin was performed to detect ubiquitinated PTEN. Anti–β-actin and GAPDH served as the loading controls.
We next determined whether PRL2 promotes proteasome-dependent PTEN degradation by increasing PTEN polyubiquitination. We discovered that PTEN is heavily ubiquitinated in the presence of PRL2 but not the catalytically inactive CS/DA mutant (Fig. 6D). Conversely, the level of ubiquitination on endogenous PTEN in PRL2-deficient mouse embryonic fibroblast (MEF) cells was markedly lower than that from WT MEF cells (SI Appendix, Fig. S6A). In addition to PRL2, PRL1 and PRL3 can also promote PTEN polyubiquitination (SI Appendix, Fig. S6B), indicating that this might be a general mechanism for all PRLs. We also established that PRL2-mediated PTEN polyubiquitination is linked through K48 (Fig. 6E), further confirming that PRL2 promotes PTEN degradation through the proteasome pathway.
Several PTEN-specific E3 ubiquitin ligases have been described, including NEDD4, WWP2, and XIAP (23–25). To determine which E3 ligase is responsible for PRL2-induced PTEN ubiquitination, we showed that PRL2, but not its CS/DA inactive mutant, is able to increase PTEN polyubiquitination (Fig. 6F), likely through endogenous E3 ligases. Strikingly, expression of NEDD4, but not XIAP and WWP2, dramatically elevated PTEN polyubiquitination in both vector control cells (containing endogenous PRL2) and cells ectopically expressing Flag-tagged PRL2 (Fig. 6F). Conversely, CRISPR-Cas9–mediated deletion of NEDD4 in HEK293 cells largely abolished PRL2-induced PTEN ubiquitination, leading to an increase in PTEN (SI Appendix, Fig. S6C), and nearly doubled the PTEN half-life (SI Appendix, Fig. S6D). Taken together, these results suggest that PRL2 down-regulates PTEN by promoting NEDD4-mediated K48-linked polyubiquitination and degradation of PTEN.
PRL2 Dephosphorylates PTEN at Y336 to Promote NEDD4-Mediated PTEN Polyubiquitination.
Given the requirement of PRL2 phosphatase activity for PTEN ubiquitination, we sought to identify the PRL2 substrate(s) in order to understand how PTEN is down-regulated by PRL. The “substrate-trapping” approach (26), by which a catalytically inactive PRL2 mutant (C101S [CS] or C101S/D69A [CS/DA]), which retains the ability to bind substrates but is unable to carry out substrate turnover, was used to capture the PRL2 substrate(s) (SI Appendix). Among the phosphorylated proteins isolated by the PRL2 substrate-trapping mutants was a 56-kDa protein, shown to be PTEN (SI Appendix, Fig. S7 A–C). To validate PTEN as a genuine PRL2 substrate, we overexpressed the vector, Flag-tagged PRL2, or the CS/DA mutant in HEK293 cells. As shown in Fig. 7A, the CS/DA mutant pulled down more tyrosine-phosphorylated proteins than did the wild-type PRL2 from the cells. Again, endogenous PTEN (∼56 kDa) was identified as one of the PRL2 substrates.
Fig. 7.
PRL2 dephosphorylates PTEN at Tyr336 to promote NEDD4-mediated PTEN polyubiquitination. (A) PTEN is a substrate of PRL2. Overexpressing control vector and Flag-PRL2 and Flag-PRL2 CS/DA mutants were transfected into HEK293 cells. Twenty-four hours after transfection, cells were treated with pervanadate for 30 min and the medium was replaced with fresh medium for another 30 min. Then the cells were lysed and immunoprecipitated by Flag beads and detected by anti-Flag, anti-PTEN, and anti-PTEN(Y336) antibodies. An asterisk indicates the potential PTEN position. PD, pull down. (B) Overexpression of PRL2 dephosphorylates endogenous PTEN at Tyr336 in HEK293 cells. PRL2 and PRL2 CS/DA mutant were transfected into HEK293 cells. Twenty-four hours after transfection, cells were treated with pervanadate for 30 min and the medium was replaced with fresh medium for another 30 min. Then the cells were lysed and immunoprecipitated with PTEN antibody and detected by anti-PTEN(Y336), 4G10(pY), pPTEN(S380/T382/T383), PTEN, Flag(PRL2), and β-actin. (C) PRL2 dephosphorylates endogenous PTEN at Tyr336 in MEFs. Control WT MEFs and Prl2 KO MEFs were treated with pervanadate for 30 min and the medium was replaced with fresh medium for another 30 min. Then the cells were lysed and immunoprecipitated with PTEN antibody and detected by anti-PTEN(Y336), 4G10(pY), pPTEN(S380/T382/T383), PTEN, Flag(PRL2), and β-actin. (D) PRL2 dephosphorylates RAK-mediated PTEN phosphorylation at Tyr336 in vitro. In vitro phosphorylation assay was performed by incubating purified His-PTEN with HA-RAK immunoprecipitated from H1299 cells for 30 min at room temperature. Then the in vitro dephosphorylation assay was performed by treating RAK-phosphorylated PTEN (10 μM) with either control buffer or purified recombinant GST-PRL2 or GST-PRL2 CS/DA (10 μM) for another 30 min at 37 °C. The level of PTEN phosphorylation was detected by 4G10(pY), pPTEN(Y336), PTEN, and PRL2 antibodies. pPTEN(Y336) levels (relative to PTEN) were measured using ImageJ (58 ± 18% in the presence of PRL2, and 96 ± 19% in the presence of PRL2/CSDA). Data are representative of three independent experiments. (E) PRL2 dephosphorylates RAK-mediated PTEN phosphorylation at Tyr336 in vivo. HA-RAK and Myc-PTEN or Myc-PTENY336F were cotransfected with or without Flag-PRL2 in HEK293 cells. Then the cells were lysed and immunoprecipitated with Myc antibody, and detected by pPTEN(Y336), 4G10(pY), Myc, HA, Flag(PRL2), and NEDD4 antibodies. (F) PRL2 enhances PTEN–NEDD4 association. HEK293 cells were cotransfected with plasmids encoding for HA-tagged ubiquitin, Flag-tagged PRL2, mutant PRL2 CS/DA, Myc-tagged PTEN, mutant PTENY336F, and V5-tagged NEDD4. The expressed PTEN was pulled down from cell lysates by Myc antibody. Subsequently, Western blotting against HA-tagged ubiquitin was performed to detect ubiquitinated PTEN. Anti–β-actin and GAPDH served as the loading controls. (G) Deletion of PRL2 decreases PTEN polyubiquitination and PTEN–NEDD4 association in mouse lymphomas. Lymphoma tissue samples were immunoprecipitated with anti-PTEN antibody and blotted with pPTEN(Y336), 4G10(pY), pPTEN (S380/T382/T383), PTEN, NEDD4, PRL1/2, GAPDH, and β-actin antibodies.
Phosphorylation of PTEN C-terminal Ser/Thr residues (S380/T382/T383) enhances PTEN stability (8). However, PRL2 displays no detectable activity toward these sites (4). Interestingly, the Src family tyrosine kinase RAK is able to block the NEDD4-mediated PTEN ubiquitination by phosphorylating PTEN at Y336 (27). We hypothesized that PRL2 promotes PTEN ubiquitination by directly dephosphorylating Y336, thereby enhancing PTEN’s binding affinity for its E3 ligase NEDD4. Using a pPTEN(Y336)-specific antibody, we showed that PTEN captured by the CS/DA mutant was indeed phosphorylated on Y336 (Fig. 7A). Furthermore, PRL2, but not the phosphatase-dead mutant, substantially reduced both the overall PTEN tyrosine phosphorylation (as measured by the 4G10 anti-pTyr antibodies) and specific pPTEN(Y336) phosphorylation (Fig. 7B). In contrast, no change in PTEN C-terminal S380/T382/T383 phosphorylation was detected upon increased PRL2 expression (Fig. 7B). Conversely, both the total tyrosine and Y336 phosphorylation in PTEN were markedly increased in Prl2−/− MEF cells, while PTEN/S380/T382/T383 phosphorylation remained unchanged upon PRL2 deletion (Fig. 7C). These data indicate that PRL2 dephosphorylates PTEN on Y336 inside the cell.
We next showed that PRL2, but not its catalytically inactive mutant, can directly dephosphorylate PTEN at Y336 using recombinant PRL2 and PTEN as well as RAK immunoprecipitated from H1299 cells (27) (Fig. 7D). To provide a quantitative assessment of PRL2’s substrate specificity for PTEN, we compared the ability of PRL2 and SHP2 (a member of the PTP family) to dephosphorylate p-nitrophenyl phosphate (pNPP), a commonly used generic small-molecule PTP substrate, the pY336-containing phosphopeptide (DKANRpY336FSPNF) derived from PTEN, and the Y336-phosphorylated PTEN protein (SI Appendix, Figs. S7 H–L and S8). The kcat/Km (a measure of catalytic efficiency and substrate specificity) values for PRL2-catalyzed hydrolysis of pNPP (0.89 ± 0.06 M−1s−1) and DKANRpY336FSPNF (0.048 ± 0.005 M−1⋅s−1) were 1,570- and 18,400-fold lower than those of the SHP2 reactions (1,400 ± 71 M−1⋅s−1 for pNPP and 885 ± 115 M−1⋅s−1 for the peptide). Strikingly, the kcat/Km for PRL2-catalyzed dephosphorylation of pPTEN(Y336) was 587 ± 76 M−1⋅s−1, whereas no appreciable dephosphorylation of pPTEN(Y336) was observed in the presence of SHP2, even after a 20-h incubation (SI Appendix, Fig. S7 H–L). Thus, even though SHP2 exhibits robust activity toward pNPP and DKANRpY336FSPNF, it fails to dephosphorylate the pPTEN(Y336) protein. In contrast, the catalytic efficiency/substrate specificity for the PRL2-catalyzed pPTEN(Y336) dephosphorylation is 660- and 12,230-fold higher than those toward pNPP and the DKANRpY336FSPNF peptide. Collectively, the results indicate that pPTEN(Y336) is a specific substrate for PRL2.
To further demonstrate that PRL2 can dephosphorylate Y336 from PTEN inside the cell, we transfected HEK293 cells with hemagglutinin (HA)-tagged RAK, Myc-tagged PTEN or PTEN/Y336F, in which Y336 was replaced with a nonphosphorylatable phenylalanine, and Flag-tagged PRL2. Consistent with the lack of phosphorylation at residue 336, the association between NEDD4 and the PTEN/Y336F mutant was considerably stronger in comparison with that between NEDD4 and wild-type PTEN, which can be phosphorylated by RAK at Y336 (Fig. 7E). As expected, overexpression of PRL2 greatly reduced RAK-mediated PTEN/Y336 phosphorylation and increased the amount of NEDD4 bound to PTEN (Fig. 7E). Collectively, these results support that PRL2 augments PTEN–NEDD4 interaction by dephosphorylating PTEN at Y336 both in vitro and in vivo. In addition, PRL1 and PRL3 could also oppose the RAK-mediated PTEN phosphorylation, suggesting that all PRL family members are able to dephosphorylate PTEN at Y336 (SI Appendix, Fig. S7D).
To furnish more evidence that PRL2-catalyzed pY336 dephosphorylation enhances PTEN binding with its E3 ligase NEDD4, we compared the effect of PRL2 on ubiquitination of wild-type PTEN and PTEN/Y336F. As observed previously (27), PTEN/Y336F displayed increased basal ubiquitination and lower protein level compared with wild-type PTEN (Fig. 7F). As noted in Fig. 6D, overexpression of PRL2, but not the CS/DA mutant, enhanced PTEN polyubiquitination (Fig. 7F). However, unlike wild-type PTEN, PRL2 failed to increase the polyubiquitination of PTEN/Y336F (Fig. 7F), suggesting that PRL2 promotes PTEN polyubiquitination through Y336 dephosphorylation. We then showed that in the presence of V5-tagged NEDD4, PTEN/Y336F displays considerably elevated ubiquitination as well as increased interaction with NEDD4 over wild-type PTEN (Fig. 7F). Moreover, the association between PTEN and NEDD4 is substantially enhanced by PRL2, whereas PTEN/Y336F and NEDD4 association remains the same with or without PRL2 (Fig. 7F). PRL2 could further augment NEDD4-mediated PTEN polyubiquitination, but has no effect on PTEN/Y336F ubiquitination (Fig. 7F). Taken together, these data indicate that PRL2-catalyzed pPTEN(Y336) dephosphorylation promotes the interaction between NEDD4 and PTEN as well as NEDD4-mediated PTEN polyubiquitination.
In line with the results in HEK293 cells, PTEN polyubiquitination was considerably decreased in tumors from Pten+/−;Prl2−/− mice compared with those from Pten+/− mice (Fig. 7G). Additionally, both Y336 phosphorylation and total tyrosine phosphorylation, but not the C-terminal S380/T382/T383 phosphorylation, were elevated in PTEN from the Pten+/−;Prl2−/− samples (Fig. 7G). Consistent with the higher PTEN level and reduced PTEN polyubiquitination, loss of PRL2 greatly decreased PTEN binding to NEDD4 in lymphomas from Pten+/−;Prl2−/− mice (Fig. 7G). Collectively, the results demonstrate that PRL2 down-regulates PTEN through direct dephosphorylation of Y336 in PTEN, leading to enhanced PTEN–NEDD4 interaction and NEDD4-mediated PTEN polyubiquitination, followed by proteasomal degradation. Consistent with this model, we determined the stoichiometry of PTEN Y336 phosphorylation in HEK293 cells (SI Appendix, Fig. S7 E–G) and found that expression of PRL2 reduced PTEN Y336 phosphorylation by 33%, in line with the observed changes in PTEN levels (25 to 40%) in vivo (Figs. 1 B and C and 7G) and up to 50% in HEK293 cells (Fig. 6C).
Discussion
PRLs garner considerable interest as novel anticancer targets (1, 2). However, the underlying mechanisms for how PRLs promote tumorigenesis are unknown. Although PRL phosphatase activity is essential for their oncogenic potential, no credible substrates have been identified for PRLs. This has greatly hampered our ability to understand their roles in cancer and devise therapeutic strategies to target these phosphatases. We previously uncovered that mice lacking PRL2 display an elevated level of PTEN and attenuated AKT activity (3–5). Given that even a subtle reduction in PTEN can result in cancer susceptibility and tumor progression (7, 15–19), we speculated that PRL2 may promote tumorigenesis by down-regulating PTEN and that PRL2 inhibition could restore PTEN, thereby blocking PTEN deficiency-induced malignancies. Here we show that deletion of Prl2 in Pten+/− mice markedly protects these animals from developing tumors and dramatically increases their overall survival. As anticipated, this is achieved through up-regulation of PTEN, leading to a reduction in AKT activity in PRL2-deficient animals. This finding is in line with early observations that PTEN overexpression in mice confers cancer resistance (28) and removal of AKT1 or hypomorphic mutation of PDK1 suppresses tumorigenesis in Pten+/− mice (29, 30). Consistent with the impairment of AKT activity, tumors from Pten+/−;Prl2−/− mice exhibit substantially decreased cell proliferation and increased apoptosis, as a consequence of diminished mTORC1/S6K and GSK3β signaling and induction of p53 and its proapoptotic target proteins PUMA and Bax. Our results provide compelling biochemical and genetic evidence that PRL2 promotes tumorigenesis by down-regulating PTEN to amplify the prooncogenic AKT pathways.
PTEN level is often decreased in cancer (7, 15–18), yet the mechanism for PTEN loss has not been well-defined. Our data indicate that PTEN itself is a substrate for PRL2. We show that PRL2 can dephosphorylate PTEN at Y336, thereby augmenting PTEN binding to NEDD4, leading to increased PTEN ubiquitination and proteasomal degradation. We find that PRL1 and PRL3 can also remove the phosphoryl group from pY336 in PTEN and promote PTEN ubiquitination. We previously reported that PRL1 and PRL2 may have functional redundancy, at least in the context of PTEN regulation during spermatogenesis (31). PRL3 expression in DLD-1 colon cancer cells could also down-regulate PTEN and activate AKT (32). Thus, the mechanism we defined for PRL2-mediated PTEN down-regulation and tumorigenesis may apply to PRL1 and PRL3 as well.
Although the rate for PRL2-catalyzed pPTEN(Y336) dephosphorylation, as measured with recombinant proteins in vitro, may seem slower than what one might have expected for an enzymatic reaction, the impact of PRL2 on PTEN half-life occurs on a timescale (Fig. 6A and SI Appendix, Fig. S6D) compatible with PRL2-mediated substrate turnover (SI Appendix, Fig. S7H). Thus, the phosphatase activity exhibited by recombinant PRL2 toward pPTEN(Y336) could be physiologically relevant when placed in the context of PRL2-mediated PTEN degradation inside the cell. Nevertheless, additional factors, such as PRL2 prenylation, trimerization, membrane colocalization with PTEN, and potential cofactors may further enhance the rate of pPTEN(Y336) dephosphorylation by PRL2 in vivo. Although our data indicate that PRL2’s phosphatase activity is required for PRL2-mediated PTEN dephosphorylation, ubiquitination, and degradation, there is still the possibility of PRL2 regulating a different phosphatase for pPTEN(Y336) that has much higher levels of PTEN phosphatase activity. Further investigation is needed to ascertain if these and other factors are involved in regulating PTEN dephosphorylation by the PRL2 phosphatase in vivo.
PRLs are also reported to play a role in modulating intracellular Mg2+ levels through direct binding interaction with the CNNM family of proteins (2). However, this finding is based primarily on experiments with cultured cell lines ectopically expressing or knocking down either PRL or CNNM. It remains to be seen whether changes in PRL expression can lead to significant perturbation in Mg2+ levels in vivo and whether such changes exert an effect on physiological and pathological processes. We found no appreciable differences in CNNM1 and CNNM3 expression in tumors from Pten+/− and Pten+/−;Prl2−/− mice (SI Appendix, Fig. S4E). We also found no appreciable differences in Mg2+ concentrations between tumor, kidney, and serum samples from Pten+/− and Pten+/−;Prl2−/− mice (SI Appendix, Fig. S4). Thus, the inhibitory effect of PRL2 ablation on PTEN heterozygosity-induced tumorigenesis is unlikely mediated by the proposed role of PRL in modulating Mg2+ homeostasis.
In addition to defining an oncogenic role for PRL2 and uncovering a regulatory mechanism for PTEN, this work also suggests a therapeutic approach for neoplasia caused by PTEN deficiency. Monoallelic PTEN loss is regularly observed in a considerable fraction of human malignancies, such as glioma, prostate, mammary, colon, and lung cancer (17, 33). PTEN expression is also frequently decreased in cancer patients even in the absence of genetic loss or mutation, and PTEN deficiency increases cancer susceptibility in a dose-dependent manner (7, 15–19). For example, low PTEN levels are observed in 70% of surgically removed prostate tumors (34). PTEN protein levels are reduced in at least 50% of breast cancers (35, 36). Indeed, even a 20% reduction in PTEN level could dramatically increase breast cancer susceptibility (15).
PTEN antagonizes the PI3K-AKT pathway by dephosphorylating PIP3. Thus, a gradual reduction in PTEN will result in a concomitant progressive dose-dependent activation of the PI3K-AKT pathway, which is inappropriately activated in many human cancers. Current efforts targeting this pathway have focused on PI3K, AKT, or mTOR (37). However, inhibitors targeting these kinases have had suboptimal therapeutic efficacy due to both on- and off-target side effects and activation of alternative signaling cascades (38). Identifying new therapeutic strategies to block PI3K-AKT pathway activation in tumors with PTEN deficiency is of paramount importance.
Our findings that removal of Prl2 in Pten+/− mice increases PTEN level and suppresses tumor progression establish that targeting PRL2 could serve as a potential PTEN restoration strategy for treating human malignancies caused by PTEN deficiency. PTEN restoration represents a direct therapeutic paradigm for PTEN deficiency-induced cancers, as it offers the exciting possibility that the full complement of PTEN functions (including protein phosphatase and nuclear function) could be restored in a straightforward manner. In addition, PTEN restoration could improve already available target therapies when administered simultaneously, since treatments with PTK inhibitors have significantly better outcome when patients still have functional PTEN (33). Indeed, adenoviral-mediated PTEN expression significantly inhibits tumor growth in multiple animal models (39–42), indicating that restoration of PTEN may be an effective strategy for tumor therapy.
In support of the therapeutic applicability of the PTEN restoration approach, overexpression of PTEN in cells with physiological PTEN levels had little adverse effect (42), and mice with systemic elevated PTEN display healthy phenotypes and are characterized by heightened resistance to cancer (28). While genetically introducing PTEN or deleting PRL2 is not currently feasible in the clinic, targeting PRL2 with small-molecule inhibitors may be an effective strategy to increase PTEN level and thereby obliterate PTEN deficiency-induced malignancies. As revealed in this study, cancers showing high PRL2 expression and low PTEN level will be prime targets for PRL2-based PTEN restoration therapy. Finally, given the potential functional redundancy for PTEN regulation by the PRLs and their universal linkage to cancer when overexpressed, a pan-PRL inhibitor may prove to be more effective than compounds that target only one PRL isoform in a clinical setting.
Materials and Methods
Please refer to SI Appendix, Materials and Methods for other materials and methods, including detailed characterization of tumors in Pten+/− and Pten+/−;Prl2−/− mice, substrate trapping, antibodies and Western blot analysis, cell culture and transfection, histology analysis and pathologic grading, immunohistochemistry, immunofluorescence, immunoprecipitation and pull-down assays, TUNEL assay, in vitro PI3K assay, effector pull-down assay, measurement of magnesium concentration, RNA isolation assay, qRT-PCR and cycloheximide chase assay, generation of NEDD4 knockout cells, Phos-tag stoichiometry measurement, and PRL2 phosphatase activity assay.
Generation of Mice.
The pure C57BL/6 mice and EIIA-Cre mice were purchased from The Jackson Laboratory. The Prl2 genetic knockout mouse (Prl2−/−) used in this study was described previously (4). The conditional Pten knockout mouse was originally developed by Lloyd C. Trotman (14). Briefly, conditional Pten knockout mice were crossed with the EIIA-Cre mouse to generate Pten+/−, and then Pten+/− mice were cross-bred with Prl2+/− mice to generate Pten+/−;Prl2+/−. The Pten+/−;Prl2+/− littermates were then bred with each other to generate Pten+/−;Prl2−/−, Prl2−/−, Pten+/−, and WT mice. All DNA used for genotyping our experimental mice was gathered from toe tissue clipped from 7- to 10-d-old pups. The tissue was then lysed in tissue lysis buffer (200 mM NaCl, 100 mM Tris, pH 8.0, 5 mM ethylenediaminetetraacetate [EDTA], 0.5% sodium dodecyl sulfate [SDS], 100 μg/mL proteinase K) overnight at 55 °C. The DNA was then isolated from solution with isopropanol and resuspended in TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA) for long-term storage. All mice were maintained in the Purdue University Animal Facility according to institutional animal care and use committee-approved protocols and kept in Thorensten units with filtered germ-free air.
TCGA Analysis.
TCGA level 3 RNAseqV2 gene expression, reverse-phase protein array (RPPA), and clinical data of primary patient clinical samples from kidney renal clear cell carcinoma (KIRC; n = 537), liver hepatocellular carcinoma (LIHC; n = 377), pancreatic adenocarcinoma (PAAD; n = 185), prostate adenocarcinoma (PRAD; n = 499), and thyroid carcinoma (THCA; n = 503) were downloaded from the Broad Institute’s Firehose (gdac.broadinstitute.org). Patients were separated into two subgroups (low PRL2 and high PRL2) based on their PRL2 mRNA expression levels using mean value as cutoff for PTEN RPPA analysis and Kaplan–Meier survival analysis. The correlation coefficient between PRL2 mRNA and PTEN protein level was measured by Spearman rank-correlation analysis.
In Vitro Substrate-Trapping Assay.
The most commonly used substrate-trapping mutant is one in which the PTP active-site nucleophilic Cys residue is replaced by a Ser (e.g., PRL2/C101S or CS). Evidence suggests that the substrate-trapping efficiency of the CS mutant can be further improved by substitution of the general acid/base Asp with an Ala (e.g., PRL2/C101S/D69A or CS/DA) (26). HEK293, Kasumi-1, or K562 cells (1 × 109) were treated with 1 mM pervanadate for 30 min and collected by centrifugation. The cell pellet was lysed with 3 mL lysis buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM iodoacetic acid, 1 mM orthovanadate, protease inhibitors). Dithiothreitol (10 mM) was added to the lysate and incubated for 15 min on ice to inactivate any unreacted iodoacetic acid and pervanadate. Supernatant was collected by centrifugation at 14,000 rpm for 15 min. Glutathione S-transferase (GST), GST-PRL2, or GST-PRL2/CS/DA, or His-tagged PRL2 or His-tagged PRL2/CS (25 μg) was coupled to GST beads or Ni-NTA beads, respectively, in lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100), and incubated at 4 °C for 1 h. Cell lysates were incubated with PRL2 proteins conjugated to beads at 4 °C for 2 h. The beads were pelleted and washed three times for 5 min with lysis buffer. Bound proteins were resuspended in 15 μL Laemmli sample buffer and boiled for 5 min, and the samples were resolved by SDS/polyacrylamide gel electrophoresis (PAGE).
In Vivo Substrate-Trapping Assay.
HEK293 cells in 15-cm dishes were transfected with Flag-PRL2 or Flag-PRL2/CS/DA. The transfected cells were treated with 300 μM pervanadate for 30 min, the medium was replaced with fresh medium for another 30 min, and the cells were collected by centrifugation. The cell pellet was lysed with 1 mL lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, supplemented with a complete protease inhibitor and phosphatase inhibitor) on ice for 1 min and then spun at 14,000 rpm at 4 °C for 30 min, and the supernatant was transferred to a fresh tube and Flag agarose beads added and incubated at 4 °C for 3 h. Beads were collected by centrifugation at 3,000 rpm for 1 min and the supernatant was removed. Beads were washed three times with 1.5 mL lysis buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100). Bound proteins were resuspended in 15 μL Laemmli sample buffer and boiled for 5 min, and the samples were resolved by SDS/PAGE.
Quantification and Statistical Analysis.
Datasets were analyzed by the Student’s t test or Mann–Whitney U test according to the experiments using GraphPad Prism software, unless otherwise described in methods or figure legends. Tumor volumes at different times and final tumor weights were compared using the Student’s t test. Survival analysis was performed according to the Kaplan–Meier method and the log-rank test. Error bars in figures indicate SD or SEM for the number of replicates, as indicated. A P value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Qi Zeng for providing antibodies against PRL1 and PRL2. This work was supported in part by NIH R01CA69202 and the Robert C. and Charlotte Anderson Chair Endowment. We also gratefully acknowledge NIH P30CA023168 for supporting Purdue University Center for Cancer Research shared resources.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. V.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002964117/-/DCSupplemental.
Data Availability.
All data discussed in this study are included in the main text and SI Appendix.
References
- 1.Frankson R. et al., Therapeutic targeting of oncogenic tyrosine phosphatases. Cancer Res. 77, 5701–5705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy S. et al., Physiological and oncogenic roles of the PRL phosphatases. FEBS J. 285, 3886–3908 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Dong Y. et al., Phosphatase of regenerating liver 2 (PRL2) deficiency impairs Kit signaling and spermatogenesis. J. Biol. Chem. 289, 3799–3810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong Y. et al., Phosphatase of regenerating liver 2 (PRL2) is essential for placental development by down-regulating PTEN (phosphatase and tensin homologue deleted on chromosome 10) and activating Akt protein. J. Biol. Chem. 287, 32172–32179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi M. et al., PRL2/PTP4A2 phosphatase is important for hematopoietic stem cell self-renewal. Stem Cells 32, 1956–1967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manning B. D., Toker A., AKT/PKB signaling: Navigating the network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollander M. C., Blumenthal G. M., Dennis P. A., PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 11, 289–301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song M. S., Salmena L., Pandolfi P. P., The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Berger A. H., Knudson A. G., Pandolfi P. P., A continuum model for tumour suppression. Nature 476, 163–169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cristofano A., Pesce B., Cordon-Cardo C., Pandolfi P. P., Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Kwabi-Addo B. et al., Haploinsufficiency of the Pten tumor suppressor gene promotes prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 98, 11563–11568 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podsypanina K. et al., Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. U.S.A. 96, 1563–1568 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki A. et al., High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 8, 1169–1178 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Trotman L. C. et al., Pten dose dictates cancer progression in the prostate. PLoS Biol. 1, E59 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alimonti A. et al., Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carracedo A., Alimonti A., Pandolfi P. P., PTEN level in tumor suppression: How much is too little? Cancer Res. 71, 629–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie N. R., Foti M., Non-genomic loss of PTEN function in cancer: Not in my genes. Trends Pharmacol. Sci. 32, 131–140 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Naguib A., Trotman L. C., PTEN plasticity: How the taming of a lethal gene can go too far. Trends Cell Biol. 23, 374–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sansal I., Sellers W. R., The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 22, 2954–2963 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Di Cristofano A. et al., Impaired Fas response and autoimmunity in Pten+/− mice. Science 285, 2122–2125 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Grzechnik A. T., Newton A. C., PHLPPing through history: A decade in the life of PHLPP phosphatases. Biochem. Soc. Trans. 44, 1675–1682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieging K. T., Attardi L. D., Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 22, 97–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddika S. et al., WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Themsche C., Leblanc V., Parent S., Asselin E., X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 284, 20462–20466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X. et al., NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agazie Y. M., Hayman M. J., Development of an efficient “substrate-trapping” mutant of Src homology phosphotyrosine phosphatase 2 and identification of the epidermal growth factor receptor, Gab1, and three other proteins as target substrates. J. Biol. Chem. 278, 13952–13958 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Yim E.-K. et al., Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 15, 304–314 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Cao I. et al., Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 149, 49–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayascas J. R., Leslie N. R., Parsons R., Fleming S., Alessi D. R., Hypomorphic mutation of PDK1 suppresses tumorigenesis in PTEN(+/−) mice. Curr. Biol. 15, 1839–1846 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Chen M. L. et al., The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/− mice. Genes Dev. 20, 1569–1574 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai Y. et al., Role of phosphatase of regenerating liver 1 (PRL1) in spermatogenesis. Sci. Rep. 6, 34211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H. et al., PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 67, 2922–2926 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Keniry M., Parsons R., The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene 27, 5477–5485 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Chen M. et al., Identification of PHLPP1 as a tumor suppressor reveals the role of feedback activation in PTEN-mutant prostate cancer progression. Cancer Cell 20, 173–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugge J., Hung M. C., Mills G. B., A new mutational AKTivation in the PI3K pathway. Cancer Cell 12, 104–107 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Stemke-Hale K. et al., An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 68, 6084–6091 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkins B. D., Parsons R. E., Molecular pathways: Intercellular PTEN and the potential of PTEN restoration therapy. Clin. Cancer Res. 20, 5379–5383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janku F., Phosphoinositide 3-kinase (PI3K) pathway inhibitors in solid tumors: From laboratory to patients. Cancer Treat. Rev. 59, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Anai S. et al., Combination of PTEN gene therapy and radiation inhibits the growth of human prostate cancer xenografts. Hum. Gene Ther. 17, 975–984 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Chen H. et al., PTEN restoration and PIK3CB knockdown synergistically suppress glioblastoma growth in vitro and in xenografts. J. Neuro Oncol. 104, 155–167 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Saito Y. et al., Adenovirus-mediated transfer of the PTEN gene inhibits human colorectal cancer growth in vitro and in vivo. Gene Ther. 10, 1961–1969 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Tanaka M., Grossman H. B., In vivo gene therapy of human bladder cancer with PTEN suppresses tumor growth, downregulates phosphorylated Akt, and increases sensitivity to doxorubicin. Gene Ther. 10, 1636–1642 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this study are included in the main text and SI Appendix.