Significance
Superresolution fluorescence microscopy (nanoscopy) has enabled the study of protein distributions in cellular organelles, albeit only down to a 3D resolution of 20 to 40 nm. By targeting single emitters with a doughnut-shaped excitation beam, the recently introduced MINFLUX nanoscopy provides single-digit nanometer 3D localizations of fluorescent labels. We show an application of multicolor MINFLUX nanoscopy to the imaging of densely packed labeled proteins, specifically of subunits of the MICOS complex, a large protein complex within the mitochondrial inner membrane. In conjunction with a mathematical framework for the identification of individual molecules and their positions, our results open the door to the analysis of the nanoscale arrangement of proteins within oligomeric protein complexes using focused visible light.
Keywords: MINFLUX, superresolution microscopy, mitochondria, MICOS, cluster analysis
Abstract
The mitochondrial contact site and cristae organizing system (MICOS) is a multisubunit protein complex that is essential for the proper architecture of the mitochondrial inner membrane. MICOS plays a key role in establishing and maintaining crista junctions, tubular or slit-like structures that connect the cristae membrane with the inner boundary membrane, thereby ensuring a contiguous inner membrane. MICOS is enriched at crista junctions, but the detailed distribution of its subunits around crista junctions is unclear because such small length scales are inaccessible with established fluorescence microscopy. By targeting individually activated fluorophores with an excitation beam featuring a central zero-intensity point, the nanoscopy method called MINFLUX delivers single-digit nanometer-scale three-dimensional (3D) resolution and localization precision. We employed MINFLUX nanoscopy to investigate the submitochondrial localization of the core MICOS subunit Mic60 in relation to two other MICOS proteins, Mic10 and Mic19. We demonstrate that dual-color 3D MINFLUX nanoscopy is applicable to the imaging of organellar substructures, yielding a 3D localization precision of ∼5 nm in human mitochondria. This isotropic precision facilitated the development of an analysis framework that assigns localization clouds to individual molecules, thus eliminating a source of bias when drawing quantitative conclusions from single-molecule localization microscopy data. MINFLUX recordings of Mic60 indicate ringlike arrangements of multiple molecules with a diameter of 40 to 50 nm, suggesting that Mic60 surrounds individual crista junctions. Statistical analysis of dual-color MINFLUX images demonstrates that Mic19 is generally in close proximity to Mic60, whereas the spatial coordination of Mic10 with Mic60 is less regular, suggesting structural heterogeneity of MICOS.
Mitochondria are essential double-membrane organelles in all eukaryotes. Their intricate inner architecture is tightly connected to their primary function, the generation of adenosine triphosphate (ATP) by oxidative phosphorylation. The highly convoluted mitochondrial inner membrane (IM) is surrounded by the smooth mitochondrial outer membrane (OM) (1). Based on its fold, the IM can be structurally subdivided into three domains: 1) the inner boundary membrane (IBM) that parallels the OM, 2) the crista membranes, which are invaginations of the IM protruding into the interior of the organelle, and 3) the crista junctions. Electron tomography revealed that crista junctions are short, round to slit-like membrane tubes that connect the IBM and the crista membrane (2).
The mitochondrial contact site and cristae organizing system (MICOS; previously also called the mitochondrial inner membrane organizing system or the mitochondrial organizing structure) (3) is a conserved hetero-oligomeric protein complex that is essential for maintaining the cristae architecture. Genetic and biochemical studies have demonstrated the crucial role of MICOS for the establishment and the maintenance of crista junctions (4–7). Several severe human diseases including Parkinson’s and Alzheimer’s disease are associated with dysfunctions of MICOS subunits (8, 9). In humans, MICOS consists of at least seven different subunits, which are part of two MICOS subcomplexes, namely, the Mic60 and Mic10 subcomplexes. The Mic60 subcomplex encompasses Mic60, Mic25, and Mic19, whereas the Mic10 subcomplex consists of Mic10, Mic13, Mic26, and Mic27 (10–12). Immunogold electron microscopy demonstrated that Mic60 is highly enriched at crista junctions (5, 13). Two-dimensional (2D) stimulated emission depletion (STED) nanoscopy suggested that Mic60 forms small distinct assemblies, which reflect the positioning of the crista junctions and which are part of an extended multiprotein network scaffolding the organelle (13–15). However, no structural data of MICOS are currently available, and the details of the distribution of the different MICOS subunits at the crista junctions are still unknown.
In this study, we employ MINFLUX nanoscopy (16) to investigate the localizations of Mic10, Mic19, and Mic60 in human mitochondria. Among all superresolution fluorescence microscopy (nanoscopy) concepts (17, 18), MINFLUX imaging stands out because of its ability to routinely attain single-digit nanometer precision in 3D (19). MINFLUX owes its unique resolving power to a combination of the specific advantages of photoactivated localization microscopy (PALM)/stochastic optical reconstruction microscopy (STORM) and those of STED nanoscopy (20). Concretely, switchable fluorophores are individually turned on and localized with a structured (doughnut-shaped) excitation beam featuring an intensity zero point that is targeted to several (3D) coordinates as close to the molecule as possible until emission ceases or localization is accomplished. While MINFLUX imaging recently enabled multicolor recordings of nuclear pores with isotropic nanometer 3D resolution by using an intensity zero confined in all dimensions (19), the promise of MINFLUX nanoscopy to study densely packed proteins within a cellular organelle remained to be demonstrated. Here, by analyzing the MINFLUX data with a dedicated clustering algorithm for the molecular assignment of localizations, we exploit the nanometer-scale isotropic localization precision offered by this method. The quantitative analysis of MINFLUX recordings of Mic60 indicates circular to elliptical multiprotein arrangements of roughly 40 to 50 nm extent, supporting the concept that a small number of Mic60 proteins surround a single crista junction. Statistical analysis of dual-color MINFLUX images demonstrates that Mic19 is generally in close proximity to Mic60, whereas the spatial coordination of Mic10 with Mic60 is less regular, suggesting structural heterogeneity of MICOS. Overall, our data demonstrate that multicolor 3D MINFLUX nanoscopy combined with an appropriate analysis approach is a powerful tool for the quantitative spatial investigation of organellar protein distributions at the molecular level.
Results
Multicolor 3D MINFLUX Nanoscopy of Mitochondrial Proteins.
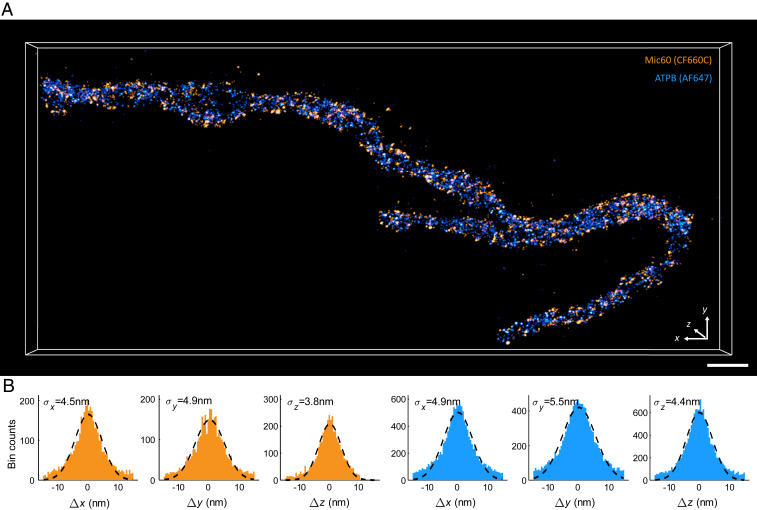
First, we established mitochondrial multicolor 3D MINFLUX imaging by labeling the -subunit of the F1FO-ATP synthase (ATPB) and the Mic60 subunit of MICOS. Whereas Mic60 forms distinct protein clusters that mark crista junctions (5, 13, 15), dimers of the F1FO-ATP synthase form extended rows which label specifically the curved rims of the cristae membrane (21). We labeled both proteins in chemically fixed primary human dermal fibroblasts (HDFa) with primary antibodies conjugated to either Alexa Fluor 647 or CF660C, organic fluorophores that are suitable for MINFLUX imaging using a conventional STORM redox buffer system. For imaging, we first selected a custom-shaped region based on a widefield fluorescence image (SI Appendix, Fig. S1) and subsequently acquired images of mitochondria using an iterative MINFLUX localization modality suitable for cellular imaging (19). In brief, we used a custom-built confocal microscopy arrangement featuring 1) fast scanning capability in all dimensions, 2) a doughnut-shaped excitation beam, 3) a regularly focused excitation beam (both 642 nm), 4) a regularly focused activation beam (405 nm), and 5) a confocal detection with two spectral channels split at a wavelength of around 685 nm. As a first step, we switched all molecules to a nonfluorescent state by scanning the regularly focused excitation beam over the region of interest. In an automated manner, the activation light was applied until the detection of emission from a single molecule, which was subsequently localized by iteratively zooming in the targeted coordinate pattern on the molecule via online position estimation. As soon as the emission burst ended, the excitation beam was switched off, and activation light was again applied until the next molecule was detected. A micrometer-sized region was covered by scanning the ensemble of activation light and the adaptable targeted coordinate pattern over the region of interest. As expected, we located Mic60 in the peripheral parts of mitochondria, whereas ATPB was more abundant at the center of the organelles (Fig. 1A, SI Appendix, Fig. S2, and Movies S1–S5). Corroborating a previous study relying on 2D STED nanoscopy (14), Mic60 clusters in HDFa cells recurrently seemed to be arranged in two opposite “distribution bands” that were occasionally twisted (Movies S2, S4, and S5).
Fig. 1.
Two-color 3D MINFLUX acquisition of a mitochondrion in primary human dermal fibroblasts. (A) Three-dimensional MINFLUX recording of Mic60 (orange) and ATPB (blue) using directly labeled primary antibodies conjugated to CF660C (orange) and Alexa Fluor 647 (blue). To keep acquisition times as short as possible, only the region close to the mitochondrion was imaged while the surroundings were padded. Individual localizations were rendered as 3D Gaussian functions with and displayed with the microscopy image analysis software Imaris. (Scale bar, 500 nm.) (B) Estimate of the localization precision in all dimensions for both molecular species. Localizations, each based on ∼2,000 photons, of all emission bursts were coaligned, and the resulting distribution was fit to a Gaussian function (dashed line) to extract the SD of the distribution . Colors are the same as in A.
We determined the experimental localization precision of our imaging approach by assessing the spread of localizations after coaligning groups of localizations obtained from individual single-molecule emission bursts. To this end, we exploited the photon efficiency of the MINFLUX localization and split the photon collection of an emission burst at a customarily chosen threshold of only 2,000 photons for each localization in postprocessing. After excluding localization groups with fewer than five localizations to avoid underestimation of the localization precision, we obtained an average of 10 localizations per emission burst, corresponding to about 20,000 to 30,000 photons (SI Appendix, Materials and Methods). One-dimensional Gaussian fits to the coaligned histograms delivered a localization precision of 3 to 6 nm (SD) in all three dimensions and for both dye species (Fig. 1B), demonstrating that MINFLUX can deliver this precision not only for sparse protein arrangements (19) but also when simultaneously imaging two complex and dense suborganellar structures in 3D.
Density-Based Clustering Assigns MINFLUX Localizations to Individual Molecules.
Numerous approaches have been established to extract quantitative biological information from single-molecule localization microscopy data, including the characterization of the clustering of proteins from point clouds of localizations (22). The broad applicability of these approaches is often challenged by a lack of high-precision information on the axial emitter position, bearing the risk of projection artifacts, as well as by the fact that several localizations are usually obtained from a given individual emitter, which can result in erroneous conclusions based on pseudoclusters. The isotropic single-digit nanometer precision provided by 3D MINFLUX opens up the possibility to tackle both of these limitations, thereby enabling the quantitative analysis of the target structure on a molecular level. To this end, we developed an approach to group all localizations that most likely originated from the same molecule so that further analysis could be based on a single precise 3D estimate of the molecular position.
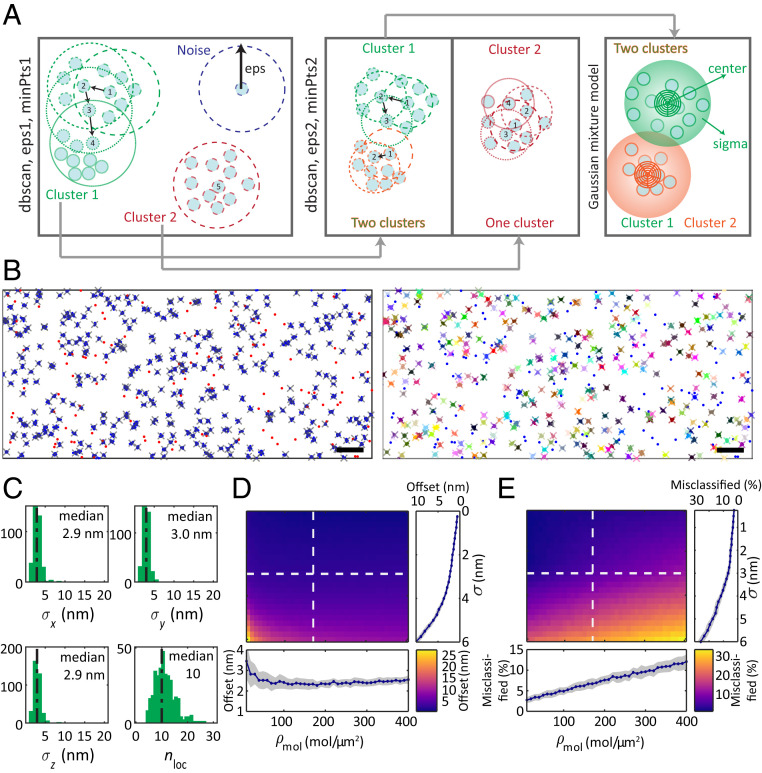
We designed a three-step clustering algorithm, combining the well-known dbscan (density-based spatial clustering of applications with noise) algorithm (23) and an expectation maximization approach using a Gaussian mixture model for the assignment of 2D or 3D localizations to fluorophore positions (Fig. 2A and SI Appendix, Materials and Methods). In the first step, we preassigned localizations and identified noise using a dbscan algorithm with a search radius much larger than the localization precision. In the second step, we identified agglomerations of molecules within the first assignment by applying a dbscan step with a reduced search radius. The result of the refined dbscan step was fed into a Gaussian mixture model for a final assignment of localizations to individual molecules. This approach was chosen as it did not require the input of the number of molecules or clusters but still exploited the Gaussian shape of individual localization clouds; thus, it delivered improved results compared to a single use of the dbscan algorithm.
Fig. 2.
Molecular assignment of localization clouds with the modified dbscan algorithm. (A) Schematic of the clustering algorithm developed in this work. In the first dbscan step, noise (blue) and isolated (red) as well as connected (green) localization clouds are distinguished. If the number of localizations in the vicinity of radius eps exceeds minPts, the localizations are assigned to the same localization cloud. The second dbscan step repeats the procedure on all localization clouds identified in the first step. If two regions of high density are found (green and orange), a Gaussian mixture model assigns the localizations to one of the two clouds. (B) Evaluation of the molecular assignment with simulated mitochondrial localizations shown as an unwrapped cylinder. Localizations (blue dots) with spread of are generated from randomly distributed molecule positions (gray crosses) at a surface density of 350 molecules/µm2 on a cylinder with a radius of 200 nm (Left). The number of localizations per molecule follows a Poisson distribution with a mean of 10. Noise localizations at a volume density of 400 localizations/µm3 (red dots) are added. The modified dbscan algorithm assigns localizations (colored dots) to molecules (identically colored crosses) or noise (blue dots) (Right). Clustering parameters eps1 = 10 nm, minPts1 = 4, eps2 = 6 nm, minPts2 = 4, and were chosen. (Scale bar, 100 nm.) (C) Spread of localizations and number of localizations for all identified molecules (histograms) and median of the distributions (dashed lines). (D) Average offset between identified molecules and the nearest-neighbor ground truth molecule position versus molecular density and 3D localization precision (Top Left). Projections along (Bottom) and (Top Right) with mean (blue line and dots) and SD (gray area) of 50 simulations. The same clustering parameters as in B were used. (E) Same as in D, but now showing the percentage of simulated molecules that were not assigned to exactly one cluster.
We optimized and tested this approach by simulating 3D localization clouds for molecules that we randomly distributed on a cylinder with a 200 nm diameter, mimicking the shape of the mitochondrion (Fig. 2B). The number of localizations per molecule was assumed to follow a Poisson distribution with a mean of 10. Applying our dbscan2 clustering algorithm to the randomly generated localizations enabled us to recover the localization precision and the mean number of localizations of the simulated data (Fig. 2C). We quantified the performance of the clustering algorithm versus the localization precision and the molecular surface density (Fig. 2 D and E), as both relate to the potential overlap of the localization clouds. The quantification was facilitated by the knowledge of the ground truth molecule positions, which we had used to simulate the localizations. We analyzed two metrics: 1) the average offset between an identified molecule and the nearest ground truth molecule, which relates assignment error to localization precision (Fig. 2D), and 2) the relative number of ground truth molecules that were assigned to either none or more than one identified molecule, which provided a measure of the misclassified fraction of molecules (Fig. 2E).
Both error metrics showed that the developed algorithm outperformed a standard dbscan clustering approach (SI Appendix, Fig. S3). Our approach achieved mean assignment errors as low as 2.3 nm and a fraction of 6.7% of wrongly assigned molecules for typical MINFLUX localizations with uncertainties of 3 nm and a surface density of 170 molecules/µm2. Both metrics strongly degraded with a decline in localization precision in the simulated data, while the influence of the molecular surface density was less severe. These results show that the fundamentally improved 3D nanometer-scale localization precision of MINFLUX extends the scope and reliability of fluorescence microscopy–based molecular quantifications.
MINFLUX Nanoscopy of Mic60 at the Level of Individual Crista Junctions.
Next, we utilized the dbscan2 clustering algorithm to investigate the distribution of Mic60, the largest subunit of MICOS. Mic60 is a highly conserved membrane protein which is essential for the formation of crista junctions (15, 24–26) and which has been demonstrated to bend membranes in vitro (27, 28). Moreover, Mic60 homologs from the fly Drosophila melanogaster and the fungus Chaetomium thermophilum have been shown to oligomerize (28, 29), giving rise to the idea that Mic60 might form structures which stabilize crista junctions. To investigate the submitochondrial distribution of Mic60, we labeled human bone osteosarcoma (U-2 OS) cells with primary anti-Mic60 antibodies that were conjugated to Alexa Fluor 647 and imaged them by MINFLUX nanoscopy. In the absence of a second fluorophore species, we observed increased signal-to-background ratio values (compared to the dual-color measurements) in conjunction with a superior localization precision of about 3 nm in all dimensions.
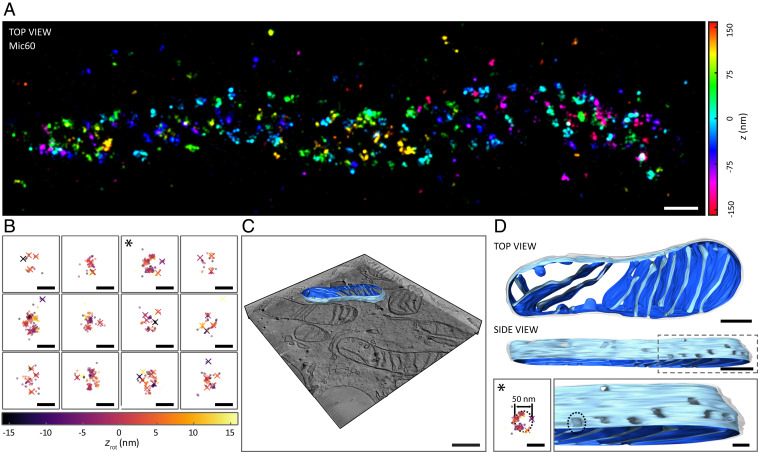
Many of the observed localizations belonged to spatially isolated agglomerations much larger than the localization precision and larger than the size of a single antibody (Fig. 3A and Movie S6). We isolated the agglomerations by applying the dbscan2 algorithm with adapted clustering parameters eps1 = 35 nm, minPts1 = 15, eps2 = 20 nm, minPts2 = 10, and . Following this feature isolation, we used the covariance matrix of the localizations to rotate the localization distributions to the paper plane. A subset of the isolated agglomerations, which contained several identified molecules, showed multiple Mic60 molecules in ring- or ellipsoid-like arrangements with a diameter of ∼40 to 50 nm (Fig. 3B). To further substantiate the visual impression of ringlike Mic60 arrangements, we compared the nearest-neighbor distribution of molecules in the MINFLUX data with simulations of molecules on a cylinder surface assuming three different models for the distribution of Mic60 molecules: 1) random, 2) a ringlike structure with a fixed number of molecules per ring, and 3) random, but preventing the appearance of molecules in circular regions that mimic crista junctions, thus representing a situation where Mic60 is randomly distributed within the IBM. We assumed a tube diameter of 200 nm, a mean number of 18 localizations per molecule, and an average molecular surface density of ∼350 detected molecules/µm2, as extracted from the measured data. The model of a ringlike distribution of molecules showed the best agreement with the experimental results (SI Appendix, Fig. S4). Considering that Mic60 is highly enriched at crista junctions (5, 13), a possible explanation is that the Mic60 arrangements observed with MINFLUX nanoscopy surround individual crista junctions. To scrutinize this explanation, we compared the spatial extent of the molecule arrangements observed with MINFLUX nanoscopy to the diameter of crista junctions using electron microscopy. Electron tomography (ET) acquisitions of the same cell line revealed individual crista junctions exhibiting circular to slit-like shapes with a size of about 20 to 25 nm in diameter (Fig. 3 C and D and Movie S7). When taking the presumed size of the Mic60 proteins as well as the antibody decoration into account, the dimensions of the crista junctions are in good agreement with the diameter of the Mic60 arrangements of ∼40 to 50 nm observed with MINFLUX nanoscopy. Therefore, our data suggest that several molecules of Mic60 are present at individual crista junctions, forming a ringlike arrangement.
Fig. 3.
Distribution of Mic60 in mitochondria of human U-2 OS cells. (A) Three-dimensional MINFLUX image of Mic60 stained with primary labeled antibodies conjugated to Alexa Fluor 647. The axial position of the individual localizations is color-coded. (Scale bar, 200 nm.) (B) Isolated and aligned assemblies of Mic60. The axial position of the localizations (dots) as well as of the estimated molecule positions (crosses, dbscan2 with eps1 = 10 nm, minPts1 = 4, eps2 = 6 nm, minPts2 = 4, and ) after rotation is color-coded. (Scale bar, 50 nm.) (C and D) ET of mitochondria. U-2 OS cells were cut into sections about 230 nm thick in parallel to the growth surface of the cells and were analyzed by electron tomography. (C) Ortho slice view of ET data with one reconstructed mitochondrion. (Scale bar, 500 nm.) (D, Top) Reconstruction of the mitochondrion highlighted in C. The OM is displayed as a transparent surface. The matrix-facing side of the IM is shown in dark blue, and the side that faces the intermembrane space is shown in light blue. (Scale bar, 200 nm.) (D, Bottom) A circular Mic60 assembly taken from B as indicated by an asterisk (Left) next to a magnified side view on individual crista junctions (Right) shown at the same scale. Dashed circles indicate the dimension of the assembly. (Scale bar, 50 nm.)
MINFLUX Nanoscopy Resolves the Distances between Different MICOS Subunits.
Genetic and biochemical data have shown that MICOS consists of a Mic10 and a Mic60 subcomplex. Although the components of the two subcomplexes are well established, the structure and spatial arrangement of the MICOS subunits are unknown. The subunits Mic19 and Mic60 are part of the Mic60 subcomplex that is stable in the absence of the Mic10 subcomplex (10–12, 15, 25). This suggests that in intact mitochondria the subunits Mic19 and Mic60 have, on average, a higher spatial proximity than Mic10 and Mic60. However, because Mic10, Mic19, and Mic60 are part of MICOS and hence in overall close proximity, this prediction is difficult to test with established (superresolution) microscopy. To assess the spatial relationship of Mic60 to Mic19 and of Mic60 to Mic10, we relied on a CRISPR/Cas9 genome-engineered Mic10-TO HeLa cell line, in which we depleted the endogenous Mic10 and inserted the coding sequence of a Mic10-FLAG-T2A-EGFP fusion protein under transcriptional control of a tetracycline-dependent promotor into the adeno-associated virus integration site 1 (AAVS1) (15, 30). Upon expression of Mic10-FLAG-T2A-EGFP, the self-cleaving T2A peptide caused the release of EGFP into the cytoplasm, whereas Mic10-FLAG was imported into the mitochondria. Induction of the expression of the Mic10-FLAG fusion protein fully rescued the wild-type cristae morphology from the aberrant cristae architecture of Mic10-depleted cells, demonstrating that Mic10-FLAG compensates for the lack of endogenous Mic10 (15). We used an anti-FLAG antibody (directly labeled with CF680) to decorate Mic10-FLAG in induced Mic10-TO cells and an anti-Mic60 antibody (directly labeled with Alexa Fluor 647) to label the endogenous Mic60. In a parallel experiment, cells of the same induced cell line were decorated for Mic19 and Mic60, also with directly labeled antibodies.
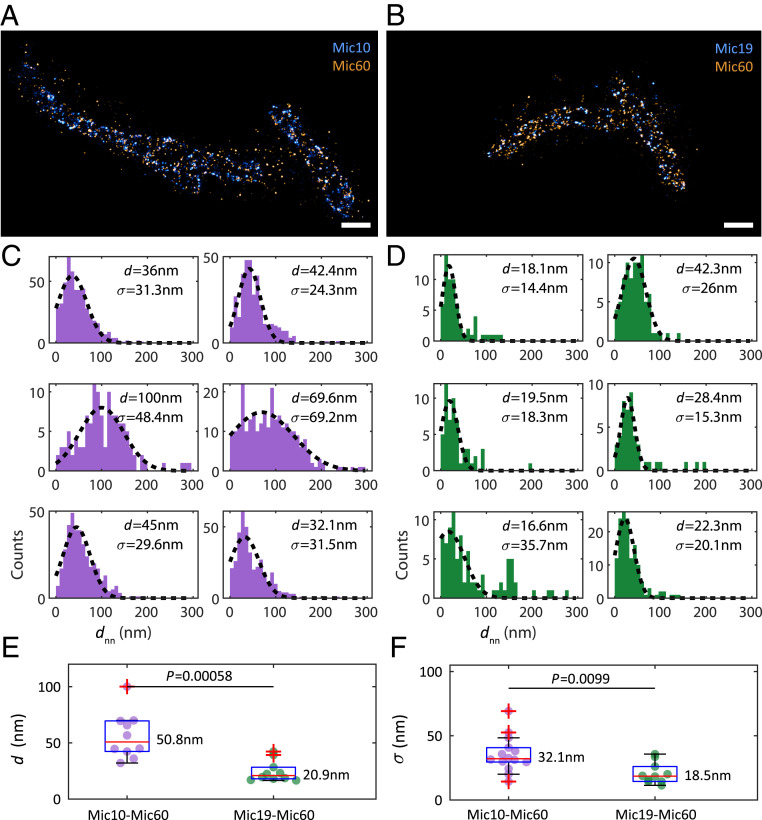
In the cells analyzed for Mic10-FLAG and Mic60, the number of observed molecules was similar in both species (Fig. 4A), whereas this number was lower for Mic19 compared to Mic60 (Fig. 4B). The difference in the number of observed molecules does not necessarily reflect the amount of the target protein present, as the observation also depends on the binding properties of the antibodies. For a quantitative comparison of the distances between Mic60 and Mic10 or Mic19 we assigned localizations to molecules using the dbscan2 algorithm with parameters eps1 = 10 nm, minPts1 = 4, eps2 = 6 nm, minPts2 = 4, and . For each molecule position of the less abundant species in a specific acquisition, we determined the distance to the nearest-neighbor molecule location of the other species (Fig. 4 C and D). Since the distributions roughly followed a Gaussian, we determined the Gaussian mean of the nearest-neighbor distance and the spread of the distances by a Gaussian fit to estimate the average intermolecular distances between Mic10 and Mic60 and between Mic19 and Mic60, as well as the variation of these distances. We excluded datasets from further analysis if the micrometer-sized acquired region contained fewer than 30 molecule positions in one of the imaged species or if the fit quality was low (coefficient of determination or negative mean value). We combined the fit values for all datasets and tested against the hypothesis that the results for the two protein combinations were the outcome of the same distribution using a Mann–Whitney U test (Matlab function ranksum). Analyzing the distribution of the Gaussian mean of nearest-neighbor distances across all measurements revealed that in the median, Mic19 was closer to Mic60 than Mic10 was to Mic60 (P = 0.00058) (Fig. 4E). Moreover, Mic10–Mic60 showed larger distance spread values compared to Mic19–Mic60 (P = 0.0099) (Fig. 4F). Taken together, this indicates that the relative localization of Mic10 to Mic60 is rather variable, whereas Mic19 and Mic60 are in a closer and more defined spatial relationship, fully in line with the finding that Mic19 and Mic60 are part of the same MICOS subcomplex (15, 25, 31).
Fig. 4.
Measurement of distances between proteins of the MICOS complex. (A) Representative 3D two-color MINFLUX acquisition of Mic10 (CF680, blue) and Mic60 (Alexa Fluor 647, orange) in Mic10-TO HeLa cells. (Scale bar, 500 nm.) (B) Representative 3D two-color MINFLUX acquisition of Mic19 (CF680, blue) and Mic60 (Alexa Fluor 647, orange) in Mic10-TO HeLa cells. (Scale bar, 500 nm.) (C) Histograms of interspecies nearest-neighbor distances between Mic10 and Mic60 for a selection of MINFLUX acquisitions. A Gaussian fit (dashed line) determines Gaussian mean distance and spread of distances . (D) Same as in C, but now for Mic19 and Mic60. (E) Gaussian mean of the nearest-neighbor distances measuring the average distance between Mic10/Mic19 and Mic60 molecules for all acquisitions for Mic10–Mic60 (purple, n = 11) and Mic19–Mic60 (green, n = 10) with 25 and 75% percentiles (blue box), the median value (red line and number), extreme values (error bar), and outliers (red crosses). A Mann–Whitney U significance test delivered a probability of P for the mean distances to result from the same distribution. (F) Same as in E, but now for the distance spreads, measuring the variation in the distance between Mic10/Mic19 and Mic60.
Discussion
This study shows that multicolor 3D MINFLUX nanoscopy is a powerful tool for imaging dense structures in complex environments like mitochondria with nanometer isotropic precision. Compared to a recent 4Pi-STORM nanoscopy study using two opposing high-aperture lenses, MINFLUX imaging achieved slightly better precision in mitochondrial imaging using just a single lens (32), which simplifies the alignment of the microscope and enhances its stability. The high and isotropic localization precision of MINFLUX extends the possibilities for drawing quantitative conclusions from microscopy data. Reassigning localization clouds to individual molecules simplifies protein counting as well as the quantitative interpretation of spatial distributions of protein arrangements.
Previous studies demonstrated that the MICOS core component Mic60 is highly enriched at crista junctions (5, 13). The MINFLUX images of Mic60 recorded in this study suggest the formation of ringlike arrangements of several Mic60 proteins around single crista junctions. Indeed, the observed heterogeneity and the measured typical diameter of the Mic60 arrangements are in accordance with the size and heterogeneity of the crista junctions, as determined by electron tomography. Ringlike arrangements of Mic60 at crista junctions would be in line with previous findings, including the observation that MICOS is a large protein complex with a molecular weight of over 1 MDa (33, 34), the observation that Mic60 is essential for crista junction stability (4–6, 15, 24), and the fact that Mic60 can bend membranes into tubular structures (27, 28). Based on this observation, we propose that the Mic60 subcomplexes are part of a pore-like structure, which stabilizes the crista junctions.
By quantitatively evaluating two-color 3D MINFLUX acquisitions, we observed that Mic60 and Mic19 were usually in close proximity to each other, whereas Mic10 and Mic60 exhibited a less defined spatial relationship (SI Appendix, Fig. S5). The observation that the localization of Mic10 is rather variable with respect to the localization of Mic60 points to substantial structural heterogeneity in the MICOS complex in mitochondria. This heterogeneity might be a consequence of the function of MICOS to control the size and distributions of crista junctions within the organelles (15). The structural flexibility of the MICOS complex should be considered in future strategies aiming at solving the stoichiometry and the atomic structure of this complex.
MINFLUX imaging allowed us to measure distances of a few nanometers as well as protein pairs several hundred nanometers apart, thus covering a dynamic range currently inaccessible to any other technique. While the interaction range of molecule pairs employed for fluorescence resonance energy transfer is usually limited to below 10 nm (35), other superresolution microscopy approaches access proteins farther apart but usually fail to resolve the 3D distances on the scale of a few nanometers in cells. We predict that future live-cell applications of MINFLUX will facilitate the analysis of the movement of proteins across these scales.
The actual limitation in accessing the exact geometry and stoichiometry of the MICOS protein distribution is the antibody-labeling approach employed, rather than the imaging technique. An insufficient labeling efficiency, possibly due to steric hindrance, renders the observed positions incomplete. Moreover, the labeling stoichiometry and the exact offset between the fluorescent reporter and the target protein (up to about 15 nm for directly labeled primary antibodies) (18) are unknown. More quantitative and accurate labeling approaches, such as Fab fragments, nanobodies, and self-labeling tags (18), will augment the power of MINFLUX nanoscopy for biological studies at the single-protein level.
Another challenge is to localize MICOS proteins in relation to the folding of the inner membrane, as most commonly used inner-membrane-specific dyes are not compatible with MINFLUX (or any stochastic) imaging. Possible solutions include the correlative acquisition of MINFLUX and electron tomography data as well as the development of novel fluorescent molecules for membrane labeling that if optimized for stochastic nanoscopy, may even allow us to distinguish the inner from the outer mitochondrial membrane in MINFLUX recordings.
The exploration of alternative fluorescent molecules with a lower photon budget but improved control of the photophysical states can additionally reduce background contributions and thus further improve the MINFLUX imaging performance. Controlled on and off switching of the emitters in combination with the low photon requirements of MINFLUX localizations can reduce molecular registration times and strongly speed up the imaging. Unlike widefield-based schemes like 4Pi-STORM nanoscopy, the current point-scanning MINFLUX acquisitions are practically restricted to regions of a few micrometers in extent because the acquisition time scales in proportion to the imaging region. Observing micrometer-sized structures can entail tens of minutes of recording time. A potential technical remedy is to develop parallelized recording schemes that are capable of addressing the molecules individually. Furthermore, moving from synthetic fluorescent molecules to photoswitchable or photoconvertible proteins not only promises a stoichiometric and spatially more accurate labeling but also has been shown to facilitate MINFLUX imaging and tracking in living cells (16, 19). Live-cell acquisitions exclude fixation artifacts and facilitate tracking of the molecules of interest over time. As the IM changes its appearance dynamically on the timescale of seconds (36, 37) and inner membrane remodeling processes are influenced by membrane-shaping proteins, including components of the MICOS complex (15), a nanometer-precision dynamic observation of MICOS proteins with 3D MINFLUX will help to shed light on the exact underlying processes.
In conclusion, we show that the single-digit nanometer resolution provided by MINFLUX nanoscopy enables the 3D mapping of the position of densely packed fluorophores with molecular precision. In conjunction with mathematical analysis, MINFLUX nanoscopy thus promises to open a new era in the stoichiometric and spatial analysis of small arrangements of proteins in (living) cells.
Materials and Methods
A detailed description of the materials and experimental methods, including the sample preparation procedures, a description of the MINFLUX microscope, the MINFLUX data acquisition, and the MINFLUX data analysis steps, and the electron microscopy protocol are provided in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ellen Rothermel for excellent technical assistance, Klaus Gwosch for providing additional expertise on the microscope software, and Peter Ilgen for help with 3D animations. This work was supported by the German Federal Ministry of Education and Research within Project 13N14122, “New fluorescence labels for protected- and multi-color-STED microscopy (STEDlabel)” (to S.W.H.), by European Research Council Advanced Grant 835102 (to S.J.), by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy with grant no. EXC 2067/1- 390729940 (to S.J. and S.W.H.), and by Sonderforschungsbereich 1190 Project P01 (to S.J.).
Footnotes
Competing interest statement: The Max Planck Society holds patents on selected embodiments and procedures of MINFLUX, benefitting S.W.H. and F.B.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009364117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Neupert W., SnapShot: Mitochondrial architecture. Cell 149, 722–722.e1 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Frey T. G., Mannella C. A., The internal structure of mitochondria. Trends Biochem. Sci. 25, 319–324 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Pfanner N. et al., Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol. 204, 1083–1086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoppins S. et al., A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J. Cell Biol. 195, 323–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harner M. et al., The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von der Malsburg K. et al., Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21, 694–707 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Alkhaja A. K. et al., MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell 23, 247–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guarani V. et al., QIL1 mutation causes MICOS disassembly and early onset fatal mitochondrial encephalopathy with liver disease. eLife 5, e17163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khosravi S., Harner M. E., The MICOS complex, a structural element of mitochondria with versatile functions. Biol. Chem. 401, 765–778 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Guarani V. et al., QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife 4, e06265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand R., Strecker V., Urbach J., Wittig I., Reichert A. S., Mic13 is essential for formation of crista junctions in mammalian cells. PLoS One 11, e0160258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman J. R., Mourier A., Yamada J., McCaffery J. M., Nunnari J., MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife 4, e07739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jans D. C. et al., STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl. Acad. Sci. U.S.A. 110, 8936–8941 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoldt S. et al., Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proc. Natl. Acad. Sci. U.S.A. 116, 9853–9858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephan T. et al., MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 39, e104105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balzarotti F. et al., Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Hell S. W., Far-field optical nanoscopy. Science 316, 1153–1158 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Sahl S. J., Hell S. W., Jakobs S., Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Gwosch K. C. et al., MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 17, 217–224 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Hell S. W., Microscopy and its focal switch. Nat. Methods 6, 24–32 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Kühlbrandt W., Structure and mechanisms of F-type ATP synthases. Annu. Rev. Biochem. 88, 515–549 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Nicovich P. R., Owen D. M., Gaus K., Turning single-molecule localization microscopy into a quantitative bioanalytical tool. Nat. Protoc. 12, 453–460 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Ester M., Kriegel H.-P., Sander J., Xu X., “A density-based algorithm for discovering clusters in large spatial databases with noise” in Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining, (AAAI Press, Menlo Park, CA, 1996), pp. 226–231. [Google Scholar]
- 24.Rabl R. et al., Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047–1063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondadi A. K. et al., Cristae undergo continuous cycles of membrane remodelling in a MICOS-dependent manner. EMBO Rep. 21, e49776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John G. B. et al., The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543–1554 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarasenko D. et al., The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J. Cell Biol. 216, 889–899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hessenberger M. et al., Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat. Commun. 8, 15258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai P.-I. et al., PINK1 phosphorylates MIC60/mitofilin to control structural plasticity of mitochondrial crista junctions. Mol. Cell 69, 744–756.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Hockemeyer D. et al., Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Laan M., Horvath S. E., Pfanner N., Mitochondrial contact site and cristae organizing system. Curr. Opin. Cell Biol. 41, 33–42 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y. et al., Nanoscale subcellular architecture revealed by multicolor three-dimensional salvaged fluorescence imaging. Nat. Methods 17, 225–231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harner M. E. et al., Aim24 and MICOS modulate respiratory function, tafazzin-related cardiolipin modification and mitochondrial architecture. eLife 3, e01684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott C. et al., Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol. 32, 1173–1188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy R., Hohng S., Ha T., A practical guide to single-molecule FRET. Nat. Methods 5, 507–516 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephan T., Roesch A., Riedel D., Jakobs S., Live-cell STED nanoscopy of mitochondrial cristae. Sci. Rep. 9, 12419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C. et al., A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 116, 15817–15822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.