Significance
Activation of hydrogen in biology is achieved by enzymes rivalling the platinum metals in catalytic activity. Attached to an electrode, hydrogenases display electrocatalytic reversibility—just the tiniest potential bias from the equilibrium value drives oxidation or production of H2 at significant rates. Hydrogenases activate H2 heterolytically; hence, reversibility, an extreme marker for evolved efficiency, is expected to depend on electron and proton transfer processes occurring in concert. Experiments in which the long-range proton-transfer pathway in two [FeFe]-hydrogenases has been mildly disrupted without affecting the electron transfer pathway show that electrocatalytic reversibility fades as electron and proton transfers become temporally decoupled. These subtlest of observations demonstrate how evolution responded to the need for concerted electron–proton transfer in optimizing catalytic efficiency.
Keywords: hydrogenase, electrocatalysis, redox enzymes, catalyst, proton-coupled electron transfer
Abstract
As paradigms for proton-coupled electron transfer in enzymes and benchmarks for a fully renewable H2 technology, [FeFe]-hydrogenases behave as highly reversible electrocatalysts when immobilized on an electrode, operating in both catalytic directions with minimal overpotential requirement. Using the [FeFe]-hydrogenases from Clostridium pasteurianum (CpI) and Chlamydomonas reinhardtii (CrHydA1) we have conducted site-directed mutagenesis and protein film electrochemistry to determine how efficient catalysis depends on the long-range coupling of electron and proton transfer steps. Importantly, the electron and proton transfer pathways in [FeFe]-hydrogenases are well separated from each other in space. Variants with conservative substitutions (glutamate to aspartate) in either of two positions in the proton-transfer pathway retain significant activity and reveal the consequences of slowing down proton transfer for both catalytic directions over a wide range of pH and potential values. Proton reduction in the variants is impaired mainly by limiting the turnover rate, which drops sharply as the pH is raised, showing that proton capture from bulk solvent becomes critical. In contrast, hydrogen oxidation is affected in two ways: by limiting the turnover rate and by a large overpotential requirement that increases as the pH is raised, consistent with the accumulation of a reduced and protonated intermediate. A unique observation having fundamental significance is made under conditions where the variants still retain sufficient catalytic activity in both directions: An inflection appears as the catalytic current switches direction at the 2H+/H2 thermodynamic potential, clearly signaling a departure from electrocatalytic reversibility as electron and proton transfers begin to be decoupled.
The [FeFe]-hydrogenases are the most efficient H2-producing catalysts found in the natural world (1–4). The active site at which H2 is oxidized or produced (known as the “H-cluster”) consists of a binuclear Fe complex (the [2FeH]-site) that is covalently linked to a [4Fe-4S]-cluster (the [4FeH]-subsite) through a shared cysteine S-atom (5–8). The two Fe atoms of the [2FeH]-site are differentiated as being proximal (Fep) and distal (Fed) according to their distance from the [4FeH]-cluster. The [2FeH]-site is further coordinated and stabilized by two CN− and three CO ligands and cross-bridged by an azadiothiolate (adt) ligand, with its central N-bridgehead overhanging Fed (Fig. 1) (9–12). The catalytic properties of the active site itself (often exceeding turnover frequencies of over 10,000 s−1) (13, 14) has led to numerous attempts by synthetic and computational chemists to mimic these extraordinary benchmarks (10, 15–20). For the native enzyme, the high turnover rates are ensured by the protein environment which provides two very distinct supply channels for electrons and protons. Electrons are transferred via accessory [FeS]-clusters (present in all [FeFe]-hydrogenases except for those from green algae) that are organized in sequence to facilitate long-range transport between the [4FeH]-subsite of the H-cluster and an external electron donor/acceptor (21). On the other hand, a long-range proton-transfer pathway (PTP) is provided by a highly conserved sequence of five amino acid side chains and a protein-bound water molecule (21–25). The proton- and electron-transfer pathways for [FeFe]-hydrogenase I (CpI) from Clostridium pasteurianum are shown in Fig. 1, together with details of the two CpI variants investigated in this study. As we stress later, it is important to note that the two pathways approach the H-cluster from opposite directions. The [FeFe]-hydrogenase CrHydA1 from the green alga Chlamydomonas reinhardtii lacks any additional [FeS]-clusters but, analogous to CpI, the inferred proton-transfer route is positioned away from the surface-exposed [4FeH]-subsite of the H-cluster, which in the algal hydrogenase represents the end of the electron-transfer path (23).
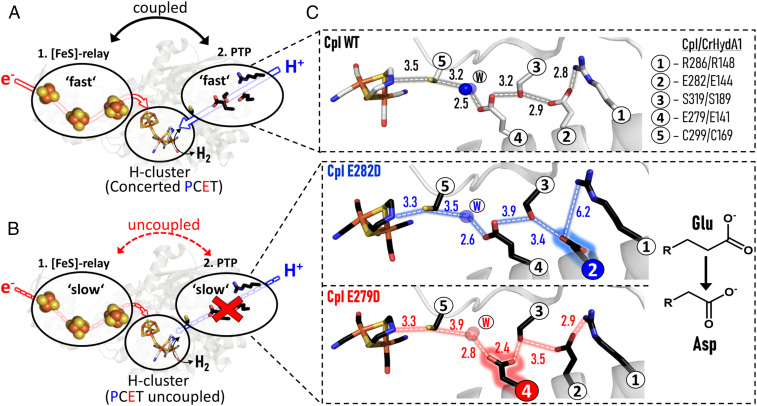
Fig. 1.
Schematic representation for electron and proton supply channels in the form of [FeS]-relay ET and PT pathways, respectively, based on the structure of CpI. As is shown in this paper, in WT enzyme both ET and PT pathways are strongly coupled, thus ensuring reversible catalysis at the H-cluster (A). If the PTP is impeded (B), ET is also impeded as PT is uncoupled from ET at the H-cluster. (C) Close-up of the PT pathway for CpI WT and exchange variants E282DCpI and E279DCpI (PDB ID codes 4XDC, 6GM2, and 6YF4, respectively). The H-bonding networks are represented by dashed lines and distances are indicated in angstroms. Color code for atoms: gray/black: carbon; red: oxygen; blue: nitrogen; yellow: sulfur; orange: iron.
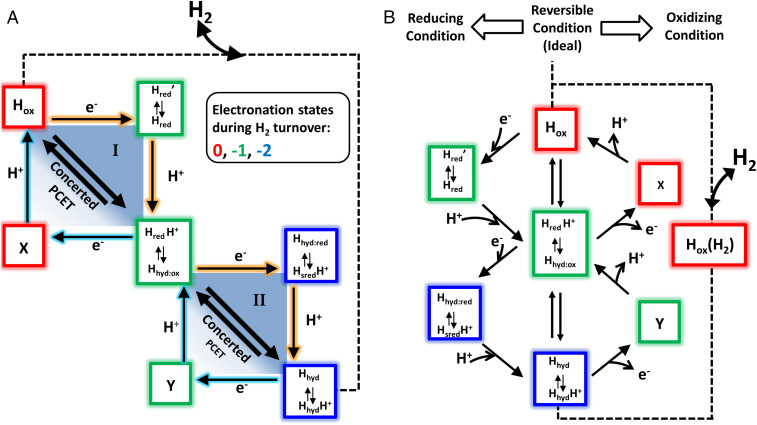
The exquisite catalytic performance of [FeFe]-hydrogenases is otherwise entirely restricted to the platinum metals (being the basis of the reversible hydrogen electrode) (26). The impossibility of developing a global hydrogen economy on such limited metal resources has stimulated research into alternative catalysts, hence the great interest in hydrogenases as examples (27). Proton-electron/hydrogen interconversion is the simplest of chemical reactions, and the underlying reason for the extraordinary catalytic activity of hydrogenases very likely lies in their ability, by virtue of their highly evolved atomic structures, to couple electron and proton transfers so tightly that, ideally, each electron transfer (ET) occurs in concert with a proton transfer (PT), see Fig. 2. There are many important theoretical papers on proton-coupled electron transfer (PCET), the overall picture being one where a concerted reaction has a significantly lower activation barrier (Fig. 2A) (28, 29).
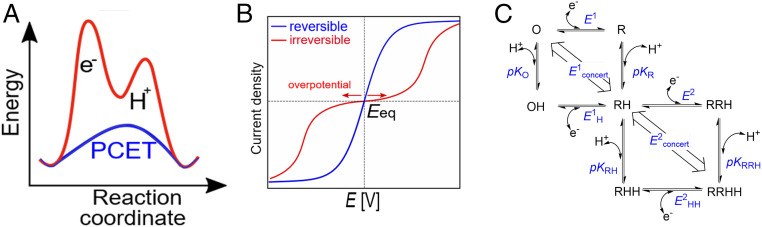
Fig. 2.
(A) General reaction scheme, showing the energetic advantage of PCET compared to uncoupled proton and ET steps. (B) Ideal waveshapes in cyclic voltammetry that are proposed to describe reversible (blue) and irreversible (red) electrocatalysts (adapted from ref. 30). (C) Square scheme in which horizontal sides are redox couples defined by electron-only reduction potentials and the diagonal lines are the redox couples defined by the reduction potential for concerted PCET. O/R: oxidized/reduced state. E1/E1H: reduction potential of the unprotonated/protonated state. E2/E2HH: reduction potential of the reduced, onefold protonated/reduced, twofold protonated state. pKO/pKR: pK of the oxidized/reduced state. pKRH/pKRRH: pK of the onefold protonated onefold/twofold reduced state.
The catalytic characteristics of hydrogenases and an increasing number of other “energy-processing” electron-transport enzymes are revealed in electrochemical experiments (protein film electrochemistry, PFE) in which the enzyme is attached directly to a suitable electrode surface (30–33). The shape of an electrocatalytic voltammogram often conveys otherwise intractable mechanistic information, as it is always possible to drive the reaction in at least one direction by applying an appropriate electrode potential. An outcome of extensive studies is that hydrogenases, with few exceptions, behave as reversible electrocatalysts, that is, they catalyze net H2 oxidation and evolution with the tiniest of overpotentials, the current (reflecting the turnover rate) cutting sharply across the zero current axis at the equilibrium potential (blue trace in Fig. 2B) (30). This reversibility, a supreme marker of efficiency, requires that high-energy intermediates or transition states are avoided, and it is associated with concerted PCET (34). Hand in hand with reversibility goes bidirectionality, often termed more quantitatively as the catalytic bias, the question addressed being that once a practical oxidizing or reducing overpotential is applied, to what degree is catalysis in one direction favored over the other (12, 26, 35–38)? Bidirectionality and reversibility should not be confused. Information on bidirectionality can be obtained from conventional steady-state solution kinetics, for example by comparing the rate of H2 oxidation vs. that of H2 evolution using different electron acceptors or donors. In contrast, the subtle degree to which a catalyst approaches reversible behavior can only be sensed and measured by dynamic electrochemistry.
For an otherwise reversible electrocatalytic system, increased decoupling of PCET should result in the emergence of an inflection (a “twist” in an otherwise regular line) at the zero-current potential as an overpotential requirement emerges in each direction. To see why the inflection appears and why oxidation and reduction activities may eventually commence at very different potentials, it is useful to consider the catalytic pathway around a square scheme that connects all possible states of a PCET process (34, 39–41). Fig. 2C shows two square schemes that are connected to represent the two consecutive 1e−/1H+ transfers that are required, stoichiometrically, for catalytic H2 oxidation or H2 production. The two squares represent a complete minimal pathway map, but at this stage, for didactic simplicity, we need only consider the behavior of one square representing the transfer of a single 1e−/1H+ pair. The two pK values of the upper square refer to oxidized (O) and one-electron reduced (R) states, and the two reduction potentials E1 and E1H relate to the couples that involve only nonprotonated and only protonated forms, respectively. At equilibrium (at the zero-current potential) the free energy changes for the four sides must sum to zero, and since pKR > pKO (as R will be a stronger base) it follows that E1 < E1H.
Under the condition that the experimental pH is below pKR (favoring protonation of R) but above pKO (favoring deprotonated O), slow PT kinetics isolate the two individual electron-only reduction potentials E1 and E1H; in contrast, fast PT occurring in concert with ET is described by the single diagonal with potential E1concert (34, 39, 40). Consequently, if reversible electrocatalysis depends on PCET’s being concerted, decoupling due to slowing down PT should result in a widening inflection, as reductive catalysis becomes controlled by E if protonation lags behind ET, and oxidative catalysis becomes controlled by E1H if the proton is trapped at the active site (39, 40). Adding or removing an electron by applying a sufficiently large overpotential helps drive the subsequent PT by rendering the process more exergonic. In each direction the expected exponential current–potential dependence eventually yields a “hockey-stick”-shaped trace which finally approaches a limiting current that depends on the rate of a step that is entirely chemical in nature, that is, for which the electrochemical driving force is no longer relevant. For simplicity, the plots shown in Fig. 2B have equal limiting currents in each direction, and it is assumed that the limiting rate is not due to a solution-based mass transport restriction such as diffusion of H2 molecules. The resulting bidirectional but irreversible electrocatalysis is represented by the red trace. The single redox wave corresponding to reversible electrocatalysis in either direction along the E1concert diagonal has been replaced by two separate waves ultimately associated with E1 and E1H. Put another way, if PT is slow, catalytic electron–proton flow will be stepwise—transfer of the electron, defined by E1 or E1H, will drive subsequent transfer of the proton. If PT is fast, concerted PCET will occur in both directions at the reversible value E1concert. In summary, the performance of each arm of the voltammogram, oxidation and reduction, is defined by an overpotential requirement at either side of the reversible potential value and by a limiting turnover rate that does not depend on ET.
For hydrogenases the single square corresponds to 50% of the catalytic cycle: Extension to include the second square does not alter the outcome as the rule remains the same, that is, at equilibrium (the zero-current potential) the free energy changes should sum to zero irrespective of pathway. The two connected squares represent the minimal electrochemical pathways linking H2, electrons, and protons, although alternative forms of each electronation/protonation state may exist and might ultimately be needed to describe the complete mechanisms for catalytic cycles. The well-known principle of microscopic reversibility dictates that for any given applied potential the same distribution of pathways must be taken in each direction.
Experimentally, it is possible to gauge the importance of proton–electron coupling by examining how systematic decoupling affects the overpotential requirement. An enzyme behaving as a reversible electrocatalyst represents a perfect starting point, as specific sites widely spaced across the macromolecule can be manipulated by site-directed mutagenesis and structures verified by X-ray crystallography. The fact that PT and ET pathways in [FeFe]-hydrogenases approach the active site in opposite directions offers a special opportunity to test the mechanistic details of remote coupling between PTs and ETs; this is achieved by examining how site-directed mutagenesis on the PTP alters the shape of the resulting electrocatalytic voltammograms while making sure that the structure of the active site and the ET pathway remain unchanged. We therefore chose conservative amino acid substitutions within the proposed catalytic PTP that have an overall minor effect on enzymatic activity, thereby still allowing electrocatalysis to be measured easily under steady-state conditions. The glutamate → aspartate substitution conserves the functional carboxylic group while shortening the residue by one methylene (-CH2-) group.
Results
Measured using methyl viologen as electron donor, exchange variants E282DCpI and E279DCpI (sites 2 and 4 in Fig. 1C) retain 53% and 22% of the in vitro H2 production activity measured for wild-type (WT) enzyme at pH 7 (SI Appendix, Fig. S1B). Protein crystallography further confirmed that the conservative amino acid substitutions leave the [FeS]-relay system and other parts of the protein structurally unaffected [see Protein Data Bank (PDB) ID codes 6GM2 for E282DCpI (25)] and 6YF4 for E279DCpI from this paper (SI Appendix, Fig. S2) compared to 4XDC for CpI WT. In contrast, there are significant increases in the H-bonding distances within the PTP network (up to 3.4 Å) which are expected to produce lower rates of PT (Fig. 1C). For CrHydA1, which is well studied kinetically but less characterized structurally (42–46), we produced the equivalent exchange variants E144DHydA1 (= E282DCpI) and E141DHydA1 (= E279DCpI) which retain 54% and 2.5% of the H2 production activity measured for WT, respectively, under the same test conditions as used for CpI (SI Appendix, Fig. S1A).
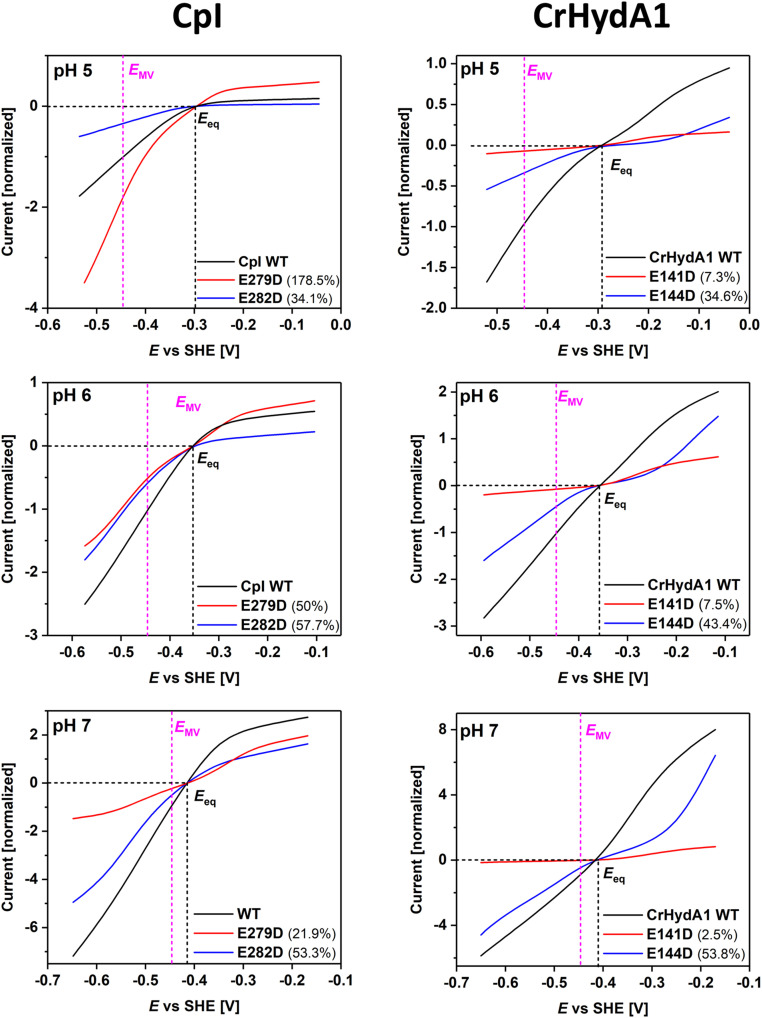
For each enzyme, a film was prepared on a rotating disk pyrolytic graphite “edge” electrode according to a well-established procedure (14, 47). Electrocatalytic voltammograms, measured in cyclic form to confirm steady-state behavior (i.e., free of hysteresis) and initiated from the negative potential limit, were recorded under 1 atmosphere of H2 over the pH range 5 to 9. Unnormalized, representative results were adjusted to set zero net current values at the appropriate pH-adjusted potentials to compensate for small instrumental offsets (SI Appendix, Fig. S3). PFE is generally not useful for obtaining absolute rates because electroactive coverage is usually unknown and variable. However, the shape of each voltammogram (the continuous current–potential dependence across the entire potential range) provides an immediate and unique visualization of bidirectionality and reversibility that is impossible to obtain by comparing separate H2 oxidation or evolution rates obtained with molecular redox partners in solution. Unlike most [NiFe]-hydrogenases, H+ reduction by [FeFe]-hydrogenases is much less inhibited by H2 (31, 48), thus allowing for a comparison between both catalytic directions under a 100% H2 atmosphere. Two procedures were adopted in order to elevate the qualitative comparisons to a more quantitative level. First, the data in SI Appendix, Fig. S3 were normalized to the current obtained at a H2 oxidation overpotential of 0.25V, an arbitrary reference point but helpful as it enabled the shapes to be compared throughout the entire pH range of 5 to 9 (Figs. 3 and 4). Note that normalization magnifies the capacitance component when catalytic activity is low. Second, the data were scaled with respect to the H2 evolution activities determined from solution assays using reduced methyl viologen (−0.446 V with negligible pH dependence) as electron donor (Fig. 5); this allowed us to estimate how the actual activity varies with potential based on the pH-dependent methyl viologen-dependent H2 evolution activities for WT enzyme and the exchange variants (SI Appendix, Fig. S1) but was restricted to the range pH 5 to 7 over which methyl viologen is thermodynamically competent to support H2 production (49).
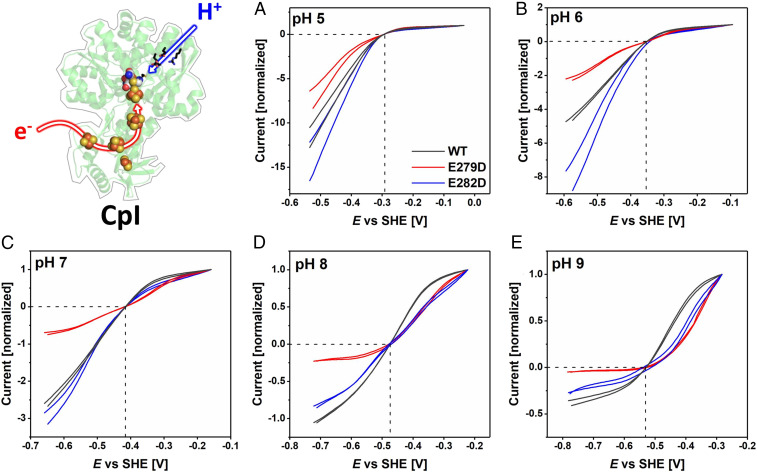
Fig. 3.
Comparisons of electrocatalytic H2 oxidation and H+ reduction for WT CpI and variants E279DCpI (red) and E282DCpI (blue) from pH 5 to pH 9 (A–E, respectively), presented after normalizing to the H2 oxidation current recorded at an H2 oxidation overpotential of 0.25 V. Note that relative current densities do not reflect absolute enzyme activities. The crossing point of the dashed lines represents the 2H+/H2 thermodynamic potential. Experimental conditions: temperature = 20 °C, 3,000 rpm electrode rotation, 5 mV/s scan rate, measured in a mixed buffer system.
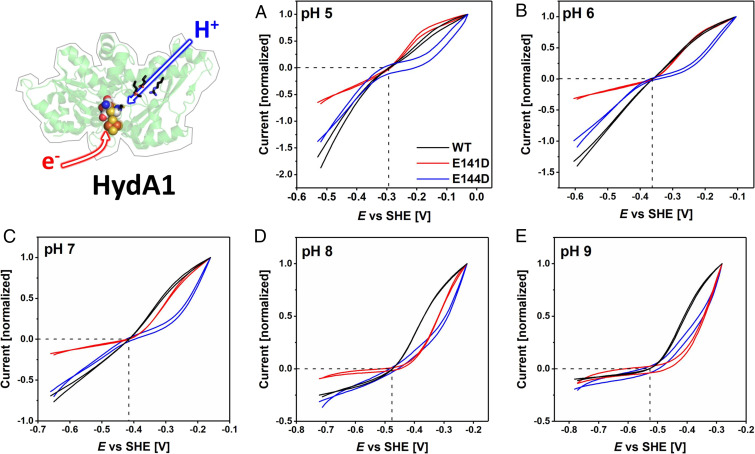
Fig. 4.
Comparisons of electrocatalytic H2 oxidation and H+ reduction for WT CrHydA1 and variants E141DCrHydA1 (red) and E144DCrHydA1 (blue) from pH 5 to pH 9 (A–E, respectively), presented after normalizing to the H2 oxidation current recorded at an H2 oxidation overpotential of 0.25 V. Note that relative current densities do not reflect absolute enzyme activities. The crossing point of the dashed lines represents the 2H+/H2 thermodynamic potential. Experimental conditions as for Fig. 3.
Fig. 5.
Comparisons of electrocatalytic voltammograms for WT CpI and CrHydA1 and their variants over the pH range 5 to 7 after scaling the currents at −0.446 V to the relative activities measured for H2 evolution using reduced methyl viologen as electron donor (SI Appendix, Fig. S1). Each trace is the average of the forward and reverse scans. The numbers shown in brackets refer to residual activities for inner and outer E → D exchange variants relative to WT enzyme. The crossing point of the dashed lines represents the 2H+/H2 thermodynamic potential (black) and the MV thermodynamic potential (magenta), respectively.
Effects of PT Pathway Disruption on Catalysis in the Direction of H+ Reduction.
The most obvious observation made for H+ reduction is that under more acidic conditions the current increases as the electrode potential is made more reducing, without any well-defined hockey-stick dependence or leveling off. Relative to WT enzymes and measured at a substantial driving force (an applied overpotential of 0.25 V), the activities of all variants decrease more sharply as the pH is raised; this was already noted from conventional solution kinetics (SI Appendix, Fig. S1) where the data were used for scaling in Fig. 5, and the observation could now be extended up to pH 9 (Figs. 3 and 4). The retardation in current evolution with increasing pH is particularly marked for E279DCpI and E141DCrHydA1 in which the innermost of the two glutamates has been exchanged for aspartate. The overall attenuation of the otherwise high turnover rate as the pH is raised reflects the decreased availability of protons in the solvent; importantly, the variants are less able to capture protons, the sole oxidizing substrate, for transfer to the active site.
Effects of PT Pathway Disruption on Catalysis in the Direction of H2 Oxidation.
Turning to H2 oxidation, experiments in which the oxidizing potential limit was extended to +0.20 V confirmed that both E141DCrHydA1 and E144DCrHydA1 undergo anaerobic oxidative inactivation similar to CrHydA1 WT enzyme, albeit showing small differences when anaerobic inactivation occurs (SI Appendix, Figs. S4 and S5). Under a 100% H2 atmosphere, the voltammograms of all variants reveal large departures from the reversible curve observed for the WT enzymes at the same pH values. First, the H2 oxidation profiles of all variants transform to a hockey-stick dependence as the pH is increased from 5 to 9 (Figs. 3 and 4). The traces for E144DCrHydA1 are markedly affected across the entire pH range, whereas those for E141DCrHydA1 only deviate above pH 7. The absolute activities of the corresponding variants of the two hydrogenases differ significantly. Whereas at pH 5 E279DCpI is even more active than WT CpI, a large deviation becomes visible at pH = 7. Second, relative to WT enzyme, the current obtained once a substantial overpotential has been applied decreases much more as the pH is raised. The H2 oxidation kinetics for the variants thus become more sluggish, despite the expectation that PT from active site to solvent should be more favored as the pH is increased.
Effects of PT Pathway Disruption on Reversibility.
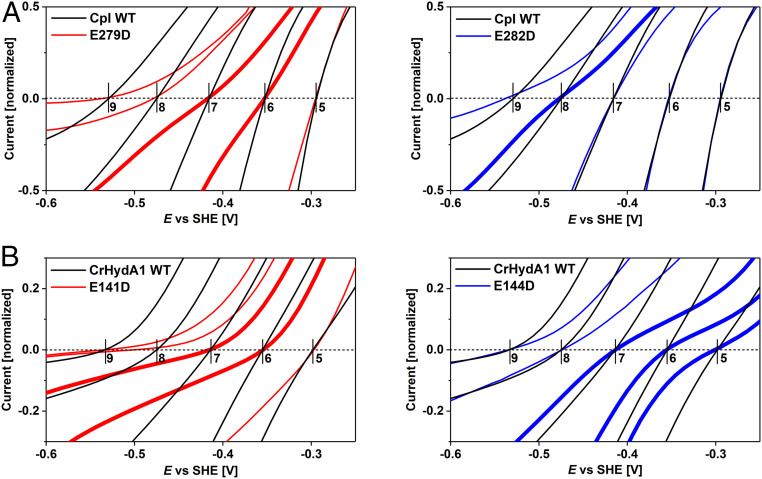
Under conditions where electrocatalysis remains clearly bidirectional, close inspection of the region of voltammograms near the equilibrium potential shows that the “clean cut” that the current normally makes through the zero-current axis is replaced by an inflection. As predicted earlier, the appearance of such an inflection signals even the slightest increase of overpotential requirement in both directions. Experiments with WT CrHydA1 normally reveal a very small inflection that has been attributed to the lack of an internal [FeS]-relay, which is present in other [FeFe]-hydrogenases to mediate electron exchange between active site and external redox partners or in this case the electrode surface (14, 26, 50). With the variants, however, the appearance of the inflection is much more profound. Fig. 6 shows expanded views of each of the voltammograms in the region of the equilibrium potential, and our attention is drawn to the results where the overall H+ reduction rate is not sufficiently suppressed to flatten the curve and obscure detail. The unmistakable “twist” in the current–potential dependence that could only originate from a disruption of the PTP provides compelling evidence that electrocatalytic reversibility requires PT and ET to occur in concert. Of particular note are the inflections observed for E279DCpI at pH 6 and 7, E282DCpI at pH 8, E141DCrHydA1 at pH 6 and 7, and E144DCrHydA1 at pH 5 to 7.
Fig. 6.
Close-up views on electrocatalytic voltammograms in the region of the reversible potential for CpI (A) and CrHydA1 (B). Traces showing the most significant inflections are highlighted by a bold line, taken from data shown in Figs. 3 and 4.
Discussion
Fourier-transform infrared (FTIR) spectroscopic evidence has been used to demonstrate that PCET is a key requirement for rapid H2 turnover by [FeFe]-hydrogenases, but the interpretation has been limited to electronic and structural features of the H-cluster (51–53). For structurally characterized CpI, the amino acid residues substituted here are well separated from the active site (∼10 Å for E279CpI and 16 Å for E282CpI) and exhibit an even longer distance to the [FeS]-relay system (∼25 to 40 Å for E279CpI and ∼29 to 45 Å for E282CpI) (SI Appendix, Fig. S6). In a recent paper in which the protonation states of several participating PT pathway residues of CrHydA1 were assigned via attenuated total reflection FTIR spectroscopy for Hox and Hred (or HredH+) states, Senger et al. (54) proposed that the PT pathway regulates both entry and exit of protons at the active site. Efficient operation of the entire PT pathway should be particularly relevant under physiological conditions, that is, between pH 7 and 8. The electrochemical results now reveal how disruptions to the PT pathway affect both the catalytic bias (bidirectionality) and the reversibility of ET.
Regarding catalytic bias, a suppression of rate in either direction could be strictly chemical in origin without requiring any step that is sensitive to the applied potential. Such rate-limiting chemical effects have already been shown to affect and tune the catalytic bias of Desulfovibrio fructosovorans (Df) [NiFe]-hydrogenase in which H2 diffusion to and/or from the active was manipulated by narrowing the gas diffusion channel (55–57). In our case, the lower limiting rate may signal a decreased ability to capture or transport solvent protons for H2 evolution, resulting in PTP variants’ being incapable of functioning as H2 producers at higher pH. Interestingly, however, any benefit that H2 oxidation might gain from a higher pH due to a more favorable proton gradient from active site to solvent fails to have an impact for the variants.
Regarding reversibility, the subtle influences of mild disruptions to the PTP that are evident by inspection of voltammograms close to the 2H+/H2 thermodynamic potential suggest that all long-range electron-transfer processes occurring in WT enzymes are tightly coupled to PT, implying that evolution strived for concerted PCET in either or both of the diagonal paths in Fig. 2C. Via electrochemistry, even minor temporal decouplings of proton and ETs are translated into a visible inflection in the current–potential curve, which is an observation that could not be made in any other way under turnover conditions. The fact that these inflections arise due to changes in the PTP rather than the ET chain shows that PT is limiting the rate of ET. As decoupling becomes more extreme, sluggish PT leads to the accumulation of redox intermediates, and thus to a temporary stalling of the catalytic cycle. This effect is evident for H2 oxidation, where the large overpotential requirement that is eventually observed for each of the variants at high pH suggests that the catalytic cycle is delayed at a reduced protonated state of the active site. The decoupling of electron release from proton removal results in a higher electrode potential being needed (corresponding to E1H or E2HH in Fig. 2C), that is, more oxidizing conditions are required to complete the catalytic cycle.
The linked two-square scheme shown in Fig. 2C was unambiguous but generic; for completeness we could have included states RR (two electrons, no protons) and OHH (no electrons, two protons), but these states (extremely strong base and acid, respectively) are highly unlikely. We are now in a position to assign the various states of Fig. 2C to specific H-cluster species that have been characterized by electron paramagnetic resonance (EPR) and vibrational spectroscopies, which is equivalent to putting place names on a map. Accordingly, in Fig. 7A we have replaced the generic states of Fig. 2C by boxes arranged in terms of their unique electronation and protonation numbers; with the exception of X and Y, each box lists the redox tautomers that have so far been identified for that state. The different species in Fig. 7 are drawn from recent literature in which there is satisfactory (although not perfect) agreement on nomenclature (52, 53, 58–61). For further illustration, the current form of the catalytic cycle (adopted from ref. 61) is shown with detailed information of each redox intermediate state (SI Appendix, Fig. S7).
Fig. 7.
Electrocatalytic pathways for the interconversion beween H2 and two electrons and two protons, based on changes in electronation and protonation level. A is a direct adaptation of Fig. 2C identifying generic states with redox states of an [FeFe]-hydrogenase that have been identified by spectroscopic measurements, either under equilibrium conditions or in fast time-resolved experiments. The horizontal shifts show (negative) changes in electronation (left to right, 0 [red], −1 [green], −2 [blue]) and the vertical shifts show changes in protonation (top to bottom 0, +1, +2). Each box shows the tautomers that have been reported and characterized for each electronation–protonation state. Boxes X and Y are highly acidic states that have not so far been reported and could evade detection and characterization. B shows a 45°-rotated/vertical-plane reflected representation of the same map to depict how different catalytic pathways can be taken for oxidation and reduction. Fast PT favors the central line that describes the reversible (most efficient), limiting case.
The central box at the intersection of the two squares in Fig. 7A houses species having electronation and protonation levels represented by −1/+1 (i.e., electronation adds a negative charge). The two species shown are HredH+ and Hhyd:ox, the latter having been shown recently through cryogenic infrared and EPR spectroscopic evidence to have an active-site electronic configuration [4FeH]2+/FeIIFeII, with an apical hydride coordinated to Fed and a nonprotonated adt bridge (61). Of particular relevance are FTIR spectroscopic data from our recent study which showed that Hhyd:ox (formally called Hhyd in that study) accumulates for all exchange variants under a 100% H2 atmosphere at pH 8 (especially in the case of E141DCrHydA1) in place of a mixture of reduced redox states (i.e., HredH+ and HsredH+) that are observed for WT CrHydA1 under the same conditions (see figure 3B on p. 5 in ref. 25). It is important to stress that under such equilibrium conditions the states observed will reflect both thermodynamic stability and catalytic lifetime; catalysis in the H2 evolution and H2 oxidation directions are each occurring at the same fast rate (14), so the fact that Hhyd:ox accumulates under equilibrium for PTP variants suggests that it is a thermodynamically more stable form of a true intermediate but does not itself lie on the main electrocatalytic pathway. The true central intermediate, plausibly the well-characterized tautomer HredH+ (52), is linked directly via concerted PCET to Hox (level 0/0) and to the box at level −2/+2 that comprises tautomers Hhyd and HhydH+. The off-diagonal states that have been characterized are tautomers Hred and Hred’ (53, 58, 62, 63) at electronation/protonation state −1/0 and tautomers Hhyd:red and HsredH+ (61) at level −2/+1. Boxes X (0/+1) and Y(−1/+2) correspond to generic states OH and RHH that might be formulated as HoxH+ Hred2H+ but which have not so far been confirmed.
In Fig. 7B the map is redrawn to depict how the electrode potential (or the reducing environment of the living cell) can influence the catalytic pathways taken in each direction, depending on whether PT is sufficiently fast to occur in concert with ET or must instead occur as a subsequent step. The map displays the possible pathways for each direction and contrasts with the conventional “clockwise–anticlockwise” way of representing a catalytic cycle. Using the WT enzymes as the starting point, the most efficient pathway, adopted at the equilibrium (exchange) potential, is described by the vertical line cutting across both squares, that is, between Hox and tautomers Hhyd/HhydH+, via concerted PCET. The concerted pathway should be regarded as the ideal limiting case. As an overpotential is applied to drive the reaction in a single net direction, all or part of the concerted pathway may well persist but the higher rate of ET will cause the stepwise pathways to become increasingly important. The pathway taken for reduction will progressively shift to include steps through intermediates at levels −1/0 (Hred and Hred′) and −2/+1 (Hhyd:red/HsredH+), whereas the oxidation pathway will shift to include steps via as-yet uncharacterized intermediates −1/+2 (Y) and 0/+1 (X). These scenarios are limiting cases; a concerted PCET in just one square may still lead to reversible catalysis if steps in the other square are fast. Disruptions to the PTP lower the probability of taking the direct concerted route, thus resulting in an inflection and ultimately a sizeable overpotential requirement. The observation of a clear and significant overpotential requirement for H2 oxidation with the PTP variants suggests the importance of acidic states X and/or Y; these would be formed upon one-electron oxidation of HhydH+ or Hhyd/HhydH+, respectively, but are probably too reactive to isolate.
Disruptions to the PTP can therefore be manifested as an apparent malfunction of the ET chain. One such malfunction has been demonstrated for Df [NiFe]-hydrogenase in a study by Dementin et al. (64). Changing the His(Cys)3 ligation of the most distal [4Fe-4S]-cluster coordination sphere to an all-Cys coordination rendered its reduction potential more negative, resulting in strongly decreased intra- and intermolecular ET rates and in a significantly affected catalytic bias (64). Moreover, the electrocatalytic waveshape observed for E279DCpI closely resembles that of C. pasteurianum [FeFe]-hydrogenase isoform II (CpII) as reported by Artz et al. (65). In their study, the authors used a simple model to explain the differences in catalytic bias observed for the different C. pasteurianum [FeFe]-hydrogenase isoforms (CpI-CpIII). For CpII they proposed that the bias toward H2 oxidation and significant overpotential requirement arose mostly from the markedly high reduction potential of the most distal [FeS]-cluster (−180 mV CpII vs. ≤−450 mV for CpI) which results in a lower internal driving force for H2 evolution (65–67). Interestingly, the Glu-to-Asp substitution of variant E279DCpI converted CpI into a “CpII-like” hydrogenase simply by impacting the PTP, while leaving the [FeS]-relay unaffected. In the light of the data presented in this paper, any model describing the catalytic bias of [FeFe]-hydrogenases solely as a function of parameters defining ET to or from the active site (65) needs to be extended by including the aspect of far-distant cooperativity between ET and PT pathways.
Conclusions
Ultimately, we have described experiments addressing both the bidirectionality and reversibility of [FeFe]-hydrogenase catalysis across a continuous range of driving force. Exploiting a very subtle observation, made possible by mildly suppressing an otherwise fast PT rate, we have adopted a more realistic and versatile way to view catalytic cycles, such as those for hydrogenases—one that emphasizes the sum of pathways for each direction. Specifically, the widely adopted representation in terms of a single circular pathway being taken in each direction is strictly true only in the unproductive, reversible limit; it breaks down, increasingly so as an overpotential is applied to drive the reaction in a net direction, and almost immediately if PT is sluggish.
The electrochemical experiments show clearly that substitutions located at a long distance from an electron-transfer relay affect catalysis differently depending on the catalytic direction (for example, proton uptake ability vs. proton release from a trapped reduced intermediate) and reveal even the most subtle breakdown of reversibility when electron and PTs are uncoupled. We conclude that the PTP acts like a “highway” for rapid proton funneling—proton-coupled electron-transfer processes at the active site are optimized over a surprisingly long range, and even innocuous amino acid substitutions within the complex proton relay network have a major impact on catalytic efficiency and directionality. Our findings provide an answer for what makes de novo catalyst design so challenging. While the critical role of pendant bases for rapid H2 turnover rates at the catalytic cofactor (i.e., the adt bridgehead ligand) is well established in the design of synthetic catalysts (68, 69), longer-range proton relay systems are now being increasingly recognized and implemented as essential attributes into the designs of synthetic Ni- or Fe-based catalysts (18, 38, 70–74). Of particular note are recent enzyme-inspired efforts by Shaw and coworkers, who modified a Ni-based Dubois-type catalyst (18) with amino acid-based proton relay systems (e.g., arginine and tyrosine). In combination with the carboxylic functional groups, the individual amino acid side chains were shown to exchange protons rapidly with the catalytically active Ni-site via a pendant amine, thus representing a minimal PTP. These features ultimately led to significantly increased H2/H+ turnover rates and a close approach to electrocatalytic reversibility (75–77). We are just beginning to understand the details necessary to ensure success in the rational design of energy-cycling electrocatalysts, the catalytic performance of which lags well behind evolutionarily optimized redox enzymes that operate efficiently under ambient conditions.
Experimental Section
Plasmid Preparation, Enzyme Expression, and Purification.
Site-directed mutagenesis and recombinant protein expression were carried out as described previously (12, 78). All enzyme preparation experiments were carried out in an anaerobic tent (Coy Labs) under a N2/H2 atmosphere (99:1). All solutions and buffers used for protein purification were supplemented with 2 mM sodium dithionite (NaDT). Recombinant proteins (strep-tagged) were purified via Strep-Tactin Superflow high-capacity cartridges (IBA Lifesciences) according to the manufacturer’s instructions. The purity of the hydrogenase isolates was monitored using sodium dodecyl sulfate polyacrylamide gel electrophoresis, and a Bradford assay (Bio-Rad) was carried out to determine the protein concentration, using bovine serum albumin as a standard (Biolabs). All protein samples were concentrated to 1 mM in 0.1 M Tris/HCl, pH 8.0, and stored at −80 °C until further use.
Preparation of [2FeH] Mimics and In Vitro Maturation of [FeFe]-Hydrogenases.
Synthesis of the [2FeH]MIM complex was carried out according to a procedure described previously (79). The complex was dissolved in 0.1 M potassium phosphate buffer (KpI) at pH 6.8 and stored at −80 °C until further use. Recombinant apo-proteins of CpI and CrHydA1 were activated with the [2Fe]MIM complex as described earlier (7) with minor modifications according to the procedure described in ref. 80.
Enzyme Activity Assays.
Hydrogen production activity assays were performed as described previously by incubating 400 ng of holo-hydrogenase in a 2 mL reaction mixture, containing buffer (0.2 M MES buffer [pH 5 to 6.5]/0.2 M Mops buffer [pH 6.5 to 7.5]/0.2 M Tris buffer [pH 8 to 9]), NaDT (0.1 M), and methyl viologen (10 mM) using sealed 8-mL vessels (Suba) (25). After purging with argon gas for 5 min, the samples were incubated for 20 min at 37 °C (100 rpm) in a shaking water bath. The resulting hydrogen production was monitored by analyzing the composition of a 400-µL sample of head space via gas chromatography (Shimadzu).
Protein Crystallography.
The exchange variant E279DCpI was crystallized by applying hanging-drop vapor diffusion as described earlier (25). A 2-µL sample of protein (15 mg/mL) plus 2 µL crystallization buffer, stemming from a 500-µL reservoir solution consisting of 0.1 M MES-NaOH (pH 6), 0.4 M MgCl2, 20% polyethylene glycol 4000, and 20% glycerol, was mixed and equilibrated against the reservoir solution. The crystals appeared within 1 wk. The full size could be achieved within another week and crystals were flash-frozen and stored in liquid N2. The X-ray data were collected under a cryo nitrogen stream at 100 K at SLS (Swiss Light Source) PXII. The diffraction data were processed using XDS. Phenix (81) was used for running molecular replacement and the refinement was assisted by Coot (82) as the visualization tool. The structure was deposited in the PDB under ID code 6YF4 (SI Appendix, Table S1).
PFE.
All PFE experiments were carried out in anaerobic chamber (Coy Labs) under a N2/H2 atmosphere (98:2). A PalmSens potentiostat (PalmSens 4) was used to control the potential, employing PalmSens software (PS trace 5.2). The gas-tight electrochemical cell was water-jacketed to control the temperature. A pyrolytic graphite “edge” rotating disk electrode was used as a working electrode and controlled by a rotator (Autolab). A nonisothermal side arm housing the reference electrode (Ag/AgCl, 3.5 M KCl) was connected to the main cell compartment by a luggin capillary. The reference potential was converted to the standard hydrogen electrode (SHE) scale, using the correction ESHE = EAg/AgCl + 0.205 V at 20 °C. Platinum wire was used as a counterelectrode. For each experiment, defined gas flow rates (Air Liquide) were adjusted, using mass flow controllers (Sierra Instruments). For enzyme film preparation, 3 µL of a 10 µM hydrogenase sample were applied onto the polished electrode surface (surface area varying between 0.06 cm2 and 0.09 cm2, depending on the electrode used). After 3 min, the electrode was rinsed thoroughly with Milli-Q water. All cyclic voltammetry experiments were performed at 20 °C and 3,000 rpm electrode rotation, under a 100% H2 atmosphere, using a varying scan rate depending on the respective experiment in a mixed buffer system (15 mM each of MES, Hepes, TAPS, CHES, and sodium acetate, as well as 0.1 M NaCl).
Supplementary Material
Acknowledgments
We gratefully acknowledge Florian Wittkamp and Ulf-Peter Apfel for synthesizing and providing the [2Fe]MIM cofactor for in vitro maturation. This work is funded by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy–EXC 2033–390677874–RESOLV. T.H. gratefully acknowledges financial support from the Volkswagen Stiftung (design of [FeS] cluster containing Metallo-DNAzymes [Az 93412]) and the DFG (HA 2555/10-1). F.A.A. gratefully acknowledges support by grants from the Biotechnology and Biological Sciences Research Council, particularly BB/M005720/1. We thank Dr. Raphael Gasper and staff in beamline SLS PXII for technical support during X-ray data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007090117/-/DCSupplemental.
Data Availability.
All data supporting the findings of this study are available within the article and/or SI Appendix. The coordinates and structure factors of exchange variant E279DCpI have been deposited in the PDB (ID code 6YF4).
References
- 1.Frey M., Hydrogenases: Hydrogen-activating enzymes. ChemBioChem 3, 153–160 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Vincent K. A., Parkin A., Armstrong F. A., Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 107, 4366–4413 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Stripp S. T., Happe T., How algae produce hydrogen–News from the photosynthetic hydrogenase. Dalton Trans. 45, 9960–9969 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Lubitz W., Ogata H., Rüdiger O., Reijerse E., Hydrogenases. Chem. Rev. 114, 4081–4148 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Silakov A., Kamp C., Reijerse E., Happe T., Lubitz W., Spectroelectrochemical characterization of the active site of the [FeFe] hydrogenase HydA1 from Chlamydomonas reinhardtii. Biochemistry 48, 7780–7786 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Berggren G. et al., Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature 499, 66–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esselborn J. et al., Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat. Chem. Biol. 9, 607–609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esselborn J. et al., A structural view of synthetic cofactor integration into [FeFe]-hydrogenases. Chem. Sci. 7, 959–968 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silakov A., Reijerse E. J., Albracht S. P. J., Hatchikian E. C., Lubitz W., The electronic structure of the H-cluster in the [FeFe]-hydrogenase from Desulfovibrio desulfuricans: A Q-band 57Fe-ENDOR and HYSCORE study. J. Am. Chem. Soc. 129, 11447–11458 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Rauchfuss T. B., Diiron azadithiolates as models for the [FeFe]-hydrogenase active site and paradigm for the role of the second coordination sphere. Acc. Chem. Res. 48, 2107–2116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silakov A., Wenk B., Reijerse E., Lubitz W., (14)N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: Evidence for a nitrogen in the dithiol bridge. Phys. Chem. Chem. Phys. 11, 6592–6599 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Lampret O. et al., Interplay between CN- ligands and the secondary coordination sphere of the H-cluster in [FeFe]-Hydrogenases. J. Am. Chem. Soc. 139, 18222–18230 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Madden C. et al., Catalytic turnover of [FeFe]-hydrogenase based on single-molecule imaging. J. Am. Chem. Soc. 134, 1577–1582 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Pandey K., Islam S. T., Happe T., Armstrong F. A., Frequency and potential dependence of reversible electrocatalytic hydrogen interconversion by [FeFe]-hydrogenases. Proc. Natl. Acad. Sci. U.S.A. 114, 3843–3848 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darensbourg M. Y., Lyon E. J., Zhao X., Georgakaki I. P., The organometallic active site of [Fe]hydrogenase: Models and entatic states. Proc. Natl. Acad. Sci. U.S.A. 100, 3683–3688 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T., Darensbourg M. Y., A mixed-valent, Fe(II)Fe(I), diiron complex reproduces the unique rotated state of the [FeFe]hydrogenase active site. J. Am. Chem. Soc. 129, 7008–7009 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Justice A. K., Rauchfuss T. B., Wilson S. R., Unsaturated, mixed-valence diiron dithiolate model for the H(ox) state of the [FeFe] hydrogenase. Angew. Chem. Int. Ed. Engl. 46, 6152–6154 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helm M. L., Stewart M. P., Bullock R. M., DuBois M. R., DuBois D. L., A synthetic nickel electrocatalyst with a turnover frequency above 100,000 s−1 for H2 production. Science 333, 863–866 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Lunsford A. M. et al., Cyanide-bridged iron complexes as biomimetics of tri-iron arrangements in maturases of the H cluster of the di-iron hydrogenase. Chem. Sci. 7, 3710–3719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding S., Ghosh P., Darensbourg M. Y., Hall M. B., Interplay of hemilability and redox activity in models of hydrogenase active sites. Proc. Natl. Acad. Sci. U.S.A. 114, E9775–E9782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vignais P. M., Billoud B., Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 107, 4206–4272 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Cornish A. J., Gärtner K., Yang H., Peters J. W., Hegg E. L., Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. J. Biol. Chem. 286, 38341–38347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler M., Esselborn J., Happe T., Molecular basis of [FeFe]-hydrogenase function: An insight into the complex interplay between protein and catalytic cofactor. Biochim. Biophys. Acta 1827, 974–985 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Gauquelin C. et al., Roles of the F-domain in [FeFe] hydrogenase. Biochim. Biophys. Acta Bioenerg. 1859, 69–77 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Duan J. et al., Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases. Nat. Commun. 9, 4726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hexter S. V., Grey F., Happe T., Climent V., Armstrong F. A., Electrocatalytic mechanism of reversible hydrogen cycling by enzymes and distinctions between the major classes of hydrogenases. Proc. Natl. Acad. Sci. U.S.A. 109, 11516–11521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazurenko I., Wang X., Poulpiquet A. d., Lojou E., H2/O2 enzymatic fuel cells: From proof-of-concept to powerful devices. Sustain. Energy Fuels 1, 1475–1501 (2017). [Google Scholar]
- 28.Migliore A., Polizzi N. F., Therien M. J., Beratan D. N., Biochemistry and theory of proton-coupled electron transfer. Chem. Rev. 114, 3381–3465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammes-Schiffer S., Proton-coupled electron transfer: Moving together and charging forward. J. Am. Chem. Soc. 137, 8860–8871 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong F. A., Hirst J., Reversibility and efficiency in electrocatalytic energy conversion and lessons from enzymes. Proc. Natl. Acad. Sci. U.S.A. 108, 14049–14054 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong F. A. et al., Guiding principles of hydrogenase catalysis instigated and clarified by protein film electrochemistry. Acc. Chem. Res. 49, 884–892 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Del Barrio M. et al., Electrochemical investigations of hydrogenases and other enzymes that produce and use solar fuels. Acc. Chem. Res. 51, 769–777 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Evans R. M. et al., The value of enzymes in solar fuels research - efficient electrocatalysts through evolution. Chem. Soc. Rev. 48, 2039–2052 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Koper M. T. M., Theory of multiple proton–electron transfer reactions and its implications for electrocatalysis. Chem. Sci. 4, 2710–2723 (2013). [Google Scholar]
- 35.Hexter S. V., Esterle T. F., Armstrong F. A., A unified model for surface electrocatalysis based on observations with enzymes. Phys. Chem. Chem. Phys. 16, 11822–11833 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Caserta G. et al., Engineering an [FeFe]-Hydrogenase: Do accessory clusters influence O2 resistance and catalytic bias? J. Am. Chem. Soc. 140, 5516–5526 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Rodríguez-Maciá P. et al., His-Ligation to the [4Fe-4S] subcluster tunes the catalytic bias of [FeFe] hydrogenase. J. Am. Chem. Soc. 141, 472–481 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Fourmond V., Wiedner E. S., Shaw W. J., Léger C., Understanding and design of bidirectional and reversible catalysts of multielectron, multistep reactions. J. Am. Chem. Soc. 141, 11269–11285 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Hirst J. et al., Kinetics and mechanism of redox-coupled, long-range proton transfer in an iron−sulfur protein. Investigation by fast-scan protein-film voltammetry. J. Am. Chem. Soc. 120, 7085–7094 (1998). [Google Scholar]
- 40.Chen K. et al., Atomically defined mechanism for proton transfer to a buried redox centre in a protein. Nature 405, 814–817 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Thammavongsy Z., Mercer I. P., Yang J. Y., Promoting proton coupled electron transfer in redox catalysts through molecular design. Chem. Commun. (Camb.) 55, 10342–10358 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Mulder D. W. et al., Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG). Nature 465, 248–251 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Mulder D. W. et al., Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure 19, 1038–1052 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Hajj V. et al., FeFe hydrogenase reductive inactivation and implication for catalysis. Energy Environ. Sci. 7, 715–719 (2014). [Google Scholar]
- 45.Fourmond V. et al., The oxidative inactivation of FeFe hydrogenase reveals the flexibility of the H-cluster. Nat. Chem. 6, 336–342 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Orain C. et al., Electrochemical measurements of the kinetics of inhibition of two FeFe hydrogenases by O2 demonstrate that the reaction is partly reversible. J. Am. Chem. Soc. 137, 12580–12587 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Armstrong F. A., Evans R. M., Megarity C. F., Protein film electrochemistry of iron-sulfur enzymes. Methods Enzymol. 599, 387–407 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Fourmond V. et al., The mechanism of inhibition by H2 of H2-evolution by hydrogenases. Chem. Commun. (Camb.) 49, 6840–6842 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Wang D., Crowe W. E., Strongin R. M., Sibrian-Vazquez M., Exploring the pH dependence of viologen reduction by alpha-carbon radicals derived from HCy and Cys. Chem. Commun. (Camb.) 14, 1876–1878 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stripp S. T. et al., How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. U.S.A. 106, 17331–17336 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulder D. W. et al., Investigations on the role of proton-coupled electron transfer in hydrogen activation by [FeFe]-hydrogenase. J. Am. Chem. Soc. 136, 15394–15402 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Sommer C. et al., Proton coupled electronic rearrangement within the H-cluster as an essential step in the catalytic cycle of [FeFe] hydrogenases. J. Am. Chem. Soc. 139, 1440–1443 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Senger M. et al., Proton-coupled reduction of the catalytic [4Fe-4S] cluster in [FeFe]-hydrogenases. Angew. Chem. Int. Ed. Engl. 56, 16503–16506 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Senger M. et al., How [FeFe]-hydrogenase facilitates bidirectional proton transfer. J. Am. Chem. Soc. 141, 17394–17403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leroux F. et al., Experimental approaches to kinetics of gas diffusion in hydrogenase. Proc. Natl. Acad. Sci. U.S.A. 105, 11188–11193 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebgott P. P. et al., Relating diffusion along the substrate tunnel and oxygen sensitivity in hydrogenase. Nat. Chem. Biol. 6, 63–70 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Abou Hamdan A. et al., Understanding and tuning the catalytic bias of hydrogenase. J. Am. Chem. Soc. 134, 8368–8371 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Katz S. et al., Vibrational spectroscopy reveals the initial steps of biological hydrogen evolution. Chem. Sci. 7, 6746–6752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ratzloff M. W. et al., CO-bridged H-cluster intermediates in the catalytic mechanism of [FeFe]-hydrogenase CaI. J. Am. Chem. Soc. 140, 7623–7628 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Sanchez M. L. K. et al., Investigating the kinetic competency of CrHydA1 [FeFe] hydrogenase intermediate states via time-resolved infrared spectroscopy. J. Am. Chem. Soc. 141, 16064–16070 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Lorent C. et al., Shedding light on proton and electron dynamics in [FeFe] hydrogenases. J. Am. Chem. Soc. 142, 5493–5497 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Adamska-Venkatesh A. et al., New redox states observed in [FeFe] hydrogenases reveal redox coupling within the H-cluster. J. Am. Chem. Soc. 136, 11339–11346 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Senger M. et al., Protonation/reduction dynamics at the [4Fe-4S] cluster of the hydrogen-forming cofactor in [FeFe]-hydrogenases. Phys. Chem. Chem. Phys. 20, 3128–3140 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Dementin S. et al., Changing the ligation of the distal [4Fe4S] cluster in NiFe hydrogenase impairs inter- and intramolecular electron transfers. J. Am. Chem. Soc. 128, 5209–5218 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Artz J. H. et al., Tuning catalytic bias of hydrogen gas producing hydrogenases. J. Am. Chem. Soc. 142, 1227–1235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams M. W., The mechanisms of H2 activation and CO binding by hydrogenase I and hydrogenase II of Clostridium pasteurianum. J. Biol. Chem. 262, 15054–15061 (1987). [PubMed] [Google Scholar]
- 67.Artz J. H. et al., Reduction potentials of [FeFe]-Hydrogenase accessory iron-sulfur clusters provide insights into the energetics of proton reduction catalysis. J. Am. Chem. Soc. 139, 9544–9550 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Henry R. M., Shoemaker R. K., DuBois D. L., DuBois M. R., Pendant bases as proton relays in iron hydride and dihydrogen complexes. J. Am. Chem. Soc. 128, 3002–3010 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Crouthers D. J., Denny J. A., Bethel R. D., Munoz D. G., Darensbourg M., Conformational mobility and pendent base effects on electrochemistry of synthetic analogues of the [FeFe]-hydrogenase active site. Organometallics 33, 4747–4755 (2014). [Google Scholar]
- 70.Das P. et al., Controlling proton movement: Electrocatalytic oxidation of hydrogen by a nickel(II) complex containing proton relays in the second and outer coordination spheres. Dalton Trans. 43, 2744–2754 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Ding S. et al., Hemilabile bridging thiolates as proton shuttles in bioinspired H2 production electrocatalysts. J. Am. Chem. Soc. 138, 12920–12927 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Ahmed M. E., Dey S., Darensbourg M. Y., Dey A., Oxygen-tolerant H2 production by [FeFe]-H2ase active site mimics aided by second sphere proton shuttle. J. Am. Chem. Soc. 140, 12457–12468 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Chu K. T. et al., A reversible proton relay process mediated by hydrogen-bonding interactions in [FeFe]hydrogenase modeling. Chemistry 21, 10978–10982 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Darmon J. M. et al., Increasing the rate of hydrogen oxidation without increasing the overpotential: A bio-inspired iron molecular electrocatalyst with an outer coordination sphere proton relay. Chem. Sci. 6, 2737–2745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dutta A., Ginovska B., Raugei S., Roberts J. A., Shaw W. J., Optimizing conditions for utilization of an H2 oxidation catalyst with outer coordination sphere functionalities. Dalton Trans. 45, 9786–9793 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Priyadarshani N. et al., Achieving reversible H2/H+ interconversion at room temperature with enzyme-inspired molecular complexes: A mechanistic study. ACS Catal. 6, 6037–6049 (2016). [Google Scholar]
- 77.Boralugodage N. P., Arachchige R. J., Dutta A., Buchko G. W., Shaw W. J., Evaluating the role of acidic, basic, and polar amino acids and dipeptides on a molecular electrocatalyst for H2 oxidation. Catal. Sci. Technol. 7, 1108–1121 (2017). [Google Scholar]
- 78.Kuchenreuther J. M. et al., High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLoS One 5, e15491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Razavet M. et al., All-iron hydrogenase: Synthesis, structure and properties of {2Fe3S}-assemblies related to the di-iron sub-site of the H-cluster. Dalton Trans. 2003, 586–595 (2003). [Google Scholar]
- 80.Lampret O. et al., The final steps of [FeFe]-hydrogenase maturation. Proc. Natl. Acad. Sci. U.S.A. 116, 15802–15810 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adams P. D. et al., PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and/or SI Appendix. The coordinates and structure factors of exchange variant E279DCpI have been deposited in the PDB (ID code 6YF4).