Significance
The major histocompatibility complex class-I (MHC-I) peptide-loading complex (PLC) translocates cytosolic degradation products to the endoplasmic reticulum to load antigenic peptides onto MHC-I molecules. Stable peptide–MHC-I complexes are presented at the cell surface to mirror cellular contents for patrolling T cells, which protect against viral infections and cancer-causing mutations by inducing apoptosis in cells that expose nonself peptides. Due to its size and dynamic nature, the atomic-level details of the PLC remained unknown. We built an all-atom model of the human MHC-I PLC by combining the recent 9.9-Å resolution cryo-EM density with microsecond molecular dynamics simulations in a membrane and water environment (1.6 million atoms). The results provide unprecedented insights into the molecular underpinnings of our adaptive immune response.
Keywords: immunity, MHC-I, peptide-loading complex, molecular dynamics simulations, protein dynamics
Abstract
The major histocompatibility complex class-I (MHC-I) peptide-loading complex (PLC) is a cornerstone of the human adaptive immune system, being responsible for processing antigens that allow killer T cells to distinguish between healthy and compromised cells. Based on a recent low-resolution cryo-electron microscopy (cryo-EM) structure of this large membrane-bound protein complex, we report an atomistic model of the PLC and study its conformational dynamics on the multimicrosecond time scale using all-atom molecular dynamics (MD) simulations in an explicit lipid bilayer and water environment (1.6 million atoms in total). The PLC has a layered structure, with two editing modules forming a flexible protein belt surrounding a stable, catalytically active core. Tapasin plays a central role in the PLC, stabilizing the MHC-I binding groove in a conformation reminiscent of antigen-loaded MHC-I. The MHC-I–linked glycan steers a tapasin loop involved in peptide editing toward the binding groove. Tapasin conformational dynamics are also affected by calreticulin through a conformational selection mechanism that facilitates MHC-I recruitment into the complex.
To protect us against cancer and intracellular pathogens, the human adaptive immune system relies on a signaling mechanism whereby antigens are exposed at the surface of cells by major histocompatibility complex class-I (MHC-I) proteins for recognition by killer T cells (1–3). These antigens are, for the most part, short peptides (8 to 12 amino acids) resulting from the degradation of intracellular proteins. Peptides exposed at the cell surface by MHC-I therefore mirror cellular contents: In healthy cells, only peptides from the “self” are exposed; in cells compromised by a virus or cancer-causing mutation, both self and “nonself” peptides, either viral or mutated, are exposed. Patrolling CD8+ T lymphocytes detect tainted cells by scanning the peptide–MHC-I (pMHC-I) complexes via T cell receptors and induce their apoptosis.
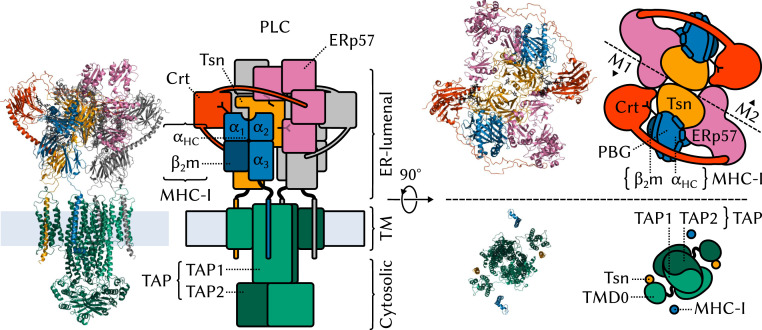
Most peptide sequences have little or no immunogenic value. Triaging the vast pool of cytosolic degradation products to find the few peptides that have a high affinity for MHC-I requires a sophisticated machinery, the peptide-loading complex (PLC) (Fig. 1) (4, 5). Anchored to the endoplasmic reticulum (ER) membrane, this large macromolecular assembly integrates several MHC-I antigen-processing functions into a single molecular machine: antigen transport to the ER, MHC-I stabilization during the loading process, and catalytic selection of high-affinity antigenic peptides (or peptide editing).
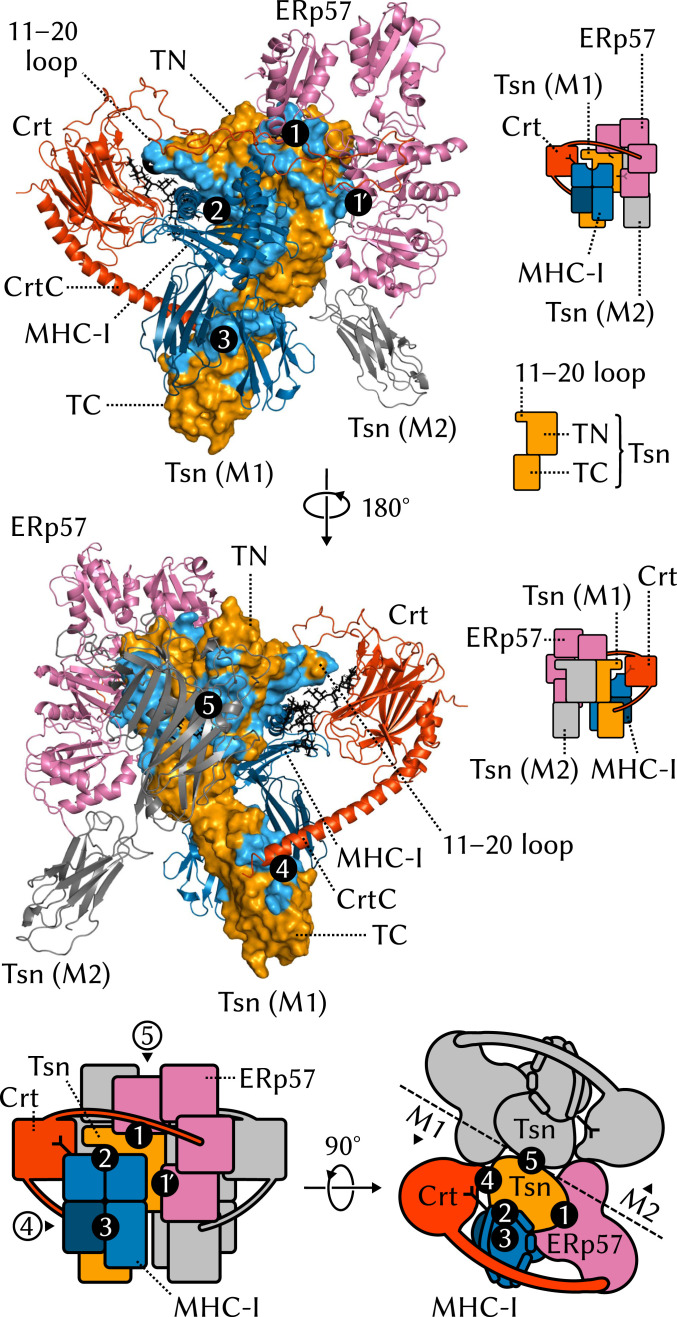
Fig. 1.
Structural overview of the MHC-I PLC. The PLC can be divided into three parts, ER-luminal, TM, and cytosolic. The ER-luminal part consists of two editing modules, M1 and M2, each of which contains one Tsn, ERp57, Crt, and MHC-I. MHC-I proteins (1–3, 6, 7) are heterodimers formed of a variable heavy chain () and an invariant, light 2 microglobulin (). comprises three soluble domains, two of which, and , form the peptide-binding groove (PBG). MHC-Is have an N-linked branched glycan that reflects their loading status: A terminal glucose allows recognition and binding by Crt and acts as a signal that MHC-I should be recruited to the PLC for peptide editing; antigen-loaded MHC-Is that exit the ER are deglucosylated. Newly synthesized “empty” MHC-I proteins (eMHC-I) are unstable; in the PLC, Tsn acts as an MHC-I chaperone. Tsn (8–10) is the central component of the editing modules. It has two ER-luminal domains: N-terminal TN and C-terminal TC. M1 and M2 are organized around a pseudosymmetry axis at the interface between the two Tsns. Tsn forms a complex with MHC-I and accelerates the off rate of low-affinity MHC-I–bound peptides to perform peptide editing (9, 11 – 13). Tsn is also disulfide bonded to ERp57 (14), a four-domain protein playing a structural role. Crt (15, 16) consists of three soluble domains, a globular lectin domain with a binding site for the monoglucosylated branch of the N-linked MHC-I glycan, a flexible P domain that extends over the PBG and contacts ERp57, and a calcium-sensing C domain with an extended -helix that contacts Tsn. The transporter associated with antigen processing (TAP) (17) is the main component of both the TM and cytosolic parts of the PLC. TAP shuttles peptides from the cytosol to the ER, providing the PLC with its substrate for antigen processing. TAP is a heterodimer of TAP-1 and TAP-2, each of which has an N-terminal four-helix TM domain, TMD0, that provides a docking site for the TM helix of Tsn. A TM helix also anchors MHC-I to the ER membrane.
The molecular structure of the PLC was only recently resolved by single-particle cryo-electron microscopy (cryo-EM) (5). The map resolution obtained for the full PLC in this study was only 9.9 Å, which could be improved to 5.8 Å by focusing on a single module of the PLC (Fig. 1). In light of the large size of the complex and the transient nature of the interactions between its components, this study represents an impressive achievement. However, such a low-resolution structure does not provide the atomic-level picture of the PLC required to fully elucidate its inner workings. A static, averaged picture like the one provided by cryo-EM does not inform us about the many dynamic events that take place in the complex, such as assembly, the roles of the various components in the formation of a stable complex, the recruitment of suboptimally loaded MHC-I, the process of peptide selection, or the release of antigen-loaded MHC-I. Understanding these events is crucial to human health since they can be hindered by immune evasion mechanisms (18, 19), rendering the PLC inoperative.
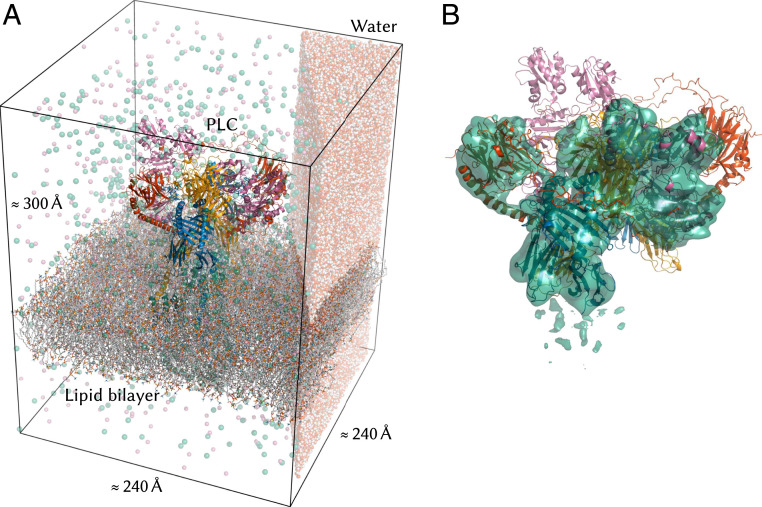
Here, we present the structure and conformational dynamics of the PLC at the atomic level. We integrated the cryo-EM density of the complex with high-resolution structural information on individual PLC components to obtain an atomistic model (Fig. 2). This atomic structure was embedded into a solvated lipid bilayer and subjected to all-atom molecular dynamics (MD) simulations, in which the motions of the complex unfold on the microsecond time scale. We also simulated alternative models of the PLC where individual domains, subunits, or even large parts of the complex were removed and compared protein dynamics in these truncated systems to what we observed in the full complex. This computational structural biology approach pinpoints the origin of dynamic events and relates them to biological functions.
Fig. 2.
Atomistic model of the MHC-I peptide-loading complex. (A) The 1.6-million-atom molecular dynamics simulation system contains the complete human MHC-I PLC (75.939 atoms), embedded in a POPC lipid bilayer (101.036 atoms) and solvated by explicit water with Na+ and Cl− ions. (Only a small slab of water is shown for clarity.) (B) The PLC model was built by fitting atomistic subunit structures to the cryo-EM density for a single editing module (EMD-3906) (5) and then duplicating this module and fitting to the cryo-EM data (EMD-3094) (5) for the pseudosymmetric assembly (cryo-EM density for a single module shown as a surface).
Our results highlight the central role of the two tapasin (Tsn) subunits in the PLC. Tsn and MHC-I form a rigid core surrounded by a flexible belt of accessory proteins, and the dual editing modules formed by Tsn are essential for global complex stability. MHC-I binding-groove width is modulated by the environment of the PLC. The MHC-I N-linked glycan modulates the conformational dynamics of a solvent-exposed Tsn loop putatively involved in peptide editing [called the “scoop loop” (20) by some authors]. Finally, the calreticulin (Crt) C domain constrains Tsn motions to facilitate the docking of MHC-I proteins to the complex, which is the last step of PLC assembly and the onset of antigen processing.
Results and Discussion
ERp57 and Crt Form a Flexible Belt around TsnMHC-I.
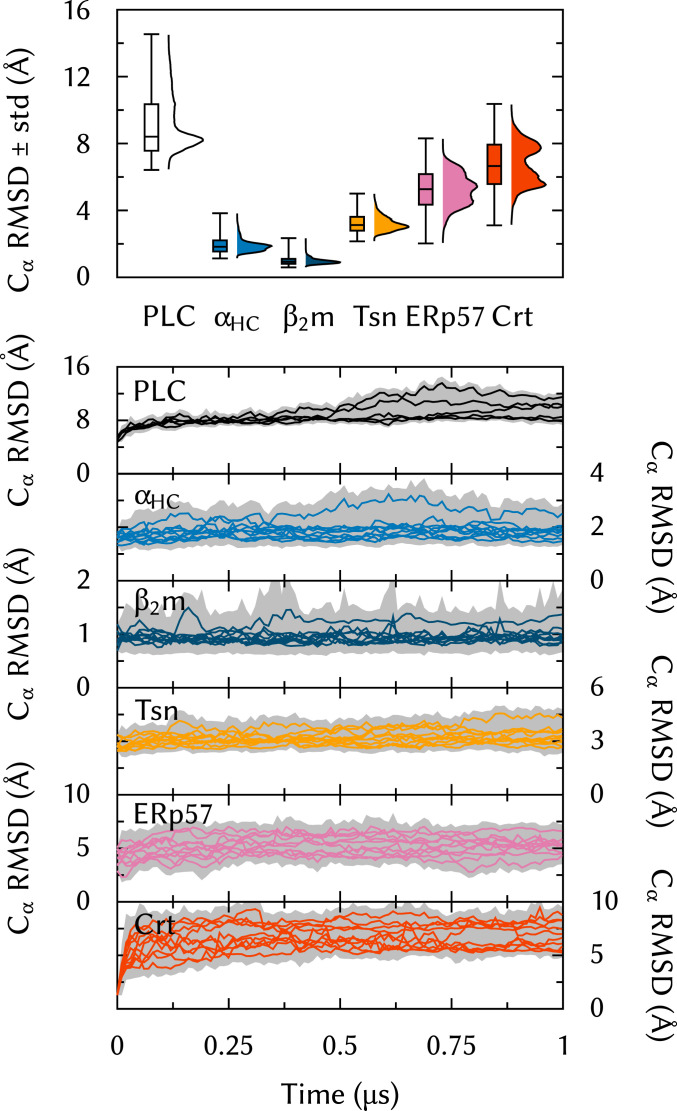
To study the structural dynamics of the human PLC at the atomic level, all-atom MD simulations of the entire PLC, embedded in a lipid bilayer and solvated by water and ions (Fig. 2), were carried out. We acquired five 1-s trajectories of this 1.6-million-atom system. To the best of our knowledge, this represents an unprecedented time scale for such a large membrane-bound protein complex. First, as a quality check, the structural integrity of the individual components was assessed in terms of the root-mean-square deviation (RMSD) of atoms from the starting structure of the simulations. Fig. 3 shows that MHC-I ( heavy chain [], β2 microglobulin []) and Tsn exhibit low rmsd values typical for highly structured proteins, whereas Crt and ERp57 are considerably more flexible with broad distributions and mean rmsd values of 6.8 and 5.3 Å, respectively. This supports the notion that Crt and ERp57 form a flexible belt of accessory proteins that encircle and stabilize a more rigid PLC core, formed by Tsn and MHC-I and dedicated to peptide editing.
Fig. 3.
rmsd from the starting structure, calculated for ER-luminal components individually and for the entire PLC.
To verify the global stability of the overall PLC, as opposed to that of individual subunits, Fig. 3 also shows the rmsd for the whole complex. The PLC is overall stable over the course of the MD simulations. Long convergence times were expected since large systems typically exhibit slow relaxation toward equilibrium.
The general architecture of the PLC allows the two Tsns to adopt a productive conformation for MHC-I binding and peptide editing, i.e., the selection of immunodominant antigens from the vast pool of cytosolic peptides. The ER-luminal part of Tsn is composed of two domains, the N-terminal TN and the C-terminal TC; the latter is anchored to the membrane by a single -helix. We reported in previous work (13) that Tsn exhibits large interdomain flexibility outside of the PLC, while structures of the homologous protein TAPBPR in complex with MHC-I (20, 21) show significant conformational changes compared to MHC-I–free Tsn (10). This raises the question of which structural elements and intercomponent contacts are essential for the formation of a stable core. To provide an answer, we truncated or removed PLC components and assessed the effects of these modifications on the global stability and on specific features of the complex.
Dual Editing Modules Stabilize the PLC.
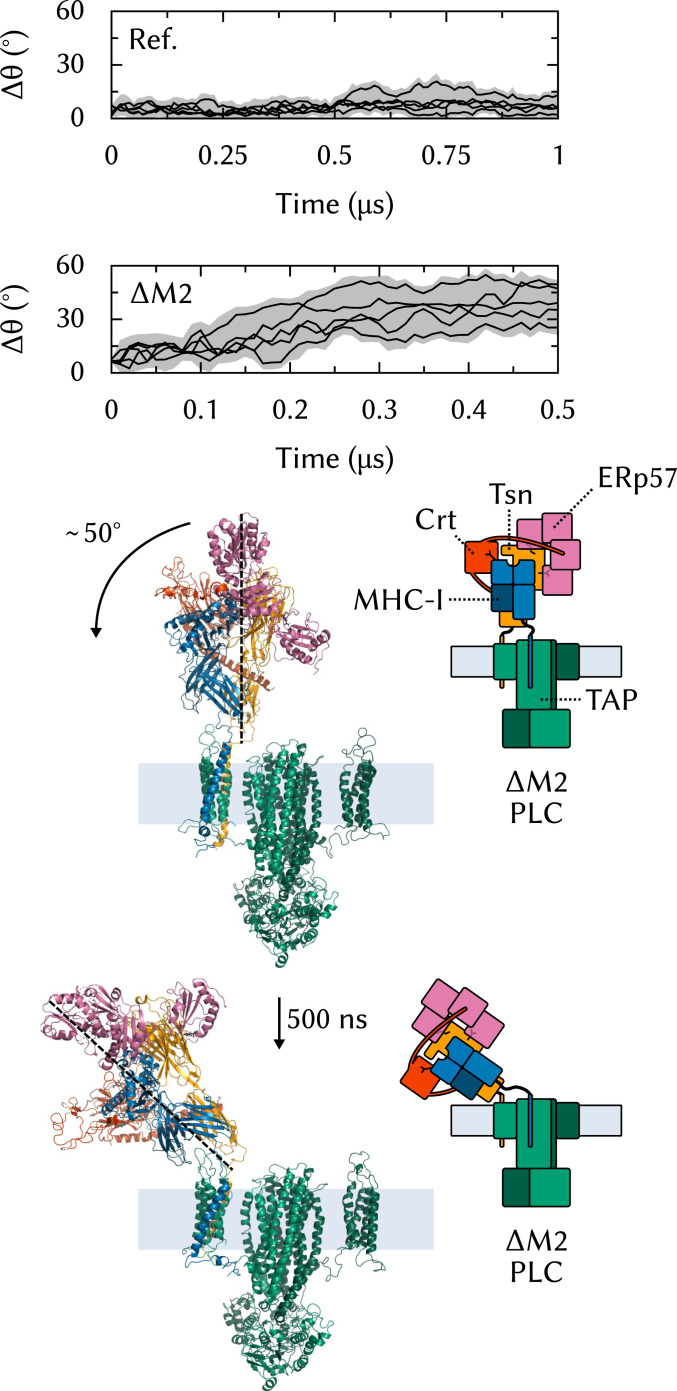
To investigate the role of the two editing modules in the formation of a stable PLC scaffold, we built a computational model of a single-module PLC () and performed five 500-ns MD simulations of this system. The removal of one MHC-I, Tsn-ERp57, and Crt leads to a pronounced deviation from the initial structure. Fig. 4 shows that in all simulations, the editing module leans increasingly outward from the complex’s center. In contrast, in the reference simulations of the full PLC, the tilt angle is stable.
Fig. 4.
Effects of on PLC structural stability. To measure the tilt of the editing module, is defined as the angle between the centers of geometry of the two soluble Tsn domains (TN and TC). is the deviation from the initial orientation; it is reported for the reference system (full PLC, Ref.) (Top) and for (Middle). (Bottom) The starting structure of and the structure with tilted editing module obtained after 500 ns of MD simulation.
If such a conformational change were to happen in vivo, it could hamper the association and dissociation of CrtMHC-I and the apo-PLC because of the very close proximity of the membrane. Therefore, it seems plausible that the assembly of two Tsn-ERp57s is required, at least in the human PLC, to form a stable scaffold for MHC-I binding. These results are consistent with the cryo-EM structure (5) and with pulldown experiments that suggest a 1:2 TAP:Tsn stoichiometry (22).
Tsn Bridges Individual PLC Components.
Tsn is a central hub around which the PLC assembles, a chaperone that stabilizes peptide-deficient MHC-I, and a catalyst for peptide editing that enables MHC-I to rapidly optimize its antigen repertoire (23). In keeping with these key functional roles, Tsn establishes extensive contacts with other components of the PLC. To map these contacts in our simulations, the buried surface area between the soluble Tsn domains and other PLC components was calculated. Five main interaction interfaces were found. First, Tsn interacts with the disulphide-conjugated ERp57 (Fig. 5, 1). Contacts involve the Tsn N-terminal domain (TN) and the first and fourth ERp57 domains (a and a′, respectively), resulting in an interface with a mean buried surface area of 2,400 ± 200 (SD) Å2. We did not observe any contact between Tsn and ERp57 in the opposite editing module, which had been cautiously suggested (5) on the basis of the proximity between these two components.
Fig. 5.
Interfaces between ER-luminal Tsn domains and other PLC components. TM regions are not shown.
In a previous computational study, (13), we showed that Tsn contacts MHC-I through two separate interfaces, a feature also present in TAPBPRMHC-I structures (20, 21) and first observed experimentally for Tsn in the low-resolution structure of the PLC (5). Tsn TN establishes an interface with the MHC-I binding-groove domain formed by and in (Fig. 5, 2; 2,600 500 Å2), while the Tsn C-terminal domain (TC) is lodged between () and (Fig. 5, 3; 1,800 500 Å2). It is worth noting that no computational study, including ours (13), predicted the specific binding interface of Tsn TC and MHC-I.
Interestingly, the N-terminal interface between Tsn and MHC-I involves the N86-linked MHC-I glycan and the Tsn 11–20 loop. This loop is suspected to play a role in peptide selection (20, 24, 25) by interacting either with the F pocket of the MHC-I binding groove or directly with the peptide C terminus which is bound at that location in properly loaded proteins. In our simulations, the residues at the apex of the loop form dynamic contacts with the polysaccharide stretch between MHC-I and the Crt glycan-binding site; effects of the MHC-I glycan on Tsn conformational dynamics are described in more detail in a later subsection.
Tsn also contacts Crt (Fig. 5, 4). The C-terminal Crt domain (C domain) is known to fluctuate between disordered and -helical states as a function of calcium concentration (26). In our simulations, the highly structured C domain contacts Tsn TC at a location directly opposite the interface with MHC-I (700 200 Å2 mean buried surface area). In previous work (13), we commented on the high flexibility between TN and TC and the conformational selection enforced by MHC-I binding. Crt could facilitate MHC-I binding by constraining TC to a specific orientation through the action of the C domain; this hypothesis is addressed in a later subsection.
Finally, the Tsn proteins in both editing modules contact each other to form a large TsnTsn interface (Fig. 5, 5; 1,900 300 Å2) that involves their N233-linked N-acetyl-D-glucosamine glycans. As discussed above, paired Tsns are essential to stabilize the PLC. This interface is strictly TNTN, with the TC domains tilted away from the complex center so that a central cavity is formed that can act as a reservoir for TAP-translocated peptides (5).
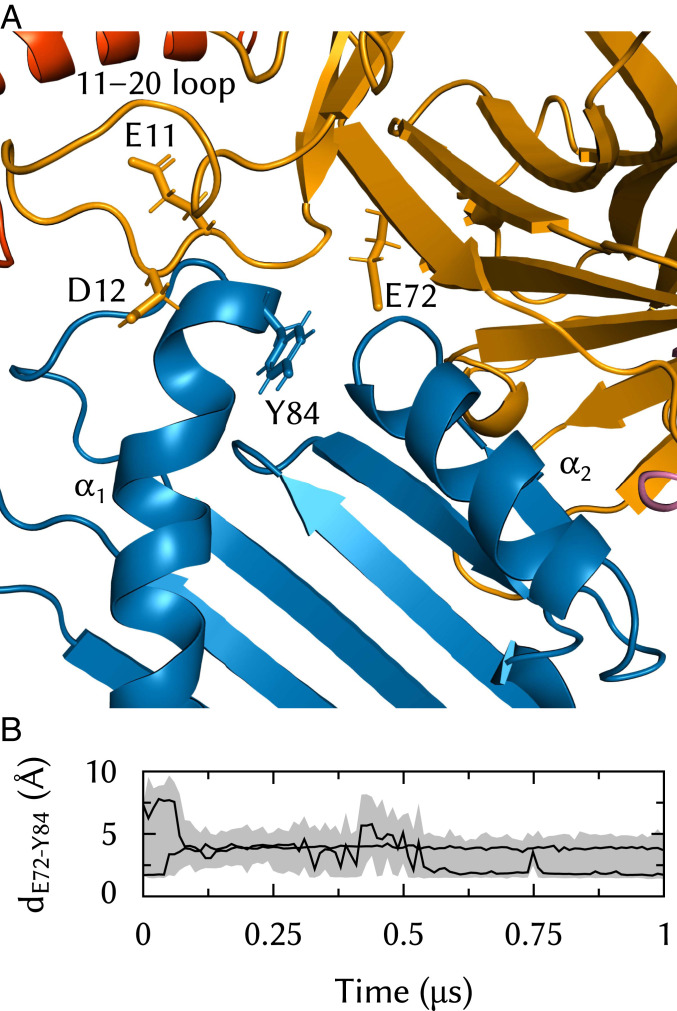
Tsn variants were used by Dong et al. (10) to map residues and features important for MHC-I association. The TN3 variant is a single-point E72K substitution that reduces Tsn peptide-loading activity to 58% of that of the wild-type (WT) protein. In our simulations, the E72 side chain is lodged between the MHC-I and α2 helices (Fig. 6A). E72 interacts with MHC-I Y84, which is known to form essential hydrogen bonds with the antigen peptide C terminus. The E72–Y84 distance fluctuates substantially over the course of a typical simulation (Fig. 6B), but the two residues stay in close proximity and establish stable interactions on the 100-ns time scale. Interactions between Y84 and the Tsn 11–20 loop were recently proposed as drivers of the peptide editing process (25). While this study was performed using only loop fragments, the TN4 variant (10), which involves loop residues E11 and D12, has only 41% of the WT activity. Thus, several Tsn elements come together to interact in the vicinity of Y84, and their synergistic effects should be studied to understand how Tsn opens up the binding groove or competes with peptide ligands.
Fig. 6.
Interactions between Tsn E72 and the MHC-I binding groove in the vicinity of the F pocket. (A) E72 is lodged between MHC-I and and contacts Y84, which is also a suggested interaction partner of the Tsn 11–20 loop. E72 and residues E11 and D12 from that loop have been shown to be important for Tsn association with MHC-I (10). Only Tsn TN and MHC-I and are shown for clarity. (B) E72 to Y84 distance time series in two trajectories.
The PLC Dampens MHC-I Dynamics.
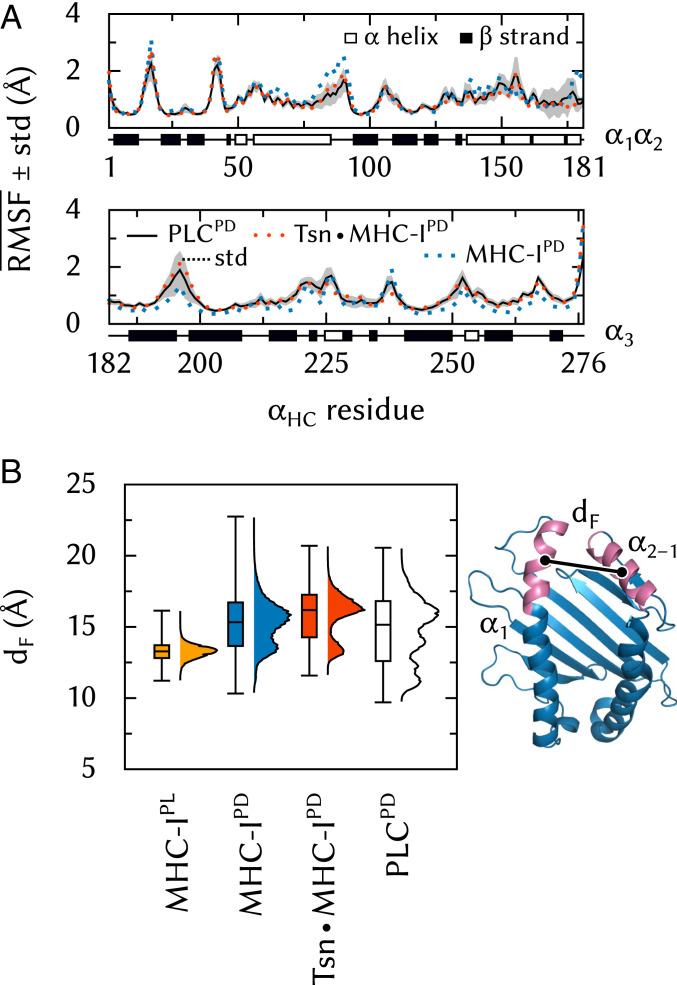
Next, to investigate whether the PLC environment affects MHC-I plasticity in terms of its structural fluctuations, we compared MHC-I dynamics in the PLC to previous results obtained for free MHC-I (27) and TsnMHC-I (13). We found that the binding groove of peptide-deficient MHC-I is affected by the PLC environment differently than by Tsn alone.
Tsn chaperones MHC-I by lowering its fluctuations, especially in the and helices of the binding groove. This effect, which we observed in previous simulations of TsnMHC-I (13), is also seen here in the full PLC (Fig. 7A). In addition, Tsn modulates the width of the binding groove near the F pocket that binds the antigen C terminus. In peptide-loaded (PL) MHC-I, this region of the binding groove is tightly packed and very stable, with an average distance (dF, Fig. 7B) of 13.3 Å between the centers of mass of the helix fragment and the opposite region in . In peptide-deficient (PD) MHC-I, however, the binding groove fluctuates between two conformations: one similar to the PL state and the other much wider (15.7 Å) and more populated. In the TsnMHC-I complex, these two open and closed conformations are still observed, but the open conformation is widened (16.4 Å) and intermediate distances are less populated. In the environment of the PLC, however, the distance distribution is broadened. Although the open conformation is still observed, about half of the dF values are closer to those seen in MHC-IPL (below the median bar in Fig. 7B).
Fig. 7.
MHC-I dynamics in the PLC. (A) Effects of Tsn and PLC binding on RMSF along the sequence. SD is shown for the PLC system. (B) Effects of peptide, Tsn, and PLC binding on binding-groove width dF, measured as the distance between the centers of mass of atoms in residues 75 to 85 () and 138 to 150 ().
While Tsn is active in vitro as a peptide editor and chaperone, the full PLC keeps the binding groove closer to the conformation of peptide-loaded MHC-I. This may optimize chaperoning during peptide loading and may also facilitate loading itself by limiting the opening of the groove so that both flanking helices can interact with incoming peptides simultaneously. The recently determined structure of an empty MHC-I protein (28) with its binding groove width constrained by a disulfide bond near dF has shown that an empty binding groove close to the peptide-loaded conformation can be loaded efficiently with a high-affinity peptide.
The MHC-I Glycan Modulates the Conformational Dynamics of the Tsn 11–20 Loop.
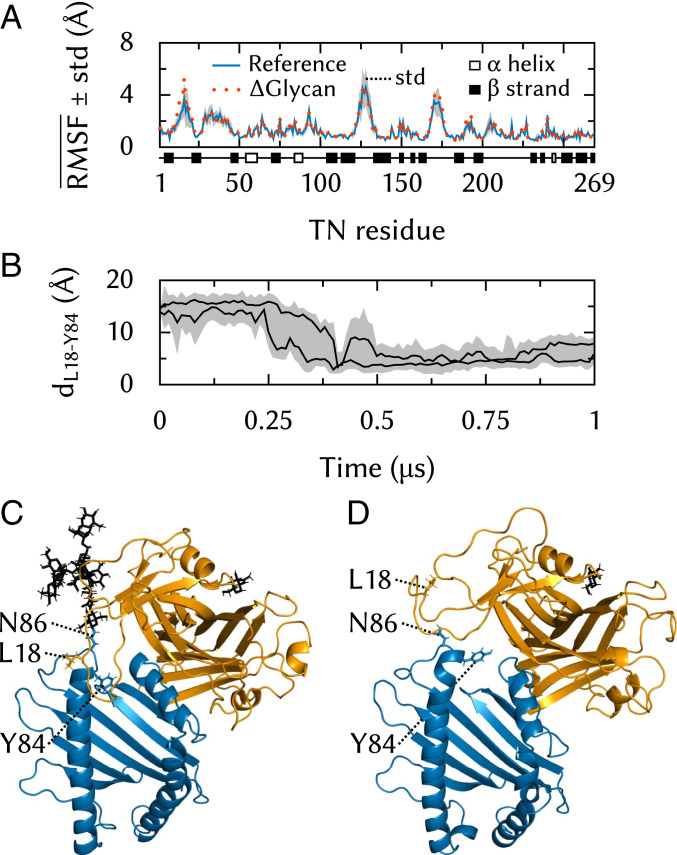
The N86-linked MHC-I glycan establishes contacts with Tsn in the vicinity of its 11–20 loop. To understand how this interaction might impact Tsn conformational dynamics, we removed the MHC-I glycan from our PLC model and performed five additional 500-ns MD simulations of this Glycan system.
The Tsn 11–20 loop exhibits similarly high fluctuations when the glycan is present on MHC-I and when it is removed (Fig. 8A). However, the conformational space explored by the loop differs. In the absence of the glycan, it fluctuates between MHC-I and the Crt glycan-binding domain (Fig. 8D). In the presence of the glycan, however, this region becomes inaccessible due to steric hindrance, and the loop is pushed toward the F-pocket region of the MHC-I binding groove (Fig. 8C). In two trajectories, the loop closes down on the groove, as can be seen from the distance (Fig. 8B) between L18, at the apex of the loop, and MHC-I Y84, which is located in the helix of the binding groove and interacts with loaded peptides at their C terminus.
Fig. 8.
Tsn 11–20 loop dynamics. (A) Effects of glycan removal on RMSF along the TN sequence. SD is shown for the reference system. (B) TN L18 to Y84 distance time series. (C) Contact between the 11–20 loop and the MHC-I binding groove in a simulation of the full PLC. (D) Typical 11–20 loop conformation in a simulation of the Glycan system.
The very large size of the systems simulated here limits the simulation times that can be reached with all-atom MD simulations. Highly flexible regions, such as the Tsn 11–20 loop, may exhibit slow dynamics on the micro- to millisecond time scale, which might be incompletely sampled in our simulations. For this reason, we can only cautiously speculate about a possible relationship between the presence of the MHC-I glycan and the motions of the loop; steric hindrance between the glycan and the loop might simply accelerate a motion that would also have been observed in the absence of the glycan, albeit at a much slower time scale. Nevertheless, the MD simulations clearly show that the two regions can establish contacts. Whether these are specific interactions that induce a structural rearrangement in TN is a hypothesis that deserves further study. Our results also show that although the Tsn 11–20 loop is much shorter than its 22–35 TAPBPR homolog, it can interact directly with the binding groove in the region of the peptide C terminus, possibly competing with ligands for binding to MHC-I during the peptide editing process.
These results are fully compatible with the partial glycan model presented with the cryo-EM PLC structure (5). However, since the cryo-EM model included only the few sugar units nearest MHC-I N86 and Crt W319, the effects of the complete, branched glycan could not be inferred in that study. Tsn L18 was identified in another work (24) as a lever that mediates peptide dissociation. Recent evidence (29) suggests instead that the 11–20 loop might act as peptide surrogate, stabilizing the empty binding groove and competing with peptides at the binding step. However, both published TAPBPRMHC-I structures (20, 21) show poor electron density in this region, in line with the pronounced conformational dynamics observed in our MD simulations. Since the Tsn and TAPBPR loops are of different lengths (11–20 in Tsn vs. 22–35 in TAPBPR), they may also have different modes of action. Clearly, more studies are needed to determine precisely the possible role of this region.
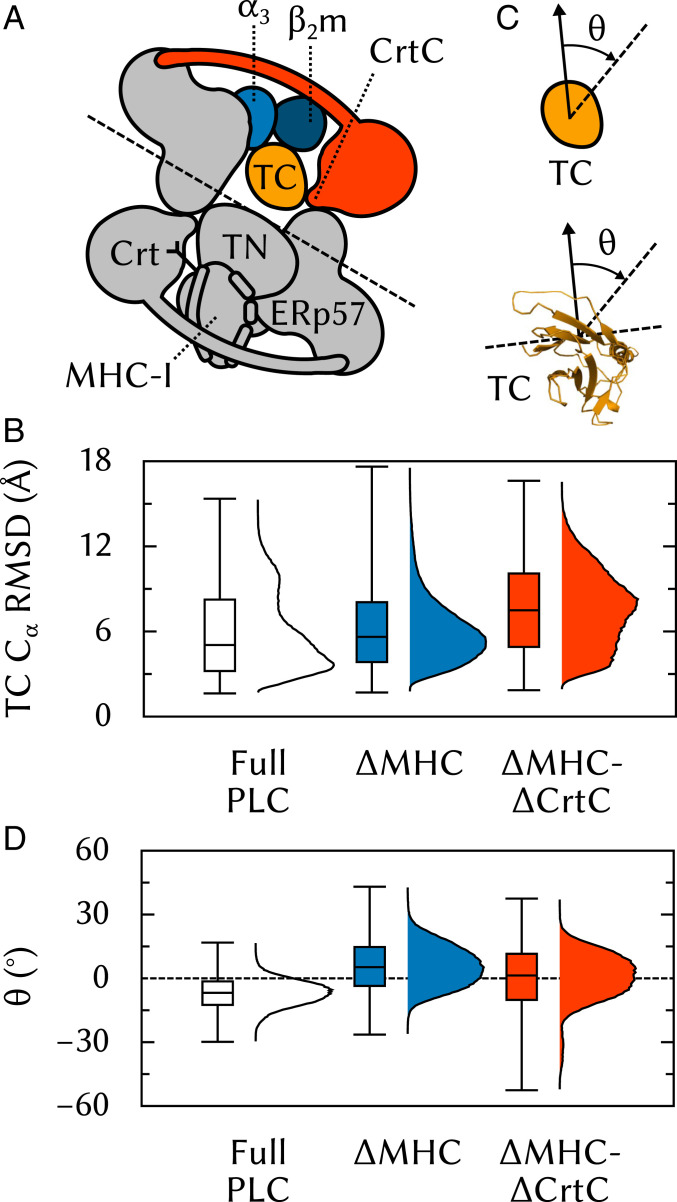
The Crt C Domain Facilitates MHC-I Binding to Tsn through Conformational Selection.
Upon assembly, TAP and two Tsn-ERp57 form an apo-PLC ready to bind suboptimally-loaded MHC-I. Crt recruit MHC-I whose glycan is glucosylated, and deliver them to the PLC (30). However, the precise sequence of events leading to the integration of these MHC-ICrt dimers into the PLC is unknown, as is the role of Crt, if any, in the assembled complex. To gain insights into the effects of MHC-I and Crt binding on the structural dynamics of the PLC, two modified PLC systems were built and subjected to five 500-ns MD simulations: one where MHC-I has been removed (MHC) and one where both MHC-I and the C domain of Crt have been removed (MHC-CrtC). It should be stressed that these two systems do not correspond to in vivo situations: Since Crt delivers MHC-I to the PLC, it would not be expected for Crt to be present in the PLC while MHC-I is absent. However, these theoretical models enable testing hypotheses that are relevant to the PLC and its assembly.
Previous work (13) showed that, in the absence of MHC-I, Tsn exhibits a significant degree of rotational flexibility between its two soluble domains, TN and TC. In the TsnMHC-I complex, this degree of freedom becomes essentially frozen. Since, in the PLC, the Crt C domain contacts TC (Fig. 5, 4) at a location opposite the TC•α3 interface (Fig. 5, 3), we posited that Crt could extend the long -helix of its C domain to constrain TC to a conformation that is conducive to MHC-I binding (Fig. 9A). To test this hypothesis, we computed TC rmsd using the TN domain as a reference frame (Fig. 9B). Using the TN domain (instead of the complete protein) for structural superposition yields a measure of the TN/TC interdomain flexibility. Fig. 9B shows that this flexibility increases slightly in the absence of MHC-I. Removal of the C domain, however, leads to a more drastic increase and a broadened distribution (Fig. 9B, MHC-CrtC). The Crt C domain thus limits TN/TC interdomain fluctuations and favors a Tsn conformation similar to the one adopted in the complex, supporting the notion that Crt facilitates MHC-I recruitment to the PLC by participating in a conformational selection mechanism at the TCMHC-I interface.
Fig. 9.
Influence of the Crt C domain on Tsn TC conformational dynamics. (A) Tsn TC is lodged between MHC-I and and also contacted by the Crt C domain. (B) rmsd of TC using TN for structural superposition. (C) The average plane of the TC -sheet region that contacts MHC-I (residues 273 to 277, 293 to 298, 327 to 329, 341 to 346) is used to measure TC rotation. (D) Angle of the -sheet to its initial orientation, viewed from the top of the ER-luminal part of the PLC toward the membrane.
Previous simulations of free Tsn (13) showed that the average relative orientations of TN and TC differ substantially between free and MHC-I–bound Tsn. To investigate the TN/TC interdomain flexibility in the PLC, we analyzed the average plane of the TC -sheet that contacts MHC-I (Fig. 9C) and measured the angle between this plane and its initial orientation in the full PLC (Fig. 9D, Full PLC). The average angle is about −7° displaced with respect to the reference value, slightly turning the -sheet toward . Interestingly, in the absence of MHC-I (Fig. 9D, MHC), is on average +9° from its initial value, turning the -sheet toward where would be. Therefore, the removal of MHC-I leads to a net TC rotation of . This observation fuels the hypothesis that Tsn could store energy, provided by thermal fluctuations, upon MHC-I binding. Such TC “spring-loading” could play a role in the subsequent dissociation of antigen-loaded MHC-I, consistent with a peptide-loading signal transduction from the binding groove to , as proposed by other researchers (31, 32). Further removal of the C domain (Fig. 9B, MHC-CrtC) does not have a significant additional impact compared to MHC, suggesting that the stabilizing effect of the Crt C domain on TC is not related to the precise orientation of the -sheet.
Conclusions
In this work, we present an atomic-resolution model of the full PLC and study its structural dynamics on the microsecond time scale. All-atom molecular dynamics simulations of such large membrane-bound protein complexes are challenging not just because of system size, but also due to the variety of macromolecules involved and, often, the low affinity of their interactions. In the case of the PLC, the system extends from a cytosolic part through the ER membrane to a 10-protein ER-luminal assembly.
We have shown that the stability of the human MHC-I PLC requires two editing modules. A single-module PLC cannot assemble the circular belt formed by ERp57 and Crt that encloses the central scaffold formed by the two TsnMHC-I dimers. Our results also highlight the crucial role of Tsn in bridging PLC components together. We found five protein–protein interfaces involving Tsn (Fig. 5), both intra- and intermodule. This complex environment modulates MHC-I dynamics, resulting in a tighter binding groove that is stabilized in a peptide-receptive conformation. Furthermore, the MHC-I N86-linked glycan was found to influence the conformational dynamics of the Tsn 11–20 loop, a region likely involved in peptide editing. Experimental studies of that loop have so far not explicitly considered the effects of this nearby glycan. Finally, the MD simulations suggest that a conformational selection mechanism involving CrtC facilitates MHC-I binding to the apo-PLC by constraining the relative orientations of the Tsn TN and TC domains.
All-atom MD simulations of this 1.6-million-atom system are computationally demanding, limiting the number of different systems that could be studied. A number of questions therefore remain open. The first one is the influence of the MHC-I peptide-loading status: How would the presence of a high-affinity antigen in the MHC-I binding groove influence the affinity and stability of the various protein–protein interfaces? Until now, most studies of TsnMHC-I have neglected the context of the PLC, focusing on smaller systems that are easier to handle. Another question that warrants closer investigation is peptide translocation through the membrane and binding to MHC-I. For instance, does the cavity atop TAP, between the two Tsn TC domains, act as a peptide reservoir, and do the dual exits of that cavity steer peptide diffusion toward the MHC-I binding groove? We expect that molecular dynamics simulations, in synergistic combination with experimental approaches, can be successfully applied to answer these questions. The present work represents a key first step in this direction and opens up the way for future work.
Materials and Methods
Modeling the TsnMHC-I Interface.
An atomic-resolution experimental structure of the catalytically active subcomplex formed by MHC-I and Tsn-ERp57 is not available, but the cryo-EM structure of the PLC provides a low-resolution approximation. Previously, we obtained a structure of this subcomplex from MD simulations (13). The recent publication of two structures of the Tsn homolog TAPBPR in complex with MHC-I (20, 21) allowed us to refine the TsnMHC-I interface. Compared to free Tsn, TAPBPR in complex with MHC-I shows twisting of the N- and C-terminal soluble domains, allowing the peptide editor to lodge its C-terminal domain between MHC-I and . A similar binding mode is observed for Tsn in the structure of the PLC.
Starting from the structure of the Tsn-ERp57 conjugate (Protein Data Bank [PDB] ID 3F8U, chains A and B, which contains only soluble domains) (10), we removed the 269–271 linker between TN and TC and fitted TN-ERp57 and TC to their homologs from the TAPBPRMHC-I complex (20) separately. The linker was then rebuilt using Modeller (version 9.19) (33), as were the unresolved Tsn loops. Human MHC-I protein HLA B*4402 (PDB ID 1M6O, which contains only soluble domains) (34) was then superposed onto the mouse MHC-I protein from the structure of the TAPBPRMHC-I complex. This allele was chosen as it is known to be PLC dependent and has been extensively studied both in experiments and in simulations. Tsn and MHC-I side chains with at least one heavy atom within 2.5 Å of another heavy atom from the binding partner were rebuilt with Modeller to remove steric clashes, followed by 100 steps of steepest-descent (SD) energy minimization.
Finally, MD simulations allowed the subcomplex to relax. The structure was solvated in a periodic rhombododecahedral box (10 Å minimal distance between protein surface and box edge) with 0.15 M Na+Cl−. The resulting system was subjected to 5 ns of MD simulation with position restraints on all atoms. This was followed by unrestrained MD simulation in the isothermal-isobaric (NpT) ensemble; protein coordinates after 45 ns were used in the next step.
Fitting to the Cryo-EM Density.
The two ER-luminal editing modules are composed of the soluble regions of Tsn-ERp57, MHC-I, and Crt. We first superposed the above-described Tsn-ERp57MHC-I subcomplex to the cryo-EM density for a single editing module (EMD-3906) (5) as a rigid body using Chimera (version 1.12) (35). This approximate positioning was then refined using DireX (version 0.7) (36). We performed 300 optimization steps using a deformable elastic network (DEN) with the recommended default parameters; the DEN was applied separately to each polypeptide chain, with no restraint between the chains. Additional harmonic distance restraints were applied to conserve hydrogen bonds in the starting structure. Hydrogen bonds were determined using the FindHBond (37) Chimera program, with donor–acceptor constraints relaxed by 0.4 Å and .
To complete the positioning of the editing modules, we took the Crt partial model from the published PLC structure (PDB ID 6ENY) (5). This model is based on structures of the Crt lectin domain (PDB ID 3O0W) (38), the tip of the Crt P domain (PDB ID 1HHN) (39), and the homologous region of the calnexin P domain (PDB ID 1JHN) (40); the C domain is modeled as an -helix. We filled in missing side chains and missing regions of the P domain with Modeller and superposed the resulting model onto the cryo-EM density as described above. We then performed another round of DireX optimization (300 steps), keeping Tsn-ERp57 and MHC-I fixed and using a DEN as described above. The complete editing module was duplicated, and the two modules were fitted, as rigid bodies, to the cryo-EM density for the pseudosymmetric assembly (EMD-3904) (5).
Finally, the MHC-I N-linked glycan was modeled to bind the Crt lectin domain. The glycan sequence is N86-GlcNAc2-Man-(Man3-Glu, Man-[Man2, Man2]), where GlcNAc is N-acetyl-D-glucosamine, Man is D-mannose and Glu is D-glucose; this corresponds to a typical glucosylated high-mannose N-glycan. Topology and initial coordinates were generated with doGlycans (source code date: 17 August 2017) (41). The four sugars of the Man3-Glu moiety were then individually repositioned to reproduce the binding mode observed in the structure of the Crt lectin domain in complex with a tetrasaccharide (38). Monte Carlo (MC) sampling was then used to find a combination of dihedral angles that would allow the GlcNAc2-Man moiety to link N86 and Man3-Glu with no steric clashes. Finally, another dihedral MC search was used to orient the Man-(Man2, Man2) moiety to avoid steric clashes.
Modeling Transmembrane and Cytosolic PLC Components.
The transmembrane (TM) and cytosolic parts of the PLC comprise TAP and the single TM helices of Tsn and MHC-I. While these are not the focus of the present work, they were included in our simulations to properly position and anchor the editing modules in the membrane.
While a cryo-EM structure of TAP in complex with the ICP47 viral inhibitor has been resolved (42), this structure shows TAP in an arrested conformation with its two nucleotide-binding domains separated. In the absence of an atomic-resolution structure of TAP that could provide suitable starting coordinates for our all-atom MD simulations, we used Modeller to generate a homology model based on the heterodimeric ABC exporter TM287/288 in the apo state (PDB ID 4Q4H) (43). The TAP model was then added to the system by rigid body superposition using the cryo-EM density of the full PLC (EMD-3905) (5); while the resolution of the density in that region is low, TAP anisotropy and the protruding nucleotide-binding domains allowed unambiguous placement. Harmonic position restraints on all atoms were then applied in the MD simulations to preserve the structural integrity of the TAP homology model. Since we do not study TAP dynamics in the present work, this is sufficient for our purposes.
The Tsn and MHC-I TM helices were modeled as ideal -helices and positioned in the plane of the membrane underneath the termini of the soluble domains they connect to, yielding a topology similar to what has been reported (5). Previously, we proposed the structure and binding mode of the TMD0 domain from TAP1 (44). Given the position of the core TAP domains and Tsn TM helix, the only location TMD0 can occupy in the membrane without steric clashes is between the Tsn and MHC-I TM helices (Fig. 1). At this position, TMD0 is close enough to the core TAP domains for the flexible linker to connect them. TMD0 and the Tsn TM helix were then oriented to reproduce the published binding mode (44). Since there is no available structure of the TMD0 domain from TAP2, we used the one from TAP1 for both editing modules; since both TMD0s are four-helix bundles and share the same binding mode (44), this should have little impact on overall PLC behavior. Membrane insertion depth was determined using the Orientation of Proteins in Membranes (OPM) Positioning of Proteins in Membrane (PPM) (45) server independently for the core TAP domains, the MHC-I TM helix, and the subcomplex formed by TMD0 and the Tsn TM helix. The three flexible linkers were built, connecting TMD0 to the core TAP domains, the Tsn TM helix to the Tsn soluble domains, and the MHC-I TM helix to the MHC-I soluble domains. The structure was finally energy minimized for 100 SD steps.
MD Simulation System Setup.
A preequilibrated 128-lipid 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer was overlaid onto the PLC structure at the previously determined insertion depth. The membrane normal and the long axis of the PLC lie in the z dimension. To accommodate the full PLC, the bilayer was replicated fourfold in the x and y dimensions and the protein complex was centered on the resulting 2,048-lipid bilayer. The water on both sides of the lipid bilayer was then extended in the z dimension by replicating a 216-molecule preequilibrated water box as needed to accommodate the full PLC and a 20-Å buffer on both sides. The membed (46) GROMACS program was then used to embed the protein structure in this orthorhombic lipid/water box. Membed proceeded over 1,000 steps of simulation and considered five TM fragments: TAP core, the two MHC-I TM helices, and the two subcomplexes formed by TMD0 and its bound Tsn TM helix. After embedding was complete, all waters whose oxygen was closer than 4.0 Å to lipid or protein heavy atoms were removed, thus avoiding trapping waters inside the membrane or in small cavities inside or between protein subunits. Na+Cl− ions were then added to a 0.15-M concentration by replacing randomly chosen water molecules. A 100 SD steps energy minimization was then performed. Finally, 50 ns of MD simulation was performed in the constant temperature and lateral area ensemble (frozen xy plane, pressure coupling applied only in the z dimension) with position restraints on all PLC heavy atoms. This allowed the box size to stabilize and the water and lipids to equilibrate around the complex. The final system (Fig. 2) contains 1.6 million atoms in an approximately 240 240 300 Å3 box.
All deletion systems were built using a similar protocol. The vacuum created by the removal of selected PLC regions was filled by water molecules, except in the TM regions. This was followed by 50 ns of MD simulation in the constant temperature and lateral area ensemble with position restraints on all PLC heavy atoms.
MD Simulations.
Simulations were carried out with GROMACS (version 5.1.2) (47). The Amber99SB*-ILDNP protein force field (48–53), the united-atom Berger lipid force field (54), the GLYCAM06h carbohydrate force field (55), and the TIP3P water model (56) were used. The SETTLE (57) and LINCS (58) algorithms were applied to constrain the internal degrees of freedom of water molecules and the bonds in other molecules, respectively. To allow for a 4-fs integration time step, we used virtual site hydrogens (59) for proteins. Since GLYCAM does not support virtual sites, we used hydrogen mass repartitioning (HMR) (60) instead to slow down vibrations involving hydrogen atoms in the carbohydrate moieties. The Berger lipids intrinsically allow for a 4-fs integration time step since they contain no explicit hydrogens. Short-range nonbonded Coulomb and Lennard-Jones 6-12 interactions were treated with a Verlet buffered pair list (61) with smoothly shifted potentials. Long-range Coulomb interactions were treated by the particle mesh Ewald (PME) method (62). Analytical dispersion corrections were applied for energy and pressure to compensate for the truncation of the Lennard-Jones interactions. The thermodynamic ensemble was NpT unless otherwise stated. Temperature was kept constant at 298 K by a velocity-rescaling thermostat with a stochastic term (63), with coupling time constant 0.1 ps. For constant 1.0-bar pressure, a semiisotropic (xy, z) Berendsen barostat (64) was used with coupling time constants of 2.0 ps and bar−1 compressibility. Isotropic pressure coupling was used instead for the Tsn-ERp57MHC-I simulations, which do not involve a bilayer.
For each system, five 500- to 1,000-ns (see Results and Discussion for details) trajectories were acquired by generating random initial velocities from a Maxwell–Boltzmann distribution at 298 K. Coordinates were recorded every 10 ps. All of these simulations were preceded by the following two-step equilibration protocol (in addition to the box equilibration performed at the end of the system generation protocol described above). First, 50 ns of simulation was carried out with position restraints on allCα atoms; this allowed protein–protein interfaces to stabilize through side-chain rotations. Second, 100 ns of simulation was performed with position restraints on Crt Cα atoms; this allowed the CrtERp57 interface, which has a low resolution in cryo-EM, to stabilize. Production trajectories were then performed without these position restraints.
To ensure that an interaction similar to the one observed in the cryo-EM structure (44) is also stably established in our MD simulations, one harmonic distance restraint for each editing module was added between the Crt W319 main chain nitrogen and C5 from the closest mannose in the MHC-I glycan (50,000 kJmol−1⋅nm−2 force constant, 4.23 Å equilibrium distance). Test simulations showed that, without this restraint, the glycan does not remain in the binding pocket of the Crt lectin domain.
Analysis.
Properties were computed after discarding the first 100 ns of each production trajectory to allow relaxation toward the equilibrium. Properties are shown as box/distribution plots; boxes extend from to SD with a median bar, while whiskers encompass all data points. Time series are shown for entire trajectories including equilibration time, with running averages over 10 ns; time series envelopes (minimum and maximum) are shown alongside the running average.
Acknowledgments
This work was funded by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy–EXC 2033–390677874–RESOLV. We gratefully acknowledge the Gauss Center for Supercomputing (GCS) e.V. (http://www.gauss-centre.eu) for funding this project by providing computing time on the GCS Supercomputer SuperMUC at Leibniz Supercomputing Center (http://www.lrz.de). We thank Robert Tampé for useful discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability.
The model of the PLC was deposited in the ModelArchive (https://www.modelarchive.org/doi/10.5452/ma-whom2). The structure, in PDB format, contains all protein subunits and glycans and incorporates protein RMSF (in angstroms) for cytosolic components, stored in the column typically used for B factors. An average structure was computed for each protein chain and used as the reference frame to compute fluctuations. Since the flexible P domain of Crt leads to a large RMSF range, the visualization of fluctuations inside a given subunit is better done by adjusting the scale for that specific subunit rather than for the whole complex; this is especially the case for the more stable MHC-I and Tsn. All study data are included in this article.
References
- 1.Neefjes J., Jongsma M. L. M., Paul P., Bakke O., Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Blum J. S., Wearsch P. A., Cresswell P., Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rock K. L., Reits E., Neefjes J., Present yourself! by MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulpke S., Tampé R., The MHC I loading complex: A multitasking machinery in adaptive immunity. Trends Biochem. Sci. 38, 412–420 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Blees A., et al. , Structure of the human MHC-I peptide-loading complex. Nature 551, 525–528 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Elliott T., van Hateren A., Protein plasticity and peptide editing in the MHC I antigen processing pathway. Biochemistry 57, 1423–1425 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Ayres C. M., et al. , Dynamically driven allostery in MHC proteins: Peptide-dependent tuning of class I MHC global flexibility. Front. Immunol. 10, 966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Momburg F., Tan P., Tapasin: The keystone of the loading complex optimizing peptide binding by MHC class I molecules in the endoplasmic reticulum. Mol. Immunol. 39, 217–233 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Chen M., Bouvier M., Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 26, 1681–1690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong G., Wearsch P. A., Peaper D R., Cresswell P., Reinisch K. M., Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity 30, 21–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams A. P., Peh C. A., Purcell A. W., McCluskey J., Elliott T., Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity 16, 509–520 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Fleischmann G., et al. , Mechanistic basis for epitope proofreading in the peptide-loading complex. J. Immunol. 195, 4503–4513 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Fisette O., Wingbermühle S., Tampé R., Schäfer L. V., Molecular mechanism of peptide editing in the tapasin–MHC I complex. Sci. Rep. 6, 19085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peaper D. R., Wearsch P. A., Cresswell P., Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 24, 3613–3623 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadasivan B., Lehner P. J., Ortmann B., Spies T., Cresswell P.. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity 5, 103–114 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Williams D. B., Beyond lectins: The calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 119, 615–623 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Trowitzsch S., Tampé R., ABC transporters in dynamic macromolecular assemblies. J. Mol. Biol. 430, 4481–4495 (2018). [DOI] [PubMed] [Google Scholar]
- 18.van de Weijer M. L., Luteijn R. D., Wiertz E. J. H. J., Viral immune evasion: Lessons in MHC class I antigen presentation. Semin. Immunol. 27, 125–137 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Praest P., Liaci A. M., Förster F., Wiertz E J. H. J., New insights into the structure of the MHC class I peptide-loading complex and mechanisms of TAP inhibition by viral immune evasion proteins. Mol. Immunol. 113, 103–114 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Thomas C., Tampé R., Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science 365, 1060–1064 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Jiang J., et al. , Crystal structure of a TAPBPR-MHC-I complex reveals the mechanism of peptide editing in antigen presentation. Science 358, 1064–1068 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panter M. S., Jain A., Leonhardt R. M., Ha T., Cresswell P., Dynamics of major histocompatibility complex class I association with the human peptide-loading complex. J. Biol. Chem. 287, 31172–31184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas C., Tampé R., MHC I chaperone complexes shaping immunity. Curr. Opin. Immunol. 58, 9–15 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Tudor Ilca F., et al. , TAPBPR mediates peptide dissociation from MHC class I using a leucine lever. eLife 7, e40126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hafstrand I., et al. , Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin. Proc. Natl. Acad. Sci. U.S.A. 116, 5055–5060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boelt S. G., et al. , Mapping the Ca(2+) induced structural change in calreticulin. J. Proteomics 142, 138–148 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Fisette O., Wingbermühle S., Schäfer L. V., Partial dissociation of truncated peptides influences the structural dynamics of the MHCI binding groove. Front. Immunol. 8, 408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anjanappa R., et al. , Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection. Nat. Commun. 11, 1314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagert L., Hennig F., Thomas C., Tampé R., A loop structure allows TAPBPR to exert its dual function as MHC I chaperone and peptide editor. eLife 9, e55326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao B., et al. , Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity 16, 99–109 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Bailey A., et al. , Selector function of MHC I molecules is determined by protein plasticity. Sci. Rep. 5, 14928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hateren A., et al. , Direct evidence for conformational dynamics in major histocompatibility complex class I molecules. J. Biol. Chem. 292, 20255–20269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eswar N., et al. , Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 5, Unit 5.6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macdonald W. A., et al. , A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198, 679–691 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen E. F., et al. , UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Wang Z., Schröder G. F., Real-space refinement with DireX: From global fitting to side-chain improvements. Biopolymers 97, 687–697 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Mills J. E. J., Dean P. M., Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput. Aided Mol. Des. 10, 607–622 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Kozlov G., et al. , Structural basis of carbohydrate recognition by calreticulin. J. Biol. Chem. 285, 38612–38620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellgaard L., et al. , NMR structure of the calreticulin P-domain. Proc. Natl. Acad. Sci. U.S.A. 98, 3133–3138 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schrag J. D., et al. , The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8, 633–644 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Danne R., et al. , doGlycans–Tools for preparing carbohydrate structures for atomistic simulations of glycoproteins, glycolipids, and carbohydrate polymers for GROMACS. J. Chem. Inf. Model. 57, 2401–2406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldham M. L., et al. , A mechanism of viral immune evasion revealed by cryo-EM analysis of the TAP transporter. Nature 529, 537–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hohl M., Briand C., Grütter M. G., Seeger M. A., Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat. Struct. Mol. Biol. 19, 395–402 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Blees A., et al. , Assembly of the MHC I peptide-loading complex determined by a conserved ionic lock-switch. Sci. Rep. 5, 17341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf M. G., Hoefling M., Aponte-Santamaría C., Grubmüller H., Groenhof G., g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 31, 2169–2174 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Pronk S., et al. , GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornell W. D., et al. , A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995). [Google Scholar]
- 49.Wang J., Cieplak P., Kollman P. A., How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules?. J. Comput. Chem. 21, 1049–1074 (2000). [Google Scholar]
- 50.Hornak V., et al. , Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Best R. B., Hummer G., Optimized molecular dynamics force fields applied to the helix-coil transition of polypeptides. J. Phys. Chem. B 113, 9004–9015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindorff-Larsen K., et al. , Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aliev A. E., et al. , Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins 82, 195–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berger O., Edholm O., Jähnig F., Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72, 2002–2013 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirschner K. N., et al. , GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 57.Miyamoto S., Kollman P. A., Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992). [Google Scholar]
- 58.Hess B., P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Feenstra K.A., Hess B., Berendsen H. J. C., Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20, 786–798 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Hopkins C. W., Le Grand S., Walker R. C., Roitberg A. E., Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Páll S., Hess B., A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 184, 2641–2650 (2013). [Google Scholar]
- 62.Essmann U., et al. , A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995). [Google Scholar]
- 63.Bussi G., Donadio D., Parrinello M., Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., DiNola A., Haak J. R., Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The model of the PLC was deposited in the ModelArchive (https://www.modelarchive.org/doi/10.5452/ma-whom2). The structure, in PDB format, contains all protein subunits and glycans and incorporates protein RMSF (in angstroms) for cytosolic components, stored in the column typically used for B factors. An average structure was computed for each protein chain and used as the reference frame to compute fluctuations. Since the flexible P domain of Crt leads to a large RMSF range, the visualization of fluctuations inside a given subunit is better done by adjusting the scale for that specific subunit rather than for the whole complex; this is especially the case for the more stable MHC-I and Tsn. All study data are included in this article.