Significance
Tissue-resident macrophages can arise from multiple origins during embryonic and adult hematopoiesis. They are important cells regulating a wide range of biological processes, such as tissue remodeling during organogenesis, tissue homeostasis in the steady state, tissue repair following injury, and immune response to pathogens. Although the origins and tissue-specific functions of resident macrophages have been extensively studied in many other tissues, they are not well characterized in skeletal muscle. Our study characterized the origins, tissue-specific gene expression, and diverse functions of skeletal muscle-resident macrophages in the steady state. Our findings provide a knowledge base for future studies of the roles of skeletal muscle-resident macrophages in various disease states.
Keywords: skeletal muscle, resident macrophage, ontogeny, transcriptome, functional subsets
Abstract
Tissue-resident macrophages can originate from embryonic or adult hematopoiesis. They play important roles in a wide range of biological processes including tissue remodeling during organogenesis, organ homeostasis, repair following injury, and immune response to pathogens. Although the origins and tissue-specific functions of resident macrophages have been extensively studied in many other tissues, they are not well characterized in skeletal muscle. In the present study, we have characterized the ontogeny of skeletal muscle-resident macrophages by lineage tracing and bone marrow transplant experiments. We demonstrate that skeletal muscle-resident macrophages originate from both embryonic hematopoietic progenitors located within the yolk sac and fetal liver as well as definitive hematopoietic stem cells located within the bone marrow of adult mice. Single-cell-based transcriptome analyses revealed that skeletal muscle-resident macrophages are distinctive from resident macrophages in other tissues as they express a distinct complement of transcription factors and are composed of functionally diverse subsets correlating to their origins. Functionally, skeletal muscle-resident macrophages appear to maintain tissue homeostasis and promote muscle growth and regeneration.
Macrophages (MPs) are heterogeneous and multifunctional cells that are critical regulators of tissue homeostasis, repair, immunity, and disease pathogenesis. Tissue MPs consist of two classes: resident MPs and infiltrating monocyte (MO)-derived MPs. In adult mammals, while resident MPs are present in all tissues, infiltrating inflammatory MPs derived from blood Ly6Chi inflammatory MOs are found in a disease state such as tissue injury. Although MPs are classically identified as innate immune cells functioning in the activation and resolution of tissue inflammation, it is now clear that they play roles in a much wider range of biological processes, such as tissue remodeling during organogenesis, tissue homeostasis in the steady state, tissue repair, and immune response to pathogens (1–5). They are critically involved in a variety of disease processes, such as chronic inflammatory diseases, tumor growth and metastasis, and tissue fibrosis (4–7). As MPs represent an attractive therapeutic target, understanding their respective origins, tissue-specific characteristics, and disease-related functions is absolutely essential to harness their therapeutic potential.
In the steady state, most tissue-resident MP populations are established prenatally from two embryonic progenitors: primitive yolk sac MPs and fetal liver MOs (i.e., fetal MOs) (3, 8–15). Primitive yolk sac MPs originate from early erythro-myeloid progenitors (EMPs) which emerge in yolk sac at embryonic day 7 (E7) in mice. The early EMPs express colony stimulating factor 1 receptor (CSF1R) but not the transcription factor (TF) c-Myb. They differentiate into primitive MPs and migrate to embryonic tissues beginning at E9.5. The c-myb+ late EMPs emerge in yolk sac at E8.5. They migrate into fetal liver and differentiate into fetal MOs at E12.5 independently of CSF1R. Recent studies also reported the existence of embryonic hematopoietic progenitors other than EMPs, which made a relatively minor contribution to the fetal MOs (8, 9), such as the Flt3+ lympho-myeloid progenitors (LMPs) (16). Fetal liver MOs seed numerous embryonic tissues except for brain (14). Within individual tissues, primitive MPs and fetal MOs differentiate into tissue-specific resident MPs. They persist into adulthood through proliferative self-renewal. Prehematopoietic stem cells (HSCs) first appear at the aorta-gonad-mesonephros at E9.5 and then seed fetal liver around E10.5, where they differentiate into mature HSCs (9). Whether HSCs contribute to fetal MOs is still in debate (8, 17). Mature HSCs migrate into nascent bone marrow (BM) at a late embryonic stage and give rise to blood MOs (adult MOs) after birth. Adult MOs contribute to resident MPs after birth in several tissues, such as intestine (18), heart (19), peritoneum (20), skin (21), and liver (22), but not brain (14). Although the origins of resident MPs have been characterized in most of the tissues, they have not been well studied in skeletal muscle.
Resident MPs express tissue-specific transcription factors (TFs) and exert tissue-specific functions in the steady state in many tissues (12, 20, 23–27). The tissue-specific TFs that are expressed by resident MPs include GATA-6 by large peritoneal MPs and C/EBPβ by lung alveolar MPs, among others. Resident MPs acquire tissue-specific functionalities via differential activation of TFs in response to specific cues within the tissue microenvironment (24, 26, 28). The tissue-specific gene expression and function of skeletal muscle-resident MPs (SMRMPs) in the steady state remains to be explored.
Previous studies by our laboratory and others have demonstrated that intramuscular MPs are important effectors and regulators of muscle inflammation, fibrosis, and regeneration associated with acute skeletal muscle injury repair and muscular dystrophy (29–32). Therapeutic manipulations of MPs hold promise for promoting muscle injury repair and improving outcomes for individuals with muscular dystrophy. In order to develop MP-targeted therapies, it is necessary to dissect the diversity, origins, and functions of intramuscular MPs. In the present study, we identified the origins, tissue-specific transcriptome, and diverse functional subsets of resident MPs in mouse skeletal muscle in the steady state.
Results
CD45+CD11b+F4/80+CD64+ MPs Are Detected in Skeletal Muscle in the Steady State.
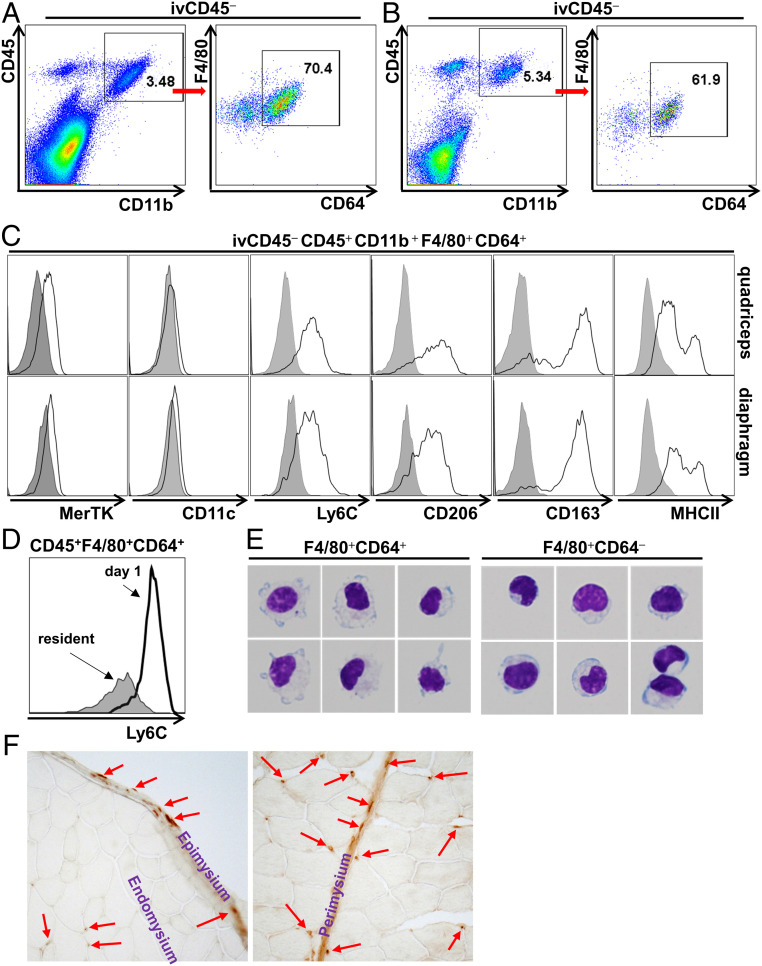
Steady-state SMRMPs were detected by flow cytometry analysis (FACS) and immunostaining in both limb muscle (quadriceps) and respiratory muscle (diaphragm) at 4 to 6 wk of age (Fig. 1). To exclude the potential contamination by immune cells within intramuscular blood vessels, we injected mice (intravenously) with a fluorescence-labeled anti-CD45 antibody. We included a separate anit-CD45 antibody conjugated to a different fluorophore in our staining panel to differentiate MPs located within the intravascular and tissue compartments. Resident MPs, gated as ivCD45−CD45+CD11b+F4/80+CD64+ cells (19, 33), were detected in both quadriceps (Fig. 1A) and diaphragm (Fig. 1B). They were MerTK+CD11c− (Fig. 1C and SI Appendix, Fig. S1A) and expressed Ly6C (Fig. 1C and SI Appendix, Fig. S1A). However, Ly6C expression level was significantly lower than that of the Ly6Chi infiltrating inflammatory MPs in injured muscle 1 d after intramuscular BaCl2 injection (Fig. 1D). We also noted an ivCD45−CD45+CD11b+F4/80+CD64− population (Fig. 1 A and B). These cells did not express Ly6G, indicating that they are not neutrophils (SI Appendix, Fig. S1B), and only a small percentage (∼30% in quadriceps and 13% in diaphragm) expressed the eosinophil marker Siglec F (SI Appendix, Fig. S1C). Giemsa staining (Fig. 1E) of sorted ivCD45−CD45+CD11b+F4/80+CD64+ and ivCD45−CD45+CD11b+F4/80+CD64− cells revealed that the CD64+ cells displayed typical MP morphology with irregular cytoplasmic border and cytoplasmic vacuoles. The CD64− population consisted of cells displaying mixed morphologies, mostly with MO-like morphology with an eccentrically placed nucleus occupying over half of the cell area. Consistent with a resident MP phenotype, ivCD45−CD45+CD11b+F4/80+CD64+ MPs expressed CD163 and CD206 (Fig. 1C and SI Appendix, Fig. S1A). They also expressed major histocompatibility complex class II (MHCII) and, based on the expression level, could be divided in to MHCIIhi and MHCIIlow subpopulations (Fig. 1C and SI Appendix, Fig. S1A). CD68 immunostaining showed that the CD68+ resident MPs were scattered in the muscle interstitial tissues, including epimysium, perimysium, and endomysium (Fig. 1F).
Fig. 1.
Resident MPs are present in adult mouse skeletal muscle in the steady state. Quadriceps and diaphragm were collected from WT C57 BL/6J mice at 4 to 6 wk of age. (A and B) FACS analysis of single-cell suspension of the quadriceps (A) and diaphragm (B) identified the existence of ivCD45−CD45+CD11b+F4/80+CD64+ resident MPs. (C) FACS analysis of the expression of MerTK, CD11c, Ly6C, CD163, CD206, and MHCII by the ivCD45−CD45+CD11b+F4/80+CD64+ MPs of the quadriceps (Upper) and diaphragm (Lower); gray area: immunoglobulin G isotype control. (D) FACS analysis comparing Ly6C expression by ivCD45−CD45+CD11b+F4/80+CD64+ resident MPs in normal muscle and infiltrating inflammatory MPs in acutely injured muscle 1 d after intramuscular injection of BaCl2. (E) Giemsa staining of sorted ivCD45−CD45+CD11b+F4/80+CD64+ cells and ivCD45−CD45+CD11b+F4/80+CD64− cells. (F) CD68 immunostaining of cryosections of quadriceps. n = 10 mice per experiment.
SMRMPs Have Both HSC and Non-HSC Origins.
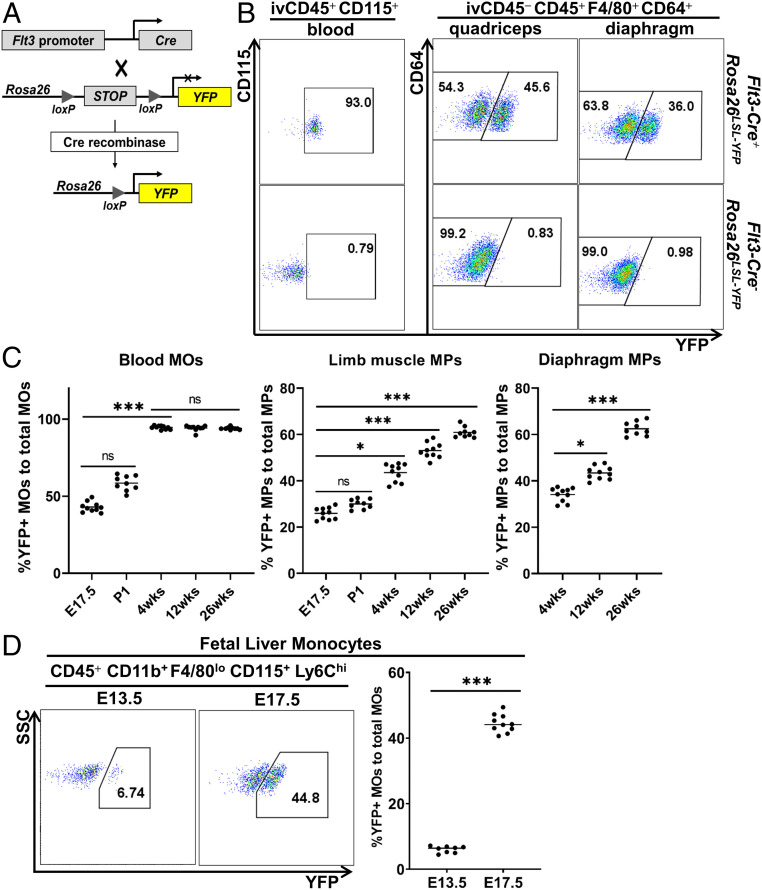
We next determined the origins of resident MPs in skeletal muscle. Since definitive HSCs transiently up-regulate growth factor fms-like tyrosine kinase 3 (Flt3) as they differentiate (17), we used Flt3Cre-Rosa26LSL-YFP mice to identify resident MPs originated from definitive HSCs (express yellow fluorescent protein, YFP) (Fig. 2A). At 4 wk of age, over 90% of blood MOs (CD45+CD115+) (34) were YFP+ (Fig. 2B). Since adult blood MOs completely originate from HSCs, incomplete Cre-lox recombination efficiency likely accounts for 10% of YFP− blood MOs (35). The percentage of the YFP+ cells in the quadriceps- or diaphragm-resident MPs was significantly lower than that in blood MOs (Fig. 2B). This finding demonstrates the existence of both HSC (YFP+) and non-HSC (YFP−) origins for SMRMPs. The percentage of the YFP+ population in both blood MOs and SMRMPs changed with age (Fig. 2C). While the YFP+ cells accounted for over 40% of the total blood MOs in embryos at E17.5, the percentage increased rapidly after birth and reached a plateau of over 90% at 4 wk. The findings suggest that definitive HSCs contribute to blood MOs, partially at the late embryonic stage but completely or nearly completely at the adult stage. In comparison, the YFP+ cells accounted for less than 30% of the total resident MPs in limb muscle at E17.5. After birth, the percentage continuously increased with age but was still significantly lower than that in blood MOs even at 26 wk of age (Fig. 2C). Similar findings were seen with the resident MPs in diaphragm from 4 to 26 wk of age (Fig. 2C). The findings demonstrate that while muscle-resident MPs of non-HSC origin persist into late adulthood, there is a continuous expansion of HSC-derived muscle-resident MPs over time.
Fig. 2.
SMRMPs arise from both HSC and non-HSC origins. Flt3Cre-Rosa26LSL-YFP mice were used for lineage tracing of HSC-derived blood MOs (ivCD45+CD115+) and SMRMPs (ivCD45−CD45+CD11b+F4/80+CD64+). (A) Scheme of Flt3Cre-Rosa26LSL-YFP lineage tracing. (B and C) Blood MOs and SMRMPs were analyzed for YFP expression by FACS at indicated ages. (B) Dot blot showing YFP expression at 4 wk of age. (C) Comparisons of the percentage of YFP+ cells in blood MOs and SMRMPs at indicated ages. (D) FACS analysis and comparison of YFP+ cells in fetal liver MOs at indicated embryonic stages. n = 8 to 10 mice per group per time point. *P < 0.05; ***P < 0.001; ns: no significance.
HSCs Contribute to Prenatal Limb Muscle-Resident MPs.
A significant percentage of YFP+ cells were present in both blood MOs and limb muscle MPs at E17.5 (Fig. 2C). These findings suggest that HSCs may contribute to fetal liver hematopoiesis, since BM hematopoiesis does not occur before birth. Consistent with this possibility, we observed a marked increase in the percentage of YFP+ cells in fetal liver MOs (CD45+CD11b+F4/80loCD115+Ly6Chi) (14, 34) from <7% at E13.5 to >40% at E17.5 (Fig. 2D). The percentages of the YFP+ MOs in fetal liver and peripheral blood were similar at E17.5 (Fig. 2 C and D). The findings suggest that HSCs contribute, in part, to the fetal liver hematopoiesis and fetal MOs at late embryonic stages, which subsequently contribute to resident MPs in prenatal limb muscle.
BM HSC-Derived Blood MOs Contribute to Postnatal SMRMPs.
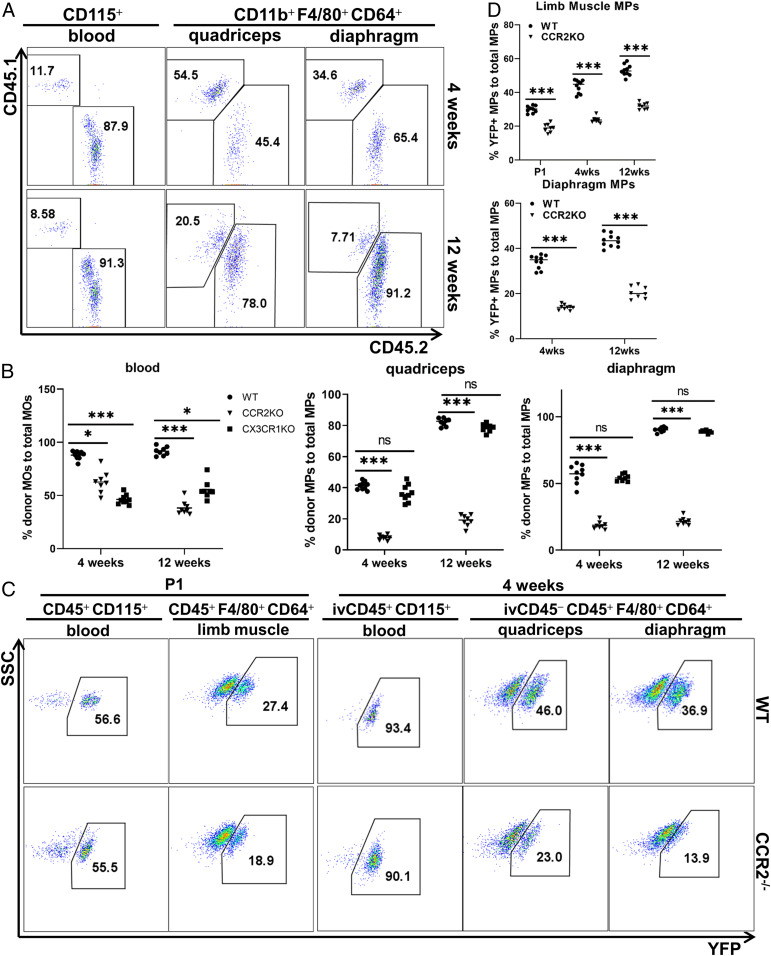
The postnatal continuous increase in the percentage of HSC-originated SMRMPs suggests that skeletal muscle in the steady state may recruit BM-derived blood MOs to replenish SMRMPs. Alternatively, the proliferation of the YFP+ MPs may be faster than that of the YFP− MPs. To address these possibilities, we first performed Ki67 staining to label intramuscular proliferative SMRMPs and found no difference in either the percentage of the Ki67+ cells or the expression level of Ki67 between YFP+ and YFP− MP subpopulations (SI Appendix, Fig. S2). We then performed BM transplant (BMT) experiments to address the recruitment of BM-derived blood MOs by postnatal skeletal muscle. BM cells from CD45.2 wild-type (WT) donor mice were transferred into sublethally irradiated CD45.1 WT recipients. A nearly complete replacement of the recipient blood MOs by the donor BM-derived MOs was achieved 4 wk after the transplantation. Donor-derived (CD45.2+) SMRMPs were detected in both quadriceps and diaphragm of the recipient mice with the percentage continuously increased until 12 wk posttransplantation (Fig. 3A). The findings indicate active recruitment of the donor BM-derived MOs to replenish SMRMPs in the recipients. The recruitment of adult HSC-derived MOs appeared faster in irradiated mice (Fig. 3A) than in nonirradiated mice (Fig. 2C). This is most likely due to the damage and/or impairment of self-renewal ability of resident MPs by irradiation, as a previous study showed that irradiation depleted resident MPs in lung and spleen, and the recovery after BMT mainly depended on the recruitment of donor-derived circulating progenitors rather than the proliferation of local MPs (3).
Fig. 3.
BM HSC-derived blood MOs contribute to postnatal SMRMPs. (A and B) BMT from indicated donors (CD45.2 background) into sublethally irradiated recipients (CD45.1 background). Blood MOs and SMRMPs were then analyzed for the expression of CD45.1 and CD45.2. (A) FACS analysis of CD45.2 expression by blood MOs and SMRMPs in recipients receiving WT CD45.2 BM cells at 4 (Upper) and 12 wk (Lower) posttransplantation. (B) Comparisons of the percentage of CD45.2+ cells in blood MOs and SMRMPs in recipients receiving BM cells from indicated donors at 4 and 12 wk posttransplantation. (C and D) FACS analysis of YFP expression by blood MOs and SMRMPs in Flt3Cre-Rosa26LSL-YFP-Ccr2−/− mice (CCR2−/−) and Flt3Cre-Rosa26LSL-YFP controls (WT) at indicated ages. (C) Dot blot showing YFP expression at P1 and 4 wk of age. (D) Comparisons of the percentage of YFP+ cells in blood MOs and SMRMPs at indicated ages. n = 8 to 10 mice per group per time point per experiment. *P < 0.05; ***P < 0.001; ns: no significance.
There are two subsets of blood MOs: CCR2+CX3CR1loLy6Chi and CCR2−CX3CR1hiLy6Clo subsets (36). C–C chemokine receptor 2 (CCR2) and CX3C chemokine receptor 1 (CX3CR1) mediate the tissue recruitment of CCR2+ and CX3CR1hi cells, respectively. To address which subset of the blood MOs is recruited to replenish SMRMPs, we performed BMT experiments using BM cells from Ccr2−/− and Cx3cr1−/− donor mice. The replenishment of SMRMPs by the donor BM-derived MOs was severely impaired by CCR2 deficiency but only marginally affected by the CX3CR1 deficiency. Notably, both Ccr2−/− and Cx3cr1−/− mice had partial reductions in blood MOs at baseline (Fig. 3B and SI Appendix, Fig. S3A). These findings indicate that skeletal muscle preferentially recruits CCR2+CX3CR1loLy6Chi blood MOs via CCR2 to replenish resident MPs.
To further address whether skeletal muscle recruits blood MOs in a true steady state without irradiation, we generated Flt3Cre-Rosa26LSL-YFP and Flt3Cre-Rosa26LSL-YFP/Ccr2−/− mice and analyzed YFP expression in SMRMPs. CCR2 deficiency did not significantly impact the number of SMRMPs in either quadriceps or diaphragm (SI Appendix, Fig. S3B). CCR2 deficiency also did not change the percentage of YFP+ blood MOs at postnatal day 1 (P1) or 4 wk of age (Fig. 3C), but it did cause a significant reduction in the percentage of YFP+ SMRMPs (Fig.3 C and D). In the Flt3Cre-Rosa26LSL-YFP control mice, the percentage of the intramuscular CCR2+ MPs, which was very small in limb muscle at P1, increased and accounted for over 30% of the YFP+ MPs. YFP− MPs were largely CCR2− in both quadriceps and diaphragm at 4 wk of age (SI Appendix, Fig. S3C). Taken together, these findings support a significant contribution of the BM HSC-derived CCR2+CX3CR1lo Ly6Chi blood MOs to the postnatal SMRMPs in the steady state.
MHCIIhiLyve1lo and MHCIIloLyve1hi Subsets of SMRMPs Derive from Different Origins Postnatally.
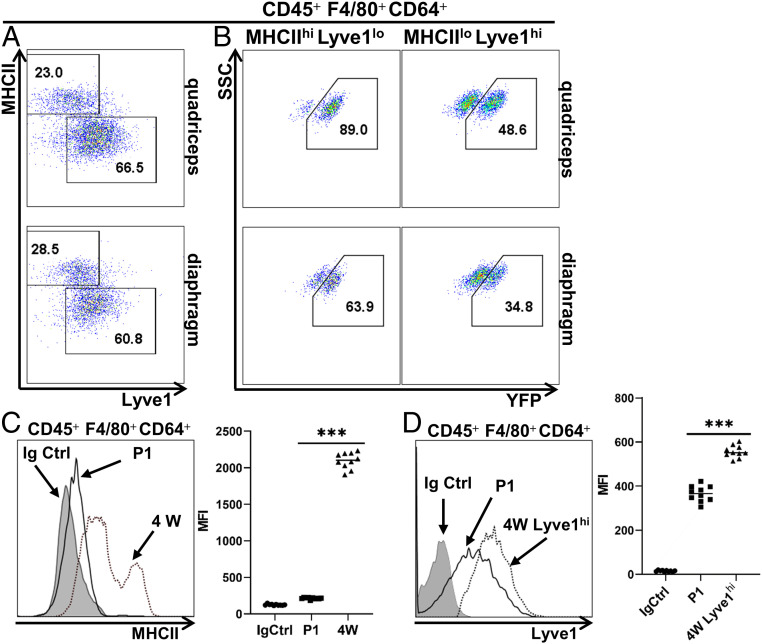
Chakarov et al. (37) reported the coexistence of two distinct interstitial MP subsets across tissues, MHCIIhiLyve1lo and MHCIIloLyve1hi MPs. In lung, both subsets were progressively replenished by blood MOs after birth (37). Using Flt3Cre-Rosa26LSL-YFP mice, we addressed whether these two subsets were also present in skeletal muscle, and whether the HSC hematopoiesis contributed to them. FACS analysis revealed the presence of both MHCIIhiLyve1lo and MHCIIloLyve1hi subsets in quadriceps and diaphragm at 4 wk of age (Fig. 4A), and the two subsets differed significantly in YFP expression (Fig. 4B). In quadriceps, 89% of the MHCIIhiLyve1lo MPs were YFP+ (Fig. 4 B, Upper Left), comparable to the percentage of the YFP+ cells in blood MOs. These data suggest that MHCIIhiLyve1lo MPs may be derived from blood MOs of HSC origin. In contrast, only about a half of the MHCIIloLyve1hi MPs were YFP+ (Fig. 4 B, Upper Right), suggesting that HSC and non-HSC origins both contributed to the MHCIIloLyve1hi MP subset. In the diaphragm, 63.9% of MHCIIhiLyve1lo MPs were YFP+ (Fig. 4 B, Lower Left), suggesting that this subset derived from both HSC and non-HSC origins. The MHCIIloLyve1hi MP subset in diaphragm contained 34.8% of YFP+ cells (Fig. 4 B, Lower Right), which was also lower than that in quadriceps (48.6%), suggesting that this subset also derived from both HSC and non-HSC origins with a preferential contribution from non-HSC progenitors.
Fig. 4.
MHCIIhiLyve1lo and MHCIIloLyve1hi subsets of SMRMPs arise from different origins postnatally. (A) FACS analysis of MHCII and Lyve1 expression by SMRMPs in quadriceps (Upper) and diaphragm (Lower) of the Flt3Cre-Rosa26LSL-YFP mice at 4 wk of age. (B) Expression of YFP by gated MHCIIhiLyve1lo and MHCIIloLyve1hi subpopulations from A. (C) Histogram comparing the MHCII expression by limb muscle resident MPs at P1 and 4 wk (4W). Scatter plot shows quantification based on mean fluorescence intensity (MFI). (D) Histogram comparing the Lyve1 expression by limb muscle-resident MPs at P1 and by the MHCIIloLyve1hi MP subpopulation in limb muscle at 4 wk. Immunoglobulin G (IgG) isotype control was done with MPs from 4W limb muscle, which was similar to P1. Scatter plot shows quantification based on MFI. n = 10 mice per time point per experiment. ***P < 0.001.
The expression of MHCII and Lyve1 by SMRMPs was also analyzed in neonatal limb muscle at P1. Surprisingly, the MHCII expression was minimally detectable (Fig. 4C). The Lyve1 expression by SMRMPs at P1 was also significantly lower than that by the quadriceps MHCIIloLyve1hi MPs at 4 wk (Fig. 4D). Therefore, the differentiation of the MHCIIhiLyve1lo and MHCIIloLyve1hi MP subsets in limb muscle appears to start postnatally.
Yolk Sac Primitive MPs and Fetal MOs of Non-HSC Origin Contribute to Adult SMRMPs.
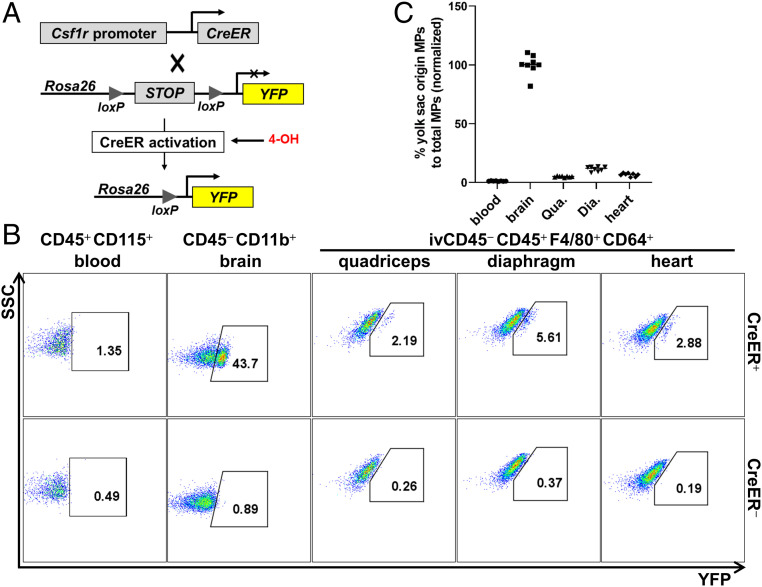
Since the YFP− SMRMPs identified in Fig. 2 could derive from yolk sac primitive MPs, non-HSC-derived fetal MOs, or both (17), we next determined the origins of these cells. We used a 4-hydroxytamoxifen (4-OH)–induced lineage tracing system (Csf1CreER+-Rosa26LSL-YFP mice) (Fig. 5A) to specifically label yolk sac primitive MPs and their progeny (see SI Appendix, Supplementary Materials and Methods for details) based on the fact that CSF1R is expressed by yolk sac early EMP and primitive MPs (11). SMRMPs were analyzed for YFP expression at 4 wk of age. Brain microglia, which completely originate from yolk sac primitive MPs (11, 15), were used as a positive control. Adult blood MOs which arise from HSCs were used as a negative control. Heart-resident MPs, known to contain yolk sac-derived resident MPs, were also included as a control (8, 9, 19). As shown in Fig. 5B, while almost no YFP+ cells were detected in blood MOs, 43.7% of microglia were YFP+. Since microglia completely arise from yolk sac primitive MPs, the observed number of YFP+ cells reflects the incomplete efficiency of 4-OH–induced Cre-lox recombination. A small percentage of quadriceps-, diaphragm-, and cardiac-resident MPs were YFP+ cells. No YFP+ cells were observed in Csf1rCreER--Rosa26LSL-YFP littermate controls (Fig. 5B). After normalizing to the percentage of YFP+ brain microglia, the estimated ratio of yolk sac-derived MPs to total resident MPs was about 5.0%, 12.8%, and 6.6% in the quadriceps, diaphragm, and heart, respectively (Fig. 5C). Since more than 50% of SMRMPs at 4 wk of age are derived from non-HSC origin (Fig. 2C), these findings may suggest that fetal MOs of non-HSC origin predominately contribute to SMRMPs at 4 wk of age. Alternatively, microglia and yolk sac-derived MPs in the skeletal muscle and heart may be specified at different time points or display preferentially labeling efficiency.
Fig. 5.
Yolk sac primitive MPs and fetal MOs of non-HSC origin contribute to adult SMRMPs. Csf1rCreER+-Rosa26LSL-YFP mice (CreER+) and Csf1rCreER--Rosa26LSL-YFP (CreER−) littermate controls born from the females with peritoneal injections of 4-OH at E8.5 were used for tracing the progeny of the yolk sac primitive MPs at age 4 wk. (A) Scheme of Csf1rCreER-Rosa26LSL-YFP lineage tracing. (B and C) Blood MOs, brain microglia, SMRMPs, and heart-resident MPs were analyzed for YFP expression by FACS: dot blot showing YFP expression (B) and scatter plot showing the percentage of YFP+ cells in blood MOs and tissue-resident MPs normalized to brain microglia cells (C). n = 8 mice per experiment.
SMRMPs Display a Tissue-Specific Gene Signature.
As resident MPs in various nonskeletal muscle tissues express tissue-specific TFs and exert tissue-specific functions in the steady state (12, 20, 23–27), we next determine whether SMRMPs have similar features. To this end, we sorted SMRMPs from both quadriceps and diaphragm and analyzed their transcriptomes by single-cell messenger RNA (mRNA) sequencing (scRNAseq). Peritoneal MPs (CD45+F4/80+MHCII−) (24) and lung alveolar MPs (CD45+CD11c+Siglec F+) (24), two well-characterized tissue-resident MPs, were analyzed simultaneously for comparison.
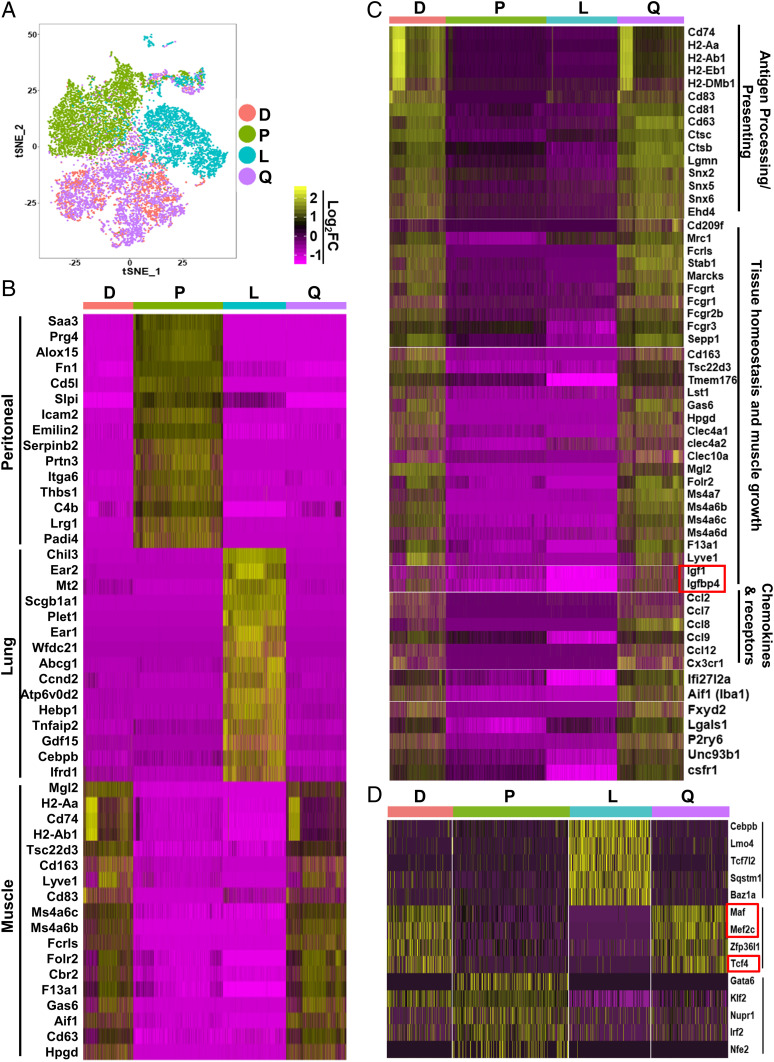
We first pooled the sequencing data from all four tissues and performed t-distributed stochastic neighbor embedding (t-SNE) dimensional reduction analysis. As shown in Fig. 6A, MPs from quadriceps (purple dots) and diaphragm (red dots) were largely grouped together. In striking contrast, MPs from peritoneum (green dots) and lung alveoli (blue dots) were each in significantly separated clusters. The results suggest that the resident MPs of different skeletal muscles are functionally similar to each other but are distinct from the resident MPs of other tissues. A heat map of top differentially expressed genes showed that the peritoneal MPs and lung alveolar MPs each expressed a higher level of a distinct set of genes (Fig. 6B), many of which were also identified by another transcriptome study comparing resident MPs in nonmuscle tissues (24). Compared to nonmuscle resident MP populations, resident MPs from quadriceps and diaphragm shared a common set of differentially expressed genes (Fig. 6B). A total of 215 genes were identified showing a significantly higher expression level (log2FC ≥ 0.5; FC: fold change) in SMRMPs (Dataset S1). Transcripts enriched by greater than or equal to twofold (log2FC ≥1) are shown as a heat map in Fig. 6C. Interestingly, many of these genes are involved in antigen processing/presentation, tissue homeostasis and muscle growth, and chemokine signaling. These findings suggest that SMRMPs have the potential to function in antigen presentation, maintenance of tissue homeostasis, and promotion of muscle growth and regeneration.
Fig. 6.
scRNAseq reveals that SMRMPs are functionally specified. Resident MPs of quadriceps (Q), diaphragm (D), peritoneum (P), and lung alveoli (L) sorted by FACS from 10 WT mice at age 10 wk were subjected to scRNAseq. (A) Sequencing data from all four samples were pooled and subjected to t-SNE dimensional reduction analysis. (B and C) Heat maps of the genes that were most differentially expressed by MPs from different tissues, the top 15 genes in B, and the genes with log2FC ≥ 1 in C. (D) Heat map of TF genes that were differentially expressed by SMRMPs, peritoneal MPs, and lung alveolar MPs.
We next performed an analysis focusing on transcriptional regulation. Several genes encoding TFs, maf, mef2c, and tcf4, were expressed at a significantly higher level (log2FC ≥ 0.5) by the muscle MPs than by the peritoneal and lung alveolar MPs (Fig. 6D). In contrast, the expression level of gata6, a signature TF of peritoneal MPs, and cebpb, a signature TF of lung alveolar MPs, was very low in muscle MPs (Fig. 6D). Therefore, like resident MPs in other tissues, SMRMPs express a combination of TFs unique to their environment. The specific roles of these TFs in the development and specification of SMRMPs will undoubtedly be a topic of future study.
SMRMPs Contain Diverse Functional Subsets.
It has been reported that resident MPs in nonskeletal muscle tissues, including lung, peritoneum, and heart, contain functionally diverse subsets correlating to their ontogenies (38–40). Since SMRMPs also showed multiple origins (Fig. 2), we hypothesized that they might also contain functionally diverse subsets. To test this hypothesis, we performed more detailed analysis of the scRNAseq data derived from the skeletal muscle tissue.
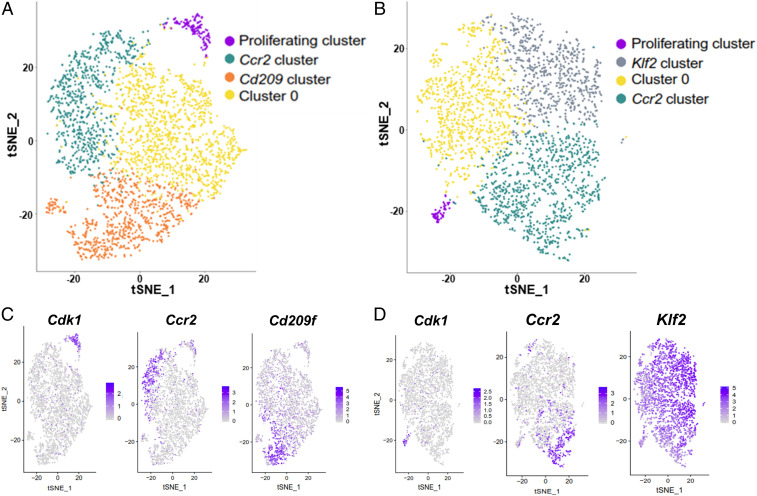
t-SNE dimensional reduction analysis of quadriceps MPs (Fig. 7A) and diaphragm MPs (Fig. 7B) identified four main clusters per population. All clusters expressed adgre1 (F4/80) and fcgr1 (CD64), confirming their MP identity (SI Appendix, Fig. S4 A and B). The top 15 genes with the highest differential expression in each individual cluster are shown as heat maps and tables in SI Appendix, Fig. S4 C and D. The four clusters within quadriceps MPs included a “proliferating cluster” (purple dots in Fig. 7A) that featured enriched expression of cell-cycle genes (Fig. 7C and SI Appendix, Fig. S4C), a “Ccr2 cluster” (green dots in Fig. 7A) with enriched expression of Ccr2 and MHCII genes, a “Cd209 cluster” (orange dots in Fig. 7A) with enriched expression of cd209 genes, and “cluster 0” (yellow dots in Fig. 7A) with no genes showing a significantly high expression level as compared with the other clusters (log2FC ≥ 0.5), including the 15 genes listed in SI Appendix, Fig. S4C.
Fig. 7.
scRNAseq analysis identifies multiple clusters within SMRMPs. scRNAseq data of quadriceps and diaphragm MPs were analyzed individually. (A and B) t-SNE dimensional reduction analysis identified four clusters in the quadriceps MPs (A) and four in the diaphragm MPs (B). (C and D) Feature plots showing single-cell expression of signature genes of each cluster: cdk1 for the proliferating cluster, Ccr2 for the Ccr2 cluster, and cd209f for the Cd209 cluster in the quadriceps MPs (C); cdk1 for the proliferating cluster, Ccr2 for the Ccr2 cluster, and Klf2 for the Klf2 cluster in the diaphragm MPs (D).
The diaphragm MPs also contained a “proliferating cluster” (purple dots in Fig. 7B) and “Ccr2 cluster” (green dots in Fig. 7B) that were almost identical to the corresponding clusters in the quadriceps MPs (Fig. 7D and SI Appendix, Fig. S4D). However, the diaphragm MPs did not contain a cluster equivalent to the “Cd209 cluster” found in the quadriceps MPs. Instead, a cluster featuring enriched expression of the genes involved in the stress response, such as klf2, Egr1, fos, and jun (Fig. 7D and SI Appendix, Fig. S4D), was identified and named the “Klf2 cluster” (gray dots in Fig. 7B). The diaphragm MPs also contained a cluster with few differentially expressed genes (SI Appendix, Fig. S4D), equivalent to the “cluster 0” in the quadriceps MPs.
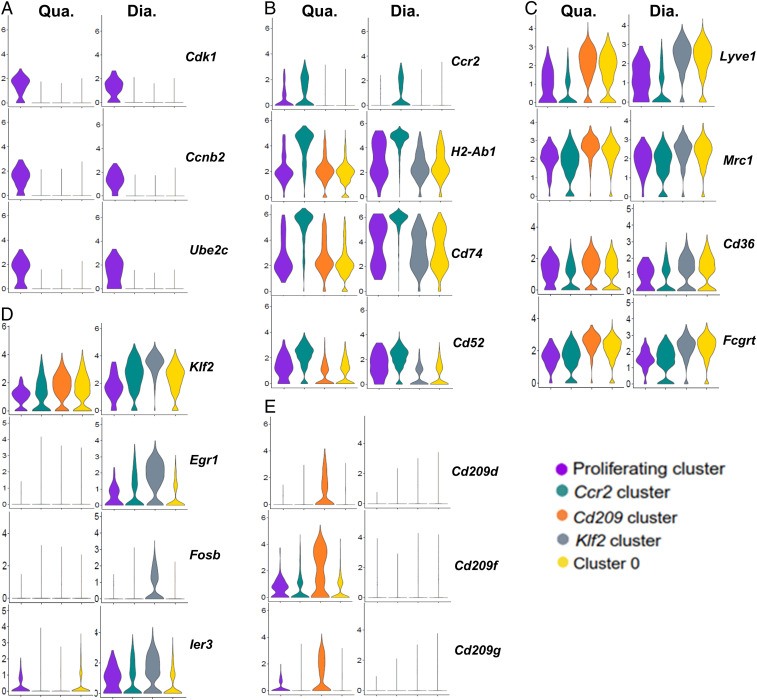
To further compare each MP cluster, we generated violin plots to visualize the distribution and probability density of individual gene expression (Fig. 8). Cell-cycle genes were only significantly expressed by the proliferating cluster in both muscle types (Fig. 8A), indicating that only the resident MPs in this cluster were undergoing active cell proliferation. The existence of the active proliferating cells in both quadriceps and diaphragm is consistent with self-renewal of SMRMPs and explains why embryonic-derived MPs are not completely replaced by blood MO-derived MPs in late adulthood. Since active cell proliferation changes the transcriptome profoundly, we excluded this cluster from the “non-Cc2 clusters” for the following comparison studies.
Fig. 8.
Clusters within SMRMPs are functionally diverse. Violin plots showing expression of (A) cell-cycle genes, (B) genes preferentially expressed by the Ccr2 cluster, (C) genes preferentially expressed by the non-Ccr2 clusters excluding the proliferating cluster, (D) genes preferentially expressed by the Klf2 cluster, and (E) genes preferentially expressed by the Cd209 cluster.
The Ccr2 Cluster and Non-Ccr2 Clusters Correspond to the MHC IIhiLyve1lo and MHCIIloLyve1hi Subsets of SMRMPs, Respectively.
In both quadriceps and diaphragm, the Ccr2 cluster showed enriched expression of Ccr2 and high expression of MHCII genes involved in antigen presentation (Fig. 8B and SI Appendix, Fig. S5A). This cluster also showed relatively low expression of “M2-like” genes when compared with the other two non-Ccr2 clusters, such as mrc1, cd36, and fcgrt (Fig. 8C and SI Appendix, Fig. S5B). Functional enrichment analysis (FEA) confirmed that, in both quadriceps and diaphragm, the genes that were differentially expressed by the Ccr2 cluster were enriched in antigen processing and presentation pathways, while the genes that were differentially expressed by the non-Ccr2 clusters were enriched in pathways related to phagocytosis and metabolism (SI Appendix, Table S1). The higher endocytic activities of the CCR2− MPs was also suggested by their morphological features, as they were larger and contained more vesicles than the CCR2+ MPs (SI Appendix, Fig. S6). In particular, the Ccr2 cluster showed a low expression level of Lyve1 (Fig. 8C) and a high expression level of the MHCII genes (Fig. 8B), strongly suggesting that this cluster corresponds to the MHCIIhiLyve1lo subset of MPs identified by FACS (Fig. 4). In contrast, the nonCcr2 clusters in both muscle types showed higher expression of Lyve1 (Fig. 8C) and a significantly lower expression of MHCII genes (Fig. 8B) compared to the Ccr2 cluster. These populations appear to correspond to the MHCIIloLyve1hi subset of MPs. Based on the results presented in Fig. 4, the Ccr2 cluster that corresponds to the MHCIIhiLyve1lo subset mainly originates from HSC-derived blood MOs, while the non-Ccr2 clusters that correspond to the MHCIIloLyve1hi subset are derived from both HSC and non-HSC origins. We further analyzed the origins of the individual clusters within the non-Ccr2 clusters corresponding to the MHCIIloLyve1hi subset in quadriceps using Flt3Cre-Rosa26LSL-YFP mice. We found that both CD209+ and CD209− MPs contained YFP+ and YFP− subsets at similar ratios (SI Appendix, Fig. S7). The finding suggests that both Cd209 cluster and cluster 0 have mixed HSC and non-HSC origins.
Diaphragm SMRMPs in the Klf2 Cluster Express a High Level of Stress-Response Genes.
The Klf2 cluster was identified only in the diaphragm, and this cluster expressed a group of stress-response genes at a significantly higher level than any other clusters (Fig. 8D and SI Appendix, Fig. S5C). FEA confirmed that the genes differentially expressed by this cluster were enriched in pathways related to stress responses (SI Appendix, Table S2). The diaphragm-resident MPs also expressed a significantly higher level of stress-response genes as compared to the quadriceps-resident MPs (Fig. 8D and SI Appendix, Fig. S5C). These findings may explain why the Ccr2 cluster demonstrated enrichment for the mRNA translation pathway in quadriceps but not in diaphragm (SI Appendix, Table S1), as the stress response could inhibit mRNA synthesis (41).
Quadriceps SMRMPs in the Cd209 Cluster Express a High Level of M2-Like Genes.
The Cd209 cluster was identified only in the quadriceps and featured a high expression of a group of genes known to be up-regulated in alternatively activated MPs (M2), such as mrc1, fcgrt, folr2, cd163, fcna, timd4, and tslp (Fig. 8C and SI Appendix, Fig. S5B) and the C-type lectin receptor (CLR) genes (Fig. 8E and SI Appendix, Fig. S5D). FEA also showed that the genes differentially expressed by this cluster were enriched in the CLR-activated pathways regulating immune responses (SI Appendix, Table S2). CLRs are pathogen recognition receptors that recognize both endogenous antigens and exogenous pathogens (42–44). They mediate the functions of myeloid cells to maintain tissue homeostasis in the steady state by detecting and clearing damaged cells and tissues for tissue repair (42, 43, 45). Therefore, the MPs in the Cd209 cluster may be more active in clearing tissue damage in quadriceps.
Discussion
We have characterized the cell surface markers, origins, and transcriptomes of SMRMPs in the steady state. SMRMPs are CD45+CD11b+F4/80+CD64+Ly6CloMerTK+CD11c−CD163+CD206+. They are located in the interstitial tissues, including epimysium, perimysium, and endomysium. Adult SMRMPs arise from both embryonic and adult hematopoiesis. The transcriptomes of resident MPs in different muscle types are highly similar to each other but are distinctively different from those in other tissues. SMRMPs express a distinct set of TFs. Functionally diverse clusters are present within resident MPs in limb and respiratory muscles, some of which are muscle type-specific.
Our lineage tracing and BMT experiments provide strong evidence that SMRMPs arise from four origins: 1) yolk sac hematopoiesis involving primitive MPs, 2) fetal liver hematopoiesis involving fetal MOs of non-HSC origin, 3) fetal liver hematopoiesis involving fetal MOs of HSC origin, and 4) postnatal BM hematopoiesis involving BM HSC-derived adult MOs. Among them, yolk sac MPs appear to have the least contribution to adult SMRMPs. Non-HSC fetal liver hematopoiesis appears to make larger contributions to adult SMRMPs. More than 50% of the SMRMPs at 4 wk of age are of non-HSC origins (YFP−). Although the Flt3Cre-Rosa26LSL-YFP fate-mapping system cannot directly distinguish cells derived from fetal liver HSC origin and adult BM HSC origin, our BMT and lineage tracing studies demonstrate that BM hematopoiesis also contributes significantly to the adult SMRMPs. The percentage of the HSC-originated SMRMPs increases with age, from 30% at P1 to over 60% at 26 wk, as a result of CCR2+CX3CR1loLy6Chi adult MO recruitment rather than proliferation of local HSC-originated MPs. These findings suggest that the recruitment of BM HSC-derived MOs contributes to SMRMPs under steady-state conditions. Embryonic-derived SMRMPs have excellent self-renewal capacity and persist into adulthood. Collectively, our data suggest that adult MO-derived and embryonic-derived SMRMPs each participate in maintaining skeletal muscle homeostasis.
It is interesting that the Flt3Cre-Rosa26LSL-YFP fate-mapping system labels a significant percentage of both fetal liver MOs and skeletal muscle MPs at a late embryonic stage, which suggests that HSCs participate in fetal liver hematopoiesis and give rise to fetal liver MOs, and, subsequently, tissue-resident MPs. This is a debated area (8, 17) as LMPs are also labeled by the Flt3Cre fate-mapping system (14). By exploiting the Rag1-cre system, one study showed that the contribution of LMPs to the fetal Mac1+ cells was reduced from around 36% at E11.5 to around 16% at E14.5 (16). However, our results showed a robust increase in the contribution of Flt3+ progenitors to fetal liver MOs from E13.5 (6.7%) to E17.5 (44.8%). It is therefore unlikely that the Flt3Cre- driven labeling is completely due to the contribution from LMPs. Gomez Perdiguero et al. (11) also reported the replacement of fetal liver myeloid cells of the yolk sac origin by cells of the HSC origin from E14.5 to E18.5. Thus, although a potential LMP contribution cannot be excluded, our findings support a significant contribution of HSCs to fetal liver MOs and prenatal SMRMPs.
SMRMPs display specific gene expression profile when comparing the transcriptomes of resident MPs from limb muscle (quadriceps), respiratory muscle (diaphragm), peritoneum, and lung alveoli. Resident MPs in different types of skeletal muscle are highly similar to each other but are distinguished from MPs in other tissues. Compared to the MPs in peritoneum and lung alveoli, SMRMPs express a relatively higher level of the genes that are involved in the regulation of immune response and skeletal muscle growth and regeneration. This may be because skeletal muscle endures constant mechanical force and therefore suffers a higher frequency of tissue microdamage than other tissues. Ongoing tissue injury may generate a microenvironment that drives resident MPs to become more active in clearing microdamage, maintaining tissue homeostasis, and promoting muscle growth and regeneration. The differential transcriptome of SMRMPs suggests the expression of tissue-specific TFs by SMRMPs. Indeed, three TF genes, maf, mef2c, and tcf4, are differentially expressed by skeletal muscle MPs, regardless of the muscle type. Whether they are responsible for the tissue-specific transcriptome of SMRMPs requires further determination by cell type-specific deletion of these genes.
Like in many other tissues, resident MPs in skeletal muscle contain functionally diverse clusters as identified by scRNAseq. The diverse subsets have been reported to correlate to their ontogenies in lung, peritoneum, and heart (38–40). We have also identified correlations of functional clusters with their ontogenies in SMRMPs. Quadriceps and diaphragm each contain four clusters, including the common proliferating cluster, Ccr2-cluster, and cluster 0, and the unique Cd209 cluster in quadriceps and Klf2 cluster in diaphragm. Based on the expression of MHCII genes and lyve1 gene, the Ccr2 cluster and non-Ccr2 clusters (including the Cd209 cluster and cluster 0 in quadriceps and the Klf2 cluster and cluster 0 in diaphragm) are correlated to the MHCIIhiLyve1lo and MHCIIloLyve1hi SMRMP subsets, respectively. The Ccr2 cluster arises mainly from adult BM HSC origin, while the non-Ccr2 clusters arise from both HSC and non-HSC origins. Within the MHCIIloLyve1hi MP subset in quadriceps of the Flt3Cre-Rosa26LSL-YFP mice, both CD209+ and CD209− MPs contain YFP+ and YFP− subpopulations, indicating that both Cd209 cluster and cluster 0 have mixed HSC and non-HSC origins. We therefore speculate that the functional differences between these two clusters may reflect the differences in the activation status rather than the origins. Whether the HSCs that contribute to the non-Ccr2 clusters in quadriceps are from fetal liver, adult BM, or both requires further investigation. The origins of the Klf2 cluster or cluster 0 in diaphragm are difficult to determine due to the lack of specific antibodies that are suitable for flow cytometry.
Compared to the Ccr2 cluster, the non-Ccr2 clusters appear more specialized in phagocytosis based on their gene expression and morphology. The Ccr2 and non-Ccr2 clusters may also play different roles in injury repair as seen in cardiac muscle (19, 46, 47). It would be interesting to explore in the future the role of different clusters of resident MPs in skeletal muscle injury repair.
Quadriceps and diaphragm contain unique clusters in addition to common clusters. It is interesting that cd209 family of genes are only expressed by MPs in quadriceps but not diaphragm, the reason for which is not clear. Diaphragm undergoes constant contraction and relaxation for breathing while quadriceps does not. Therefore, diaphragm is likely to endure more mechanical stress than quadriceps. This may explain why the Klf2 cluster with enriched expression of stress response genes is present in diaphragm but not quadriceps. Many myopathies, including those with prominent muscle inflammation, show differential involvement of limb and respiratory muscles. It is therefore tempting to explore in the future whether the differences in resident MPs contribute, in part, to the differential muscle involvement.
Collectively, our findings build a knowledge base for future studies of resident MPs in muscle development, injury repair, and myopathies with prominent muscle inflammation. This line of research will likely provide key insights into methodologies that target skeletal muscle MPs in muscle inflammation and regeneration.
Materials and Methods
Detailed animal information of strains, genetic background, sources, and breeding strategies, and protocols of BM transplant and lineage tracing can be found in SI Appendix, Supplementary Materials and Methods. Our study protocols were approved by the Institutional Animal Care and Use Committees at the Boston University Medical Center and the University of Texas Southwestern Medical Center. To study SMRMPs, skeletal muscles as well as control tissues were collected for immunostaining or flow cytometry analysis using the protocols described in SI Appendix, Supplementary Materials and Methods and the antibodies listed in SI Appendix, Table S3. Flow-sorted MPs were used for scRNAseq, which is described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the NIH under Award 1R01AR074428 (L.Z.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915950117/-/DCSupplemental.
Data Availability.
The scRNAseq data of resident MPs from mouse quadriceps, diaphragm, lung alveoli, and peritoneum have been deposited and are available at the Gene Expression Omnibus repository under accession no. GSE142480.
References
- 1.Wynn T. A., Chawla A., Pollard J. W., Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kierdorf K., Prinz M., Geissmann F., Gomez Perdiguero E., Development and function of tissue resident macrophages in mice. Semin. Immunol. 27, 369–378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashimoto D. et al., Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wynn T. A., Vannella K. M., Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginhoux F., Schultze J. L., Murray P. J., Ochando J., Biswas S. K., New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17, 34–40 (2016). [DOI] [PubMed] [Google Scholar]
- 6.McNelis J. C., Olefsky J. M., Macrophages, immunity, and metabolic disease. Immunity 41, 36–48 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Noy R., Pollard J. W., Tumor-associated macrophages: From mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoeffel G., Ginhoux F., Fetal monocytes and the origins of tissue-resident macrophages. Cell. Immunol. 330, 5–15 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Hoeffel G., Ginhoux F., Ontogeny of tissue-resident macrophages. Front. Immunol. 6, 486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginhoux F., Jung S., Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14, 392–404 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Gomez Perdiguero E. et al., Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mass E. et al., Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz C. et al., A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Hoeffel G. et al., C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–678 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginhoux F. et al., Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Böiers C. et al., Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13, 535–548 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Boyer S. W., Schroeder A. V., Smith-Berdan S., Forsberg E. C., All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell 9, 64–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bain C. C. et al., Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15, 929–937 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epelman S. et al., Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 40, 91–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bain C. C. et al., Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 7, ncomms11852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamoutounour S. et al., Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Scott C. L. et al., Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies L. C., Jenkins S. J., Allen J. E., Taylor P. R., Tissue-resident macrophages. Nat. Immunol. 14, 986–995 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier E. L. et al.; Immunological Genome Consortium , Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T. et al., Pulmonary macrophage transplantation therapy. Nature 514, 450–454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gosselin D. et al., Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Squarzoni P. et al., Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8, 1271–1279 (2014). [DOI] [PubMed] [Google Scholar]
- 28.T’Jonck W., Guilliams M., Bonnardel J., Niche signals and transcription factors involved in tissue-resident macrophage development. Cell. Immunol. 330, 43–53 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H. et al., Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J. 25, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X., Zhao W., Ransohoff R. M., Zhou L., Infiltrating macrophages are broadly activated at the early stage to support acute skeletal muscle injury repair. J. Neuroimmunol. 317, 55–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W., Wang X., Ransohoff R. M., Zhou L., CCR2 deficiency does not provide sustained improvement of muscular dystrophy in mdx5cv mice. FASEB J. 31, 35–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhao W., Ransohoff R. M., Zhou L., Identification and function of fibrocytes in skeletal muscle injury repair and muscular dystrophy. J. Immunol. 197, 4750–4761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dick S. A. et al., Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20, 29–39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breslin W. L., Strohacker K., Carpenter K. C., Haviland D. L., McFarlin B. K., Mouse blood monocytes: Standardizing their identification and analysis using CD115. J. Immunol. Methods 390, 1–8 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Zheng B., Sage M., Sheppeard E. A., Jurecic V., Bradley A., Engineering mouse chromosomes with Cre-loxP: Range, efficiency, and somatic applications. Mol. Cell. Biol. 20, 648–655 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geissmann F., Jung S., Littman D. R., Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Chakarov S. et al., Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Epelman S., Lavine K. J., Randolph G. J., Origin and functions of tissue macrophages. Immunity 41, 21–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassado Ados. A., D’Império Lima M. R., Bortoluci K. R., Revisiting mouse peritoneal macrophages: Heterogeneity, development, and function. Front. Immunol. 6, 225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schyns J., Bureau F., Marichal T., Lung interstitial macrophages: Past, present, and future. J. Immunol. Res. 2018, 5160794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryoo H. D., Vasudevan D., Two distinct nodes of translational inhibition in the integrated stress response. BMB Rep. 50, 539–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancho D., Reis e Sousa C., Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 30, 491–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez F. O., Helming L., Gordon S., Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 27, 451–483 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Murray P. J. et al., Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiffoleau E., C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front. Immunol. 9, 227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W. et al., Heart-resident CCR2+ macrophages promote neutrophil extravasation through TLR9/MyD88/CXCL5 signaling. JCI Insight 1, e87315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bajpai G. et al., Tissue resident CCR2- and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ. Res. 124, 263–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNAseq data of resident MPs from mouse quadriceps, diaphragm, lung alveoli, and peritoneum have been deposited and are available at the Gene Expression Omnibus repository under accession no. GSE142480.