Significance
The fibroblast growth factor–extracellular signal-regulated kinase (FGF-ERK) signaling pathway drives embryonic stem cells (ESCs) toward lineage commitment. How FGF-ERK signaling is regulated during this developmental window is poorly understood. Here we show that Profilin-2 (PFN2), an actin/dynamin-binding protein, controls FGF-ERK signaling in ESCs by regulating endocytosis. Members of the developmentally regulated miR-290 microRNA family suppress PFN2, enabling maximal FGF signaling, ESC proliferation, and ESC differentiation. These findings define a previously unknown axis of post-transcriptional control, endocytosis, and signal transduction important for ESC biology.
Keywords: FGF/ERK signaling, embryonic stem cells, microRNAs, PFN2, endocytosis
Abstract
Profilin2 (PFN2) is a target of the embryonic stem cell (ESC)-enriched miR-290 family of microRNAs (miRNAs) and an actin/dynamin-binding protein implicated in endocytosis. Here we show that the miR-290-PFN2 pathway regulates many aspects of ESC biology. In the absence of miRNAs, PFN2 is up-regulated in ESCs, with a resulting decrease in endocytosis. Reintroduction of miR-290, knockout of Pfn2, or disruption of the PFN2–dynamin interaction domain in miRNA-deficient cells reverses the endocytosis defect. The reduced endocytosis is associated with impaired extracellular signal-regulated kinase (ERK) signaling, delayed ESC cell cycle progression, and repressed ESC differentiation. Mutagenesis of the single canonical conserved 3′ UTR miR-290–binding site of Pfn2 or overexpression of the Pfn2 open reading frame alone in otherwise wild-type cells largely recapitulates these phenotypes. Taken together, these findings define an axis of posttranscriptional control, endocytosis, and signal transduction that is important for ESC proliferation and differentiation.
MicroRNAs (miRNAs) arising from the miR-290 family are highly enriched in pluripotent stem cells (1–4) and are active over much of early mammalian embryonic development (5, 6). The miR-290 miRNA family also enhances the promotion of somatic cell reprogramming to induced pluripotency (7–9). A number of confirmed miR-290 targets are preferentially suppressed in pluripotency and on knockdown also enhance somatic cell reprogramming to induced pluripotency (7, 8). One of these targets, Profilin2 (PFN2), is an evolutionarily highly conserved regulator of actin cytoskeletal dynamics (10) that has been shown to act as a negative regulator of endocytosis (11–13). Here we study the role of miRNA regulation of PFN2 in pluripotency and early embryonic stem cell (ESC) differentiation.
Results
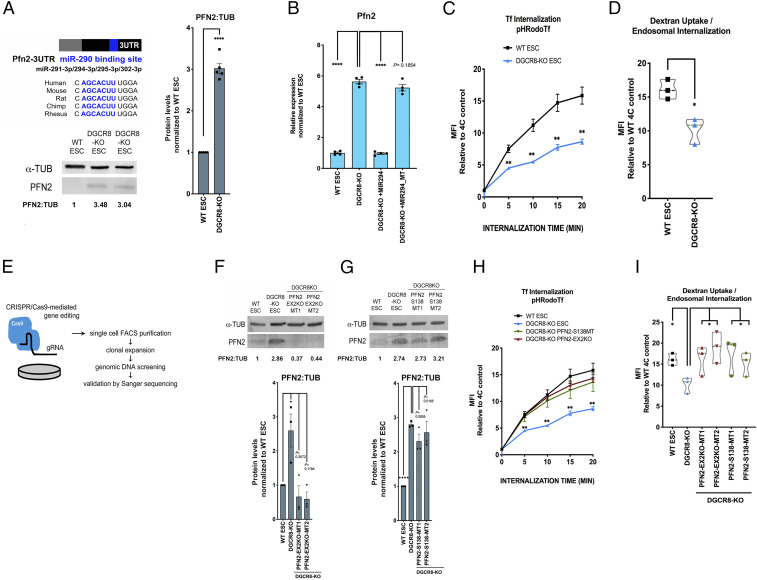
To confirm miRNA-mediated repression of PFN2 in pluripotent stem cells, we first examined endogenous PFN2 in wild-type (WT) vs. miRNA-deficient Dgcr8-KO mouse ESCs and found significantly increased PFN2 in Dgcr8-KO ESCs at the protein and messenger RNA (mRNA) levels (Fig. 1 A and B). Addback of the miR-290 family member miR-294 into Dgcr8-KO ESCs by mimic transfection returned Pfn2 expression to WT levels, while addback of miR-294 mimic with a mutant seed sequence did not (Fig. 1B).
Fig. 1.
MicroRNAs regulate endocytosis through their inhibition of Pfn2 in pluripotent stem cells. (A, Upper Left) Schematic depicting the Pfn2 3′ UTR with the conserved miR-290–binding site highlighted in blue. (A, Lower Left) Western blot analysis of endogenous PFN2 in v6.5 (WT) and Dgcr8-KO ESCs. A representative image is shown. (A, Right) Summary quantification of five independent Western blot analyses. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (B) qRT-PCR analysis of Pfn2 expression in WT ESCs, Dgcr8-KO ESCs, and Dgcr8-KO ESCs with addback of miR-294 mimic (+MIR294) or miR-294 mimic with mutant seed (MIR294_MT). n = 4 independent experiments. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (C) pH-labeled transferrin endosomal uptake assay in WT and Dgcr8-KO ESCs over the indicated times, as measured by the change in mean fluorescence intensity (MFI) relative to control. n = 3 independent experiments. Medium, serum+LIF. Error bars represent SEM. **P < 0.01, unpaired two-tailed t test. (D) pH-labeled dextran endosomal uptake assay in WT and Dgcr8-KO ESCs, measured by the change in MFI relative to control. n = 3 independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. *P < 0.05, unpaired two-tailed t test. (E) Schematic representation of CRISPR/Cas9-mediated gene editing strategy to generate Pfn2 mutant ESC lines. (F, Top) Western blot analysis of PFN2 expression in WT ESCs, Dgcr8-KO ESCs, and two clones (MT1 and MT2) of Pfn2-EX2KO (in Dgcr8-KO background) ESCs. A representative image is shown. (F, Bottom) Summary quantification of Western blot data. n = 3 or more independent experiments. (G, Top) Western blot analysis of PFN2 expression in WT ESCs, Dgcr8-KO ESCs, and two clones (MT1 and MT2) of Pfn2-S138 (in Dgcr8-KO background) ESCs. A representative image is shown. (G, Bottom) Summary quantification of Western blot data. n = 3 or more independent experiments. (H) pH-labeled transferrin endosomal uptake assay in WT, Dgcr8-KO +/− Pfn2-EX2KO or Pfn2-S138 ESCs over the indicated times, as measured by the change in MFI relative to control. n = 3 independent experiments. Medium, serum+LIF. Error bars represent SEM. **P < 0.01, unpaired two-tailed t test. (I) pH-labeled dextran endosomal uptake assay in WT, Dgcr8-KO +/− Pfn2-EX2KO, and Pfn2-S138 ESCs as measured by change in MFI relative to control. n = 3 independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. *P < 0.05, unpaired two-tailed t test. Scanned images of unprocessed blots are shown in SI Appendix, Fig. S6.
Given PFN2’s reported role in endocytosis, we next asked whether miRNAs are required for normal endocytosis in ESCs. To test this, a canonical assay for receptor-mediated endocytosis, uptake of a pH-sensitive labeled transferrin, was performed in WT vs. Dgcr8-KO ESCs (Fig. 1C). Dgcr8-KO ESCs demonstrated significantly decreased transferrin uptake, and addback of miR-294 mimic increased transferrin uptake to WT levels (SI Appendix, Fig. S1A). To study non–receptor-mediated dynamin-dependent endocytosis, endosomal uptake of pH-sensitive labeled nonspecific dextran was also performed, and Dgcr8-KO ESCs demonstrated significantly decreased dextran uptake (Fig. 1D). These data demonstrate that a miRNA-dependent mechanism regulates endocytosis in pluripotent stem cells.
PFN2 has been shown to interact directly with the proline-rich domain of dynamin (11) and inhibit endocytosis in neuronal and nonneuronal contexts (11, 12, 14). Previous studies have shown that the dynamin proline-rich domain is important for colocalization with clathrin, as deletion of the proline-rich region abolishes colocalization of dynamin with clathrin (15). Importantly, disruption of PFN2 at S138 abrogates the PFN2–dynamin interaction and abrogates the inhibition of endocytosis (11).
To study the potential role for PFN2 up-regulation in the endocytosis defect seen with miRNA loss, we used CRISPR genomic editing to create both a total Pfn2 knockout and a specific mutation to the S138 region in the Dgcr8-KO background. To produce the Pfn2 knockout, the second exon was disrupted (Pfn2-EX2KO) (Fig. 1E and SI Appendix, Fig. S1B), resulting in a dramatic reduction in mRNA and loss of protein detectable by Western blot analysis (Fig. 1F). In contrast, specific mutagenesis of the Pfn2 S138 region (SI Appendix, Fig. S1C) resulted in WT mRNA and protein levels (Fig. 1G and SI Appendix, Fig. S1D). These Dgcr8;Pfn2 double mutants were then evaluated for changes in endocytosis. Both receptor-mediated endocytosis and non–receptor-mediated endocytosis were significantly enhanced in the double mutants relative to Dgcr8-KO alone, reaching levels similar to those seen in WT cells (Fig. 1 H and I). These data suggest that up-regulation of PFN2 and the interaction of PFN2 with dynamin account for much of the endocytosis defect seen in the miRNA-deficient cells.
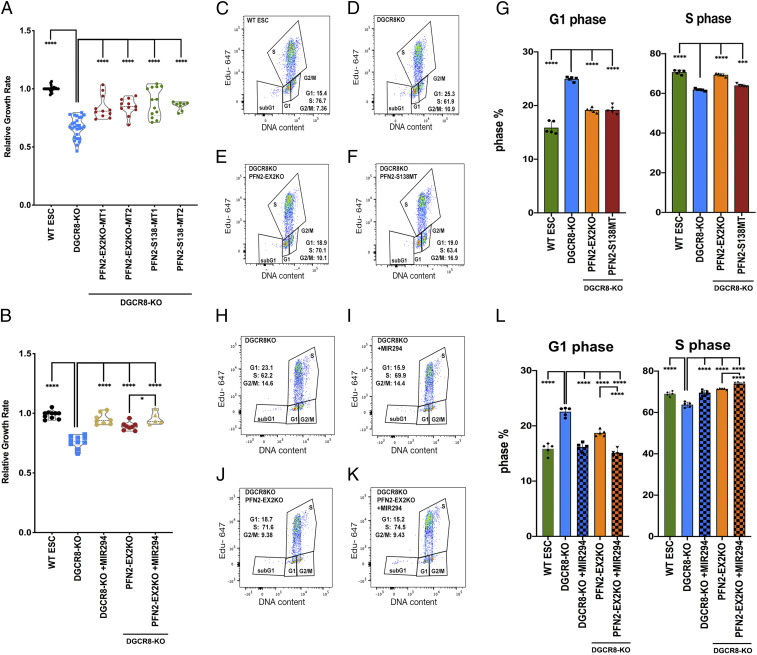
We next sought to determine the significance of PFN2-regulated endocytosis in pluripotent stem cell biology. Since the miR-290 family miRNAs play a role in the proliferation (16–18), unique cell cycle structure (19), and differentiation of ESCs (16, 18), we looked at the potential contribution of PFN2 levels in each of these essential miRNA-mediated processes. As described previously, the loss of Dgcr8 led to a decreased proliferation rate of ESCs (Fig. 2A). The loss of PFN2 protein or the PFN2–dynamin interaction domain in the Dgcr8KO background partially rescued this defect (Fig. 2A). The addition of a miR-294 mimic produced a further increase in the rate, suggesting additional targets for the miRNA in growth control (Fig. 2B). Moreover, as described previously, miRNA-deficient Dgcr8-KO ESCs showed an accumulation of cells in G1 relative to that seen in WT cells (19, 20) (Fig. 2 C and D). Loss of PFN2 protein or the PFN2–dynamin interaction in the miRNA-deficient Dgcr8-KO ESCs shifted the cell cycle structure to one closer to WT ESCs (Fig. 2 E–G). Addback of miR-294 in the Dgcr8;Pfn2 double KOs showed a further, albeit slight, correction in the cell cycle structure of the cells (Fig. 2 H–L). These data show that the loss of PFN2 function can largely reverse the proliferation and cell cycle defects seen with the loss of miRNAs.
Fig. 2.
Elevated PFN2 in miRNA-deficient cells suppresses growth rate and inhibits G1/S transition in ESCs. (A) Growth rates of indicated ESCs relative to WT ESCs. n = 8 or more independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. ****P < 0.0001, unpaired two-tailed t test. (B) Growth rate of indicated ESCs relative to WT ESCs. n = 5 or more independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. Error bars represent SEM. ****P < 0.0001; ***P < 0.001, unpaired two-tailed t test. (C–F) Representative dot plot depicting dual Click-iT–based EdU and FxCycle DNA content analysis of the indicated ESC sample from five independent experiments. (G) Summary statistics of G1 phase distribution (Left) and S-phase distribution (Right) from five independent experiments. (H–K) Representative dot plot depicting dual Click-iT–based EdU and FxCycle DNA content analysis of the indicated ESC sample from five independent experiments. (L) Summary statistics of G1 phase distribution (Left) and S-phase distribution (Right) from five independent experiments. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test.
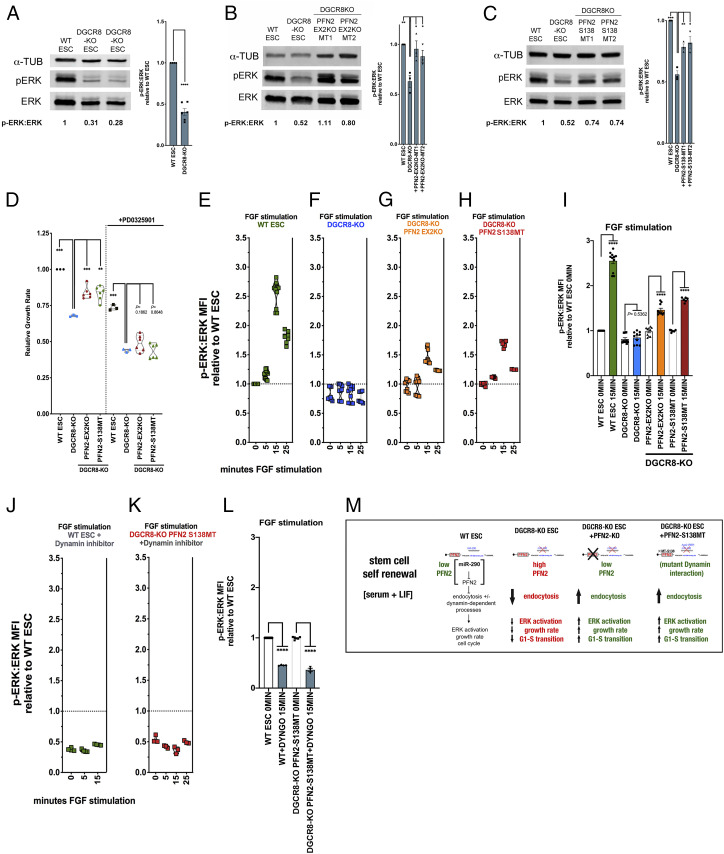
In pluripotent stem cells, the MAPK/ERK signaling pathway has been suggested to regulate proliferation and cell cycle progression in serum plus leukemia inhibitory factor (LIF) (21, 22). Therefore, we asked whether a miRNA-PFN2 axis regulates ERK signaling in ESCs. ERK is activated by phosphorylation (23). Basal phosphorylated ERK (pERK) levels were decreased in Dgcr8-KO ESCs compared with WT ESCs grown in serum+LIF (Fig. 3A). This decrease was largely reversed with the simultaneous loss of PFN2 and partially reversed with deletion of the PFN2–dynamin interaction domain (Fig. 3 B and C).
Fig. 3.
Elevated PFN2 in miRNA-deficient ESCs regulates ERK activity in a dynamin-dependent manner. (A, Left) Western blot analysis of ERK and phospho-ERK expression in WT and Dgcr8-KO ESCs. A representative image is shown. (A, Right) Summary quantification of Western blot data of at least three independent experiments. (B, Left) Western blot analysis of ERK and phospho-ERK expression in WT ESCs, Dgcr8-KO ESCs, and two clones (MT1 and MT2) of Pfn2-EX2 (in Dgcr8-KO background) ESCs. A representative image is shown. (B, Right) Summary quantification of Western blot data of at least three independent experiments. (C, Left) Western blot analysis of ERK and phospho-ERK expression in WT ESCs, Dgcr8-KO ESCs, and two clones (MT1 and MT2) of Pfn2-S138 (in Dgcr8-KO background) ESCs. A representative image is shown. (C, Right) Summary quantification of Western blot data of at least three independent experiments. (D) Growth rates of indicated ESCs relative to WT ESCs in the presence or absence of MEK inhibitor PD0325901. n = 3 independent experiments. Medium, serum+LIF, with addition of indicated inhibitor. Data are shown as median ± quartiles. ***P < 0.001; **P < 0.01, unpaired two-tailed t test. (E–H) Ratio of intracellular phospho-ERK to ERK protein levels in ESCs of the indicated genetic background and at the indicated time points following FGF stimulation relative to WT at time 0. n = 3 independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. (I) Summary statistics from the FGF stimulation assay at the minimum (0 min) and maximum (15 min) time points shown in E–H. ****P < 0.0001, unpaired two-tailed t test. (J and K) FGF stimulation assay ratio of intracellular phospho-ERK to ERK protein levels from indicated ESC samples with addition of the dynamin inhibitor Dyngo4a, relative to WT ESC basal levels over the indicated time course. n = 3 independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. (L) Summary statistics from the FGF stimulation assay at the 0 and 15 min time points shown in J and K. ****P < 0.0001, unpaired two-tailed t test. Scanned images of unprocessed blots are shown in SI Appendix, Fig. S6. (M) Schematic summarizing the identified roles of PFN2 level in endocytosis and other aspects of stem cell biology in the indicated genetic backgrounds during stem cell self-renewal conditions.
We next asked whether this role for the miRNA-PFN2 axis in ERK signaling is associated with the proliferation defect associated with Dgcr8 loss. The MEK inhibitor PD0325901 suppressed growth in all genetic backgrounds, including Dgcr8-KO and Pfn2 mutant Dgcr8-KO ESCs (Fig. 3D). Importantly, however, the positive impact of the knockout of Pfn2 or the mutagenesis of the PFN2–dynamin interaction domain on the growth of Dgcr8-KO cells was lost in the presence of the MEK inhibitor.
To capture the range of in vitro pluripotency conditions, we also cultured the pluripotent stem cells with GSK3 inhibition (CHIR99021) with and without added MEK inhibition and performed growth rate studies. Again, inhibition of ERK activation extinguished the increased growth rate seen in Pfn2-mutant Dgcr8-KO ESCs (SI Appendix, Fig. S2A). These results indicate that ERK-dependent signaling acts downstream of PFN2-regulated endocytosis to promote the rapid growth rate of ESCs grown in serum+LIF conditions.
The MAPK/ERK signaling pathway is also an important regulator in the transition from pluripotency to early lineage commitment (24). In pluripotency, LIF overrides the autoinductive capacity of fibroblast growth factor (FGF)-induced MAPK/ERK pathway activation necessary for exiting pluripotency (24). To directly interrogate the effect of PFN2 on ERK activation under serum+LIF conditions, we performed FGF stimulation assays in WT, Dgcr8-KO, and Pfn2-mutant Dgcr8-KO ESCs at single-cell resolution. To achieve this, the cells were subjected to short-term FGF4-stimulation, a physiologically relevant cue that induces MAPK/ERK signaling and ERK activation in pluripotent stem cells (24, 25), with simultaneous p90 ribosomal S6 kinase (RSK) inhibition to block negative feedback regulation. ERK activation was measured by intracellular staining and flow cytometry. As expected, ERK activation was induced in WT ESCs (Fig. 3E); however, this induction was absent in Dgcr8-KO ESCs (Fig. 3F and SI Appendix, Fig. S2 B and C). Knockout of Pfn2 or mutation of its dynamin interaction domain in Dgcr8-KO ESCs partially rescued ERK activation (Fig. 3 G–I and SI Appendix, Fig. S1 B–D).
To further evaluate the role of dynamin-dependent endocytosis in FGF signaling, we treated cells with the dynamin inhibitor Dyngo-4a. As expected, the inhibitor blocked receptor-mediated endocytosis (SI Appendix, Fig. S2E). Furthermore, the inhibitor reduced basal ERK activity by half and blocked FGF stimulation of phospho-ERK levels in WT cells (Fig. 3J). Mutation of the PFN2–dynamin interaction domain did not rescue the effect of this inhibitor (Fig. 3 K and L). These data place dynamin-mediated endocytosis downstream of PFN2 and upstream of FGF-mediated ERK activation.
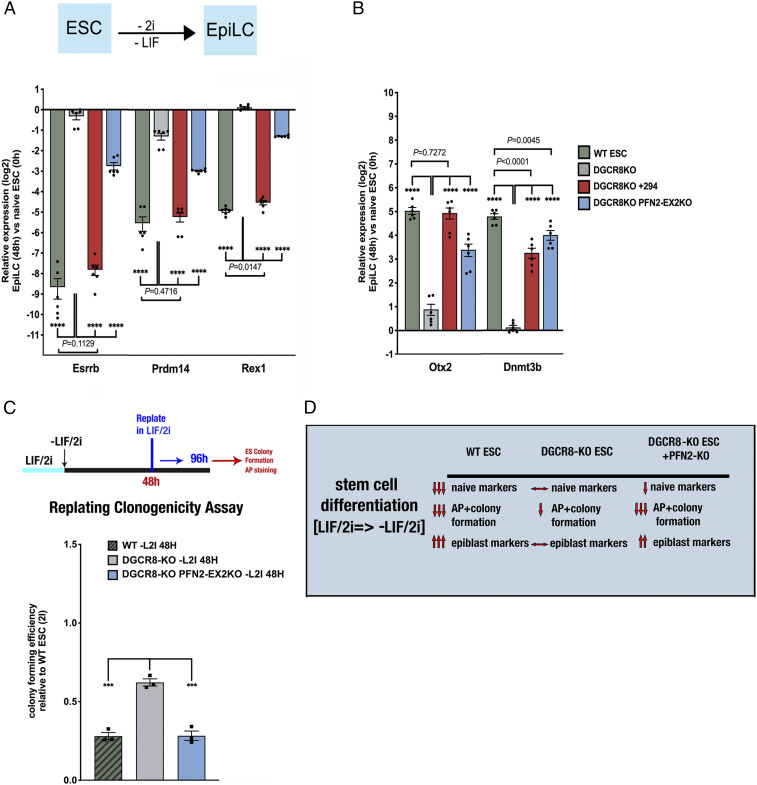
Along with regulating ESC proliferation, the miR-290 miRNA family is required for the transition of naïve ESCs to postimplantation epiblast-like cells (EpiLCs) (26). This transition requires an early increase in ERK activation (24, 25). Therefore, we wondered whether the up-regulation of PFN2 in Dgcr8-KO cells could help explain their differentiation defect. ESCs were grown in serum+LIF/2i to mimic the naïve state (2i = CHIR99021 plus PD0325901; ref 27), followed by LIF/2i removal, allowing for induction of ERK activation in the absence of LIF. These conditions induce a transition to EpiLCs in 48 to 72 h (28, 29). Indeed, WT ESCs down-regulated naïve markers (e.g., Esrrb, Prdm14, Rex1) and up-regulated EpiLC markers (e.g., Otx2, Dnmt3b) over a 48-h time course (Fig. 4 A and B and SI Appendix, Fig. S3A). In contrast, however, Dgcr8-KO cells showed a dramatic reduction in both the down-regulation of the naïve markers and the up-regulation of EpiLC markers. The introduction of miR-294 mimic into the Dgcr8-KO cells largely rescued the defect (SI Appendix, Fig. S3 A and B). Loss of PFN2 largely rescued the up-regulation of EpiLC markers while having less (but still significant) effect on the down-regulation of the naïve markers (Fig. 4 A and B and SI Appendix, Fig. S3A).
Fig. 4.
miR-294 addback and PFN2 depletion rescue pluripotency transition defects in Dgcr8-KO ESCs. (A, Top) Schematic ESC to EpiLC transition strategy. (A, Bottom) qRT-PCR analysis of naive factors Esrrb, Prdm14, and Rex1 expression after transition to EpiLC (at 48 h) relative to naïve state (0 h) in WT ESCs, Dgcr8-KO ESCs, and Dgcr8-KO with addback of miR-294 mimic (+294) or Pfn2-EX2KO (in Dgcr8-KO background) ESCs. n = 3 independent experiments. Medium, serum+LIF/2i with indicated changes. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (B) qRT-PCR analysis of expression levels of EpiLC markers Otx2 and Dnmt3b after transition to EpiLC (at 48 h) relative to the naïve state (0 h) in WT ESCs, Dgcr8-KO ESCs, and Dgcr8-KO with addback of miR-294 mimic (+294) or Pfn2-EX2KO (in Dgcr8-KO background) ESCs. n = 3 independent experiments. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (C, Top) Schematic replating assay strategy. (C, Bottom) Colony-forming efficiency assay of AP-positive ESC colonies in control, Dgcr8-KO, and Pfn2-EX2KO (in Dgcr8-KO background) ESCs. n = 3 independent experiments. Error bars represent SEM. ***P < 0.001, unpaired two-tailed t test. (D) Schematic summarizing the role of PFN2 levels in endocytosis and pluripotency transition in Dgcr8-KO ESCs.
An important functional measure of ESC differentiation is the loss of the cells’ ability to reform alkaline phosphatase (AP)-positive colonies when returned to ESC culture conditions. At 48 h after LIF/2i removal, WT ESCs showed a 72% reduction in colony-forming capacity. Similar to previous findings (16), Dgcr8-KO cells showed a lower reduction (38%) in colony formation after 48 h, consistent with suppressed differentiation. Loss of PFN2 rescued this block (Fig. 4C). These data show a role for the miR-290 miRNA family-dependent down-regulation of PFN2 in enabling multiple aspects of ESC differentiation (Fig. 4D).
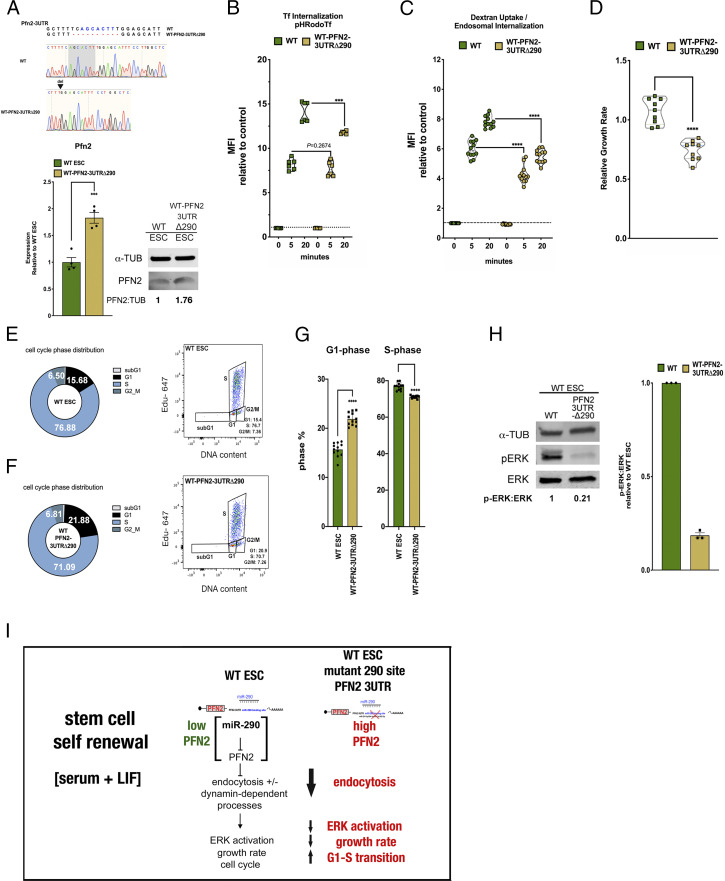
As PFN2 has one canonical miR-290 target site in its 3′ UTR (30), we sought to characterize the singular effect of this site on the phenotypes described above. Using CRISPR-mediated genomic editing, we mutated the region of the Pfn2 3′ UTR that complements the seed sequence for the miR-290 family (Fig. 5A). These WT-Pfn2-3UTRΔ290 mutant ESCs showed elevated Pfn2 RNA levels relative to WT ESCs, although not to the same degree as the loss of Dgcr8 (compare Figs. 1B and 5A). Nonetheless, the mutation of this target site led to decreased receptor-mediated and non–receptor-mediated endocytosis (Fig. 5 B and C), decreased growth rate, increased fraction of cells in G1 phase in serum+LIF conditions (Fig. 5 D–G), and decreased basal phosphorylated ERK (pERK) (Fig. 5H). Thus, manipulation of PFN2 levels by either altering miR-290 expression or targeting the canonical miR-290 target site in the Pfn2 3′ UTR leads to reciprocal changes in endocytosis, ERK activation, growth rate, and the cell cycle (Fig. 5I).
Fig. 5.
Deletion of a single miR-290 seed site in ESCs recapitulates defects seen in Dgcr8-KO ESCs. (A, Top) Sanger sequencing results showing CRISPR-induced mutations to the Pfn2 3′ UTR miR-290 seed site in WT ESCs. (A, Bottom Left) qRT-PCR analysis of Pfn2 expression in WT and WT-Pfn2-3UTRΔ290 mutant ESCs. n = 4 independent experiments. Error bars represent SEM. ***P < 0.001, unpaired two-tailed t test. (A, Bottom Right) Western blot analysis of protein levels in WT and WT-Pfn2-3UTRΔ290 mutant ESCs. (B) pH-labeled transferrin endosomal uptake assay in WT and WT-Pfn2-3UTRΔ290 mutant ESCs over the indicated times, as measured by change in MFI relative to control. n = 3 independent experiments. Medium, serum+LIF. Error bars represent SEM. ***P < 0.001, unpaired two-tailed t test. (C) pH-labeled dextran endosomal uptake assay in WT and WT-Pfn2-3UTRΔ290 mutant ESCs, measured by change in MFI relative to control. n = 3 independent experiments. Medium,serum + LIF. Data are shown as median ± quartiles. ****P < 0.0001, unpaired two-tailed t test. (D) Growth rates of indicated WT-Pfn2-3UTRΔ290 mutant ESCs relative to WT ESCs under the indicated conditions. n = 3 independent experiments. Medium, serum+LIF. Data are shown as median ± quartiles. ****P < 0.0001, unpaired two-tailed t test. (E and F) Mean cell cycle phase distribution of the indicated ESC sample (Left) and representative dot plot depicting dual Click-iT–based EdU and FxCycle DNA content analysis of the indicated ESC sample (Right) from three independent experiments. (G) Summary statistics of G1 phase distribution (Left) and S-phase distribution (Right) from three independent experiments. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (H) Intracellular phospho-ERK protein levels relative to ERK protein levels in WT and WT-Pfn2-3UTRΔ290 mutant ESCs relative to naïve conditions over the indicated time course. n = 3 independent experiments. Data are shown as median ± quartiles. Error bars represent SEM. *P < 0.05; ****P < 0.0001, unpaired two-tailed t test. (I) Schematic summarizing the effect of miR-294–dependent target PFN2 on stem cell self-renewal state in WT and Dgcr8-KO ESCs. Scanned images of unprocessed blots are shown in SI Appendix, Fig. S6.
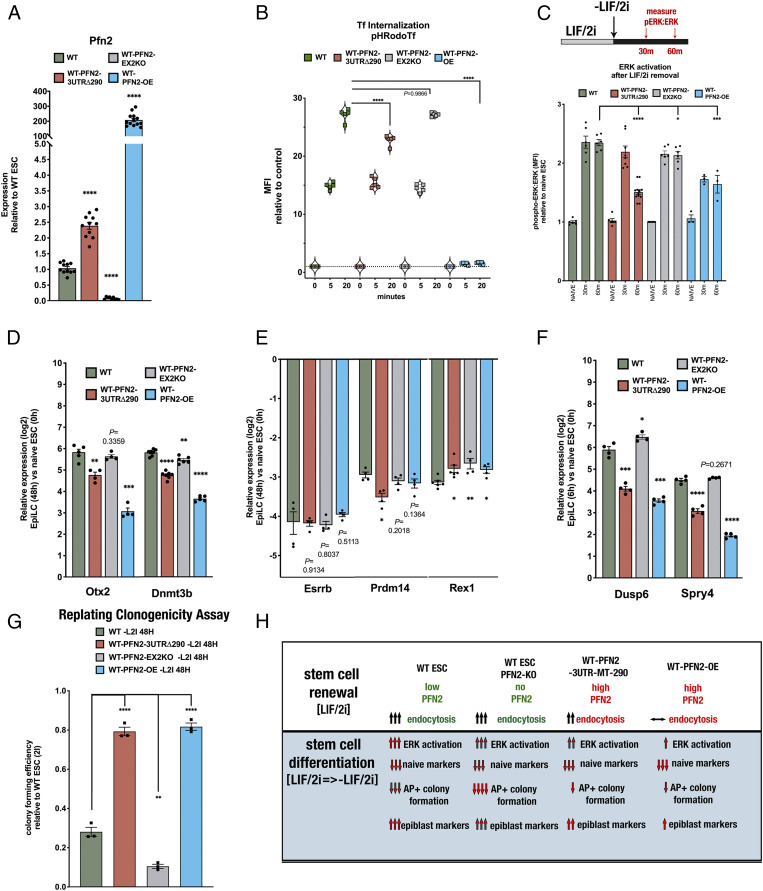
To further study the specific contribution of PFN2 in the context of unaltered miRNA expression, we generated additional mutant cell lines featuring loss of PFN2 (WT-Pfn2-EX2KO) or ectopic overexpression of PFN2 (WT-Pfn2-OE) (Fig. 6A and SI Appendix, Fig. S4) in the otherwise WT ESC background. Loss of PFN2 in an otherwise WT background had no significant impact on endocytosis (as measured by transferrin uptake), while PFN2 overexpression fully blocked endocytosis (Fig. 6B). Notably, the block with PFN2 overexpression is much greater than the mutation of the single miRNA site in the 3′ UTR, consistent with the much higher levels of PFN2 in the overexpression line (Fig. 6 A and B). We then compared the impact of the mutant Pfn2Δ290 3′ UTR, PFN2 loss, and PFN2 overexpression on activation of FGF/ERK signaling with ESC differentiation (Fig. 6C). ERK activation was measured at 30 min and 60 min following removal of LIF/2i, matching the time window of the reported maximum increase in ERK activation during the naïve to formative transition (25). WT cells showed a greater than twofold increase in activated ERK at 30 to 60 min (Fig. 6C and SI Appendix, Fig. S5). While Pfn2-3UTRΔ290 mutant cells showed a similar increase at 30 min, it was not sustained at 60 min. Loss of PFN2 showed levels close to those of WT at all time points, while overexpression of PFN2 suppressed ERK activation at both 30 min and 60 min. These changes in ERK activation were associated with changes in the activation of differentiation markers (e.g., Otx2, Dnmt3b) at 48 h postdifferentiation (Fig. 6D), with Pfn2-OE cells showing a more severe defect than the Pfn2-3UTRΔ290 mutant cells. In contrast, there was little to no change in the down-regulation of expression of naïve markers (e.g., Esrrb, Prdm14, Rex1) across the different genotypes (Fig. 6E). Consistent with changes in activated ERK, expression levels of the ERK target genes Dusp6 and Spry4 were decreased in both the Pfn2-3UTRΔ290 and Pfn2-OE cells at 6 h after removal of LIF/2i (Fig. 6F).
Fig. 6.
PFN2 mutants alter pluripotency transition-associated ERK activation and early differentiation in WT ESCs. (A) qRT-PCR analysis of Pfn2 expression in WT, WT-Pfn2-3UTRΔ290 mutant, WT-Pfn2-EX2KO mutant, and WT-Pfn2-OE ESCs. n = 3 independent experiments. Medium, serum + LIF/2i. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (B) pH-labeled transferrin endosomal uptake assay in WT, WT-Pfn2-3UTRΔ290 mutant, WT-Pfn2-EX2KO mutant, and WT-Pfn2-OE ESCs over the indicated times, measured by the change in MFI relative to control. n = 3 independent experiments. Medium, serum+LIF/2i. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (C) Intracellular phospho-ERK protein levels relative to ERK protein levels in indicated ESC samples relative to naïve conditions over the indicated time course. Statistics show comparisons at the 60 min time point after LIF/2i removal. n = 3 independent experiments. *P < 0.05; ***P < 0.001; ****P < 0.0001, unpaired two-tailed t test. (D) qRT-PCR analysis of EpiLC markers Otx2 and Dnmt3b after transition to EpiLC (at 48 h) relative to naïve state (0 h) in indicated ESC samples. n = 3 independent experiments. Error bars represent SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001, unpaired two-tailed t test. (E) qRT-PCR analysis of expression levels of naive factors Esrrb, Prdm14, and Rex1 after transition to EpiLC (at 48 h) relative to naïve state (0 h) in indicated ESC samples. *P < 0.05; **P < 0.01, unpaired two-tailed t test. (F) qRT-PCR analysis of expression levels of direct ERK substrates targets Dusp6 and Spry4 during transition to EpiLC (at 6 h) relative to naïve state (0 h) in indicated ESC samples. *P < 0.05; ***P < 0.001; ****P < 0.0001, unpaired two-tailed t test. (G, Top) Schematic replating assay strategy. (G, Bottom) Colony-forming efficiency assay of AP-positive ESC colonies in indicated samples. n = 3 independent experiments. Error bars represent SEM. **P < 0.01;****P < 0.0001, unpaired two-tailed t test. (H) Summary schematic reviewing effects of Pfn2 mutant ESCs in stem cell renewal and early pluripotency transition.
We next evaluated the impact of these different genotypes on AP-positive colony formation during ESC differentiation. Both Pfn2-3UTRΔ290 and PFN2 OE showed a retention in colony-forming ability relative to WT after 48 h of differentiation (Fig. 6G). In contrast, the loss of PFN2 showed a slight but significant reduction in colony formation, suggesting enhanced differentiation in the absence of PFN2. These data identify miRNA control of PFN2 as critical for normal endocytosis levels, ERK signaling, and ESC differentiation (Fig. 6H).
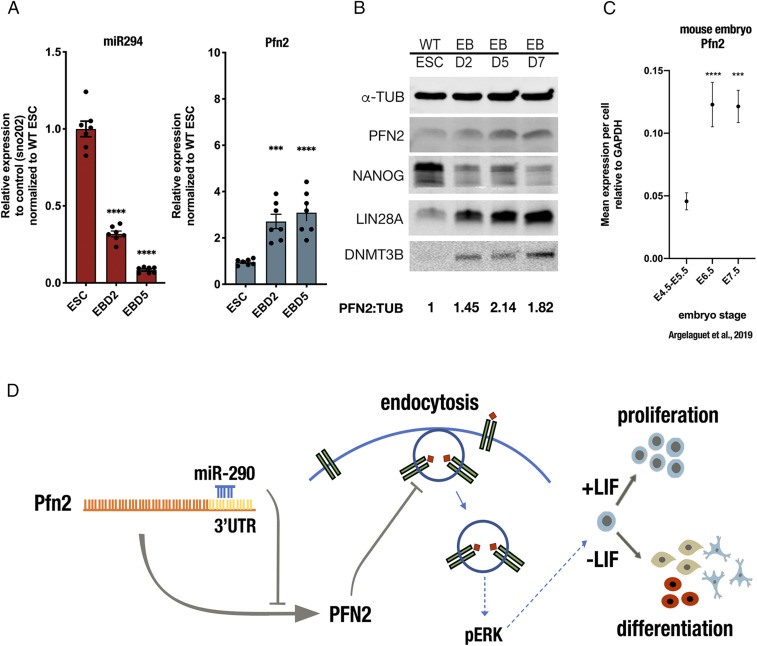
We then asked whether PFN2 expression changes during normal development. Embryoid body differentiation represents an in vitro model of early development with high transcriptional similarity to normal in vivo development through the gastrulation stage (31). Expression level analysis showed that miR-294 and Pfn2 mRNA levels were anticorrelated with decreasing miR-294 as Pfn2 increases (Fig. 7A). Similarly, PFN2 protein levels increased together with the late epiblast-enriched proteins DNMT3b and LIN28, while the early epiblast marker NANOG decreased (Fig. 7B). Analysis of recently published single-cell data from developing early mouse embryos also showed an increase in Pfn2 mRNA with the transition from the early to late epiblast, with levels remaining high during gastrulation (31) (Fig. 7C). These data are consistent with a model in which the PFN2 level is kept low in the early epiblast to enable the activation of ERK, which in turn initiates the transition to late epiblast.
Fig. 7.
PFN2 levels increase on ESC differentiation and in early embryonic development. (A) miR-294 and Pfn2 expression at indicated time points during embryoid body differentiation. Error bars represent SEM. ****P < 0.0001, unpaired two-tailed t test. (B) Western blot analysis of PFN2 protein levels in WT ESCs at indicated time points during embryoid body differentiation. A representative image is shown. (C) Pfn2 expression in mouse embryo at indicated stages (31). (D) Model illustrating the role of miR-290 control of PFN2 in regulation of endocytosis, ERK signaling, and ESC proliferation and differentiation. Note that multiple inputs occur at each step, with only one set of events diagrammed here. Error bars represent SEM. ***P < 0.001; ****P < 0.0001, unpaired two-tailed t test. Scanned images of unprocessed blots are shown in SI Appendix, Fig. S6.
Discussion
Our findings uncover a crucial role for miRNA repression of PFN2 in fine-tuning the levels of endocytosis in ESCs which in turn is essential for normal FGF/ERK signaling, ESC proliferation, and differentiation (Fig. 7D). We show that both basal and FGF-induced ERK activation in ESCs are dependent on dynamin-regulated endocytosis. It has long been proposed that endosomal membranes may serve as platforms for the formation of functional MAPK/ERK pathway signaling complexes (32–35). The most comprehensive studies to date have centered almost exclusively on the epidermal growth factor (EGF)/EGF receptor-stimulated MAPK/ERK pathway (36, 37). However, EGF signaling is not active in ESCs; instead, the ERK pathway is driven through autocrine signaling by FGF4 through FGF receptors (24). Intriguingly, clathrin-mediated dynamin-dependent endocytosis (38) has been reported to play a crucial role in maintaining a fine balance between antagonistic signaling pathways in ESC pluripotency, suggesting an additional layer of regulation for the pluripotent state (39). Future studies examining the spatiotemporal localization of the PFN2-dynamin-FGF receptor relationship in relation to ERK activation over the early phases of ESC differentiation could help further clarify the possibility of a contributory endosomal signaling cascade and other dynamin-dependent processes in pluripotent state transitions.
The results of this study demonstrate clear biological effects of the posttranscriptional regulation of PFN2 on ESC cell cycle, proliferation, and differentiation kinetics. Removal of a single canonical miR-290 site in the Pfn2 3′ UTR resulted in elevated PFN2 levels, reduced endocytosis, decreased ERK activation, altered cell cycle, and slowed growth. However, data from miRNA-deficient ESCs (Dgcr8 loss) with addback of miR-294 demonstrated a more robust effect on Pfn2 expression and associated downstream phenotypes, suggesting that 1) other miRNA-mediated sites in the PFN2 3′ UTR, including possible noncanonical miR-290 sites, likely coordinately control PFN2 mRNA stability, and/or 2) other miRNA-mediated targets may secondarily affect PFN2 levels. In addition, while overexpression of PFN2 was able to fully block endocytosis, it reduced but did not fully block ERK signaling. In contrast, Dgcr8 fully blocked ERK signaling, suggesting roles for miRNAs outside of PFN2 regulation and endocytosis in the regulation of this pathway.
These data are consistent with the idea that each step in the pathway is one node of a complex network of regulation. Further determination of the precise mechanisms by which miR-290–regulated targets contribute to the overall effect of endocytic mechanisms, cell cycle structure, cell growth rates, and ERK activation will continue to be an important goal in dissection of the molecular and signaling dynamics underlying pluripotency and pluripotent state transitions.
Deletion of the canonical miR-290 site in the Pfn2 3′ UTR also results in defects in differentiation, as evidenced by reduced up-regulation of late epiblast markers and retention of ESC colony formation. Again, these defects are associated with reduced ERK activation. The FGF-induced MAPK/ERK signaling pathway is an essential regulator of exit of ESC from pluripotency (24, 40–43), with multiple lines of evidence suggesting that the pathway is under tight temporal control (25, 44). The regulation of endocytosis could provide a rapid and highly tunable means of regulating FGF signaling during these early stages. Interestingly, similar to our findings, a recent preprint described a role for endocytosis in the promotion of ERK signaling that regulates the transition of ESCs from naïve to formative pluripotency (45). The authors describe a role for a change in membrane tension as promoting endocytosis and FGF-driven ERK activation during the pluripotency transition. Of note, in addition to regulating dynamin-driven endocytosis, PFN2 regulates actin dynamics (11). It will be interesting to determine whether PFN2 regulates membrane tension tying our results with those of these recent publications.
The miR-290 family of miRNAs diminishes with ESC differentiation and in the developing embryo (27, 46). We find a reciprocal increase in PFN2 levels both in an in vitro development model (embryoid bodies) and in vivo from early epiblast to gastrulation. This increase may play an important role in the dampening of specific signaling pathways during early development. It will be important to see how PFN2 levels are controlled in various developmental contexts and how its regulation is associated with controlling signaling and cell fate. In this study, we specifically uncover links among miR-290, PFN2, endocytosis, ERK signaling, and ESC self-renewal and differentiation.
Materials and Methods
Cell Culture and ESC Monolayer Differentiation.
V6.5 ESCs were maintained on 0.2% gelatin-coated plates, cultured in ESC medium (custom DMEM) supplemented with 15% FBS (Corning Mediatech), LIF (1000 U/mL, custom) and, when indicated, 2i, 1 uM MEK inhibitor (PD0325901; Axon Medchem), and 3 uM GSK3 inhibitor (CHIR99021; Axon Medchem). Differential states of pluripotency from naïve to EpiLC transition were generated by removal of LIF and 2i as described previously (28). In brief, ESCs were plated in ESC medium. To initiate differentiation, LIF and 2i were removed approximately 24 h after seeding. Cells were collected at the indicated time points after removal of LIF and 2i.
ESC Transition Assays and Embryoid Body Generation and Differentiation.
For ESC colony formation assays, colonies were trypsinized to a single-cell suspension and replated in LIF/2i conditions at 200 cells per well, then scored by AP staining according to the manufacturer’s (Abcam) protocols for ESC-like (AP-positive) colonies after 4 d. For embryoid body (EB) generation, ESCs were plated in rotary suspension culture on a low attachment plate to allow for self-aggregation and regular reproducible spontaneous differentiation over a course of 7 to 10 d. EB differentiation was evaluated by qRT-PCR and immunoblot analysis.
miRNA Mimic Transfections.
Mimic transfections were performed in indicated ESCs using miRIDIAN miRNA mimics and DharmaFECT 1 transfection reagent (Dharmacon, Thermo Fisher Scientific) according to the manufacturer’s protocols. Control transfections to evaluate transfection efficiency were performed with every mimic transfection experiment using Dy547-labeled miRNA mimic (Dharmacon, Thermo Fisher Scientific) and evaluated by flow cytometry.
Generation of PFN2 ESC Mutant Lines by CRISPR-CAS9 Gene Editing.
PFN2-deleted mutant Dgcr8-KO ESC lines were generated using clustered regularly interspaced short palindromic repeats (CRISPR) technology, as described previously (47). Pairs of guide RNAs were designed against exon 2 of PFN2 using the CRISPR Design Tool (http://crispr.mit.edu/). Details are provided in SI Appendix. Guide sequences are listed in SI Appendix, Table S2.
Endosomal Uptake Assays.
Endosomal uptake assays were performed with pHrodo-conjugated transferrin and dextran, which are nonfluorescent at neutral pH and increasingly fluorescent with the decreasing pH of the maturing endosomes. Details are provided in SI Appendix.
Immunoblot Analysis.
Cells were lysed with ice-cold RIPA lysis buffer containing EDTA-free protease inhibitor mixture and PhosSTOP phosphatase inhibitor (Roche), with DNA shearing by a 21-gauge needle. Lysates were incubated at 4 °C for 15 min and centrifuged at 20,000 × g at 4 °C on a table-top centrifuge. Protein was quantified using a Bio-Rad protein assay, with 30 μg of protein resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. After electrophoresis, proteins were transferred onto an Immobilon-FL PVDF membrane (Millipore) and processed for immunodetection. Further details are provided in SI Appendix.
qRT-PCR.
RNA for all qRT-PCR analyses was prepared using TRIzol (Invitrogen) and quantified on a Nanodrop spectrophotometer (Thermo Fisher Scientific). qRT-PCR was performed with an ABI 7900HT PCR system (Thermo Fisher Scientific). Details are provided in SI Appendix. Primer sets are listed in SI Appendix, Table S1.
Growth Rate Assay.
Counts were typically performed over 3 d after plating (on day 0), with day 1 and day 3 counts used to determine the population growth rate. The growth rate was calculated using the equation [(log2 (Yf) – log2 (Yi)]/t, where Yi is the initial count and Yf is the final count over time period t. Unless indicated otherwise, comparisons were performed relative to WT ESCs cultured at the same time under the same culture conditions.
Cell Cycle Analysis.
Cell cycle phase distribution analysis was conducted using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific) and FxCycle Violet stain (Thermo Fisher Scientific) according to the manufacturer’s protocol. Details are provided in SI Appendix.
FGF Stimulation and ERK Activation Assay.
Cells were pretreated with 3 μM BI‐D1870 RSK inhibitor (Selleck Chemical) to potentiate FGF-stimulated ERK activation by blocking negative feedback regulation. For FGF stimulation, cells were treated with 100 ng/mL human recombinant FGF4 (R&D Systems; 235-F4) in the presence of 10 μg/mL heparin sulfate and 3 μM BI‐D1870 RSK inhibitor at 37 °C, then washed immediately with ice-cold PBS and fixed with paraformaldehyde. Cells were permeabilized with 0.1% Triton-X and stained for intracellular phospho‐p44/42 Erk1/2 Thr202/Tyr204 at 1:700 dilution (Cell Signaling Technology; 4370) and p44/42 Erk1/2 at 1:700 dilution (Cell Signaling Technology; 4696) for 30 min at 4 °C, followed by incubation with species-specific secondary antibodies Alexa Fluor 568 or 594 goat anti-mouse IgG at 1:200 dilution (Invitrogen; A11032) and Alexa Fluor 647 goat anti-rabbit IgG (Invitrogen; A21244) at 1:200 dilution for 30 min at 4 °C. Finally, cells were washed with cold PBS and prepared for analysis by flow cytometry using an LSR II flow cytometer (BD Biosciences).
Flow Cytometry.
Unless indicated otherwise, events were collected with FACS Diva software (BD Biosciences) using a hierarchical gating strategy selecting for a homogeneous cell population (forward-scatter area vs. side-scatter area) of single cells (forward-scatter width vs. forward-scatter area). Dot plots and mean fluorescence intensities for gated events were generated using FlowJo analysis software.
Statistical Analysis.
Statistical details of experiments are provided in the figure legends, including the statistical test used, meaning of error bars, and P values. For all figures, the value of n represents the number of independent experiments, and unless specified otherwise, each dot indicates the number of biological replicates, defined as distinct dishes of cultured cells. Statistical analyses were performed using GraphPad Prism software.
Supplementary Material
Acknowledgments
This work was supported by NIH/ Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant F32HD088051 (to C.S.) and NIH/National Institute of General Medical Sciences Grants R01 GM125089 and R01 GM122439 (to R.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002750117/-/DCSupplemental.
Data Availability Statement.
All pertinent data for this study are provided in the main text or SI Appendix.
References
- 1.Houbaviy H. B., Murray M. F., Sharp P. A., Embryonic stem cell-specific microRNAs. Dev. Cell 5, 351–358 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Stadler B. et al., Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 19, 935–950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jouneau A. et al., Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA 18, 253–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suh M.-R. et al., Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Parchem R. J. et al., miR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 12, 760–773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marson A. et al., Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Judson R. L., Babiarz J. E., Venere M., Blelloch R., Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Judson R. L., Greve T. S., Parchem R. J., Blelloch R., MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat. Struct. Mol. Biol. 20, 1227–1235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anokye-Danso F. et al., Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8, 376–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witke W., The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14, 461–469 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Gareus R., Di Nardo A., Rybin V., Witke W., Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J. Biol. Chem. 281, 2803–2811 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Pilo Boyl P. et al., Profilin2 contributes to synaptic vesicle exocytosis, neuronal excitability, and novelty-seeking behavior. EMBO J. 26, 2991–3002 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luscieti S. et al., The actin-binding protein profilin 2 is a novel regulator of iron homeostasis. Blood 130, 1934–1945 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Witke W. et al., In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 17, 967–976 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto P. M., Herskovits J. S., Vallee R. B., Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J. Biol. Chem. 272, 11629–11635 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R., DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murchison E. P., Partridge J. F., Tam O. H., Cheloufi S., Hannon G. J., Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 102, 12135–12140 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanellopoulou C. et al., Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y. et al., Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 40, 1478–1483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y. et al., miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep. 4, 99–109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H. et al., Erk signaling is indispensable for genomic stability and self-renewal of mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 112, E5936–E5943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberger L., Ayyash M., Novershtern N., Hanna J. H., Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155–169 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Boulton T. G. et al., ERKs: A family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65, 663–675 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Kunath T. et al., FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Yang P. et al., Multi-omic profiling reveals dynamics of the phased progression of pluripotency. Cell Syst. 8, 427–445.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu K.-L. et al., Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 26, 350–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying Q. L. et al., The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnakumar R. et al., FOXD3 regulates pluripotent stem cell potential by simultaneously initiating and repressing enhancer activity. Cell Stem Cell 18, 104–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen A. F. et al., GRHL2-dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell 23, 226–238.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam J.-W. et al., Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 53, 1031–1043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Argelaguet R. et al., Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teis D., Wunderlich W., Huber L. A., Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3, 803–814 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Rizzo M. A., Kraft C. A., Watkins S. C., Levitan E. S., Romero G., Agonist-dependent traffic of raft-associated Ras and Raf-1 is required for activation of the mitogen-activated protein kinase cascade. J. Biol. Chem. 276, 34928–34933 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Vieira A. V., Lamaze C., Schmid S. L., Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274, 2086–2089 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Lefkowitz R. J., Shenoy S. K., Transduction of receptor signals by β-arrestins. Science 308, 512–517 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Sorkin A., Von Zastrow M., Signal transduction and endocytosis: Close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 3, 600–614 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Villaseñor R., Kalaidzidis Y., Zerial M., Signal processing by the endosomal system. Curr. Opin. Cell Biol. 39, 53–60 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Mettlen M., Chen P. H., Srinivasan S., Danuser G., Schmid S. L., Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayana Y. V., Gadgil C., Mote R. D., Rajan R., Subramanyam D., Clathrin-mediated endocytosis regulates a balance between opposing signals to maintain the pluripotent state of embryonic stem cells. Stem Cell Reports 12, 152–164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X., Chen H., Chen L., A dual role of Erk signaling in embryonic stem cells. Exp. Hematol. 44, 151–156 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Stavridis M. P., Lunn J. S., Collins B. J., Storey K. G., A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889–2894 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Hamilton W. B. et al., Dynamic lineage priming is driven via direct enhancer regulation by ERK. Nature 575, 355–360 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Whistler J. L., von Zastrow M., Dissociation of functional roles of dynamin in receptor-mediated endocytosis and mitogenic signal transduction. J. Biol. Chem. 274, 24575–24578 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Nett I. R., Mulas C., Gatto L., Lilley K. S., Smith A., Negative feedback via RSK modulates Erk-dependent progression from naïve pluripotency. EMBO Rep. 19, e45642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Belly H., Jones P. H., Paluch E. K., Chalut K. J., Membrane tension mediated mechanotransduction drives fate choice in embryonic stem cells. bioRxiv. 10.1101/798959 (2019).
- 46.Parchem R. J. et al., Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell 14, 617–631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran F. A. et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All pertinent data for this study are provided in the main text or SI Appendix.