Significance
Oncogenic transcription factors, such as the CIC-DUX4 fusion protein, constitute cancer-specific but highly challenging therapeutic targets. Consequently, pharmacologic targeting of transcription factor fusions has relied on identifying downstream actionable targets that relay fusion protein function. While informative, this approach has been limited by the divergent pleotropic effects of transcription factor fusion output. To overcome this, we have developed a mechanism-based strategy to directly degrade the CIC-DUX4 oncoprotein through ligand-dependent or pharmacologic MAPK activation. Using small-molecule drugs that block negative regulators of MAPK-ERK signaling, such as DUSP6, we reveal MAPK-mediated CIC-DUX4 degradation as an innovative therapeutic approach to targeting CIC-fused sarcoma.
Keywords: Capicua, oncoprotein, CIC-DUX4
Abstract
Transcription factor fusions (TFFs) are present in ∼30% of soft-tissue sarcomas. TFFs are not readily “druggable” in a direct pharmacologic manner and thus have proven difficult to target in the clinic. A prime example is the CIC-DUX4 oncoprotein, which fuses Capicua (CIC) to the double homeobox 4 gene, DUX4. CIC-DUX4 sarcoma is a highly aggressive and lethal subtype of small round cell sarcoma found predominantly in adolescents and young adults. To identify new therapeutic targets in CIC-DUX4 sarcoma, we performed chromatin immunoprecipitation sequencing analysis using patient-derived CIC-DUX4 cells. We uncovered multiple CIC-DUX4 targets that negatively regulate MAPK-ERK signaling. Mechanistically, CIC-DUX4 transcriptionally up-regulates these negative regulators of MAPK to dampen ERK activity, leading to sustained CIC-DUX4 expression. Genetic and pharmacologic MAPK-ERK activation through DUSP6 inhibition leads to CIC-DUX4 degradation and apoptotic induction. Collectively, we reveal a mechanism-based approach to therapeutically degrade the CIC-DUX4 oncoprotein and provide a precision-based strategy to combat this lethal cancer.
Transcription factors (TFs) are frequently altered in the pathogenesis of human cancer (1). In ∼30% of soft-tissue sarcoma (STS), transcriptional regulatory genes are altered through chromosomal rearrangements that lead to transcriptional dependence on a specific fusion oncoprotein (2). While these cancer-specific TF fusions create highly attractive therapeutic targets, the majority of these oncogenic fusions remain “undruggable” in a direct pharmacologic manner.
The CIC-DUX4 oncoprotein, which fuses Capicua (CIC) to the double homeobox 4 gene, DUX4, is a prototypical transcription factor fusion that leads to a highly aggressive and chemotherapy-resistant undifferentiated round cell sarcoma (3–5). Moreover, there are no known effective therapeutic strategies to combat CIC-fused sarcomas (3), leading to dismal clinical outcomes. Recent evidence suggests that blocking distinct downstream transcriptional nodes that regulate cell-cycle progression and metastasis limits the growth and spread of CIC-DUX4–driven sarcoma (6). While mechanistically insightful, combinatorial strategies to therapeutically target these distinct transcriptional repertoires are not clinically feasible.
The CIC-DUX4 fusion protein structurally retains >90% of native CIC, including its DNA and putative extracellular signal-regulated kinase (ERK)-binding domains, yet it functions as a transcriptional activator instead of as a repressor (6–9). Mechanistically, mitogen-activated protein kinase (MAPK) regulates wild-type (WT) CIC protein expression through direct ERK-mediated degradation (10–12); thus, we reasoned that enhanced ERK activity potentially leads to CIC-DUX4 fusion protein degradation. Leveraging this understanding, we used rare patient-derived CIC-DUX4 cell lines and primary tumors to perform a coordinated transcriptomic and pharmacologic analysis aimed at directly degrading the CIC-DUX4 fusion oncoprotein.
Results
CIC-DUX4 Binds Negative Regulators of MAPK Activity.
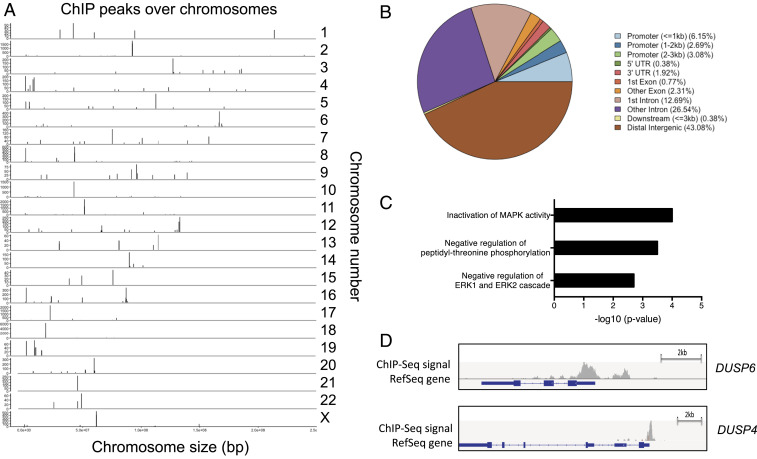
To identify endogenous CIC-DUX4–binding sites, we performed chromatin immunoprecipitation sequencing (ChIP-seq) in a well-validated CIC-DUX4 bearing cell line, NCC_CDS_X1 (6, 13). Using model-based analysis for ChIP-seq (MACS) (14, 15), we identified 260 CIC-DUX4 binding peaks present at a high level of significance (P < 0.05 and false discovery rate [FDR] <0.01) (Fig. 1A). Eighty-two percent of the binding peaks were located in either intergenic (43%) or intronic (39%) sites, and 12% were within 3 kb of the transcriptional start site (TSS), defined as the promoter region (Fig. 1B). Among the genes identified were known CIC-DUX4 targets including ETV1, ETV4, and ETV5 (Dataset S1). Similar to WT CIC, functional annotation revealed multiple negative regulators of MAPK activity as putative CIC-DUX4 target genes (16, 17) (Fig. 1C and Datasets S1 and S2).
Fig. 1.
The CIC-DUX4 binding landscape in undifferentiated sarcoma. (A) Genome-wide annotation of endogenous CIC-DUX4 ChIP-seq peak intensity in sarcoma cell line NCC_CDS_X1. Vertical bars represent CIC-DUX4 oncoprotein binding peaks on labeled chromosomes (corresponding chromosome number on the right). The x-axis corresponds to the genomic location, and the y-axis quantifies the number of reads within each peak. (B) Relative frequency of CIC-DUX4 binding sites in annotated genomic regions. (C) Functional enrichment analysis of putative CIC-DUX4 target genes. The significance is shown on the x-axis as −log10(P value). (D) Integrative Genomic Viewer showing CIC-DUX4 binding peaks upstream of the TSSs at genes DUSP6 and DUSP4.
Intriguingly, in the context of WT CIC, we and others have found that MAPK-ERK activation results in decreased WT-CIC expression through increased proteasome-mediated degradation (11, 18). Since CIC-DUX4 retains >90% of WT-CIC, including its ERK-binding interface (6, 7, 10), we hypothesized that direct transcriptional activation of these negative MAPK regulators would enable CIC-DUX4 expression. Thus, to identify the critical CIC-DUX4 target genes that sustain CIC-DUX4 expression, we focused on direct negative regulators of ERK activity, including the dual specificity phosphatases DUSP6 (ERK-selective) and DUSP4 (multiple potential substrates, including ERK) (19), which contain binding peaks in upstream regulatory elements (Fig. 1D).
Negative MAPK Regulators Are Conserved CIC-Fusion Targets.
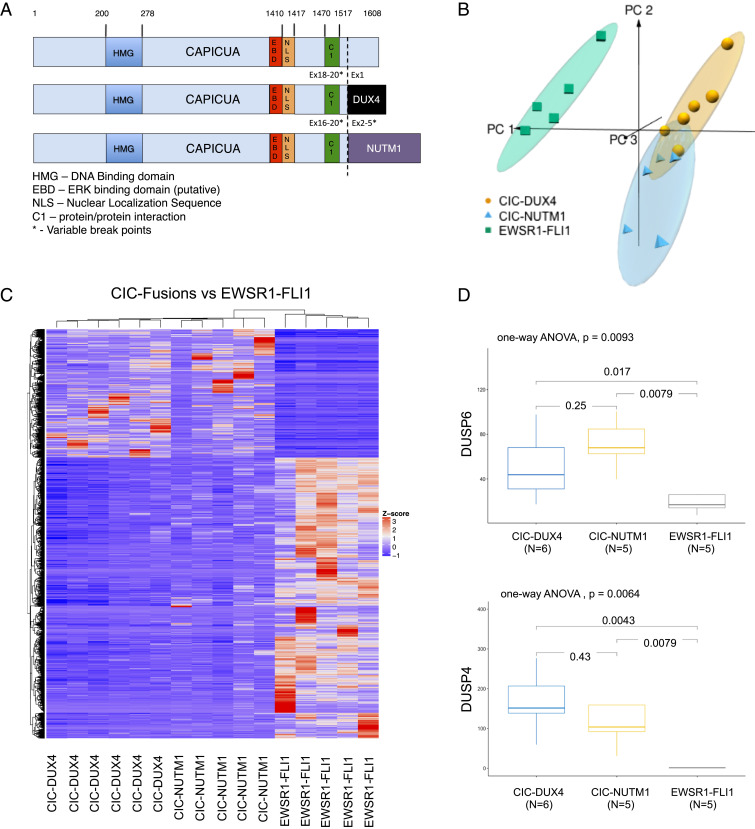
To broaden our findings beyond CIC-DUX4 fusions, we leveraged a validated transcriptomic dataset (9) to perform a comparative analysis with other known CIC-fusion oncoproteins, including CIC-NUTM1. Similar to CIC-DUX4, CIC-NUTM1 fusions retain the highly conserved CIC DNA-binding motif (7, 8) while adding the NUTM1 moiety to the C terminus (Fig. 2A) (20). These structural findings suggest that CIC-DUX4 and CIC-NUTM1 fusions transcriptionally activate the same target genes that WT-CIC repress. To test this, we performed a comparative transcriptional analysis among molecularly defined small round blue cell sarcomas, including CIC-DUX4 (n = 6), CIC-NUTM1 (n = 5), and the well-characterized EWSR1-FLI1 (n = 5) oncoprotein. This analysis revealed that CIC-DUX4 and CIC-NUTM1 fusion-positive sarcomas exhibit similar transcriptional profiles that are distinct from EWSR1-FLI1 tumors (the principal component analysis [PCA] in Fig. 2B and hierarchical clustering in Fig. 2C). Consistent with known CIC target genes, we observed increased expression of ETV1, ETV4, and ETV5 in CIC-DUX4 and CIC-NUTM1 tumors compared with EWSR1-FLI1 tumors (Dataset S3). In concordance with our ChIP-Seq findings in CIC-DUX4–bearing cells (NCC_CDS_X1), we observed increased expression of negative MAPK regulators (DUSP6 and DUSP4) in CIC-NUTM1 tumors compared with EWSR1-FLI1 tumors (Fig. 2D). These findings suggest that CIC-DUX4 and CIC-NUTM1 fusions may operate through a similar molecular mechanism to dampen MAPK-ERK flux and enable oncoprotein expression.
Fig. 2.
CIC fusions transcriptionally control negative regulators of MAPK signaling. (A) Structural alignment of WT CIC, CIC-DUX4, and CIC-NUTM1 demonstrating conserved functional domains. (B) Unsupervised PCA discriminates sarcoma samples harboring CIC-DUX4 (n = 6), CIC-NUTM1 (n = 5), or EWSR1-FLI1 (n = 5) oncoprotein-driven transcriptional signatures. (C) Hierarchical clustering analysis was conducted using 1,170 differentially expressed genes (absolute log2[fold change] ≥1 and FDR <0.05) in sarcoma samples harboring CIC-DUX4, CIC-NUTM1, or EWSR1-FLI1 fusion oncoproteins. The color key for gene expression levels (z-score) is shown to the right of the heat map. (D) Boxplots demonstrating the expression of negative MAPK regulators DUSP6 and DUSP4 in tumors bearing CIC-DUX4 (blue), CIC-NUTM1 (yellow), or EWSR1-FLI1 (gray) oncoproteins. The distribution of expression (TPM; normalized reads from RNA-seq) in each group is displayed as median (horizontal line inside box), 25th percentile (bottom line of the box), and 75th percentile (top line of the box). The whisker vertical line shows minimum and maximum expression level. Statistical significance for the difference was evaluated by one-way ANOVA.
DUSP6 Is a CIC-DUX4 Target Gene that Controls MAPK-ERK Activity.
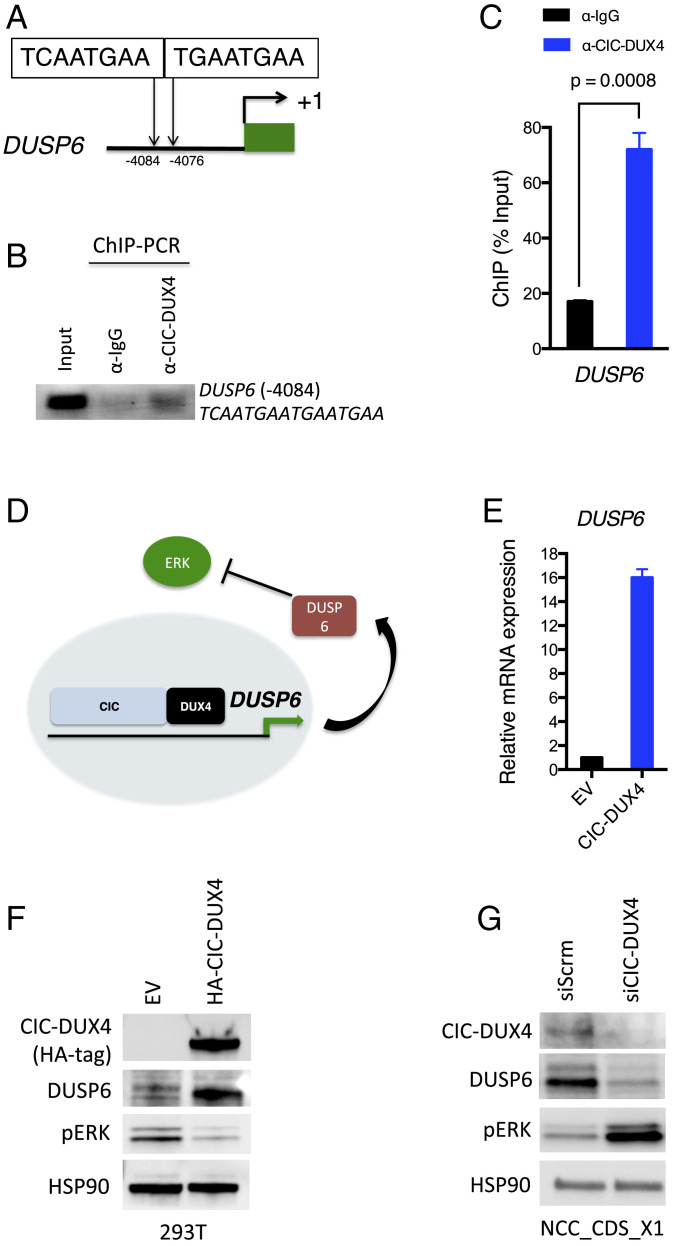
In order to demonstrate direct transcriptional regulation of DUSP family members by CIC-DUX4, we initially searched for the highly conserved CIC-binding motif (TG/CAATGA/GA) in the regulatory elements of DUSP6 and DUSP4. Indeed, we identified tandem CIC-binding sequences (TCAATGAATGAATGAA) in the upstream regulatory region of DUSP6 and a single CIC-binding motif (TGAATGGA) upstream of DUSP4 (Fig. 3A and SI Appendix, Fig. S1A). ChIP-PCR was performed to confirm CIC-DUX4 occupancy of these DUSP6 and DUSP4 upstream regulatory elements (Fig. 3 B and C and SI Appendix, Fig. S1B). DUSP6 and DUSP4 belong to different subgroups of the dual specificity family of phosphatases (21). Since DUSP6 has selective substrate specificity for ERK (22) while DUSP4 dephosphorylates multiple MAPK substrates in a context-dependent manner (23), we first aimed to identify whether DUSP4 could modulate ERK activity in the context of CIC-DUX4 sarcoma. Thus, we genetically silenced DUSP4 in NCC_CDS_X1 (endogenous CIC-DUX4 without WT-CIC expression) cells but did not consistently observe increased ERK activity (SI Appendix, Fig. S1C).
Fig. 3.
DUSP6 is a direct transcriptional target of the CIC-DUX4 fusion oncoprotein. (A). Putative tandem CIC canonical binding sites (−4,084 ∼ −4,076 bp to the TSS) in the DUSP6 regulatory domain. (B and C) ChIP-PCR demonstrating CIC-DUX4 occupancy in the regulatory region of the DUSP6 gene. IgG antibody pulldown served as a negative control (B), and the quantification of ChIP-PCR–amplified DNA bands was normalized to input DNA (C). P values were calculated using Student’s t test. Error bars indicate SEM. (D) The hypothetical model of CIC-DUX4 transcriptionally up-regulates DUSP6 expression, which dampens ERK activity to sustain CIC-DUX4 expression. (E) DUSP6 mRNA expression in 293T cells expressing exogenous CIC-DUX4 compared with empty vector (EV) control. P = 0.0001, Student’s t test. Error bars indicate SEM. (F) Immunoblot of CIC-DUX4 (HA-Tag), DUSP6, and phosphorylated-ERK from 293T cells expressing CIC-DUX4 compared with EV control. (G) Immunoblot of CIC-DUX4, DUSP6, and phosphorylated-ERK from NCC_CDS_X1 (endogenous CIC-DUX4) cells with CIC-DUX4 knockdown (siCIC) compared with scramble control (siScrm).
Given these findings, we next wanted to experimentally test whether CIC-DUX4 could directly regulate DUSP6 expression to silence ERK activity and potentially enable CIC-DUX4 oncoprotein stability (Fig. 3D). To explore this, we tested whether CIC-DUX4 expression could regulate DUSP6 protein expression to control ERK activity. Indeed, we observed increased DUSP6 expression on CIC-DUX4 expression in 293T cells, which resulted in decreased ERK phosphorylation (pERK) (Fig. 3 E and F). Moreover, when we genetically silenced CIC-DUX4 using siRNAs in NCC_CDS_X1 cells, we observed decreased DUSP6 expression and increased pERK activity (Fig. 3G).
To mitigate potential off-target effects of RNAi-mediated knockdown, we used a validated CRISPR single guide RNA (sgRNA) targeting CIC (24). Stable selection of sgCIC-DUX4 expressing NCC_CDS_X1 cells resulted in a significant decrease in viability, limiting protein expression analysis (SI Appendix, Fig. S2A). To overcome this, we next transiently expressed sgCIC-DUX4 and observed decreased DUSP6 expression and reduced growth capacity of NCC_CDS_X1 cells (SI Appendix, Fig. S2 B and C). We then engineered a CRISPR-resistant CIC-DUX4 cDNA, termed CIC-DUX4PAM (silent mutation in the PAM recognition sequence of CIC-DUX4), that retained target gene specificity and rescued the decreased DUSP6 expression and growth-suppressive effect of sgCIC-DUX4 in NCC_CDS_X1 cells (SI Appendix, Fig. S2 B–D). These findings suggest that CIC-DUX4 regulates DUSP6 through direct transcriptional control, which modulates ERK activity and thus CIC-DUX4 expression (Fig. 3D).
ERK Interacts with CIC-DUX4 to Modulate Oncoprotein Expression.
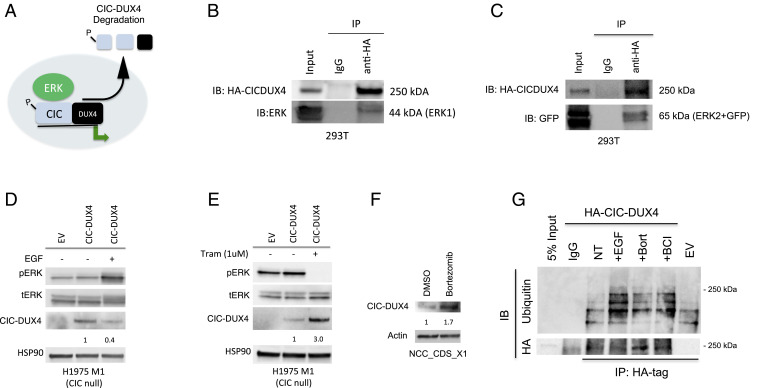
Our data indicate that DUSP6 is a direct transcriptional target of CIC-DUX4 that controls ERK activity. In the context of WT-CIC, ERK physically interacts with CIC, resulting in decreased protein stability and enhanced proteasome-mediated degradation (10–12, 18). Thus, we hypothesized that ERK physically interacts with CIC-DUX4 to control oncoprotein expression (Fig. 4A). To directly test this, we first performed coimmunoprecipitation (co-IP) experiments with HA-tagged CIC-DUX4 and human ERK (hERK) in 293T cells and observed a physical interaction between CIC-DUX4 and both hERK1 and hERK2 (Fig. 4 B and C).
Fig. 4.
ERK1 and ERK2 bind CIC-DUX4 to regulate its expression. (A) Diagram of how MAPK-ERK regulates CIC-DUX4 oncoprotein degradation. (B and C) Co-IP demonstrating ERK1 (B) and ERK2 (C) binding to HA-tagged CIC-DUX4. (D) Immunoblot of CIC (WT) null (H1975 M1) cells expressing HA-tagged CIC-DUX4, stimulated with EGF (100 ng/mL) for 6 h. (E) Immunoblot of CIC (WT) null (H1975 M1) cells expressing HA-tagged CIC-DUX4 treated with trametinib (1 μM) for 6 h. (F) Endogenous CIC-DUX4 expression in NCC_CDS_X1 cells treated with bortezomib (1 μM) for 6 h. (G) Co-IP of HA-tagged CIC-DUX4 and endogenous ubiquitin following treatment with EGF (100 ng/mL), bortezomib (proteasome inhibitor, 1 μM), and BCI (DUSP6 inhibitor, 1 μM) compared with no treatment in 293T cells.
We next assessed whether ERK activity could modulate CIC-DUX4 expression. To explore this, we introduced exogenous CIC-DUX4 into CIC-null cells (H1975 M1) (11) and activated MAPK signaling with epidermal growth factor (EGF) in serum-starved conditions for 6 h. We observed decreased CIC-DUX4 expression on ERK activation (Fig. 4D). In contrast, blocking ERK activity with the MEK inhibitor trametinib for 6 h increased CIC-DUX4 expression (Fig. 4E). Notably, endogenous CIC-DUX4 transcript levels did not change in response to EGF stimulation or MEK blockade with trametinib, further suggesting that the observed changes in protein expression were not related to changes in transcriptional output (SI Appendix, Fig. S3). These findings demonstrate that CIC-DUX4 expression is regulated in part by ERK activity.
We next assessed whether the decreased expression that we observed on ERK activation was proteasome-mediated. To do so, we first treated NCC_CDS_X1 cells with the proteasome inhibitor bortezomib and observed increased expression of endogenous CIC-DUX4 (Fig. 4F). To further assess how CIC-DUX4 ubiquitination changes on ERK activation, we performed co-IPs to compare endogenous ubiquitination of CIC-DUX4 in 293T cells treated with bortezomib (control), EGF (MAPK-ERK activator), or BCI (DUSP6 inhibitor that increases ERK activity) and compared this with no treatment (Fig. 4G). These experiments revealed a selective increase in CIC-DUX4 ubiquitination in response to ligand-dependent (EGF) or pharmacologic (BCI, DUSP6 inhibitor) ERK activation that was comparable to bortezomib-mediated proteasome inhibition. Collectively, these data suggest that ERK activation can decrease CIC-DUX4 expression through enhanced proteasome-mediated degradation.
DUSP6 Inhibition Increases ERK Activity to Degrade the CIC-DUX4 Oncoprotein.
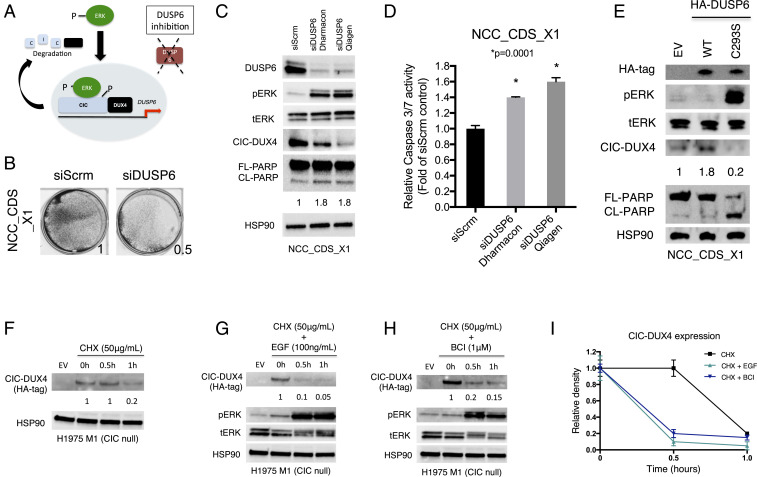
CIC-DUX4 directly controls DUSP6 expression to dampen ERK activity, enabling oncoprotein expression (Fig. 3D). We hypothesized that inhibition of DUSP6 could increase ERK activity, leading to CIC-DUX4 degradation and apoptosis (Fig. 5A). To test this, we genetically silenced DUSP6 with siRNA, shRNA, and CRISPR-based approaches and observed decreased viability of CIC-DUX4–bearing cells (NCC_CDS_X1) (Fig. 5B and SI Appendix, Fig. S4 A and B). Moreover, genetic inhibition of DUSP6 using two independent siRNAs (Dharmacon-8 siDUSP6 targeting the 3′ coding sequence and Qiagen siDUSP6 targeting the 5′ coding sequence of DUSP6) induced apoptosis, as measured by cleaved-PARP and caspase 3/7 activity (Fig. 5 C and D). In contrast, we did not consistently observe increased ERK activity or apoptosis in NCC_CDS_X1 cells with DUSP4 knockdown (SI Appendix, Fig. S4 B and C).
Fig. 5.
DUSP6 inhibition decreases CIC-DUX4 expression. (A) Diagram showing how DUSP6 inhibition increases ERK activity to degrade CIC-DUX4. (B) Crystal violet assay for cell viability in patient-derived CIC-DUX4–expressed sarcoma cell line NCC_CDS_X1 with either genetic inhibition of DUSP6 (siDUSP6) or scramble control (siScrm). (C) Immunoblot of NCC_CDS_X1 cells expressing siDUSP6 (Dharmacon 8), siDUSP6 (Qiagen), or scramble control. (D) Relative caspase 3/7 activity in NCC_CDS_X1 cells expressing siDUSP6 (Dharmacon 8), siDUSP6 (Qiagen), or scramble control. P values were calculated using Student’s t test. (E) Immunoblot of NCC_CDS_X1 cells expressing either HA-tagged WT DUSP6 or HA-tagged dominant negative DUSP6C293S. (F) CHX chase assay of WT CIC null cells expressing CIC-DUX4. (G) CHX chase assay of WT CIC null cells expressing CIC-DUX4 and stimulated with EGF (100 ng/mL) over the designated times. (H) CHX chase assay of WT CIC null cells expressing CIC-DUX4 and treated with DUSP6 inhibitor BCI (1 μM) over time. (I) Relative CIC-DUX4 expression comparing CHX alone, CHX + EGF, and CHX + BCI. The x-axis represent the period of indicated treatment, and the y-axis is the intensity of relative CIC-DUX4 expression. Error bars indicate SEM.
To further demonstrate the impact of DUSP6 on endogenous CIC-DUX4 expression, we next expressed a well-characterized dominant negative form of DUSP6 (DUSP6C293S) (25) in NCC_CDS_X1 cells and observed decreased CIC-DUX4 expression and increased PARP cleavage compared with WT-DUSP6 and EV control cells (Fig. 5E).
To further explore the temporal dynamics of CIC-DUX4 oncoprotein stability, we next performed cycloheximide (CHX) chase assays in CIC-null cells engineered to express HA-tagged CIC-DUX4. We observed decreased CIC-DUX4 half-life on ERK activation with either EGF or the DUSP6 inhibitor BCI compared with control (26) (Fig. 5 F–I). These data indicate that CIC-DUX4–mediated tumor cell survival depends in part on DUSP6 expression. Consequently, we identify DUSP6 as a potential therapeutic vulnerability in CIC-DUX4–driven sarcoma.
DUSP6 Is a Therapeutic Target in CIC-DUX4 Sarcoma.
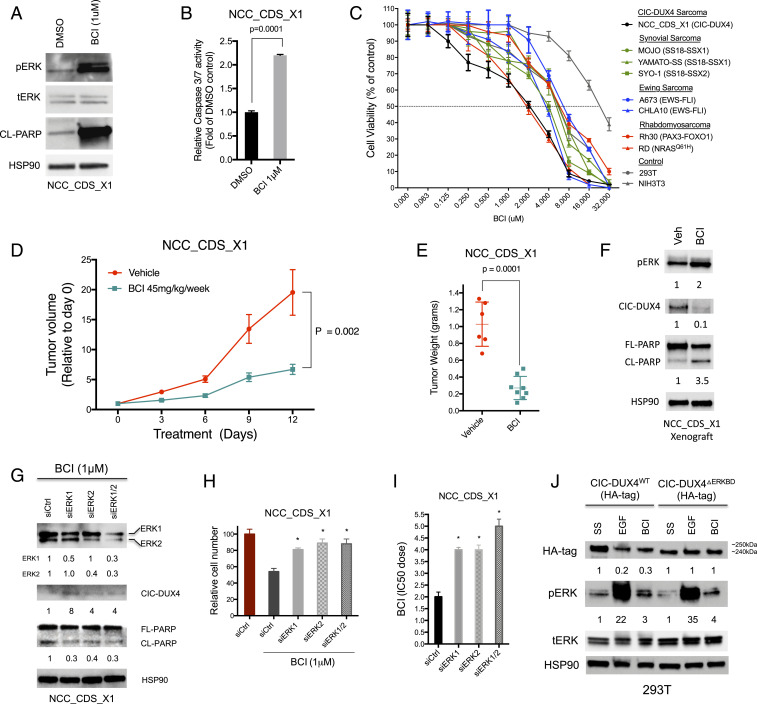
To increase the translational impact of our findings, we next explored whether pharmacologic inhibition of DUSP6 with BCI could induce apoptosis in CIC-DUX4–expressing cells. Indeed, we observed rapid apoptotic induction on BCI treatment, as measured by PARP cleavage and caspase 3/7 activity in NCC_CDS_X1 cells compared with control (Fig. 6 A and B). Moreover, BCI or genetic silencing of DUSP6 decreased the viability of CIC-DUX4–bearing cells (NCC_CDS_X1) compared with a panel of genetically well-defined fusion-positive sarcoma cell lines: synovial sarcoma, Ewing sarcoma, and rhabdomyosarcoma (Fig. 6C and SI Appendix, Fig. S5 A and B). Notably, the viability of a fusion-negative rhabdomyosarcoma cell line (RD) was similarly impacted by pharmacologic and genetic DUSP6 inhibition (Fig. 6C). Intriguingly, RD rhabdomyosarcoma cells are known to harbor an oncogenic NRASQ61H mutation (27). These findings are consistent with recent reports of DUSP6 inhibitor-induced cellular toxicity in other RAS mutant cancers (28).
Fig. 6.
DUSP6 is a pharmacologic target in CIC-DUX4 sarcoma. (A) Immunoblot of NCC_CDS_X1 cells treated with DUSP6 inhibitor BCI (1 μM). (B) Relative caspase 3/7 activity in NCC_CDS_X1 cells treated with BCI (1 μM) or DMSO control. P = 0.0001, Student’s t test. Error bars indicate SEM. (C) CellTiter-Glo assay for cell viability in NCC_CDS_X1, MOJO, Yamato-SS, SYO-1, A673, CHLA10, Rh30, RD, 293T, and NIH 3T3 cells treated with BCI for 72 h, performed in triplicate. Error bars indicate SEM. (D) Relative NCC_CDS_X1 tumor volume in mice treated with BCI (45 mg/kg/wk) (teal line) compared with vehicle control (red line) over time. P values were calculated using Student’s t test. Error bars indicate SEM. (E) Individual weights from tumor explants derived from mice in D. P = 0.0001, Student’s t test. Error bars indicate SEM. (F) Immunoblot of tumor xenografts derived from mice treated with either BCI or vehicle control. (G) Immunoblot of BCI-treated NCC_CDS_X1 cells expressing siCtrl, siERK1, siERK2, or both siERK1 and siERK2. (H) Relative number of BCI (1 μM)-treated NCC_CDS_X1 cells expressing siCtrl, siERK1, siERK2, or both siERK1 and siERK2 compared with DMSO-treated siCtrl. *P < 0.01, one-way ANOVA. Error bars indicate SEM. (I) BCI IC50 dose of NCC_CDS_X1 cells expressing siCtrl, siERK1, siERK2, or both siERK1 and siERK2. *P < 0.001, one-way ANOVA. Error bars indicate SEM. (J) Immunoblot of serum-starved, EGF, or BCI-treated 293T cells expressing either HA-tagged WT CIC-DUX4 (CIC-DUX4WT) or HA-tagged CIC-DUX4 lacking the ERK-binding domain (CIC-DUX4ΔERKBD).
To test the therapeutic impact of DUSP6 inhibition in vivo, we treated mice bearing NCC_CDS_X1 tumor xenografts with BCI (45 mg/kg/wk) weekly and noted a reduction in tumor growth in the BCI-treated mice compared to vehicle-treated control mice (Fig. 6 D and E). Further analysis of tumor explants revealed increased PARP cleavage and decreased CIC-DUX4 expression in NCC_CDS_X1 tumor xenografts treated with BCI relative to control (Fig. 6F). Similar to previous studies, BCI therapy did not result in overt toxicity in immunodeficient mice (29, 30) (SI Appendix, Fig. S6).
While these in vivo findings corroborate our in vitro studies and highlight the critical dependence on DUSP6 expression in CIC-DUX4 sarcoma, we cannot entirely rule out the possibility of unanticipated “off-target” effects of BCI. Thus, these findings should be approached with caution and may encourage the development of more clinically advanced DUSP6-specific inhibitors.
To demonstrate that the decreased viability observed in BCI-treated CIC-DUX4–expressing cells was the result of ERK activation, we genetically silenced ERK1 and/or ERK2 in NCC_CDS_X1 cells. Genetic inhibition of ERK1 and/or ERK2 increased CIC-DUX4 expression and rescued BCI-mediated apoptosis, as measured by PARP cleavage in NCC_CDS_X1 cells compared with scramble control (Fig. 6G). Moreover, we observed increased viability and a higher BCI IC50 dose in BCI-treated NCC_CDS_X1 cells expressing siERK1, siERK2, and siERK1 + siERK2 compared with siCtrl (Fig. 6 H and I). Notably, ERK1 and/or ERK2 knockdown did not impact NCC_CDS_X1 viability in the absence of BCI therapy (SI Appendix, Fig. S7 A and B).
To mechanistically link ERK activity to CIC-DUX4 degradation, we leveraged previous studies that localized and validated an ERK-binding domain in WT CIC (ERK-CIC binding interface) (10, 18). We identified the ERK-CIC binding interface, a highly conserved 75-aa sequence in CIC-DUX4, and genetically engineered a CIC-DUX4 variant that lacked this region, termed CIC-DUX4ΔERKBD (SI Appendix, Fig. S8A). Notably, CIC-DUX4ΔERKBD retained its target gene specificity and transcriptional activity (SI Appendix, Fig. S8 B and C). In contrast to WT CIC-DUX4 (CIC-DUX4WT), when CIC-DUX4ΔERKBD was expressed in 293T cells, CIC-DUX4ΔERKBD protein levels did not decrease following EGF or BCI treatment compared with serum-starved conditions (Fig. 6J). Moreover, we did not observe a decrease in CIC-DUX4ΔERKBD expression in 293T cells following genetic inhibition of DUSP6 (SI Appendix, Fig. S8D). Collectively, these findings reveal that BCI-mediated apoptotic induction in CIC-DUX4–expressing cells is ERK-dependent.
Discussion
Oncogenic transcription factors, such as the CIC-DUX4 fusion protein, constitute cancer-specific but highly challenging therapeutic targets. Consequently, pharmacologic targeting of transcription factor fusions has relied on identifying downstream actionable molecular targets that relay fusion protein function. Unfortunately, this strategy has been hampered by the functional diversification and pleotropic effects of downstream transcriptional repertoires, which limit the efficacy of single-agent pharmacologic approaches.
A deeper understanding of the mechanistic properties that enable CIC-DUX4 fusion oncoprotein stability revealed a dependence on negative regulators of ERK activity. Leveraging this mechanistic insight, we have uncovered a direct pharmacologic approach to degrading the CIC-DUX4 fusion oncoprotein. Specifically, we demonstrate that CIC-DUX4 transcriptionally controls DUSP6 expression to dampen ERK activity, which enables sustained fusion oncoprotein stability. DUSP6 inhibition in CIC-DUX4–bearing cells leads to decreased viability and enhances apoptosis both in vitro and in vivo. Thus, we identify DUSP6 as a therapeutic vulnerability in CIC-DUX4 sarcoma.
The role of DUSP6 in human cancer remains controversial, with a tumor-suppressive effect (via negative feedback on ERK1/2) in certain solid tumors and a protumorigenic role in other subsets of human cancers (19, 21). Thus, the biochemical and biological roles of DUSP6 in different histological subtypes of cancer are likely to be distinct. Therefore, mechanistically exploring and identifying the correct subset of cancers that may respond to DUSP6 inhibition remain of paramount importance. To this end, others have recently reported that specific targeting of negative regulators of the RAS-RAF-MEK-ERK pathway, including DUSP6, can lead to cellular toxicity (28). Moreover, DUSP6 inhibition has been shown to suppress tumor growth and induce apoptosis in another sarcoma subset, malignant peripheral nerve sheath tumors (31). These findings are consistent with our observations that the growth of fusion-negative, NRASQ61H mutant rhabdomyosarocma cells is suppressed with chemical or genetic DUSP6 inhibition. These results, coupled with our findings that mechanistically reveal a specific dependence on DUSP6 in CIC-DUX4 tumors, should encourage further clinical development of DUSP6 inhibitors in these currently undruggable subsets of human cancer.
In addition, future studies may reveal a broader role for targeting additional negative regulators of MAPK signaling, including SPROUTY and other DUSP family members. Notably, our ChIP-seq analysis revealed CIC-DUX4–binding peaks in SPRY2, SPRED1, and SPRED2 (Dataset S1), components of the SPROUTY family that may also impact MAPK-ERK signaling through the regulation of receptor tyrosine kinase adapter proteins or upstream substrates (e.g., RAF). Future studies are needed to elucidate how these potential CIC-DUX4 targets influence MAPK-ERK signaling and whether they represent potential therapeutic targets in CIC-fused and/or RAS mutant cancers.
The potential impact of our findings may extend beyond CIC-DUX4, as we observe increased levels of DUSP6 in other CIC-fused sarcomas, including CIC-NUTM1. While we were not able to directly test the therapeutic efficacy of DUSP6 inhibition in CIC-NUTM1 sarcomas, the clinical and transcriptional findings mimic CIC-DUX4 sarcomas. Thus, DUSP6 inhibition potentially may be an effective therapeutic strategy in CIC-NUTM1–driven tumors. We have uncovered a DUSP6-ERK-dependent molecular circuit that enables CIC-fusion oncoprotein stability. More broadly, our findings reveal a precision-based therapeutic strategy to target CIC-fused sarcomas.
Experimental Procedures
Tumor Xenograft Assays.
Six- to 8-wk-old female SCID mice were purchased from Taconic Biosciences. For s.c. xenotransplantation, 3.0 × 106 NCC_CDS1_X1 cells were resuspended in 50% PBS/50% Matrigel matrix and injected s.c. into the flanks of immunodeficient mice. Mice were observed postprocedure for 1 to 2 h, and body weight and wound healing were monitored weekly in accordance with IACUC protocol.
Cell Lines, Drug, and Reagents.
Cell lines were cultured as recommended by the American Type Culture Collection (ATCC). NIH 3T3, 293T, A673, CHLA10, RD, and RH30 cells were obtained from ATCC. MOJO was a kind gift from Kevin Jones (University of Utah, Salt Lake City, Utah), Yamato-SS was a gift from Norifumi Naka (Osaka International Cancer Institute, Osaka, Japan), and SYO-1 was a gift from Akira Kawai (National Cancer Center Hospital, Tokyo, Japan). NCC_CDS_X1 was obtained from Tadashi Kondo at the National Cancer Center in Tokyo, Tokyo, Japan. The presence of the CIC-DUX4 fusion in NCC_CDS_X1 cells was confirmed through RNAseq analysis using the “grep” command. H1975 M1 (CIC null) cells were derived from parental H1975 cells. All cell lines were maintained at 37 °C in a humidified atmosphere at 5% CO2 and grown in RPMI 1640 medium supplemented with 10% FBS, 100 IU/mL penicillin, and 100 ug/mL streptomycin. (E/Z)-BCI hydrochloride (B4313), recombinant hEGF (E9644), and cycloheximide (01810) were purchased from Sigma-Aldrich. Trametinib and bortezomib were purchased from SelleckChem.
Gene Knockdown and Overexpression Assays.
ON-TARGET plus Scramble, CIC, ERK1, ERK2, DUSP4, and DUSP6 siRNAs were obtained from GE Dharmacon, and transfection was performed with Dharmafect transfection reagent. Individual DUSP6 siRNAs were obtained from Dharmacon (DUSP6-8 #J-003964-08-0005) with target sequence GGCATTAGCCGCTCAGTCA (nucleotides 1,348 to 1,366 in the DUSP6 coding sequence) and Qiagen (DUSP6 FlexiTube SI03106404) with target sequence GTCGGAAATGGCGATCAGCAA (nucleotides 495 to 515 in the DUSP6 coding sequence). DUSP6 and LacZ control CRISPR plasmids were a gift from Arun Unni (Weill Cornell Medical College, New York, NY) with the following sequences: sgRNA_lacZ (control) guide sequence, GAGCGAACGCGTAACGCGAA; sgRNA_DUSP6-1 guide sequence, GTGCGCGCGCTCTTCACGCG (nucleotides 679 to 698 in the DUSP6 coding sequence); sgRNA_DUSP6-2 guide sequence, ACTCGTATAGCTCCTGCGGC (nucleotides 582 to 601 in the DUSP6 coding sequence). shRNAs targeting DUSP6 were obtained from Sigma-Aldrich. Sequences were as follows: TRCN0000002436, CCGGACTTGGACGTGTTGGAGGAATCTCGAGATTCCTCCAACACGTCCAAGTTTTTT; TRCN0000233474, CCGGTCTAATCCAAAGGGTATATTTCTCGAGAAATATACCCTTTGGATTAGATTTTTG.
The HA-tagged CIC-DUX4 plasmid was obtained from Takuro Nakamura (The Cancer Institute of JFCR, Tokyo, Japan). Sequence verification was performed using Sanger sequencing. DUSP6 WT (27975) and delta DUSP6 (C293S) (27977) were gifts from Igor Astasturov and purchased from Addgene. pFLAG-CMV-hErk1 plasmid was a gift from Melanie Cobb (Addgene 49328), and ERK2-GFP was purchased from Genecopoeia (EX-A0354-M03). The Cas9 + sgCICDUX4 plasmid was a gift from William Hahn (Addgene 74959). Fugene 6 transfection reagent was used for all transfections.
Mutagenesis.
The Q5 Site-Directed Mutagenesis Kit (New England BioLabs; E0554) was used according to the manufacturer’s protocol. The HA-tagged CIC-DUX4 plasmid was obtained from Takuro Nakamura, Tokyo, Japan. The CIC-DUX4ΔERKBD mutant was generated using the following primers: forward, 5′- CTGGATTCAGCACCCGAGGACC; reverse, 5′-CGCCTCCTTGCGCTCCGG. The annealing temperature was set at 72 °C. The CIC-DUX4PAM mutant was generated using the following primers: forward, 5′-AAGGCTCCGGAGAGCAGCTCC; reverse, 5′-GGTGTCTGAGCTCAGGAGTG. The annealing temperature was set at 68 °C.
ChIP-Seq and PCR.
CIC-DUX4 immunoprecipitation was performed with NCC_CDS_X1 cells using the SimpleCHIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) with IgG (Cell Signaling Technology) and CIC (Acris-Origene) antibodies in accordance with the manufacturer’s protocol. Paired-end 150-bp (PE150) sequencing on an Illumina HiSeq platform was then performed. ChIP-Seq peak calls were identified through MACS.
For DUSP6 ChIP-PCR validation, primers were designed to flank a tandem TCAATGAA/TGAATGAA motif at positions −4,084 and −4,076. The primer sequences were as follows: DUSP6_F, GGACTGTCCTCATTAGAATG; DUSP6_R, CTGTCACTCAGTAGTAGTCTGG. For DUSP4 ChIP-PCR validation, primers were designed to flank a TGAATGGA sequence at position−5,094 upstream of the TSS. The primer sequences were as follows: DUSP4_F, CCGTTCCATTCCGGGCCCCG; DUSP4_R, CACAAAGAGCGGAGTAAACAG.
Western Blot Analysis.
All immunoblots represent at least two independent experiments. Adherent cells were washed and lysed with RIPA buffer supplemented with proteinase and phosphatase inhibitors. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes, and blotted with antibodies recognizing HA-tag (Cell Signaling Technology), DUSP6 (Cell Signaling Technology), total-ERK (Cell Signaling Technology), phospho-ERK (Cell Signaling Technology), HSP90 (Cell Signaling Technology), PARP (Cell Signaling Technology), Actin (Sigma-Aldrich), CIC (Acris-Origene), Ubiquitin–FK2 clone (Millipore), and IgG (Cell Signaling Technology). Band densities were quantified using ImageJ software.
Xenograft Tumors.
Subcutaneous xenografts were explanted on day 4 of treatment. Tumor explants were immediately immersed in liquid nitrogen and stored at −80°. Tumors were disrupted with a mortar and pestle, followed by sonication in RIPA buffer supplemented with proteinase and phosphatase inhibitors. Proteins were separated as above. Antibodies to PARP, total-ERK, phosphorylated-ERK, and HSP90 were obtained from Cell Signaling Technology. Quantification of immunoblots was performed using ImageJ software.
Co-IP.
Here 293T cells were cotransfected with HA-CIC-DUX4 and hERK (ERK1 or ERK2) for 48 h, lysed, quantified, and incubated with either IgG (Cell Signaling Technology; 2729) fused to Dynabeads Protein G (Thermo Fisher Scientific; 10004D) or anti-HA magnetic beads (Thermo Fisher Scientific; 88836) overnight at 4 °C. Proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, and blotted with antibodies recognizing total ERK and HA-Tag.
Ubiquitination Experiments.
Here 293T cells were transfected with HA-tagged CIC-DUX4 and serum-starved for 6 h, followed by stimulation with EGF (100 ng/mL) for 30 min, bortezomib (1 uM) for 6 h, and BCI (1 uM) for 30 min. Cells were lysed and compared with EV control and CIC-DUX4–transfected 293T cells without treatment.
Real-Time qPCR.
Isolation and purification of RNA was performed using the RNeasy Mini Kit (Qiagen). Here 500 ng of total RNA was used in a reverse-transcriptase reaction with the SuperScript III First-Strand synthesis system (Invitrogen). qPCR included three replicates per cDNA sample. Human CIC, DUSP6, and GAPDH were amplified with Taqman gene expression assays (Applied Biosystems). Expression data were acquired using an ABI Prism 7900HT Sequence Detection System (Thermo Fisher Scientific). Expression of each target was calculated using the 2−ΔΔCt method and expressed as relative mRNA expression.
CIC-DUX4, CIC-NUTM1, and EWSR1-FLI1 Transcriptome Analysis: EGAD00001003121 Dataset.
A publicly curated RNAseq dataset (EGAD00001003121) of sarcoma tissues with CIC-DUX4 fusion (n = 6), CIC-NUTM1fusion (n = 5), and EWSR1-FLI1fusion (n = 5) was downloaded, the fastq files were mapped to hg19 using STAR (version 2.4) algorithm, and transcript expressions were quantified using RSEM (version 1.2.29) algorithm. The normalized transcript reads (TPM) were used for downstream analysis. The expression levels of 15,526 transcripts were scaled and used to generate the PCA (pca3d package in R) plots based on different fusion attributes.
The differential analysis was performed using the R edgeR package. There were 1,170 significant differentially expressed genes identified among the individual CIC-DUX4, CIC-NUTM1, and EWSR1-FLI1 tumors based on the criteria of absolute log2 fold change ≥1 and FDR <0.05. We subsequently performed hierarchical clustering using the Heatmap function from the ComplexHeatmap package in R.
To compare individual gene (DUSP4 and DUSP6) expression in CIC-DUX4, CIC-NUTM1, and EWSR1-FLI tumors, we retrieved normalized transcript reads of DUSP4 and DUSP6 across samples. The expression of these two genes in the samples with three different chimeric fusion proteins were displayed in boxplots. Three-group comparisons were done using a nonparametric statistical method (Kruskal–Wallis).
ChIP-Seq Analysis.
The reads from ChIP-seq were mapped to reference genome hg19. The MACS2 algorithm (version 2.2.1) (PMID: 18798982) was used for the peak calling. The significant peaks between the protein of interest and background IgG binding were identified based on an adjusted P <0.05. The peaks were further annotated including functional enrichment analysis of target genes using the R package ChIPseeker (PMID: 25765347) algorithm.
Viability and Apoptosis Assays.
Crystal violet assays were performed 72 h after drug treatments with either DMSO or BCI. CellTiter Glo and Caspase 3/7 Glo (Promega) experiments were performed according to the manufacturer’s protocol. In brief, cells were plated in a 96-well plate, treated with indicated drug or control, and analyzed on a Spectramax microplate reader (Molecular Devices) after 72 h of treatment.
CHX Chase Assays.
H1975 M1 cells were transfected with HA-CIC-DUX4, serum-starved for 6 h, and treated with CHX (50 ug/mL) alone or with either EGF (100 ng/mL) or BCI (1 μM) for the indicated times. The relative density was determined using ImageJ software.
Statistical Analysis.
Experimental data are presented as mean ± SEM. P values derived for all in vitro experiments were calculated using the two-tailed Student’s t test or one-way analysis of variance (ANOVA).
Study approval.
For tumor xenograft studies, specific pathogen-free conditions and facilities were approved by the American Association for Accreditation of Laboratory Animal Care. Surgical procedures were reviewed and approved by the University of California, San Francisco Institutional Animal Care and Use Committee (IACUC), protocol #AN178670-02.
Supplementary Material
Acknowledgments
We thank T. Kondo for the NCC-CDS_X1 cells, T. Nakamura for the HA-tag CIC-DUX plasmid, A. Unni for the sgLacZ and sgDUSP6 plasmids, and K. Jones, N. Naka, and A. Kawai for the synovial sarcoma cell lines. R.A.O. was supported by a grant from the National Cancer Institute (1K08CA222625-02).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009137117/-/DCSupplemental.
Data Availability.
The data supporting the findings of this study are openly available in the European Genome-Phenome Archive at https://ega-archive.org/datasets/EGAD00001003121 (ID no. EGAD00001003121).
References
- 1.Bhagwat A. S., Vakoc C. R., Targeting transcription factors in cancer. Trends Cancer 1, 53–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden E. C. et al., Soft tissue sarcomas of adults. Clin. Cancer Res. 9, 1941–1956 (2003). [PubMed] [Google Scholar]
- 3.Antonescu C. R. et al., Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 cases. Am. J. Surg. Pathol. 41, 941–949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshimoto T. et al., CIC-DUX4 induces small round cell sarcomas distinct from Ewing sarcoma. Cancer Res. 77, 2927–2937 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Specht K. et al., Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer 53, 622–633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okimoto R. A. et al., CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J. Clin. Invest. 129, 3401–3406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamura-Saito M. et al., Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 15, 2125–2137 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Simón-Carrasco L., Jiménez G., Barbacid M., Drosten M., The Capicua tumor suppressor: A gatekeeper of Ras signaling in development and cancer. Cell Cycle 17, 702–711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson S. et al., Transcriptomic definition of molecular subgroups of small round cell sarcomas. J. Pathol. 245, 29–40 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Futran A. S., Kyin S., Shvartsman S. Y., Link A. J., Mapping the binding interface of ERK and transcriptional repressor Capicua using photocrosslinking. Proc. Natl. Acad. Sci. U.S.A. 112, 8590–8595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okimoto R. A. et al., Inactivation of Capicua drives cancer metastasis. Nat. Genet. 49, 87–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm O. et al., Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139, 3962–3968 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oyama R. et al., Generation of novel patient-derived CIC- DUX4 sarcoma xenografts and cell lines. Sci. Rep. 7, 4712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. et al., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong D. et al., Transcriptomic analysis of CIC and ATXN1L reveal a functional relationship exploited by cancer. Oncogene 38, 273–290 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Weissmann S. et al., The tumor suppressor CIC directly regulates MAPK pathway genes via histone deacetylation. Cancer Res. 78, 4114–4125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunda S. et al., CIC protein instability contributes to tumorigenesis in glioblastoma. Nat. Commun. 10, 661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens D. M., Keyse S. M., Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26, 3203–3213 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Schaefer I.-M. et al., CIC-NUTM1 fusion: A case which expands the spectrum of NUT-rearranged epithelioid malignancies. Genes Chromosomes Cancer 57, 446–451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidger A. M., Keyse S. M., The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs). Semin. Cell Dev. Biol. 50, 125–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad M. K., Abdollah N. A., Shafie N. H., Yusof N. M., Razak S. R. A., Dual-specificity phosphatase 6 (DUSP6): A review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 15, 14–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes-Xavier C. et al., Dual-specificity MAP kinase phosphatases as targets of cancer treatment. Anticancer. Agents Med. Chem. 11, 109–132 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Wang B. et al., ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep. 18, 1543–1557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagnyukova T. V. et al., DUSP6 regulates drug sensitivity by modulating DNA damage response. Br. J. Cancer 109, 1063–1071 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina G. et al., Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat. Chem. Biol. 5, 680–687 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinson A. R. P. et al., Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: Utility and pitfalls. Front. Oncol. 3, 183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unni A. M. et al., Hyperactivation of ERK by multiple mechanisms is toxic to RTK-RAS mutation-driven lung adenocarcinoma cells. eLife 7, e33718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Q.-N. et al., Pharmacological inhibition of DUSP6 suppresses gastric cancer growth and metastasis and overcomes cisplatin resistance. Cancer Lett. 412, 243–255 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Shojaee S. et al., Erk negative feedback control enables pre-B cell transformation and represents a therapeutic target in acute lymphoblastic leukemia. Cancer Cell 28, 114–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramkissoon A. et al., Targeted inhibition of the dual specificity phosphatases DUSP1 and DUSP6 suppress MPNST growth via JNK. Clin. Cancer Res. 25, 4117–4127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are openly available in the European Genome-Phenome Archive at https://ega-archive.org/datasets/EGAD00001003121 (ID no. EGAD00001003121).