Abstract
The limited available treatment options for patients with chronic lung diseases, such as fibrosis, lead to poor prognosis after diagnosis and short survival rates. An exciting new bioengineering approach utilizes de- and recellularization of lung tissue to potentially overcome donor organ shortage and immune reactions toward the received transplant. The goal of decellularization is to create a scaffold which contains the necessary framework for stability and functionality for regenerating lung tissue while removing immunomodulatory factors by removal of cells. After decellularization, the scaffold could be re-functionalized by repopulation with the patient’s own stem/progenitor cells to create a fully functional organ or can be used as ex vivo models of disease. In this chapter the decellularization of lung tissue from multiple species (i.e., rodents, pigs, and humans) as well as disease states such as fibrosis is described. We discuss and describe the various quality control measures which should be used to characterize decellularized scaffolds, methods for protein analysis of the remaining scaffold, and methods for recellularization of scaffolds.
Keywords: Biomaterial, Decellularization, Lung, Mass spectrometry, Proteomics, Scaffold, Tissue engineering
1. Introduction
Pulmonary fibrosis is a devastating disease characterized by a marked increase in lung stiffness, mainly due to excessive deposition of collagen and loss of alveolar surface area, and thereby leads to declining lung function. While two pharmaceutical treatments have recently become available for the treatment of pulmonary fibrosis, they only serve to halt disease progression and cannot reverse disease pathogenesis. Thus, at end-stage disease, the only option is lung transplantation.
While mouse models can be used to study certain aspects of fibrosis, they incompletely recapitulate the heterogeneity seen in human disease. The most common mouse model is the intratracheal instillation of bleomycin which leads to fibrosis development after a short period of time (about 14 days) but resolves after longer periods (about 28–56 days) [1–5]. Other models include irradiation, intratracheal administration of fluorescein isothiocyanate (FITC), instillation of mineral fibers like silica, as well as diverse transgenic and viral induced fibrosis but all of them are only capable of recapitulating some of the features of the human disease [1]. New models are thus needed to help close the gap between promising in vitro and in vivo studies and the clinic. An exciting new bioengineering approach utilizes de- and recellularization of lung tissue to generate three-dimensional scaffolds.
The initial goal of decellularization technologies was to use these scaffolds to overcome donor organ shortage and provide a scaffold purported to be devoid of immune reactions in the transplant recipient. After initial proof-of-concept studies in rodents [6, 7], several groups scaled these processes up to more clinically relevant lung tissue from pigs, nonhuman primates, and human [8–14]. Although clinical application of recellularized lungs is still many years away, the use of decellularized preclinical and patient tissue has provided a powerful tool for expanding our knowledge of human disease onset and pathogenesis. Lungs from both normal and diseased murine and human lungs have been successfully decellularized. Emphysematous lungs from murine and human chronic obstructive pulmonary disease (COPD) patients can be decellularized and retain pathologic appearances such as distal airspace enlargement. Strikingly, immortalized murine epithelial cells recellularized onto these scaffolds did not survive as long as those recellularized on normal lung scaffolds [3]. In an emphysematous model with aging superimposed, cell survival was further reduced. In a follow-up study, it was found that epithelial cells seeded onto scaffolds derived from aged mice have decreased laminin expression, and thus this might be one reason for changes in epithelial cell survival and proliferation [15]. Interestingly, we observed comparable trends of decreased cell survival on human scaffolds derived from patients with emphysema and COPD as compared to healthy patients, indicating that there may be something fundamentally deranged with emphysematous scaffolds [10].
Murine lungs derived from bleomycin-treated animals were found to retain their characteristic fibrotic appearance by histological assessment as well as have an increase in a range of extracellular matrix (ECM) components as compared to murine lungs decellularized from healthy mice [3]. While their ECM composition seemed to differ, there were no detectable differences in cell survival or proliferation by any of the endpoints utilized for two different cell types (immortalized distal epithelial cells and murine bone marrow-derived mesenchymal stem cells).
With regard to human studies related to fibrosis, recellularization of decellularized fibrotic human lung tissue with healthy human fibroblasts led to activation to myofibroblasts with increased smooth muscle actin expression, a feature typically seen in fibrotic tissue [8]. This indicates that the composition of the ECM and environment is a critical factor and might be critically involved in disease progression. In an additional study, cultivation of primary lung fibroblasts from healthy and idiopathic pulmonary fibrosis (IPF) patients on decellularized lungs from healthy and IPF patients resulted in significant differential expression of pro-fibrotic genes. Notably, the origin of the scaffold had a greater effect on gene expression than did origin of the cells [16]. Another study identified chitinase 3-like 1 (CHI3L1), a prototypic chitinase-like protein that is elevated in IPF patients, to be sufficient to induce myofibroblast transformation in human fibroblasts when cultured on decellularized normal lung tissue [17]. De- and recellularization approaches have also been used to study the effect of the ECM of scleroderma patients on the differentiation capacity of peripheral blood mononuclear cells into fibrocytes [5]. A netrin-1-dependent pathway was identified which is involved in bleomycin-induced fibrosis in mice and may therefore be a possible treatment target [5]. These examples all demonstrate good correlation with hallmarks of the human disease and point out the important knowledge that can be gained by studying the ECM composition of and cell behavior on diseased decellularized lungs. Further analyses of the protein composition of normal vs. diseased decellularized lungs with advanced proteomic methods such as mass spectrometry will provide valuable additional insight into specific pathogenic changes in the ECM and help with the identification of novel treatment candidates. In summary, decellularized scaffolds have been invaluable tools for furthering our understanding of lung biology and disease.
In this chapter the induction of fibrosis in a rodent model and a protocol for decellularization of lung tissue from multiple species like rodents, pigs, and humans is described. Further, quality controls and methods for protein analysis of the remaining scaffold are discussed, including mass spectrometry. In addition, recellularization techniques for high-throughput studies from single murine lungs or single human patients are discussed. These tools can be used as new technologies for researchers to deepen understanding of human fibrosis pathology.
2. Materials
2.1. Induction of Fibrosis in Mice via Bleomycin
Adult mice (usually 8 weeks of age).
Anesthesia: a mixture of medetomidine (0.5 mg/kg mouse), midazolam (5.0 mg/kg mouse), and fentanyl (0.05 mg/kg mouse) (MMF).
Anesthesia antagonist: a mixture of atipamezole (2.5 mg/kg mouse), flumazenil (0.5 mg/kg mouse), and naloxone (1.2 mg/kg mouse).
Bleomycin (3.6 U/kg mouse), prepared in 75 μl of sterile 1X PBS.
Scale capable of weighing in a range of 10–100 g.
Respiratory assessment system to evaluate lung function and determine fibrosis severity (we use FlexiVent from SCIREQ).
2.2. Solutions and Materials for Decellularization
Prepare solutions sterile or sterile filter (0.22 μm) before usage. All solutions should be prepared with deionized (di) or ultrapure water. An overview of the amount of the solutions needed for different species on each day and steps of the decellularization protocol is shown in Table 1. The following solutions and materials are used in the decellularization process:
Sterile environment (cell culture hood) and dissection utensils (sterilized, i.e., autoclaved or rinsed with 70% ethanol—forceps, scissors, petri dishes, beakers, plastic bins, syringes, and cannulas).
Wash solution: 5X penicillin/streptomycin (PS), i.e., 500 U/ml penicillin, 500 μg/ml streptomycin in diH2O.
Triton solution: 0.1% (w/v) Triton X-100, 5X PS.
Sodium deoxycholate (SDC): 2% SDC (w/v) (see Note 1).
NaCl solution: 1 M NaCl, 5X PS.
DNAse solution: 30 μg/ml porcine pancreatic DNAse, 1.3 mM MgSO4, 2 mM CaCl2, 5X PS (see Note 2).
Peracetic acid (PAA) solution: 0.1% peracetic acid, 4% ethanol.
Storage solution: 5X PS, 2.5 μg/ml amphotericin-B, 50 μg/ml gentamicin, in PBS.
Roller pump (i.e., Stockert Shigley) capable of a flow rate of 2 l/min.
Table 1.
Details of the decellularization protocol for different species
| Mouse | Rat | Pig | Human | |||
|---|---|---|---|---|---|---|
| Each via trachea and right ventricle | Each via trachea and right ventricle | Each via trachea/main bronchus and pulmonary artery/main vessel | Each via trachea/main bronchus and pulmonary artery/main vessel | |||
| Day 1 | Manually | Manually | Perfusion pump 2 l/min | Perfusion pump 2 l/min | ||
| Solution | Incubation time | Amount (ml) × times | Amount (ml) × times | Amount (l) × times | Amount (l) × times | |
| diH2O 5X PS | Washing | – | 3, 5X | 20–30, 5X | 2–3, 3X | 2–3, 3X |
| 0.1% Triton | Washing | – | – | – | 2–3 | 2–3 |
| 0.1% Triton | Filling | 24 h, 4 °C, shaking | 3 | 20–30 | 2–3 | 2–3 |
| Day 2 | ||||||
| diH2O 5X PS | Washing | – | 3, 5X | 20–30, 5X | 2–3, 3X | 2–3, 3X |
| 2% SDC | Washing | – | – | – | 2–3 | 2–3 |
| 2% SDC | Filling | 24 h, 4 °C, shaking | 3 | 20–30 | 2–3 | 2–3 |
| Day 3 | ||||||
| diH2O 1X PS | Washing | – | 3, 5X | 20–30, 5X | 2–3, 3X | 2–3, 3X |
| 1 M NaCl | Washing | – | – | – | 2–3 | 2–3 |
| 1 M NaCl | Filling | 1 h, RT, shaking | 5 | 20–30 | 2–3 | 2–3 |
| diH2O 1X PS | Washing | – | 3, 5X | 20–30, 5X | 2–3, 3X | 2–3, 3X |
| DNAse | Washing | – | – | – | 2–3 | 2–3 |
| DNAse | Filling | 1 h, RT, shaking | 3 | 20–30 | 2–3 | 2–3 |
| diH2O 1X PS | Washing | – | – | – | 2–3, 3X | 2–3, 3X |
| Peracetic acid | Washing | – | – | – | 2–3 | 2–3 |
| Peracetic acid | Filling | 1 h, RT, shaking | – | – | 2–3 | 2–3 |
| diH2O 1X PS | Washing | – | 3, 5X | 20–30, 5X | 2–3, 3X | 2–3, 3X |
| Storage solution | Washing | – | – | – | 2–3, 2X | 2–3, 2X |
| Storage solution | Filling | Storage, 4 °C | 3 | 20–30 | 2–3 | 2–3 |
2.3. Solutions and Materials for Mass Spectrometry
4X lysis buffer: 250 mM Tris, pH 6.8, 8% (w/v) sodium dodecyl sulfate (SDS), 400 mM dithiothreitol (DTT).
Running buffer for SDS-PAGE: 50 mM Tris base, pH 8.3, 380 mM glycine, 0.22% (w/v) SDS.
100 mM NH4HCO3.
50 mM NH4HCO3 in 50% (v/v) acetonitrile (ACN).
100% (v/v) ACN.
10 mM DTT in 100 mM NH4HCO3.
55 mM (10 mg/ml) iodoacetamide (IAA) in 100 mM NH4HCO3.
Trypsin solution: 6–12 ng/μl trypsin, 5% (v/v) ACN in 40 mM NH4HCO3.
4 mM NH4HCO3.
5% (v/v) formic acid (FA) in 50% (v/v) ACN.
Loading solution: 2.5% (v/v) ACN in 2.5% (v/v) FA.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE): To clean the protein lysate from salts and detergents of the lysis buffer, it should be run on a denaturing gel (see Note 3) permitting the sample to run until it is located in the separation gel. The bands should not separate in order to minimize the amount of gel to be excised for trypsin digestion. In our setup, we have found that running at 80 V for around 10–15 min is sufficient with the use of 20–50 mg per lane.
Sample homogenizer (e.g., Polytron PT2100 from Kinematica; Ball mill such as Mikro-Dismembrator S from Sartorius).
Centrifuge.
Protein assay reagents such as BCA, Bradford, etc. (we use the DC detergent compatible protein detection Kit from Bio-Rad).
70% (v/v) ethanol.
Razorblades to excise protein bands.
Microcentrifuge tubes.
Vortex.
Speed vac.
Heat block for incubations at 56 °C.
Conventional ultrahigh-pressure liquid chromatography (UHPLC) or high-pressure liquid chromatography (HPLC) equipment.
Capillary fused silica column (12 cm × 100 μm inner diameter) packed with HALO C18 2.7 μm particle size, 90 A (from Michrom Bioresources).
Gradient of 0–35% ACN/0.1%FA (Fisher Chemical, Optima, LC/MS grade), 35–100% ACN/0.1% FA, 100% ACN, and 0.1% FA in H2O.
Mass spectrometer (we use a Q-Exactive mass spectrometer from Thermo Fisher Scientific).
Proteome Discoverer 1.4 software (Thermo Fisher Scientific).
2.4. Lung Histology
4% paraformaldehyde (20 cm H2O pressure).
Xylene.
Graded ethanol series.
Paraffin.
Glass slides.
Cover slips.
Microtome.
Deparaffinization station.
Histological stain as needed (e.g., hematoxylin and eosin, DAPI, Verhoeff’s Van Gieson, Masson’s Trichrome, Alcian Blue).
Normal light or fluorescence microscope.
2.5. Electron Microscopy
0.1 M cacodylate buffer, pH 7.2.
Karnovsky’s fixative: 2.5% (w/v) glutaraldehyde, 1.0% (w/v) paraformaldehyde in 0.1 M cacodylate buffer, pH 7.2.
1% (w/v) osmium tetroxide.
Graded ethanol series.
Propylene oxide.
Spurr’s epoxy resin.
Glass knife or diamond knife on an ultracut microtome.
Methylene blue-azure II.
200 mesh thin bar nickel grids.
Uranyl acetate: 2% (w/v) uranyl acetate in 50% ethanol.
Lead citrate.
Transmission electron microscope (TEM, we use a JEOL 1400 from JEOL USA Inc.).
2.6. Immunohistochemistry (IHC)
Standard deparaffinization station.
Sodium citrate buffer: 10 mM citrate buffer pH 6 (we use the one from Dako).
0.1% (w/v) Triton X-100.
Wash solution: 1% (w/v) BSA in PBS.
Blocking solution: 10% goat serum.
Primary antibody. Antibodies routinely used in our lab include anti-fibronectin (#610077 from BD Transduction Laboratories, used at 1:100), anti-laminin (#ab11575 from Abcam, used at 1:100), anti-alpha elastin (#ab21607 from Abcam, used at 1:100), anti-smooth muscle myosin heavy chain 2 (#ab53219 from Abcam, used at 1:100), anti-collagen I (#ab292 from Abcam, used at 1:100), anti-proliferation marker Ki67 (#ab16667 from Abcam, used at 1:50), anti-cleaved caspase-3 (#Asp175 from Cell Signaling Technology, used at 1:100), antihuman actin (#1A4 from Dako, used at 1:10,000).
Secondary antibody. Secondary antibodies routinely used in our lab include Alexa Fluor 568 goat anti-rabbit IgG (H + L) (1:500, Invitrogen), Alexa Fluor 568 F(ab’)2 fragment of goat anti-mouse IgG (H + L) (1:500, Invitrogen) [10, 12, 18–20].
4’,6-diamidino-2-phenylindole (DAPI) nuclear stain.
Mounting medium (we use Aqua-Poly/Mount from Polysciences Inc.)
2.7. Assessment of Residual DNA
Tissue paper (e.g., Kimwipe®).
DNA extracting kit (we use the DNeasy Blood & Tissue Kit from Qiagen).
0.8% (w/v) agarose gel and agarose gel running apparatus.
SYBR Safe DNA Gel Stain.
UV light or molecular imager (we use a Versa Doc from Bio-Rad).
100 bp ladder and salmon sperm DNA.
dsDNA quantitation assay (we use the Quant-iT™ PicoGreen® dsDNA Assay kit from Thermo Fisher Scientific)
2.8. Gelatinase Assay
Gelatinase activity measuring kit (we use the EnzChek Gelatinase Assay from Molecular Probes) as previously described [21].
Tris-buffered saline (TBS): 50 mM Tris–HCl, pH 7.4, 150 mM NaCl.
Gelatin standard (we use DQ™ gelatin from Life Technologies).
2.9. Assessment of Residual Detergent
0.0125% (w/v) methylene blue (MB) in diH2O.
Chloroform.
Vortex.
Spectrophotometer to read absorbance at 630 nm (platereader is recommended).
Polypropylene 96-well plates.
Pure diH2O or PBS for blanks.
Triton, SDC, NaCl, or DNAse for standard curve preparation [22].
2.10. Cell Survival
Cell counting chamber or viability assays like WST/MTT or LDH release.
2.11. Cells and Cultivation Media for Recellularization
A variety of different cell types can be used for recellularization studies, depending on the experimental questions. We have also found that different cell types have different thresholds for detergent-induced cytotoxicity, and therefore testing of different cell types is advantageous in assessing cytocompatibility of acellular scaffolds [22]. Although in disease models or diseased human tissue, assessment of cytotoxicity may be complicated to interpret due to potential differences in residual proteins or toxic components.
- For recellularization of rodent, nonhuman primate, pig, and human scaffolds, we have generally utilized four different cell lines (see Note 4):
- Human mesenchymal stem cells (i.e., hMSC, University of Minnesota, NHLBI Production Assistance for Cell Therapy program [23]).
- Human lung fibroblasts (HLF) (ATCC, CCL 171).
- Human bronchial epithelial cells (HBE, courtesy of Albert van der Vliet, University of Vermont, originally from Dr. J. Yankaskas [24]).
Cell culture-treated plastic (i.e., petri dishes, T-flasks, multiwall plates).
Incubator capable of 37 °C and 5% CO2.
hMSC medium: Modification of Eagle’s Medium-Earle’s Balanced Salt Solution (MEM-EBSS), 20% fetal bovine serum, 100 IU/ml penicillin/100 μg/ml streptomycin, 2 mM l-glutamine (see Note 5).
HLF medium: DMEM/F-12 (1:1, v/v) with 10% (v/v) fetal bovine serum, 100 IU/ml penicillin/100 μg/ml streptomycin, 2 mM l-glutamine.
HBE medium: DMEM/F-12 (1:1, v/v) with 10 ng/ml cholera toxin, 10 ng/ml epidermal growth factor, 5 μg/ml insulin, 5 μg/ml transferrin, 0.1 μM dexamethasone, 15 μg/ml bovine pituitary extract, 0.5 mg/ml bovine serum albumin, and 100 IU/ml penicillin/100 μg/ml streptomycin.
CBF medium: EGM-2 (Lonza), 5% fetal bovine serum, 0.04% hydrocortisone, 0.4% hFGF-B, 0.1% VEGF, 0.1% R3-IGF-1, 0.1% ascorbic acid, 0.1% hEGF, 0.1% gentamicin sulfate amphotericin-B, 100 IU/ml penicillin/100 μg/ml streptomycin. These cells are grown and expanded on collagen type I-coated tissue culture surfaces.
3. Methods
3.1. Induction of Fibrosis in Mice via Bleomycin
There are several different models for the induction of fibrosis in mice. One common method is intratracheal administration of bleomycin (see Chap. 2 for detailed protocols and tips). Other mouse models of fibrosis such as radiation, silicosis, or adeno-TGFβ could also be used.
Anesthetize mice (usually 8 weeks of age) according to local IACUC protocols.
Instill 3.6 U/kg of bleomycin by oropharyngeal or intratracheal administration (see Note 6).
Allow mice to recover from anesthesia or antagonize the narcosis.
Monitor mouse weight and health during the next 14 days. Anticipated weight loss is approximately 5–10%. If the mice lose too much weight or exhibit other symptoms of distress, follow your local protocols for euthanasia.
Harvest heart-lung blocs 14 days post-bleomycin instillation and subsequently decellularize as described below [1, 3, 5, 26].
3.2. Decellularization of Rodent Lungs
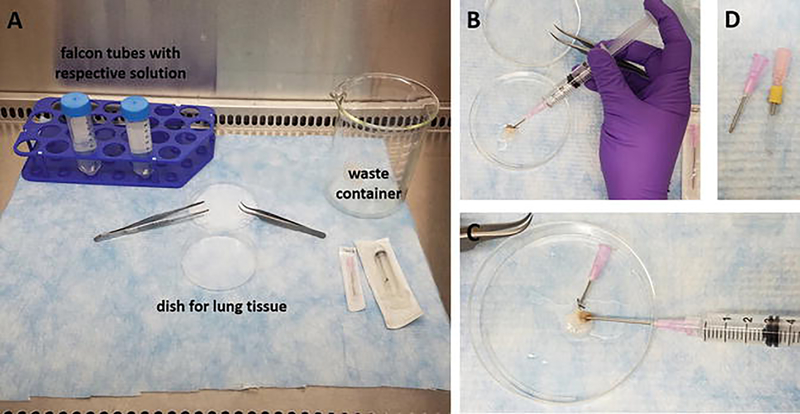
Perform all works in a sterile environment (cell cultivation hood) using sterilized utensils. A typical hood setup is shown in Fig. 1.
Euthanize the rodents according to your local animal regulations.
Extract the heart and lung en bloc.
Cannulate the trachea with a blunted 18 gauge cannula with (for rats) or without silicone tubing (for mice) to account for trachea size (Fig. 1d), and immediately clear the lung vasculature of blood by injecting 10 ml wash solution into the right ventricle.
Flush the lungs slowly and carefully using a syringe; flush five times, each alternating via the trachea and right ventricle with the amounts indicated in Table 1 [3, 18, 19, 25]. Between each filling the lung should be cleared of the respective solution as much as possible by passive drainage due to elastic recoil of the lung (see Note 7). At the conclusion of each filling and draining cycle (immediately before the next filling), take a sample to evaluate effluent contents, such as detergent or proteins. We have found that proper removal of detergents, in particular anionic detergents, is critical for producing cytocompatible acellular scaffolds [22].
At the conclusion of the decellularization protocol, take samples/biopsies as desired (see Note 8). Store the decellularized lung tissue in a sterile container filled with storage solution, covered and placed at 4 °C until further processing or usage for reseeding (see Note 9).
Fig. 1.
Rodent lung decellularization. (a) Hood setup with indication of the dish for the rodent lung tissue, waste container, and conical polypropylene tubes with solutions used during the decellularization; (b) flushing of a mouse lung via the tracheal cannula; (c) flushing of a mouse lung vasculature via the right ventricle; (d) cannulas used for rodent lung decellularization; left, cannula for mouse lungs, 18 gauge cannula with blunted edge and groove for attachment of the surgical thread; right, cannula for rat lungs, a silicone tube is used over a blunted 18 gauge cannula to increase diameter of the cannula and fit rat trachea size
3.3. Decellularization of Pig and Human Lungs
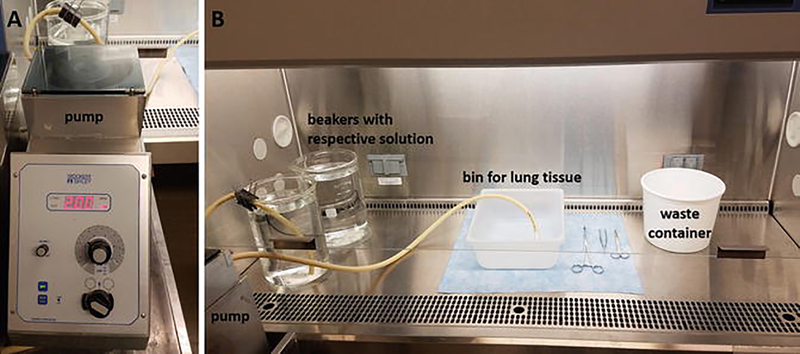
Perform all works in a sterile environment (cell cultivation hood) using sterilized (autoclaved or rinsed with 70% ethanol) plastic basins/4 l-pyrex beakers, tubings, forceps, and scissors. A typical hood setup is shown in Fig. 2.
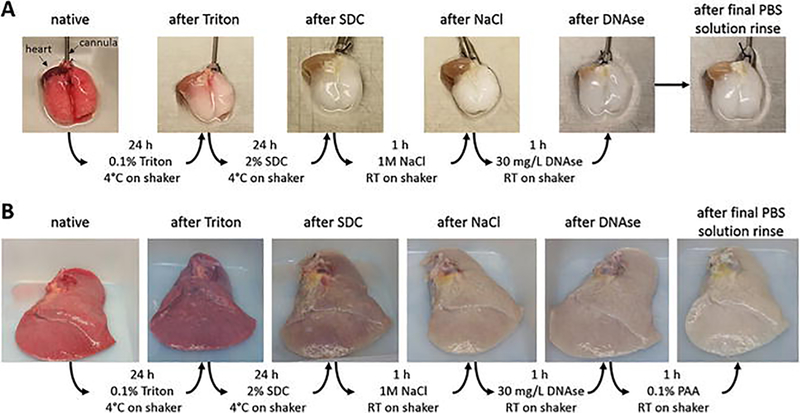
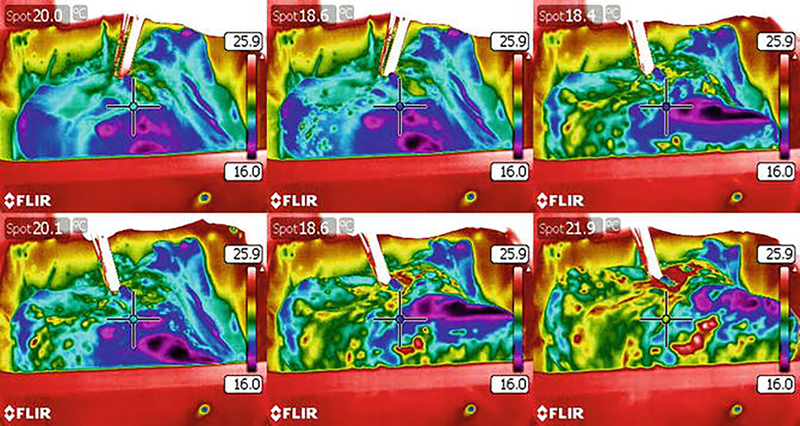
Take photographs of the lungs after each decellularization step to document loss of coloration and gross morphology. During decellularization the lungs should lose red coloration and should result in a glossy, white looking tissue (Fig. 3). Thermography, a method that is used to study heat distribution in structures or regions with the help of an infrared camera, can also be used to assess the distribution of perfused fluids during the decellularization protocol. We have found by the use of thermography that decellularization solutions are not evenly distributed in IPF lungs [10] (Fig. 4). This is most likely due to the heterogeneity in the lungs in fibrotic versus normal areas and the derangement of the normal perfusion pathways.
On day 1, dissect a single lobe by retaining as much of the main bronchi and vasculature as possible.
Flush the lungs by using a roller pump at a flow rate of 2 l/min with wash solution alternating via trachea/main bronchi and pulmonary artery/main vessel (see Note 10) [3, 12, 14, 18, 19, 25]. During the vascular flushing, clamp the trachea/main bronchus to allow for tissue inflation. Fluids will flow out of both the pulmonary artery and veins by passive drainage. At the conclusion of each filling and draining cycle (immediately before the next filling), take a sample of the effluent to evaluate effluent contents, such as detergent or protein content [22].
After the tracheal flushing is completed and the liquid is drained from the tissue (see below), connect the pump and tubing to the vasculature.
Typical volumes and the order of the solutions used for decellularization can be found in Table 1. Inflate lungs carefully (see Note 11). The inflation volume required for full inflation depends on the size of the lung/lobe and underlying disease. Between each filling, clear the lung from the respective solution as much as possible by a combination of elastic recoil and light manual manipulation, (i.e., gently kneading) to help drain the liquid (see Note 7).
Since fibrotic tissue is denser and not as accessible for decellularization reagents as healthy tissue due to obliterated vasculature and airways, a second cycle or decellularization via immersion of excised smaller segments may be necessary [12]. For this, repeat all the steps described above. Immerse tissue in the respective decellularization agent (detergent, NaCl, DNAse, PAA) for the time periods indicated in Table 1 with 3 diH2O wash steps of 10 min each (instead of the diH2O flushes) under agitation to facilitate mixing and distribution of the solutions.
At the conclusion of the decellularization, take samples/biopsies (see Note 8). Store the decellularized lung tissue covered with storage solution in a sterile container and at 4 °C until further processing or usage for reseeding. Change the storage solution every 3 weeks to minimize the risk of contamination (see Note 9).
Fig. 2.
Human and porcine lung decellularization. (a) Pump used for volume-controlled decellularization; (b) hood setup with indication of the pump, bin for the lung tissue, waste container, and beakers with solutions used during the decellularization
Fig. 3.
Overview of loss of tissue coloration during decellularization. (a) Murine lungs at the different steps of the protocol; (b) human lung lobe at the different steps of the protocol
Fig. 4.
Thermographic imaging of the liquid distribution during decellularization of a human idiopathic pulmonary fibrosis (IPF) lung lobe. Representative still images from thermographic analysis demonstrate nonhomogenous liquid distribution during decellularization of a human IPF lung lobe indicated by color changes. Regions that were limited or not accessible remained blue/violet. The human lobe was chilled at 4 °C in PBS after completion of the decellularization protocol. diH2O was warmed to 37 °C to create a thermal gradient (temperature scale bar shown) for detection by the FLIR camera
3.4. Scaffold Evaluation
3.4.1. Evaluation of Fibrosis Severity
It is known that the bleomycin model for fibrosis in rodents resolves over time and that fibrotic change peaks are established by day 14 after intratracheal bleomycin instillation with a peak around days 21–28 [27]. Lung function generally declines during fibrosis development, and a reduction in lung compliance and increase in lung elastance are seen and can be assessed using a device like the flexiVent [4]. Lung fibrosis is further characterized by excessive deposition of ECM proteins (like collagen, fibronectin, alpha smooth muscle actin, elastin, and tenascin C) [1, 28]. These changes can be evaluated in native tissue either by qPCR; quantitative protein assays such as Sircol™ (see Chap. 22 for detailed protocol and tips), Fastin™ Elastin, BioColor, etc.; Western blotting; histology (e.g., Masson Trichrome staining); or immunohistochemistry. After establishing and evaluating the preferred model of fibrosis in your laboratory, these lungs can then be decellularized and evaluated for fibrotic markers using the quantitative protein assays above and histological, immunofluorescence, or mass spectrometric proteomics assessment to evaluate changes as compared to healthy decellularized lungs. As the cells have been removed, normalization for quantitative Western blotting with cell-associated loading controls cannot be performed. Alternatively, prior to beginning decellularization, the right lungs can be removed after ligating the right bronchus. The right lung lobes can be used for traditional quantitative readouts as to the extent of fibrosis, and the left lobe could be used for decellularization. However, volumes would need to be adjusted as just the left lobe would be filled.
Lung Histology
3.4.2. Tissue Morphology and Integrity
Fix native or decellularized rodent lungs by gravity in 4% paraformaldehyde (20 cm H2O pressure) for at least 3 h at room temperature. Fix tissue samples of native and decellularized pig and human lungs (about 1 cm3) by immersion in 4% paraformaldehyde for at least 3 h at room temperature.
Embed samples in paraffin, and cut 5 μm sections on glass slides.
Deparaffinize and stain sections with any standard histological stain.
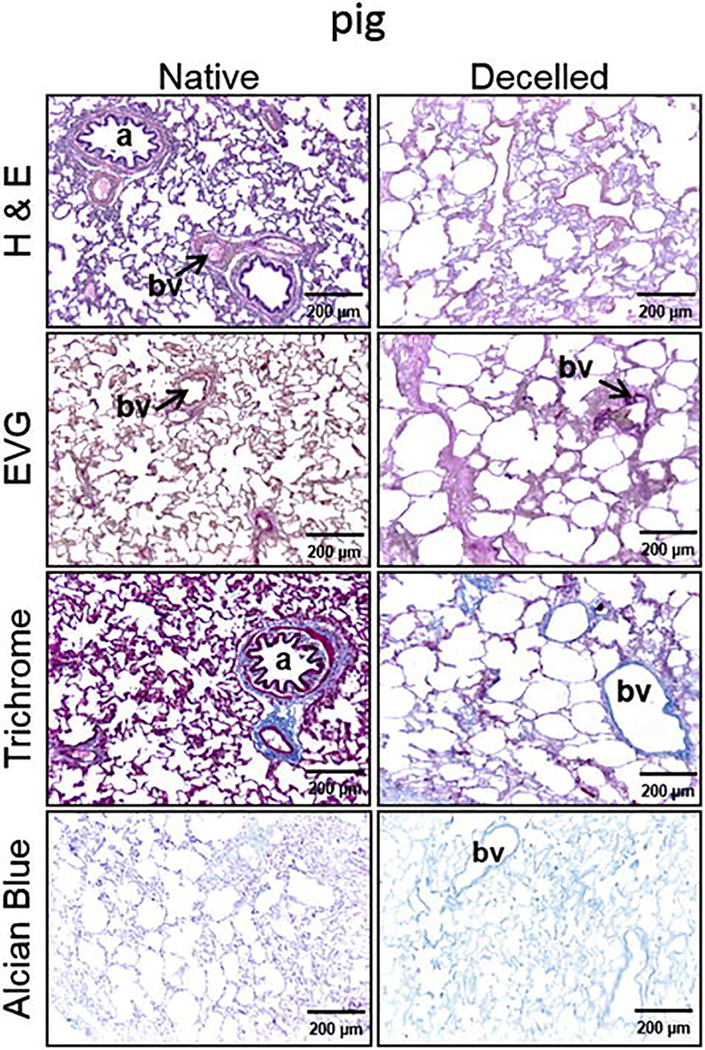
Assess sections using normal light or fluorescence microscopy (Fig. 5). Use H&E staining to determine overall tissue integrity.
Assess proper decellularization by the absence of nuclear material in the H&E and loss of DAPI staining [29]. The presence of visible red cellular material or “ghost cells” in H&E or in Masson’s Trichrome staining indicates incomplete cell removal.
Assess the retention/loss of elastin and proteoglycans by Verhoeff’s Van Gieson and Alcian Blue staining.
Fig. 5.
Evaluation of lung morphology using histology. Hematoxylin and eosin (H&E), Masson Trichrome, Verhoeff’s Van Gieson (EVG), and Alcian Blue staining of a native and decellularized pig lung lobe (Reprinted with permission from Ref. 14)
Electron Microscopy
For electron microscopic analyses, small segments (about 10 mm3) of native or decellularized lung tissue can be processed as follows:
Fix tissue overnight at 4 °C in Karnovsky’s fixative.
Rinse in cacodylate buffer.
Mince the tissue into approximately 1 mm3 pieces.
Fix in osmium tetroxide for 2 h at 4 °C.
Rinse the pieces again in cacodylate buffer.
Dehydrate through a graded ethanol series.
Clear in propylene oxide.
Embed in Spurr’s epoxy resin.
Cut semi-thin sections (1 μm) using glass knives on a Reichert ultracut microtome.
Stain with methylene blue-azure II and then evaluate for areas of interest (proximal and distal alveolar septa, large/small airways, blood vessels).
Cut ultrathin sections (60–80 nm) with a diamond knife.
Retrieve on 200 mesh thin bar nickel grids.
Contrast with uranyl acetate and lead citrate.
Examine with a TEM operating at 60 kV [12].
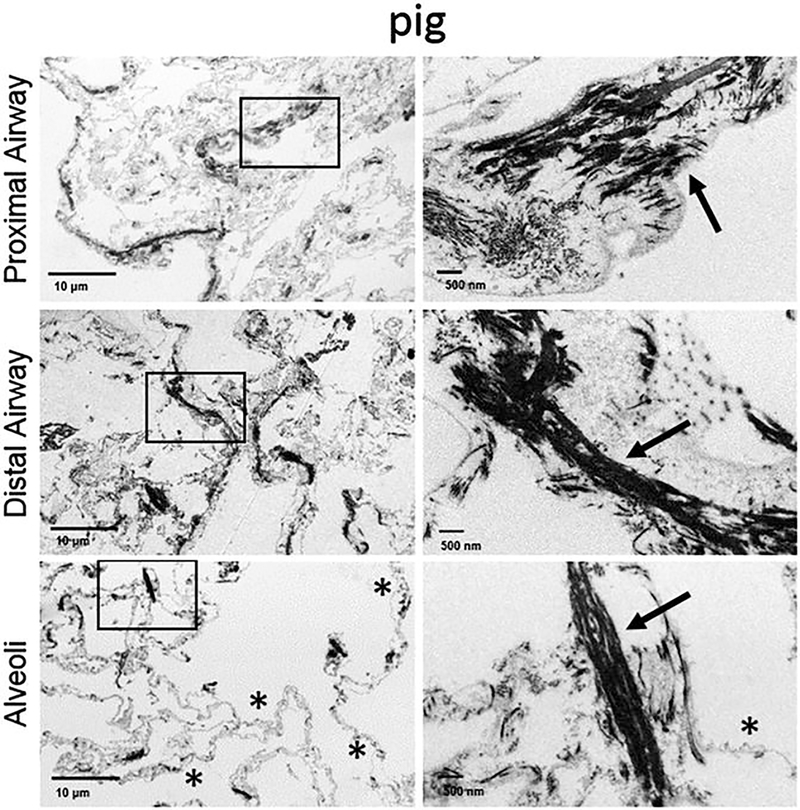
Use electron microscopy images to compare the native and decellularized tissue morphologies, assess the integrity of the alveolar-capillary barrier, and determine whether the ultrastructure of fibers like collagen and elastin is retained (Fig. 6).
Fig. 6.
Scanning electron microscopy images from a decellularized pig lung lobe. Collagen fibrils are pointed out by arrows and intact alveolar barrier can be observed next to the stars (Modified with permission from Ref. 14)
Immunohistochemistry (IHC)
Perform standard deparaffinization with three separate 10 min incubations in xylene.
Rehydrate in a descending series of ethanol and finally in water.
Perform antigen retrieval by heating tissue in 1X sodium citrate buffer at 98 °C for 20 min followed by a brief 20 min cool at room temperature.
Permeabilize tissue sections in Triton X-100 solution for 15 min.
Remove Triton X-100 with two 10 min washes in wash solution.
Block with blocking solution for 60 min.
After blocking, add primary antibody and incubate tissue sections overnight at 4 °C in a humidified chamber.
Wash tissues three times with wash solution for 5 min each.
Add secondary antibody and incubate for 60 min at room temperature in a dark humidified chamber.
Wash tissues again three times in wash solution for 5 min each in the dark.
Add DAPI nuclear stain for 5 min at room temperature in the dark followed by two washes in wash solution for 5 min each.
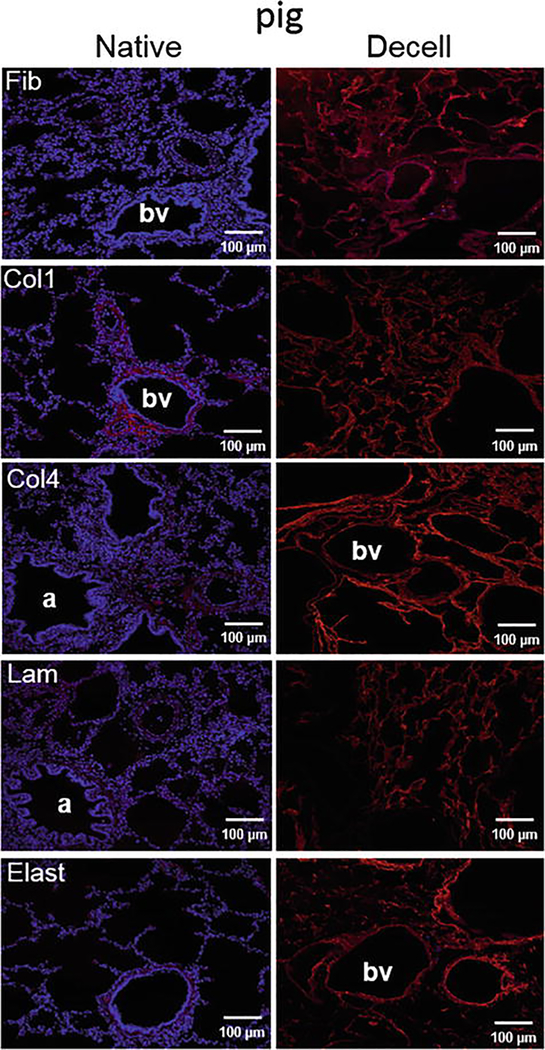
Mount the sections in Aqua-Poly/Mount and image with a fluorescence microscope. Typical staining is shown in Fig. 7.
Fig. 7.
Decellularization preserves major extracellular matrix (ECM) proteins in wild-type pig lungs. Representative photomicrographs are depicted and demonstrate qualitative retention of ECM proteins. Nuclear DAPI staining is depicted in blue and the stains of interest are depicted in red. Original magnifications: 100X. Col1, type I collagen; Col4, type 4 collagen; Elast, elastin; Fib, fibronectin; Lam, laminin (Reprinted with permission from Ref. 14)
3.4.3. Assessment of Residual DNA
Dry native and decellularized lung tissue on a tissue paper until no more liquid is visibly released and weigh the tissue.
Extract DNA by using a DNA extraction kit following the instructions provided by the manufacturer.
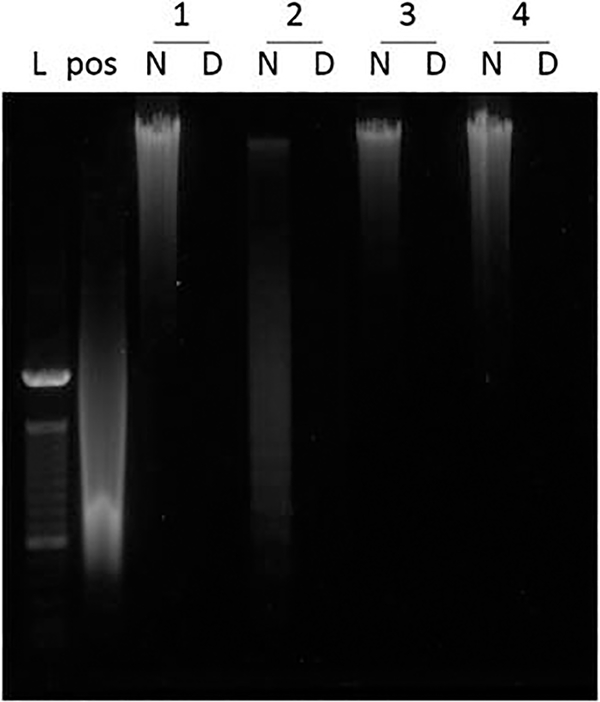
Run the isolated DNA on a 0.8% agarose gel and visualized under UV light or molecular imager with SYBR Safe DNA Gel stain dsDNA quantitation assay.
Use a 100 bp ladder and salmon sperm DNA as DNA size marker and positive control (Fig. 8). Ideally no DNA fragments will be visible on the gel.
Quantify the amount of double stranded (ds) DNA using a dsDNA quantitation assay following the instructions of the manufacturer. The threshold for adequate decellularization is less than 50 ng/mg dry tissue weight [29] (see Note 12).
Fig. 8.
Agarose gel for visualization of residual DNA in the decellularized lung tissue. Bands from left to right. L, 100 bp ladder; pos, salmon sperm DNA used as positive control; N, DNA from native human tissue; D, DNA from decellularized human tissue; 1–4, sample number
3.4.4. Gelatinase Assay
Determine net gelatinolytic activity using a gelatinase activity measuring kit as previously described [21].
Homogenize the decellularized lung tissue in TBS and report gelatinolytic activity (in 80 mg protein) as the rate of fluorescence increase over 4 h normalized to assay reagent containing DQTM gelatin alone [18].
Use native lung tissue as a control.
3.5. Scaffold Cytotoxicity
3.5.1. Assessment of Residual Detergent
Concentrations of SDC in wash effluents can be determined using a modified MB assay we recently developed [22]:
Mix effluent samples with MB at a ratio of 1:10 (w/v) and vortex.
Add chloroform at a ratio of 1:2 (sample: chloroform, v/v) and vortex for 1 min.
Incubate 30 min at room temperature.
Extract 150 μl from the bottom chloroform layer and collect in a polypropylene 96-well plate (see Note 13).
Add wells with diH2O or PBS containing no detergent for blanks.
Add SDC standards (concentration range—0–5 mg/ml, i.e., 0–0.5% (w/v)) prepared in either diH2O (for Triton, SDC, NaCl, and DNAse effluents) or storage solution (for PBS effluents).
Read the absorbance at 630 nm with a plate reader.
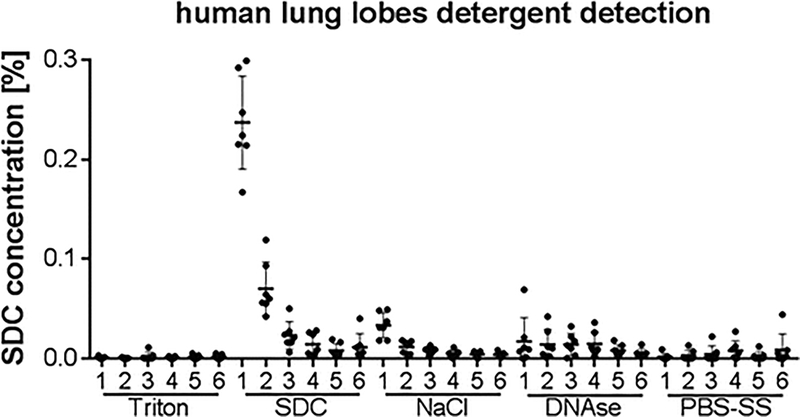
Calculate SDC concentration based on SDC standard curve. An example graph from several human lung lobe decellularizations is shown in Fig. 9.
Fig. 9.
Detergent detection in human lung lobes shows removal of detergent with consecutive diH2O washes. Calculated sodium deoxycholate (SDC) concentration is displayed from seven human lung lobe decellularizations after detergent quantification in the respective effluents. Data are displayed as mean ± standard deviation
3.5.2. Cell Survival
Determine the survival of cells on acellular lung scaffolds via indirect or direct contact with the scaffold by simple cell counting or viability assays like WST/MTT or LDH release in accordance to the ISO 10993–5:2009(E) standard.
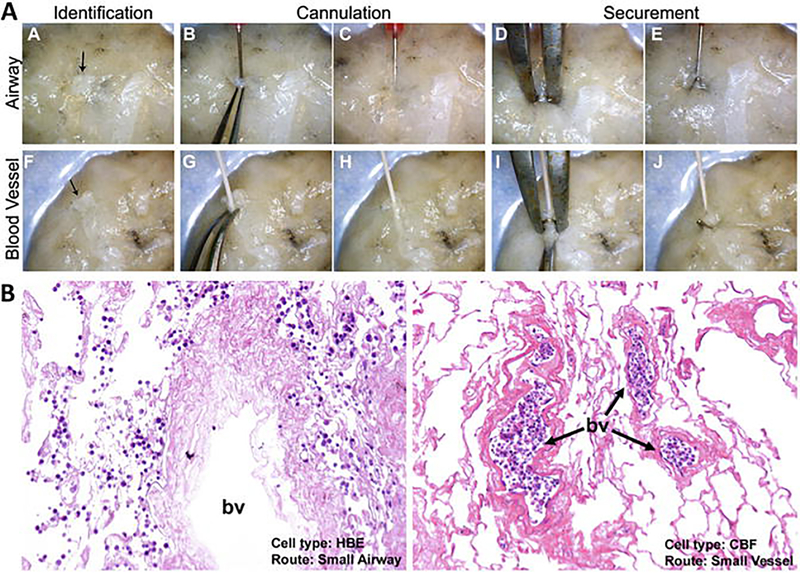
For indirect determination incubate small pieces of the tissue in PBS for a fixed period of time (e.g., 24 h) to generate effluents and treat different cell lines with it for 24 h before evaluating cell survival. For direct contact seed the cells into/onto tissue pieces/slices (see Note 14) and evaluate cell survival 24 h later. Both methods were used in a recently published study from our group, and details about the assays can be found there [22]. In general we suggest the use of multiple different cell types rather than just one as each cell type may have different thresholds for detergent-induced cytotoxicity (see Subheading 2.4). An example of the seeding of cells into the different compartments (airway and vasculature) can be seen in Fig. 10.
Fig. 10.
Recellularization of different compartments of decellularized human lung lobes. Excised segments of acellular human lung were selectively seeded through airways or vasculature with the use of a calcium alginate coating. (a) Identification of small airways and vessels, cannulation and securement with surgical clips for controlled cellular inoculation. (b) Human bronchial epithelial (HBE) and pulmonary endothelial colony-forming (CBF) cells were retained in their respective compartment after 24 h of culture (Reprinted with permission from Ref. 12)
3.6. Protein Analysis
3.6.1. General Considerations
Residual proteins in the decellularized tissue can be analyzed in multiple ways. An easy qualitative method is immunohistochemical stainings for specific proteins. Standard Western blotting [18, 19] can also be used to quantitate differences; however normalization in decellularized scaffolds can be difficult since there is loss of cellular proteins normally used for quantification and normalization. Both methods are limited to the availability and expense of antibodies and can only detect a small number of proteins at once. Proteins of interest need to be identified beforehand for these techniques, and thus techniques such as mass spectrometry proteomics offer a more unbiased and general approach to determine the protein composition of the decellularized scaffold. There are both targeted and nontargeted approaches as well as quantitative and semiquantitative approaches. Targeted approaches offer the benefit of being able to quantitate differences between native and decellularized lungs or between disease states, but standards must be generated beforehand. Thus these methods, while powerful, can be costly and are limited to known proteins of interest. In nontargeted, semiquantitative approaches, the composition of the scaffolds, including previously unknown proteins, has the potential to be detected. Thus, there are pros and cons for each approach depending on the experimental questions. Here, we present our approach for nontargeted, semiquantitative mass spectrometry proteomics.
3.6.2. Semiquantitative Mass Spectrometry Proteomics
Sample Preparation
Obtain three samples of about 125 mg wet weight from different locations of the decellularized lung tissue. In the case of murine lungs, we typically take one single lobe from each animal following decellularization.
Homogenize each sample for 30 s using a sample homogenizer at maximal speed in 800 μl of 1X lysis buffer.
Centrifuge for 5 min at 15,000 g at 4 °C.
Separate supernatant and pellet.
Determine the protein content of the supernatant with a protein assay kit.
Run the protein lysate (20–50 mg per lane) on an SDS-PAGE gel to clean it from salts and detergents (see Note 3) letting the sample run until it is located in the separation gel. The bands should not separate in order to keep the amount of gel for the trypsin in-gel digestion small. At 80 V this will take about 10–15 min.
Submit the whole band of each sample to trypsin in-gel digestion.
Trypsin In-Gel Digestion
Clean all surfaces and tools with 70% ethanol to avoid sample contaminations.
Excise protein spot/band and cut into small cubes (~1 mm3) with razorblades. Transfer to a clean microcentrifuge tube and incubate with 200 μl water for at least 15 min (see Note 15).
Remove water and destain three times by adding 50 mM NH4HCO3 in 50% ACN, incubate for 15–30 min with occasional vortexing, and discard the liquid with each wash (see Note 16).
Remove destaining solution and add enough 100% ACN to fully cover gel piece, incubate for 5 min, and discard ACN (see Note 17).
Dry gel pieces for 5 min using a SpeedVac (see Note 18).
Add 10 mM DTT in 100 mM NH4HCO3 to the dried gel piece and incubate for 1 h at 56 °C (see Note 19).
Let the gel pieces cool to room temperature and discard DTT solution. Add 55 mM IAA in 100 mM NH4HCO3 until gel pieces are covered. Incubate gel pieces in the dark for 45 min at room temperature (see Note 20).
Remove IAA/NH4HCO3 solution and discard to waste.
Add 100 mM NH4HCO3 and incubate for 5 min at room temperature.
Remove and discard solution.
Dehydrate with 100% ACN. Remove and discard solution when gel pieces are white/opaque. Repeat this step to ensure gel pieces are fully dehydrated.
Re-swell in 100 mM NH4HCO3 for 5 min. Remove and discard solution.
Dehydrate with 100% ACN. Remove and discard solution when gel pieces are white/opaque. Repeat this step to ensure gel pieces fully dehydrated.
Evaporate remaining solvents in a SpeedVac for 5 min.
Add 30 μl of trypsin solution to re-swell gel pieces completely at 4 °C for 30 min. If after 30 min, gel pieces are uncovered, add more NH4HCO3 buffer to cover gel pieces (see Note 21).
Digest 16–18 h at 37 °C in trypsin solution (see Note 22).
Transfer the solution to a new vial and add 5% FA in 50% ACN to the gel pieces; incubate for 1 h at room temperature (see Note 23).
Transfer the solution to the same vial as in step 17.
Add ACN and dehydrate. Transfer the solution to the same vial as step 17 (see Note 24).
Dry the solution using a SpeedVac. and store at −80 °C until used for mass spectrometry.
Mass Spectrometry
Add 50 μl loading solution and load 5 μl of sample onto a capillary fused silica column packed with HALO C18 at a flow rate of 300 nl/min.
Separate peptides by a gradient of 0–35% ACN/0.1% FA over 120 min, 35–100% ACN/0.1% FA for 1 min, and a hold of 100% ACN for 8 min, followed by an equilibration 0.1% FA in H2O for 21 min.
Introduce the peptides to the mass spectrometer via a nano-spray ionization source and a laser pulsed ~3 μm orifice with a spray voltage of 2.2 kV.
Acquire mass spectrometry data in a data-dependent “Top 10” acquisition mode, in which a survey scan from m/z 360–1600 is followed by ten higher-energy collisional dissociation tandem mass spectrometry (MS/MS) scans of the most abundant ions. Acquire MS/MS scans with the following parameters: isolation width = 1.6 m/z and normalized collision energy = 26.
Search product ion spectra using the SEQUEST HT engine implemented on the Proteome Discoverer 1.4 software against the respective species-specific UniProt/NCBI protein database. Search parameters are as follows: (1) full trypsin enzymatic activity, (2) two missed cleavages, (3) peptides between the MW of 350–5000, (4) mass tolerance at 20 ppm for precursor ions and 0.02 Da for fragment ions, (5) dynamic modifications on methionine (+15.9949 Da: oxidation), (6) three maximum dynamic modifications allowed per peptide, and (7) static modification on cysteine (+57.0215 Da: carbamidomethylation). Filter the combined data set to contain less than 1% false positive (with the Target Decoy PSM Validator node).
Assign proteins positively identified with two or more distinct peptide hits manually to one of six groups: ECM, cytoplasm, cytoskeletal, nuclear, membrane-associated, secreted, and uncharacterized in case no subcellular location is specified.
Generate heatmaps with the log2 transformation of peptide hits from each positively identified protein [3, 10, 12]. We have found that the log2 transformation is beneficial due to the non-normality of the data. If any of the proteins is matched to more than one category, choose its predominant subcellular location for functional grouping.
Table 2.
Properties of different detergents commonly used in whole-organ decellularization
| Agent | CMC (mM) | HLB | Properties |
|---|---|---|---|
| Triton X-100 | 0.2–0.9 | 13.5 | Nonionic detergent used to solubilize proteins; mild non-denaturing detergent |
| Sodium deoxycholate (SDC) | 4–8 | 16 | Water-soluble anionic detergent used for disrupting and dissociated protein interaction |
| Sodium dodecyl sulfate (SDS) | 7–10 | 40 | Anionic surfactant used for lysing cells and unraveling proteins |
| 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) | 6–10 | – | Non-denaturing zwitterionic detergent used to solubilize proteins |
CMC critical micelle concentration, HLB hydrophile-lipophile balance
Acknowledgments
The authors are grateful to Douglas Taatjes Ph.D. and Nicole Bishop of the UVM Microscopy and Imaging Center for assistance with IHC staining, transmission electron microscopy imaging, and image interpretations. We would like to thank Caroline Andrews, Nicholas Bonenfant, Zachary Borg, Amy Coffey, Amanda Daly, Jacob Dearborn, Nathan Gasek, Ethan Griswold, Zachary Phillips, Joseph Platz, Alexander Riveron, Dino Sokocevic, John Wallis, Sean Wrenn, and Basa Zvorova for the assistance with lung decellularizations, microscopy, and analysis of the detergent effluents. Studies were supported by the NIH RO1 HL127144-01 (DJW), Vermont Lung Center CoBRE grant (P20RR15557), NIH PACT program (contract HHSN268201000008C), NHBLI Lung Biology Training grant T32 HL076122, NIH Institutional Development Award (IDeA) NIGMS grant P20GM103449, and NCRR grant P40RR017447.
Footnotes
SDC solution should not be stored long term at 4 °C since this may cause precipitation or gelation, resulting in a weakening of the SDC solution. We have also found that the SDC must be at neutral or basic conditions as acidic conditions lead to the formation of precipitates. Other groups have reported the usage of the ionic detergents SDS or CHAPS as decellularization agents [11, 13]. The hydrophile-lipophile balance (HLB) is a value used to determine the hydrophilic character of a detergent. While a detergent with a low value is regarded as a water in oil emulsifier, detergents with higher values (>16) are regarded as solubilizer [30]. SDC (ionic detergent) and Triton X-100 (nonionic detergent) both have an HLB in the non-denaturing range, while SDS is a harsher solubilizer. CHAPS is a zwitterionic, non-denaturing detergent [31] (Table 2). Different decellularization protocols and detergents have been previously compared by different groups in different species (mouse and rat) analyzing differences in ECM composition, residual enzyme activity (e.g., matrix metalloproteinases), and initial viability of cells seeded onto the scaffolds. While matrix composition, total peptide amounts detected via mass spectrometry, and residual enzyme activity varied, no obvious differences were observed in initial recellularization studies [11, 18]. To date, there has been no comparison of the method of decellularization in fibrotic tissue. Due to the differences in cross-linking and composition of fibrotic tissue, usage of different decellularization techniques may also preferentially alter the remaining scaffold:
DNAse solution should always be either prepared fresh or stored in aliquots at −20 °C. The magnesium and calcium in the solution are necessary for the activity of the enzyme.
Gels for SDS-PAGE should be in the range of 10–15% polyacrylamide.
Other investigators have used a range of other cells and cell types including human umbilical vein endothelial cells [11, 32], small airway epithelial cells [11], pulmonary alveolar epithelial cells [11], rat distal lung epithelial cells [33], induced pluripotent stem cells [32, 34, 35], mouse A9 fibroblast cells [36], human lung fibroblasts [8], human peripheral blood mononuclear cells (PBMC) [5], and bone marrow-derived mesenchymal stem cells [37].
hMSCs have previously been extensively characterized for cell surface marker expression and differentiation capacity [23] and should not be used past passage 7.
Amounts of bleomycin administration vary between laboratories. Usually a single dose of 1.25–4 U/kg is administered [1, 3–5, 17]. It should be noted that different mouse strains have different susceptibilities to bleomycin [1]. Chapter 2 of this volume provides ample details for this method.
The lung volume should decrease significantly before the next filling. For mouse and rat lungs, this takes up to 1 min. Pig and human lungs need manual handling which means gently kneading the lung tissue in order to drain the liquid. Having the trachea/main bronchi at the lowest point of the tissue increases draining efficiency. Especially when the lung is filled, handling of the tissue should be done carefully. Kneading needs to be done more carefully with the pig lungs due to the lack of alveolar interconnectivity (lack of pores of Kohn). Therefore pig lungs are more likely to form pleural blebs which will significantly reduce decellularization efficiency and should be avoided.
Samples for quality control from pig and human lungs should be taken from at least three different locations of the tissue to account for local variations. Quality control samples include but are not limited to pieces for histology/IHC and DNA determination.
Based on our experience, acellular lungs should not be stored longer than 3 months due to degradation and loss of biological activity [25].
Pump-aided inflation (pig and human lungs) should start with a low flow rate slowly increasing to 2 l/min. A lower flow rate was shown to not properly clear the lungs from debris, while a higher flow rate negatively impacted tissue integrity [12]. Other groups reported different flow rates and the usage of pressure-controlled decellularization protocols [7, 11, 38–40]. An overview of different protocols used for decellularization is given in [41]. We believe that the decellularization is not only based on the membrane-disrupting action of the detergents to lyse the cells but also on the shear stresses imposed by flushing the tissue. For this reason a pressure-controlled decellularization protocol may need higher volumes to achieve a similar clearance of cellular debris.
Do not under- or overinflate the lungs. Underinflation will lead to improper decellularization and can be noticed by visual and manual examination. If the tissue feels soft at the edges or is still expanding while filling it is not filled completely. Proper filling volume should be recorded with the initial wash solution washes and be used as a guide for later fillings. During the decellularization process, the lungs will become leaky due to the loss of cells, and inflation volumes may need to be increased at later stages to achieve full tissue inflation and distribution of solutions to the alveoli and capillaries. Great care should be taken to not overinflate since this will result in damage and may lead to pleural blebbing (dislocation of the pleura from the lung tissue with fluid entrapment and bubble formation) especially in pig lungs. As soon as all parts of the lung are filled, you can proceed with draining/manual manipulation of the lung to empty the tissue from the fluid.
Absorbance measurements (e.g., using a Nanodrop) can be used to evaluate DNA content, but it is not as accurate as methods like the Quant-iT™ PicoGreen® dsDNA assay since it will additionally detect single-stranded DNA as well as RNA. It was found that this overestimates the amount of dsDNA in the sample since the amount of dsDNA detected by PicoGreen was found to be in the range of 25–40% of the amount determined via Nanodrop [42].
Transfer only eight samples at once and measure the absorbance in order to prevent inaccuracy due to chloroform evaporation.
To seed the cells into/onto the tissue, slices of about 1 mm thickness can be cut with sterile razorblades and trimmed to a diameter of 6 mm with a standard biopsy punch to fit the tissue into a 96-well plate. Cells can then directly be seeded onto the tissue for incubation and viability evaluation.
Ensure that gel pieces are completely covered with each reagent.
Destain until gel pieces are clear (~30 min or overnight if the dye has not diffused out).
ACN shrinks the gel pieces and dehydrates them by extracting H2O. Gel pieces should be white/opaque and shrunken after incubation with acetonitrile. Repeat ACN step if gel pieces are not completely white.
When using the SpeedVac, it is important to first turn on the centrifuge before turning on the vacuum in order to not lose the sample. After drying, first turn off the vacuum and then the centrifuge.
DTT reduces the disulfide bonds and makes the protein linear. Make sure all the gel pieces are covered.
IAA leads to alkylation by binding covalently with the thiol group of cysteine so the protein cannot form disulfide bonds.
Trypsin should be 6 ng/μl for silver stain and 12 ng/μl for Coomassie blue stained gels. The trypsin to protein ratio should be 1:20–1:100 (w/w).
Do not digest longer than 18 h. Trypsin cuts at lysine (Lys) and arginine (Arg) locations. It is important to have predictable peptide fragments for proper identification in mass spectrometry proteomic databases. Proteins are trapped in the gel, but when you introduce the enzyme, they are cleaved into peptides and diffuse out of the gel as peptides ending in Lys or Arg.
This step will extract the peptides from the gel pieces.
This step may have to be done twice to fully dehydrate the gel pieces.
References
- 1.Moore BB, Hogaboam CM (2008) Murine models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 294(2):L152–L160. doi: 10.1152/ajplung.00313.2007 [DOI] [PubMed] [Google Scholar]
- 2.Mouratis MA, Aidinis V (2011) Modeling pulmonary fibrosis with bleomycin. Curr Opin Pulm Med 17(5):355–361. doi: 10.1097/MCP.0b013e328349ac2b [DOI] [PubMed] [Google Scholar]
- 3.Sokocevic D, Bonenfant NR, Wagner DE et al. (2013) The effect of age and emphysematous and fibrotic injury on the re-cellularization of de-cellularized lungs. Biomaterials 34(13):3256–3269. doi: 10.1016/j.biomaterials.2013.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller HB, Fernandez IE, Burgstaller G et al. (2015) Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol Syst Biol 11(7):819. doi: 10.15252/msb.20156123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun H, Zhu Y, Pan H et al. (2016) Netrin-1 regulates fibrocyte accumulation in the decellularized fibrotic sclerodermatous lung microenvironment and in bleomycin-induced pulmonary fibrosis. Arthritis Rheumatol 68(5):1251–1261. doi: 10.1002/art.39575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price AP, England KA, Matson AM et al. (2010) Development of a decellularized lung bioreactor system for bioengineering the lung: the matrix reloaded. Tissue Eng Part A 16(8):2581–2591. doi: 10.1089/ten.TEA.2009.0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen TH, Calle EA, Zhao L et al. (2010) Tissue-engineered lungs for in vivo implantation. Science 329(5991):538–541. doi: 10.1126/science.1189345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Booth AJ, Hadley R, Cornett AM et al. (2012) Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med 186(9):866–876. doi: 10.1164/rccm.201204-0754OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilpin SE, Guyette JP, Ren X et al. (2013) Up-scaling decellularization and whole organ culture for human lung regeneration. J Heart Lung Transpl 32(4):S69–S70. doi: 10.1016/j.healun.2013.01.172 [DOI] [Google Scholar]
- 10.Wagner DE, Bonenfant NR, Parsons CS et al. (2014) Comparative decellularization and recellularization of normal versus emphysematous human lungs. Biomaterials 35(10):3281–3297. doi: 10.1016/j.biomaterials.2013.12.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilpin SE, Guyette JP, Gonzalez G et al. (2014) Perfusion decellularization of human and porcine lungs: bringing the matrix to clinical scale. J Heart Lung Transplant 33(3):298–308. doi: 10.1016/j.healun.2013.10.030 [DOI] [PubMed] [Google Scholar]
- 12.Wagner DE, Bonenfant NR, Sokocevic D et al. (2014) Three-dimensional scaffolds of acellular human and porcine lungs for high throughput studies of lung disease and regeneration. Biomaterials 35(9):2664–2679. doi: 10.1016/j.biomaterials.2013.11.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balestrini JL, Gard AL, Liu A et al. (2015) Production of decellularized porcine lung scaffolds for use in tissue engineering. Integr Biol (Camb) 7(12):1598–1610. doi: 10.1039/c5ib00063g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platz J, Bonenfant NR, Uhl FE et al. (2016) Comparative study to the use of decellularized alpha-Gal KO pig lungs for xenogeneic lung transplantation. Tissue Eng Part C Methods. doi: 10.1089/ten.TEC.2016.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godin LM, Sandri BJ, Wagner DE et al. (2016) Decreased laminin expression by human lung epithelial cells and fibroblasts cultured in acellular lung scaffolds from aged mice. PLoS One 11(3):e0150966. doi: 10.1371/journal.pone.0150966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker MW, Rossi D, Peterson M et al. (2014) Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest 124(4):1622–1635. doi: 10.1172/jci71386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Peng H, Sun H et al. (2014) Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med 6(240):240ra276. doi: 10.1126/scitranslmed.3007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallis JM, Borg ZD, Daly AB et al. (2012) Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng Part C Methods 18(6):420–432. doi: 10.1089/ten.TEC.2011.0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daly AB, Wallis JM, Borg ZD et al. (2012) Initial binding and recellularization of decellularized mouse lung scaffolds with bone marrow-derived mesenchymal stromal cells. Tissue Eng Part A 18(1–2):1–16. doi: 10.1089/ten.TEA.2011.0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonvillain RW, Danchuk S, Sullivan DE et al. (2012) A nonhuman primate model of lung regeneration: detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells. Tissue Eng Part A 18(23–24):2437–2452. doi: 10.1089/ten.TEA.2011.0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lluri G, Langlois GD, McClellan B et al. (2006) Tissue inhibitor of metalloproteinase-2 (TIMP-2) regulates neuromuscular junction development via a beta1 integrin-mediated mechanism. J Neurobiol 66(12):1365–1377. doi: 10.1002/neu.20315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zvarova B, Uhl FE, Uriarte JJ et al. (2016) Residual detergent detection method for nondestructive cytocompatibility evaluation of decellularized whole lung scaffolds. Tissue Eng Part C Methods 22(5):418–428. doi: 10.1089/ten.TEC.2015.0439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed W, Noga SJ, Gee AP et al. (2009) Production Assistance for Cellular Therapies (PACT): four-year experience from the United States National Heart, Lung, and Blood Institute (NHLBI) contract research program in cell and tissue therapies. Transfusion 49(4):786–796. doi: 10.1111/j.1537-2995.2008.02027.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yankaskas JR, Haizlip JE, Conrad M et al. (1993) Papilloma virus immortalized tracheal epithelial cells retain a well-differentiated phenotype. Am J Phys 264(5 Pt 1):C1219–C1230 [DOI] [PubMed] [Google Scholar]
- 25.Bonenfant NR, Sokocevic D, Wagner DE et al. (2013) The effects of storage and sterilization on de-cellularized and re-cellularized whole lung. Biomaterials 34(13):3231–3245. doi: 10.1016/j.biomaterials.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo E, Cardenes N, Garreta E et al. (2014) Inhomogeneity of local stiffness in the extracellular matrix scaffold of fibrotic mouse lungs. J Mech Behav Biomed Mater 37:186–195. doi: 10.1016/j.jmbbm.2014.05.019 [DOI] [PubMed] [Google Scholar]
- 27.Moeller A, Ask K, Warburton D et al. (2008) The bleomycin animal model: a useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int J Biochem Cell Biol 40(3):362–382. doi: 10.1016/j.biocel.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez IE, Amarie OV, Mutze K et al. (2016) Systematic phenotyping and correlation of biomarkers with lung function and histology in lung fibrosis. Am J Physiol Lung Cell Mol Physiol 310(10):L919–L927. doi: 10.1152/ajplung.00183.2015 [DOI] [PubMed] [Google Scholar]
- 29.Crapo PM, Gilbert TW, Badylak SF (2011) An overview of tissue and whole organ decellularization processes. Biomaterials 32(12):3233–3243. doi: 10.1016/j.biomaterials.2011.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffin WC (1954) Calculation of HLB values of non-ionic surfactants. J Soc Cosmet Chem 5:249–256 [Google Scholar]
- 31.Hjelmeland LM (1980) A nondenaturing zwitterionic detergent for membrane biochemistry: design and synthesis. Proc Natl Acad Sci U S A 77(11):6368–6370. doi: 10.1073/pnas.77.11.6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren X, Moser PT, Gilpin SE et al. (2015) Engineering pulmonary vasculature in decellularized rat and human lungs. Nat Biotechnol 33(10):1097–1102. doi: 10.1038/nbt.3354 [DOI] [PubMed] [Google Scholar]
- 33.Calle EA, Mendez JJ, Ghaedi M et al. (2015) Fate of distal lung epithelium cultured in a decellularized lung extracellular matrix. Tissue Eng Part A 21(11–12):1916–1928. doi: 10.1089/ten.TEA.2014.0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghaedi M, Calle EA, Mendez JJ et al. (2013) Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J Clin Invest 123(11):4950–4962. doi: 10.1172/JCI68793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilpin SE, Ren X, Okamoto T et al. (2014) Enhanced lung epithelial specification of human induced pluripotent stem cells on decellularized lung matrix. Ann Thorac Surg 98(5):1721–1729. doi: 10.1016/j.athoracsur.2014.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun H, Calle E, Chen X et al. (2014) Fibroblast engraftment in the decellularized mouse lung occurs via a beta1-integrin-dependent, FAKdependent pathway that is mediated by ERK and opposed by AKT. Am J Physiol Lung Cell Mol Physiol 306(6):L463–L475. doi: 10.1152/ajplung.00100.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z, Gong X, Zhu H et al. (2014) Inhibition of Wnt/beta-catenin signaling promotes engraftment of mesenchymal stem cells to repair lung injury. J Cell Physiol 229(2):213–224. doi: 10.1002/jcp.24436 [DOI] [PubMed] [Google Scholar]
- 38.Ott HC, Clippinger B, Conrad C et al. (2010) Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med 16(8):927–933. doi: 10.1038/nm.2193 [DOI] [PubMed] [Google Scholar]
- 39.Nichols JE, Niles J, Riddle M et al. (2013) Production and assessment of decellularized pig and human lung scaffolds. Tissue Eng Part A 19(17–18):2045–2062. doi: 10.1089/ten.TEA.2012.0250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AP, Godin LM, Domek A et al. (2015) Automated decellularization of intact, human-sized lungs for tissue engineering. Tissue Eng Part C Methods 21(1):94–103. doi: 10.1089/ten.TEC.2013.0756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner DE, Uhl FE, Weiss DJ (2015) Acellular lung scaffolds in lung bioengineering In: Bertoncello I (ed) Stem cells in the lung: development. Repair and Regeneration. Springer International Publishing, Cham, pp 309–347. doi: 10.1007/978-3-319-21082-7_18 [DOI] [Google Scholar]
- 42.Huleihel L, Hussey GS, Naranjo JD et al. (2016) Matrix-bound nanovesicles within ECM bioscaffolds. Sci Adv 2(6):e1600502. doi: 10.1126/sciadv.1600502 [DOI] [PMC free article] [PubMed] [Google Scholar]