Significance
Cytotoxic CD8+ T cells control infectious diseases as well as cancer. Memory T cells are a subset of CD8+ T cells that have previously encountered and rapidly responded to a specific antigen. Here we identified the epigenetic writer DOT1L as a central regulator of T cell physiology. In the absence of DOT1L, which methylates histone H3K79 at active genes, cytotoxic T cells prematurely acquired memory features without having encountered an antigen. However, loss of DOT1L in the T cell lineage led to a compromised immune response. Our findings highlight the importance of epigenetic signals in regulating T cell differentiation. Understanding these epigenetic signals is important for developing strategies to modulate T cell immunity.
Keywords: T cell, epigenetics, DOT1L, H3K79me2, virtual memory
Abstract
Cytotoxic T cell differentiation is guided by epigenome adaptations, but how epigenetic mechanisms control lymphocyte development has not been well defined. Here we show that the histone methyltransferase DOT1L, which marks the nucleosome core on active genes, safeguards normal differentiation of CD8+ T cells. T cell-specific ablation of Dot1L resulted in loss of naïve CD8+ T cells and premature differentiation toward a memory-like state, independent of antigen exposure and in a cell-intrinsic manner. Mechanistically, DOT1L controlled CD8+ T cell differentiation by ensuring normal T cell receptor density and signaling. DOT1L also maintained epigenetic identity, in part by indirectly supporting the repression of developmentally regulated genes. Finally, deletion of Dot1L in T cells resulted in an impaired immune response. Through our study, DOT1L is emerging as a central player in physiology of CD8+ T cells, acting as a barrier to prevent premature differentiation and controlling epigenetic integrity.
Lymphocyte development and differentiation are tightly regulated and provide the basis for a functional adaptive immune system. Development of mature T cells initiates in the thymus with progenitor T cells that have to pass two key checkpoints: T cell receptor (TCR) β selection and positive selection, both of which are controlled by intricate signaling pathways involving the pre-TCR/CD3 and αβTCR/CD3 complexes, respectively (1). Upon positive selection, mature thymocytes are licensed to emigrate and populate peripheral lymphatic organs as naïve T cells (TN). Further differentiation of naïve T cells into effector or memory T cells normally depends on TCR-mediated antigen recognition and stimulation. However, it has become evident that a substantial fraction of mature CD8+ T cells acquires memory-like features independent of exposure to foreign antigens. The origin and functionality of these unconventional memory cells in mice and humans, also referred to as innate or virtual memory cells, are only just being uncovered (2–4).
The dynamic transitions during development and differentiation of CD8+ T cells are governed by transcriptional and epigenetic changes, including histone modifications that are controlled by chromatin modifiers. Well-established histone marks are mono- and trimethylation of histone H3K4 at enhancers (H3K4me1) and promoters (H3K4me3), H3K27me3 at repressed promoters, and H3K9me2/3 in heterochromatin (5–10). Although epigenetic “programming” is known to play a key role in T cell development and differentiation, the causal role of epigenetic modulators in T cell differentiation is still poorly understood, especially for chromatin modifiers associated with active chromatin (5).
One of the histone modifications positively associated with gene activity is mono-, di-, and trimethylation of histone H3K79 mediated by DOT1L. This evolutionarily conserved histone methyltransferase methylates H3K79 in transcribed promoter-proximal regions of active genes (11, 12). Although the association with gene activity is strong, how H3K79 methylation affects transcription is still unclear and repressive functions have also been proposed (11, 13). DOT1L has been linked to several critical cellular functions, including embryonic development, DNA damage response, and meiotic checkpoint control (14) and DOT1L has also been shown to function as a barrier for cellular reprogramming in generating induced pluripotent stem cells (15). DOT1L gained wide attention as a specific drug target in the treatment of MLL-rearranged leukemia, where MLL fusion proteins aberrantly recruit DOT1L to MLL target genes leading to their enhanced expression (16). A similar dependency on DOT1L activity and sensitivity to DOT1L inhibitors was recently observed in thymic lymphoma (17). Interestingly, inhibition of DOT1L activity in human T cells attenuates graft-versus-host disease in adoptive cell transfer models (18) and it regulates CD4+ T cell differentiation (19).
Given the emerging role of DOT1L in epigenetic reprogramming and T cell malignancies, we investigated the role of DOT1L in normal T cell physiology using a mouse model in which Dot1L was selectively deleted in the T cell lineage. Our results suggest a model in which DOT1L plays a central role in CD8+ T cell differentiation, acting as a barrier to prevent premature antigen-independent differentiation and maintaining epigenetic integrity.
Results
DOT1L Prohibits Premature Differentiation toward Memory-Like CD8+ T Cells.
Given the essential role of DOT1L in embryonic development (20), we determined the role of DOT1L in T cell development and differentiation by employing a conditional knockout (KO) mouse model in which Dot1L is deleted in the T cell lineage by combining floxed Dot1L with a Cre-recombinase under the control of the Lck promoter. This leads to deletion of exon 2 of Dot1L during early thymocyte development (SI Appendix, Fig. S1A) (17). The observed global loss of H3K79me2 in T cells in Lck-Cre+/−;Dot1Lfl/fl mice, as confirmed by immunohistochemistry on fixed thymus tissue (SI Appendix, Fig. S1B), agreed with the notion that DOT1L is the sole methyltransferase for H3K79 (11, 17, 20, 21).
To validate the efficacy of Dot1L deletion at the single-cell level, we developed an intracellular staining protocol for H3K79me2. Histone dilution by replication-dependent and -independent means has been suggested to be the main mechanisms of losing methylated H3K79 (22). Flow-cytometric analyses of thymocyte subsets from Lck-Cre+/−;Dot1Lfl/fl mice (hereafter, KO) revealed that double-negative (DN, CD4−CD8−) thymocytes started losing H3K79me2. From the subsequent immature single-positive state (ISP) onward, all of the thymocytes had lost DOT1L-mediated H3K79me2 (SI Appendix, Fig. S1C). This confirmed that upon early deletion of Dot1L, successive rounds of replication in the thymus allowed for loss of methylated H3K79.
No changes in H3K79 methylation levels were found in T lineage cells of Lck-Cre+/−;Dot1Lwt/wt control mice (hereafter, wild type [WT]).
Ablation of Dot1L resulted in a reduction of overall thymic cellularity, mainly caused by reduced numbers of double-positive (DP), mature single-positive (SP) CD4+ and CD8+ thymocytes (1.4-, 2.2-, and 1.8-fold, respectively) (SI Appendix, Fig. S1 D and E). Further analysis of the CD8+ SP subset revealed that Dot1L-KO mice harbored fewer CD8+ SP thymocytes expressing CD69, a marker of recent positive selection (23) (SI Appendix, Fig. S1F). In line with the lower percentage of SP CD8+CD69+ cells, the transition of semimature to mature stage was also affected in Dot1L-KO mice. The percentage of mature SP CD8+ T cells, as characterized by surface expression of Qa2 and CD24, was reduced in Dot1L-KO (SI Appendix, Fig. S1G) (24, 25). Together this suggests a role of DOT1L in controlling intrathymic T cell selection and maturation of CD8+ SP T cells.
In the spleen, overall cellularity was not affected, but within the T cell compartment, CD4+ T cells were drastically reduced (3.2-fold), whereas CD8+ T cells were increased (1.7-fold) in number (SI Appendix, Fig. S1 E and H). However, while flow cytometry of H3K79me2-stained splenic T cells confirmed the lack of DOT1L activity in CD8+ T cells and CD44−CD62L+ CD4+ T cells, CD44+CD62L− CD4+ T cells showed a partial loss of H3K79me2 and CD4+CD25+ regulatory T cells (Treg) remained H3K79me2 positive (SI Appendix, Fig. S1I). Since earlier in development, CD4-expressing cells in the thymus were mostly H3K79me2 negative, this suggests that a strong selection occurred for the maintenance of DOT1L for the development of Tregs in this mouse model. Indeed, partial deletion of Dot1L in CD4+ cells was confirmed by PCR analysis (SI Appendix, Fig. S1J). Here, we focused our study on defining the role of DOT1L in the cytotoxic T cell compartment in which efficient deletion of Dot1L and loss of H3K79me2 was found in both the thymus and the periphery.
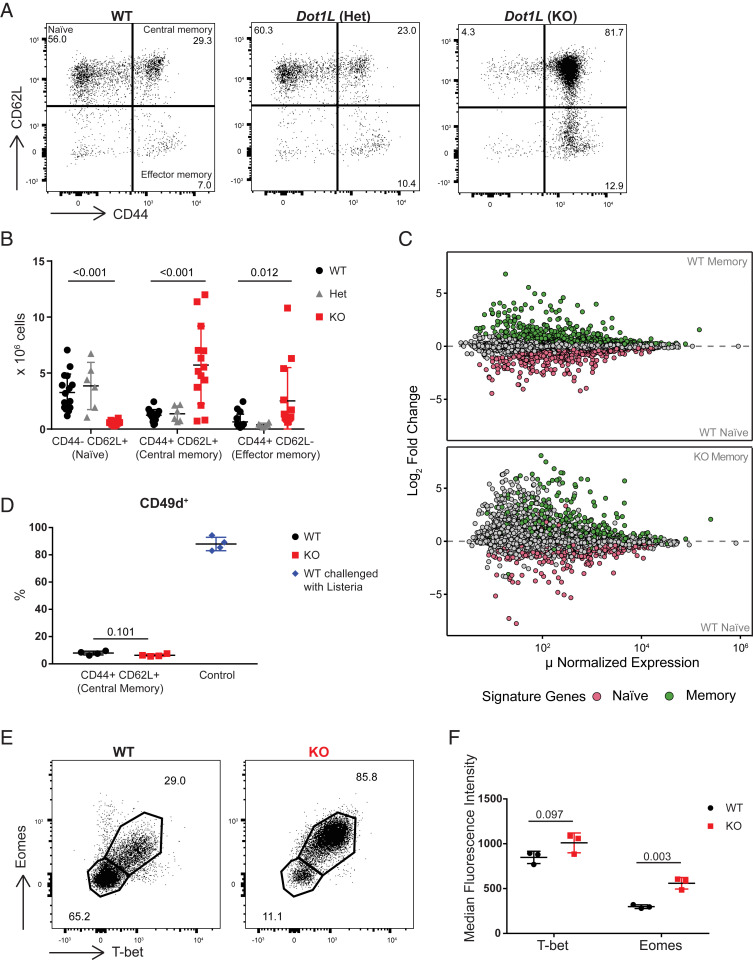
CD8+ T cell differentiation was strongly affected by the absence of DOT1L. Analysis of CD8+ T cell subsets in the spleen revealed that Dot1L-KO mice showed a severe loss of naïve (CD44−CD62L+) CD8+ T (TN) cells and a massive gain of the CD44+CD62L+ phenotype, a feature of central memory T cells (TCM; Fig. 1 A and B). In Dot1L-KO, the cells in the naïve gate did not form a distinct population as in WT, but rather were a tail of the TCM population. This is in line with the observed aberrant thymic maturation in Dot1L-KO. In Lck-Cre+/−;Dot1Lfl/wt heterozygous knockout mice (Het), CD8+ T cells did not show any phenotypic differences compared to WT (Fig. 1 A and B). The lack of haploinsufficiency was further confirmed by principal component analysis of RNA sequencing (RNA-Seq) data indicating that WT and Het CD8+ T cells were similar, but were dissimilar from the KO CD8+ T cells, excluding gene-dosage effects (SI Appendix, Fig. S1K). Therefore, we restricted our further studies to the comparison of the KO and WT mice. The strong shift toward a CD8+ memory phenotype in Dot1L-KO was unexpected because Dot1L-KO mice were housed under the same conditions as their WT controls and the mice had not been specifically immunologically challenged.
Fig. 1.
Ablation of Dot1L results in CD44+CD62L+ memory-like CD8+ T cells. (A) Flow-cytometry analysis of CD8+ T cell subsets in the spleen based on CD44 and CD62L expression in WT and heterozygous and homozygous Dot1L-KO mice. Subsets are indicated in the Top Left. (B) Quantification of CD8+ T cell subsets in the spleen indicated in the representative plots in A. Data from three to five individual experiments with three to four mice per genotype per experiment, shown as mean ± SD. (C) Mean average (MA) plot of RNA-Seq data from FACS-sorted CD44+CD62L+ (TCM) and CD44−CD62L+ (TN) CD8+ T cells from four mice per indicated genotype. Naïve and memory signatures were defined based on differentially expressed genes (false discovery rate [FDR] < 0.01) between WT TN and WT TCM cells. (D) Percentage of CD49d+ cells in CD44+CD62L+ CD8+ T cells from unchallenged mice and in CD44+CD62L− CD8+ T cells from WT mice challenged with L. monocytogenes for 7 d. Data are from one experiment with four mice per genotype, represented as mean ± SD. (E) Representative flow-cytometry plots of T-bet and Eomes expression in CD8+ T cells from the spleen. (F) Median fluorescence intensity (MFI) of T-bet and Eomes in CD44+CD62L+ CD8+ T cells from the spleen. Data are from one experiment with three mice per genotype, represented as mean ± SD.
To unravel the molecular identity of the CD44+CD62L+ Dot1L-KO cells in more detail we performed RNA-Seq analysis on sorted CD44−CD62L+ (TN) and CD44+CD62L+ (TCM) CD8+ T cell subsets from WT and KO mice. Based on differential gene expression between TN and TCM CD8+ cells from WT mice, naïve and memory gene signatures were defined. Interestingly, overlay of these signatures on WT CD44−CD62L+ (TN) and KO CD44+CD62L+ (TCM) cells showed that differential expression between TN and TCM cells was mostly preserved when Dot1L was ablated. (Fig. 1C), although there was misregulation of other genes as well (see below). These data suggest that in the absence of DOT1L, CD8+ T cells acquire, prematurely and in the absence of any overt immunological challenge, a transcriptome of memory-like CD8+ T cells.
Dot1L-KO Memory-Like Cells Are Antigen Inexperienced.
Although WT and KO mice were exposed to the same environment, it cannot be excluded that KO mice responded differentially to antigens in the environment. If this is the case, one expects skewing in the clonality of the TCRβ gene usage. In order to investigate this possibility, we examined the TCRβ repertoire. Tcrb sequencing revealed no difference in productive clonality scores between WT and KO CD8+ T cells (SI Appendix, Table S1). Also, CDR3 length as well as Tcrb-V and Tcrb-J gene usage were unaffected (SI Appendix, Fig. S2 A–C). These data, together with the nearly complete loss of naïve CD8+ T cells, argued against any antigen-mediated bias in the selection for CD8+ T cells and indicated that CD44+CD62L+ memory-like CD8+ T cells in KO mice were polyclonal and arose by antigen-independent differentiation of TN cells.
Antigen-independently differentiated memory-like CD8+ T cells have already been described in the literature and their origins and functions are the subject of ongoing studies (2–4, 26). Depending on their origin and cytokine dependency they are referred to as “virtual” or “innate” memory cells. Virtual memory CD8+ T cells have been suggested to arise in the periphery from cells that are CD5high, related to high TCR affinity, and require IL-15 (4, 27). In contrast, innate memory CD8+ T cells develop in the thymus and their generation and survival are generally considered to be dependent on IL-4 signaling (3). We here collectively refer to them as antigen-independent memory-like CD8+ T cells (TAIM). A common feature of these TAIM cells is reduced expression of CD49d (28), a marker that is normally up-regulated after antigen exposure. In addition, they express high levels of T-bet and Eomes, encoding two memory/effector transcription factors (29, 30). In both Dot1L-KO and WT, the majority of the CD44+CD62L+ (TCM) cells were CD49d negative. As a control, CD44+CD62L− effector T cells (TEFF) from WT mice challenged with Listeria monocytogenes were mostly CD49d positive (Fig. 1D). This further indicates that the generation of CD44+CD62L+ memory-like T cells in KO mice is independent of antigen exposure. Of note, the percentage of CD49d-negative CD44+CD62L+ cells that we observed in WT mice corresponds to the percentage of TAIM cells reported in WT C57BL/6 mice (31). Regarding the expression of T-bet and Eomes, most of the KO CD8+ T cells coexpressed T-bet and Eomes. Furthermore, Eomes was expressed at a higher level in KO CD8+ T cells as compared to their WT counterpart (Fig. 1 E and F). Together these characteristics are all in agreement with antigen-independent differentiation of naïve CD8+ T cells in the absence of DOT1L.
The TAIM Phenotype in Dot1L-KO Initiates in the Thymus and Is Cell Intrinsic.
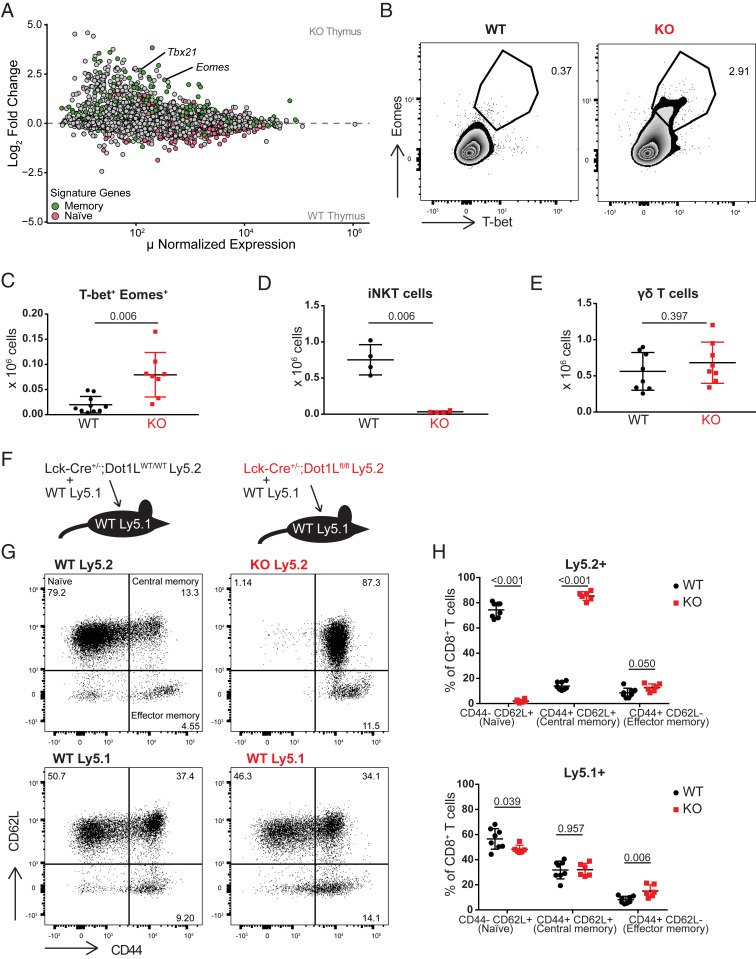
To determine whether peripheral TAIM cells observed in the Dot1L-KO setting originate intrathymically, as reported previously for IL-4-dependent innate memory T cells (3), we compared RNA-Seq data from SP CD8+ thymocytes from KO and WT mice. Analyzing the relative distribution of memory and naïve signature genes revealed that memory genes were among the genes up-regulated in KO SP CD8+ thymocytes (Fig. 2A). Importantly, like in peripheral CD8+ T cells the expression of T-bet and Eomes was up-regulated in KO SP CD8+ thymocytes. This transcriptional up-regulation was corroborated by flow-cytometric analysis of protein expression. Intracellular staining for the transcription factors showed that among KO SP CD8+ thymocytes a small but substantial subset was double positive for T-bet and Eomes (T-bet+ Eomes+) (Fig. 2 B and C). This subset was nearly absent in WT SP CD8+ thymocytes. Regarding T cell maturity as characterized by CD69 and MHC-I surface expression, the T-bet+ Eomes+ subset behaved differently from the T-bet− Eomes− subset. The T-bet+ Eomes+ subset had a more mature phenotype (M2; CD69−MHC-I+) compared to T-bet−Eomes−CD8+ KO SP T cells that were more semimatured (SM; CD69+MHC-I−) (32), suggesting that in Dot1L-KO CD8+ SP thymocytes started to acquire the TAIM phenotype just before emigrating from the thymus (SI Appendix, Fig. S2 D and E). Another possible scenario that could have explained the gain of intrathymic memory-phenotype cells (thymic TAIM cells) in KO was their recirculation from the periphery into the thymus. However, as compared to the spleen, the T-bet+Eomes+CD8+ SP subset in the thymus had a lower fraction of memory-phenotype cells. This observation is inconsistent with the possibility that thymic TAIM cells originated from recirculating peripheral T cells (SI Appendix, Fig. S2F). Together with the unperturbed TCRβ repertoire, this further supports the notion that differentiation of Dot1L-KO CD8+ T cells toward memory-like cells initiates intrathymically in an antigen-independent manner.
Fig. 2.
The TAIM phenotype in Dot1L-KO initiates in the thymus and is cell intrinsic. (A) MA plot of RNA-Seq data from sorted CD4−CD8+CD3+ thymocytes from three WT and four KO mice. Naïve and memory signature genes were defined as described in Fig. 1C. (B) Representative plot and (C) quantification of T-bet and Eomes expression on CD4−CD8+CD3+ thymocytes; data are of three individual experiments with two to four mice per genotype, represented as mean ± SD. (D) Quantification of iNKT (CD1d-PBS57+TCRβ+) cells in total thymus. Data are from one experiment with four mice per genotype, represented as mean ± SD. (E) Absolute number of γδTCR+ cells in spleen; data are of two individual experiments with four mice per genotype, represented as mean ± SD. (F) Outline of the mixed bone-marrow chimeras. (G and H) Representative flow-cytometry plots and quantification of CD44 and CD62L expression on Ly5.1+ and Ly5.2+ CD8+ splenocytes from mixed bone-marrow chimeras 4 mo after irradiation and reconstitution. All recipient mice were Ly5.1+ and transplanted with a mixture of either WT Ly5.1+ and Ly5.2+ Lck-Cre+/−;Dot1Lwt/wt (WT; Left) or WT Ly5.1+ and Ly5.2+ Lck-Cre+/−;Dot1Lfl/fl (KO; Right). Data are from one experiment with six or eight mice per genotype, represented as mean ± SD.
Innate memory cells have been suggested to arise in the thymus in response to an increase in IL-4-producing PLZFhigh invariant NKT (iNKT) cells or γδ T cells (3). However, iNKT cells (CD1d-PBS57+TCRβ+) were nearly absent in the thymus of Dot1L-KO mice (Fig. 2D and SI Appendix, Fig. S2G). Furthermore, the number of γδ T cells (γδTCR+) did not differ significantly between WT and KO mice (Fig. 2E). In addition, introduction of the transgenic OT-I TCR, a condition under which the number of iNKT and γδ T cells is strongly reduced (33–36), did not affect the memory phenotype of Dot1L-KO CD8+ T cells (SI Appendix, Fig. S2H). Together, these findings indicate that the intrathymic differentiation of TAIM CD8+ cells in the absence of DOT1L did not depend on an excess of IL-4-producing cells in the thymic microenvironment as reported for innate memory T cells. To test whether the differentiation toward TAIM cells in Dot1L-KO occurs by cell-intrinsic mechanisms, we generated mixed bone-marrow chimeras. Bone-marrow cells from WT Ly5.1+ mice were mixed with Ly5.2+;Lck-Cre+/−;Dot1Lwt/wt (WT) or Ly5.2+;Lck-Cre+/−;Dot1Lfl/fl (KO) bone-marrow cells and transplanted into lethally irradiated Ly5.1+ recipient mice (Fig. 2F). While T cells derived from WT Ly5.2+ hematopoietic stem cells gave rise to both naïve and memory-phenotype CD8+ T cells, KO Ly5.2+CD8+ T cells were predominantly of memory phenotype. This indicated that the Dot1L-KO TAIM phenotype is cell intrinsic and not dictated by environmental stimuli (Fig. 2 G and H). In addition, the memory phenotype of the WT Ly5.1+ cells remained indistinguishable between the two mixed bone-marrow chimera conditions. This indicated that within the same environment, Dot1L-KO T cells do not instruct WT T cells to gain a memory phenotype and have no effect on WT T cells as shown by the differentiation of Ly5.1+ WT cells (Fig. 2 G and H). The increased percentage of memory-phenotype Ly5.1+ cells is most likely a result of homeostatic proliferation of the T cells of the recipient after irradiation (Fig. 2H) (37, 38). Together, the results from the mixed bone-marrow chimeras further indicated that the TAIM phenotype is cell intrinsic for Dot1L-KO CD8+ T cells.
Dot1L Ablation Impairs TCR/CD3 Expression.
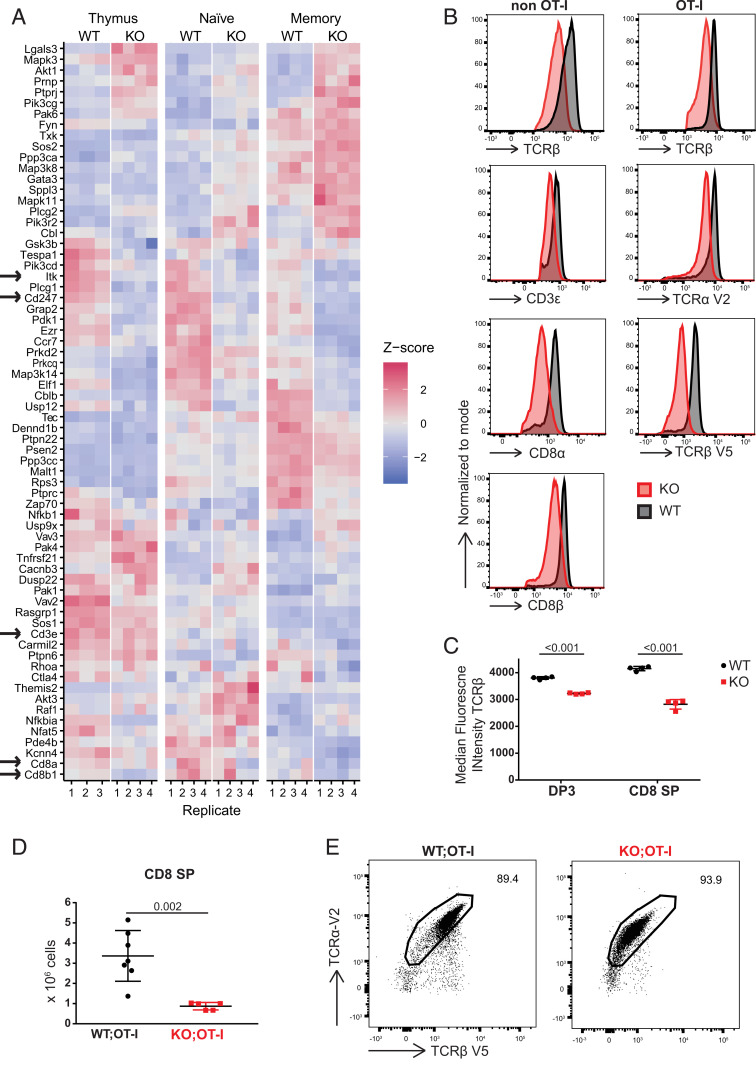
One of the cell-intrinsic mechanisms reported to be involved in the formation of TAIM cells is aberrant TCR signaling (2, 27, 39, 40). Furthermore, treatment of human T cells with DOT1L inhibitor impaired TCR sensitivity and attenuated low avidity T cell responses (18). This led us to investigate the expression of genes encoding TCR signaling components in the absence of DOT1L. RNA-Seq analyses confirmed that many TCR signaling genes were differentially expressed between WT and KO SP CD8+ thymocytes (Fig. 3A). Importantly, CD3ζ (Cd247), a critical rate-limiting factor in controlling the transport of fully assembled TCR/CD3 complexes to the cell surface, was down-regulated in Dot1L-KO T cells (1, 41, 42). In addition, other components of the TCR/CD3 complex like Cd3e and its associated coreceptor Cd8a/b were also down-regulated in KO T cells. H3K79me2 chromatin immunoprecipitation sequencing (ChIP-Seq) showed that these genes contained H3K79me2 in WT mice and might therefore be directly regulated by DOT1L (SI Appendix, Fig. S3 A–C). As a consequence, one expects TCR/CD3 and CD8αβ to be reduced at the cell surface of TAIM cells, which we confirmed by flow cytometry (Fig. 3B). Reduced expression of TCR in KO peripheral CD8+ T cells can be a consequence of T cell activation and might not be directly influenced by DOT1L activity. However, when analyzing DP3 (TCRβhighCD5int) and CD8+ SP thymocytes, the stages where T cell activation can be excluded, TCR levels were reduced in Dot1L-KO (Fig. 3C). This suggested that down-regulation of TCR expression in KO is most likely a direct effect of Dot1L ablation. In addition to the CD3/TCR complex, we observed down-regulation of Itk, a key TCR signaling molecule reported to be involved in innate memory CD8+ T cell formation (43). To exclude that the impaired TCR signaling in KO T cells could be compensated for by the selection of thymocytes expressing TCRs with altered affinity, we kept the TCR affinity identical by crossing the OT-I TCR transgene into our system. Thymocytes expressing OT-I are positively selected in the presence of MHC class I (H-2Kb), mainly generating CD8+ SP T cells expressing the exogenous OT-I TCR, with concomitant reduction of the CD4+ lineage (44). If DOT1L deficiency impairs TCR surface density and signaling, positive selection of conventional OT-I CD8+ thymocytes is expected to be compromised. In Dot1L-KO mice expressing OT-I, the number of SP CD8+ thymocytes was decreased (3.8-fold) compared to WT mice expressing OT-I (Fig. 3D). This revealed that, similar to the situation in mice with endogenous TCR, early intrathymic ablation of Dot1L in the T cell lineage prohibits positive selection of conventional OT-I CD8+ T cells, but yet supports the generation and selection of TAIM cells (SI Appendix, Fig. S2F) of which the vast majority expressed both exogenous TCR chains (TCRαV2 and TCRβV5) (Fig. 3E and SI Appendix, Fig. S3D). Consistent with the low surface expression of TCR/CD3 and CD8 in the KO condition, the surface expression of OT-I TCR was also lower, as determined by SIINFEKL/H-2Kb tetramer staining (SI Appendix, Fig. S3E). This was further validated by staining with antibodies for the transgenic TCR chains TCRαV2 and TCRβV5 which indicated two-fold reduction in KO (Fig. 3 B and E). The TCRαV2 element of the OT-I TCR was under the control of an exogenous promoter, suggesting that the reduced surface expression of OT-I TCR on KO T cells did not relate to transcriptional silencing of native TCR gene promoters. Instead, the observed lower TCR surface levels in KO T cells likely relate to the reduced CD3ζ expression (42). In conclusion, DOT1L regulates the levels of TCR complex and signaling molecules independent of the selected TCR. In the absence of these regulatory mechanisms, the identity of naïve CD8+ T cells cannot be maintained, contributing to premature T cell differentiation.
Fig. 3.
Dot1L ablation impairs TCR/CD3 expression. (A) Heatmap showing RNA expression of TCR signaling genes, defined by differential expression between WT and Dot1L-KO, in any of the sorted CD4−CD8+CD3+ thymocytes (thymus), CD44−CD62L+ (naïve) CD8+ T cells, and CD44+CD62L+ (memory) CD8+ T cells; four mice per genotype except for WT thymus where there are three mice. Genes are clustered based on Z-score. Arrows indicate genes involved in the TCR complex and Itk. Z-score is calculated row-wise within genes between samples. (B) Expression of TCRβ, CD3ε, TCRα V2, TCRβ V5, CD8α, and CD8β on CD8+ T cells in spleen from Lck-Cre;Dot1L and Lck-Cre;Dot1L;OT-I mice. (C) Median fluorescence intensity for TCRβ on DP3 and CD8+ SP thymocytes; data are from one experiment with four mice per genotype, represented as mean ± SD. (D) Absolute number of CD4−CD8+CD3+ thymocytes in WT and KO mice from Lck-Cre;Dot1L;OT-I background; data are from two individual experiments with three to four mice per genotype, represented as mean ± SD. (E) Representative plot of TCRα V2 and TCRβ V5 expression on CD8+ T cells from spleen.
DOT1L Is Required for Maintenance of Epigenetic Integrity of CD8+ T Cells.
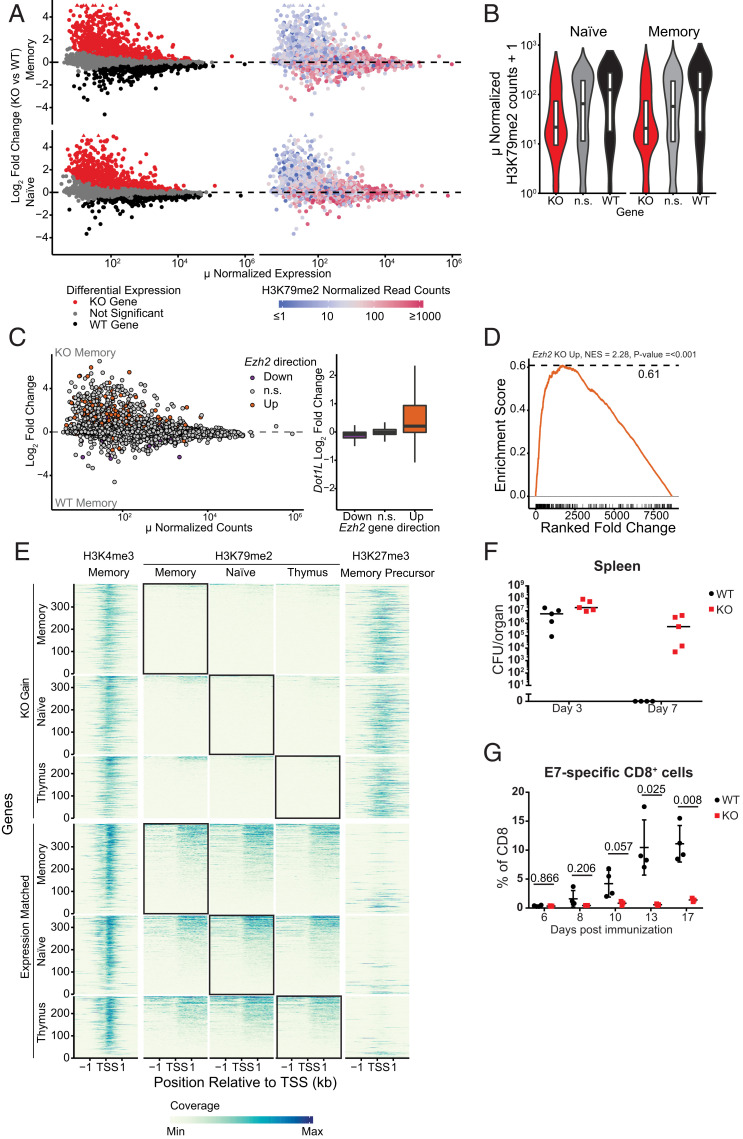
In order to mechanistically understand how the aberrant differentiation of Dot1L-KO CD8+ T cells in vivo relates to the epigenome, we performed H3K79me2 ChIP-Seq on sorted WT CD8+ T cell populations and compared it to the RNA-Seq data of sorted WT and KO CD44−CD62L+ (TN) and CD44+CD62L+ (TCM) populations. RNA-Seq analyses from CD44− TN and TCM cells revealed that genes that were up-regulated in Dot1L-KO were biased toward being lowly expressed, whereas down-regulated genes tended to have a higher expression level (Fig. 4 A, Left). To assess how the transcriptome changes in T cells and the transcriptional bias are related to the chromatin-modifying function of DOT1L, we compared the level of H3K79me2 at the 5′ end of genes in WT cells with the mRNA expression changes caused by the loss of DOT1L. H3K79me2 was enriched from the transcription start site into the first internal intron of transcribed genes (SI Appendix, Fig. S4A), as reported for human cells (45). Further analysis showed that most of the lowly expressed genes up-regulated in Dot1L-KO contained no or very low H3K79me2 in WT cells (Fig. 4 A, Right and B). Therefore, these genes are unlikely direct targets of DOT1L and are likely to be indirectly controlled by DOT1L. In contrast, most of the more highly expressed genes down-regulated in Dot1L-KO were marked by H3K79me2 (Fig. 4 A, Right and B). This demonstrates that in normal CD8+ T cells DOT1L-mediated H3K79 methylation generally marks expressed genes, but only a subset of the methylated genes needs H3K79 methylation for maintaining full expression levels. Thus, in normal T cells DOT1L does not act as a transcriptional switch but rather seems to be required for transcription maintenance of a subset of H3K79-methylated genes that are already expressed and it indirectly promotes repression of genes. Dot1L mRNA expression levels were not different between naïve, true memory, and virtual memory cells (SI Appendix, Fig. S4B), suggesting that not the global expression level, but rather differential translation, differential methylation of DOT1L targets, or differential “reading” of the H3K79me modifications may play an important role in preventing premature TAIM differentiation.
Fig. 4.
DOT1L is required for maintenance of the epigenetic identity of CD8+ T cells. (A) MA plot indicating differentially expressed genes between Dot1L-KO and WT in sorted CD44−CD62L+ (naïve) CD8+ T cells and CD44+CD62L+ (memory) CD8+ T cells (Left). Genes significantly up-regulated (red) and down-regulated (black) in Dot1L-KO are indicated. Genes were considered differentially expressed when FDR <0.01. Genes outside the limit of the y axis are indicated with triangles. Right shows H3K79me2 normalized read counts at the transcription start site (TSS) ± 2 kb of these genes. Genes with low H3K79me2 read counts are blue, genes with high H3K79me2 are red. (B) Violin plots of H3K79me2 read counts around 4 kb of the TSS of genes up in KO (KO), down in KO (WT), and nonsignificant (n.s.) between KO and WT. (C) MA plot of gene expression between WT and Dot1L-KO memory CD8+ T cells, with genes differential between activated WT and activated Ezh2-KO CD8+ T cells indicated in orange (derepressed in Ezh2 KO) and blue (down-regulated in Ezh2 KO) (49). Box plot notes effect size in Dot1L-KO vs. WT RNA-Seq data based on the indicated differential gene expression category in Ezh2 KO data. (D) Gene Set Enrichment Analysis (GSEA) of genes up-regulated in Ezh2-KO in Dot1L-KO and WT memory CD8+ cells. (E) H3K4me3, H3K79me2, and H3K27me3 ChIP-Seq data around transcription start sites from genes that are up-regulated in Dot1L-KO (KO Gain) and genes that did not change expression (Expression Matched). Coverage was calculated as reads per genomic content, cutoff at the 0.995th quantile per sample and rescaled to a maximum of 1. (F) Clearance of L. monocytogenes in spleen defined by the number of CFUs per organ. Lines indicate median. Data are from one experiment with four to five mice per genotype. (G) Percentage of E7-specific CD8+ cells in blood after vaccination with HPV-E7. Data are from one experiment with three or four mice per genotype, represented as mean ± SD.
To search for candidate direct target genes that could explain the prominent gene derepression in Dot1L-KO cells, we analyzed the relatively few differentially expressed genes marked with H3K79me2. To this end we selected genes that were significantly down-regulated in KO and had harbored H3K79me2 at the 5′ end of the gene, both in TN and TCM. We further narrowed the list down to genes that were annotated as “negative regulator of transcription by RNA polymerase II,” resulting in 14 transcriptional regulators. Among those, Ezh2 emerged as a potentially relevant target of DOT1L that could explain part of the derepression of genes in TN and TCM Dot1L-KO cells (SI Appendix, Table S2). EZH2 is part of the Polycomb-repressive complex 2 (PRC2), which deposits H3K27me3 (46, 47), a mark involved in repression of developmentally regulated genes and in switching off naïve and memory genes during terminal differentiation of effector CD8+ T cells (34, 48). While the change in Ezh2 mRNA expression was modest between WT and KO (SI Appendix, Fig. S4C), the Ezh2 gene was H3K79me2 methylated and therefore possibly affected by Dot1L loss (SI Appendix, Fig. S4D). EZH2, being a catalytic unit of master transcriptional regulator PRC2, can greatly affect the transcriptional landscape. To further investigate the idea that misregulation of PRC2 targets could be one of the downstream consequences of loss of DOT1L in CD8+ T cells, we compared the gene expression changes in an Ezh2-KO model (49) with those seen in Dot1L-KO CD8+ T cells. This revealed substantial overlap between the derepressed genes in the two models (Fig. 4 C and D), suggesting a functional connection between two seemingly opposing epigenetic pathways. Furthermore, we determined H3K27me3 scores based on previous ChIP-Seq studies (50) and compared them with the gene expression in WT and Dot1L-KO SP CD8+ thymocytes and peripheral CD8+ T cells. This analysis showed that the genes that were up-regulated in Dot1L-KO lack H3K79me2 in WT and were strongly enriched for H3K27me3 in WT memory precursor CD8+ T cells (Fig. 4E). As a control, expression matched nondifferentially expressed genes were not enriched for H3K27me3, indicating that the enrichment is not a consequence of low expression (Fig. 4E). Taken together, these findings suggest that one of the consequences of loss of DOT1L-mediated H3K79me2 is derepression of a subset of PRC2 targets that are actively repressed in WT. This, together with the other transcriptional changes likely contributes to the perturbation of the epigenetic identity of CD8+ TAIM cells.
Deletion of Dot1L in T Cells Leads to an Impaired Immune Response.
Our Lck-Cre+/−;Dot1Lfl/fl mouse model showed that deletion of Dot1L in the T cell lineage leads to a reduction in CD4+ and iNKT cells. Furthermore, lower TCR surface levels and aberrant TCR signaling in Dot1L-KO CD8+ cells was associated with antigen-independent differentiation toward memory-type cells that also show loss of epigenetic integrity. These distinct changes in the T cell compartment of this mouse model suggested an altered immune response. To determine the immune responsiveness of Dot1L-KO T cells in vivo, mice were challenged with a sublethal dose of L. monocytogenes. At day 3 and day 7 postinjection, mice were killed and spleen and liver were used to determine clearance of the Listeria by counting colony-forming units (CFUs). From the 14 Dot1L-KO mice that were analyzed 7 d after infection, 3 mice were excluded from further analysis as either they died at day 5, were sick at day 7, or had almost no live T cells. In WT mice, no adverse effects were observed. At day 7 complete clearance was observed in the spleen from WT mice, which is in accordance with literature (51); however, the Dot1L-KO mice failed to clear Listeria (Fig. 4F and SI Appendix, Fig. S4E). Furthermore, in WT mice, the peak of activated (CD44+CD62L−) CD8+ T cells was at day 7, whereas in Dot1L-KO mice the response was very heterogeneous with some KO mice showing the peak of CD44+CD62L− cells at day 3 and others at day 7 (SI Appendix, Fig. S4 F and G). To test whether the failure to respond is a general feature of KO T cells or specific for L. monocytogenes, we made use of the HPV-E7 DNA vaccination system (52). One advantage of this system is the possibility to monitor the expansion capacity and differentiation of antigen-specific CD8+ T cells. In WT mice, E7-specific CD8+ T cells increased over time, whereas in Dot1L-KO the cells did not expand, as measured in the blood (Fig. 4G). Taken together, two independent immunization challenges indicated a compromised response upon challenges in vivo.
Also in vitro Dot1L-KO TAIM cells showed a compromised effector response. A hallmark of memory T cells is that they produce IFNγ rapidly upon stimulation compared to naïve T cells (53). Likewise, innate and virtual memory T cells rapidly produce IFNγ upon TCR stimulation (31, 40). We activated B cell-depleted splenocytes in vitro with anti-CD3 and anti-CD28 antibodies. Dot1L-KO CD8+ T cells rapidly became activated (CD44+CD62L−, SI Appendix, Fig. S4 H and I). However, in contrast to WT, KO CD8+ T cells failed to produce IFNγ (SI Appendix, Fig. S4 J and K), and expressed lower levels of CD69, a marker of activation (23) (SI Appendix, Fig. S4L). This indicates a functional impairment of Dot1L-KO T cells. To test their intrinsic competence to produce IFNγ, Dot1L-KO T cells were stimulated in a TCR-independent manner with phorbol myristate acetate (PMA) and ionomycin, which act downstream of the TCR, thereby bypassing membrane-proximal events. Intracellular staining for IFNγ revealed that the percentage of stimulated CD8+ T cells producing IFNγ was 1.9-fold higher in KO as compared to WT (SI Appendix, Fig. S4 M and N). This suggests that although Dot1L-KO CD8+ T cells intrinsically do have the capacity to produce IFNγ when exposed to nonphysiological stimuli, they only partially respond to physiological, TCR-mediated stimulation, possibly due to their aberrant TCR signaling. Taken together, these results show that deletion of Dot1L in the T cell lineage in the thymus leads to an impaired immune response.
Discussion
The histone methyltransferase DOT1L has emerged as a druggable target in MLL-rearranged leukemia and additional roles in cancer have been suggested (16, 54–57). This in combination with the availability of highly specific DOT1L inhibitors make DOT1L a potential target for cancer therapy. However, the role of DOT1L in normal lymphocyte physiology has remained unknown. Here we show that DOT1L plays a central role in ensuring normal T cell differentiation.
T cell differentiation is intimately linked to epigenetic programming, but the mechanistic role of epigenetic marks in steering T cell differentiation (34, 48, 49, 58, 59) and thymic selection (34, 48, 49, 58–61) has only recently become clearer. Here we observed that ablation of Dot1L in the T cell lineage differentially affected CD4+ and CD8+ T cells. Following deletion of Dot1L by Lck-Cre during early thymocyte development, CD4+ T cells were strongly reduced, while the number of CD8+ T cells was increased. The fact that a substantial fraction of the remaining CD4+ T cells had not lost DOT1L activity indicates that the CD4+ compartment and especially regulatory T cells depend on DOT1L for their normal development. Further research exploring the dependency of CD4+ T cells on DOT1L may provide novel strategies for immune modulation and treatment of CD4+ T cell malignancies.
In CD8+ T cells, loss of DOT1L resulted in a massive gain of CD44+CD62L+ memory-like cells. These cells start to acquire memory features intrathymically, express a diverse unskewed TCRβ repertoire, and lack expression of CD49d. Taken together, this suggests that these memory-like CD8+ T cells arose independently of foreign antigens, leading us to designate these cells as antigen-independent memory-like TAIM CD8+ cells. Importantly, such unconventional memory-phenotype cells constitute a substantial (15 to 25%) fraction of the peripheral CD8+ T cell compartment in WT mice and humans (4, 28, 40, 62, 63). The fraction of unconventional memory-phenotype T cells further increases with age (64, 65). The biological role of antigen-inexperienced memory CD8+ T cells is still not completely understood, but these cells have been observed to respond more rapidly to TCR activation than TN cells and they have been suggested to provide bystander protection against infection in an antigen-independent way (27, 40). However, in older mice, virtual memory cells have been reported to lose their proliferative potential and acquire characteristics of senescence (66). The mechanism of unconventional memory-phenotype differentiation is also poorly understood (2). In some mouse models CD8+ T cells differentiate upon excess production of IL-4 by iNKT or γδ T cells (3, 33, 67–70). In contrast, other mouse models report that antigen-inexperienced memory CD8+ T cell differentiation is cell intrinsic (71, 72). Here we show that DOT1L plays an important role in TAIM CD8+ cell differentiation by cell-intrinsic mechanisms.
It has been established in several independent mouse models that the quality of TCR signaling closely relates to the formation of TAIM cells (27, 39, 40). Our results show that loss of DOT1L leads to reduced surface expression of the CD3/TCR complex and coreceptors. This phenotype is likely related to the reduced expression of CD3ζ (Cd247), a target of DOT1L and a rate-limiting molecule for assembly and transport of TCR/CD3 complexes to the cell surface (1, 41, 42). The failure to up-regulate the TCR/CD3 complex upon positive selection likely prohibits differentiation of conventional TN and supports formation of TAIM cells. In addition, Dot1L ablation perturbed expression of TCR signaling genes, including Itk (IL-2-inducible T cell kinase; a member of the Tec kinase family). Disruption of ITK signaling has also been reported to lead to antigen-independent T-cell differentiation (43). Together, this suggests that one of the key functions of DOT1L in T cells is to ensure adequate TCR surface expression and signaling to maintain naïvety and prevent TAIM cell differentiation. The discovery of DOT1L as a key player in preventing premature antigen-independent differentiation toward memory-type cells warrants further investigation and will aid in further uncovering the origin and regulation of this emerging and intriguing subset of the immune system. It will also be important to further dissect the cause of the compromised immune response in mice lacking DOT1L in the T cell lineage. Specifically, given the TAIM phenotype, the reduced TCR levels and aberrant TCR signaling, as well as the epigenetic changes in Dot1L-KO CD8+ T cells, it will be relevant to determine the intrinsic cytotoxic potential of antigen-specific T cells lacking H3K79 methylation and understand the role that this modification plays in effector differentiation, specifically in the presence of an uncompromised CD4+ T cell compartment.
How does DOT1L affect T cell differentiation at the chromatin level? Inspection of the transcriptome and epigenome provided evidence that DOT1L methylates transcriptionally active genes in T cells and positively affects gene expression. However, only a subset of the targets required DOT1L for maintenance of normal expression levels, which agrees with previous observations (73, 74). Why some genes depend on DOT1L/H3K79 methylation and others do not is not known yet, although a recent study indicates that in MLL-rearranged leukemic cell lines, some genes harbor a 3′ enhancer located in the H3K79me2/3 marked genic region, which can make them more sensitive to loss of DOT1L (73). One of the genes that was H3K79 methylated and dependent on DOT1L in normal peripheral CD8+ T cells was Ezh2. Of note, Ezh2 expression was not reduced in Dot1L-KO CD8+ SP thymocytes. Importantly, analysis of data from Kagoya et al. (18) showed that Ezh2 expression is also reduced in human T cells in which DOT1L was inactivated, not by deletion but by treatment with a DOT1L inhibitor (SI Appendix, Fig. S4O). This suggests that the epigenetic cross-talk that we uncovered in mice is evolutionarily conserved. However, the exact mechanisms by which DOT1L affects EZH2/PRC2 activity are still unknown.
Derepression of some of the targets of PRC2 is just one of the consequences of loss of DOT1L. Besides Ezh2, DOT1L affects the expression of other genes, including several additional candidate transcription regulators (SI Appendix, Table S2). In the future, it will be important to determine other mechanisms by which DOT1L affects the CD8+ T cell transcriptome to fully understand its central role in CD8+ T cell biology. Furthermore, we cannot exclude that DOT1L has additional methylation targets besides H3K79 that contribute to the role of DOT1L in safeguarding T cell differentiation and effector functions. Although understanding the mechanisms in more detail will require further studies, the role of DOT1L in preventing premature differentiation and safeguarding the epigenetic identity is conserved in other lymphocyte subsets. In an independent study (75), we observed that loss of Dot1L in B cells also led to premature differentiation, perturbed repression of PRC2 targets, and a compromised humoral immune response. Therefore, DOT1L is emerging as a central epigenetic regulator of lymphocyte differentiation and functionality.
In conclusion, we identify H3K79 methylation by DOT1L as an activating epigenetic mark critical for CD8+ T cell differentiation and maintenance of epigenetic identity. Further investigation into the central role of the druggable epigenetic writer DOT1L in lymphocytes is likely to provide novel strategies for immune modulations and disease intervention (76).
Methods
Mice.
Lck-Cre;Dot1Lfl/fl mice have been described previously (17). OT-I (B6J) mice were a kind gift from the Ton Schumacher group, originally from The Jackson Laboratory, Bar Harbor, ME. Details of animal breeding, crossing and selection, and genotyping are described in SI Appendix, Materials and Methods. Mice used for experiments were between 6 wk and 8 mo old and of both genders. For each individual experiment mice were matched for age and gender. All experiments were approved by the Animal Ethics Committee of the Netherlands Cancer Institute (NKI) and performed in accordance with institutional, national, and European guidelines for animal care and use.
Flow Cytometry.
Single-cell suspensions were made from spleen and thymus. Erylysis was performed on blood and spleen samples. Cells were stained with fluorescently labeled antibodies in a 1:200 dilution unless otherwise indicated (SI Appendix, Table S3). Details of the antibodies used are provided in SI Appendix, Materials and Methods. For H3K79me2 staining, cells were first stained with surface markers and fixed and permeabilized. After fixation and permeabilization, cells were washed with Perm/Wash buffer containing 0.25% sodium dodecyl sulfate (SDS) in order to expose the epitope. Flow cytometry was performed using the LSR Fortessa (BD Biosciences) and data were analyzed with FlowJo software (Tree Star, Inc.). Histograms were smoothed.
In Vitro Stimulation.
To determine cytokine production upon in vitro stimulation, splenocytes were stimulated with 20 ng/mL PMA (Sigma) and 0.5 μg/mL ionomcyin (Sigma), and incubated with 1 μL/mL Golgi Plug protein transport inhibitor (Becton Dickinson) for 4 h. Cells were stained as described above. To determine proliferation and IFNγ production upon in vitro TCR-mediated stimulation splenocytes were enriched for T cells using CD19 microbeads depletion (Miltenyi Biotec) or pan-T cell Isolation Kit II (Miltenyi Biotec) on LS columns (Miltenyi Biotec). Cells were plated in a 96-well plate coated with anti-CD3 (145-2C11) (BD) and anti-CD28 (37.51) (BD) was added to the medium.
In Vivo Immunization and Vaccination.
L. monocytogenes strain LM-OVA was a gift from Ton Schumacher, NKI, Amsterdam, The Netherlands. A sublethal dose (10,000 CFUs) of L. monocytogenes in Hank’s balanced salt solution (HBSS) was injected i.v. into the mice. The Help-E7SH vaccination was performed as described in ref. 52.
Mixed Bone-Marrow Chimera.
Whole bone-marrow cells were isolated from femurs of the indicated mice. Single-cell suspensions from Ly5.1+ and Ly5.2+ were mixed in a 1:1 ratio in HBSS. A total of 106 cells were injected i.v. into Ly5.1+ lethally irradiated recipients. After 20 wk, mice were killed and spleens isolated for further analysis.
RNA-Seq and ChIP-Seq Sample Collection, Preparation, and Analysis.
Flow cytometrically purified cell populations were prepared as described in SI Appendix, Materials and Methods. For RNA-Seq, strand-specific cDNA libraries were generated using the TruSeq Stranded mRNA sample preparation kit (Illumina) according to the manufacturer’s protocol. For ChIP-Seq, chromatin was cross-linked with paraformaldehyde. Antibodies against H3K79me2 (NL59, Merck Millipore) or H3K4me3 (ab8580, Abcam) were added to purified chromatin and incubated overnight at 4 °C. Protein G Dynabeads (Life Technologies) were added to the IP and beads with bound immune complexes were subsequently washed and libraries were prepared using the KAPA LTP Library preparation kit (Roche). All samples were sequenced as 65 base single reads on a HiSeq2500 (Illumina). Detailed description of the methods and analysis can be found in SI Appendix, Materials and Methods.
Statistics.
Statistical analyses were performed using Excel. Variance was determined using a F test, and an unpaired Student’s t test with two-tailed distribution was used for statistical analyses. Data are presented as means ± SD unless otherwise indicated in the figure legends. For Fig. 1B and SI Appendix, Fig. S1 D, E, and H, a Student’s t test with Bonferroni correction was performed in R. A P value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank the NKI animal pathology facility for histology and immunohistochemistry, as well as advice; the NKI Genomics Core Facility for library preparations and sequencing; the NKI Research High Performance Computing (RHPC) facility for providing computational resources; the NKI Flow Cytometry facility for assistance; the NKI Animal Laboratory Intervention Unit for performing immunization experiments; and the caretakers of the NKI Animal Laboratory facility for assistance and excellent animal care. The following reagents were obtained through the NIH Tetramer Core Facility: CD1d-PBS57 and CD1d-unloaded tetramer. We thank Paul van den Berk for advice and Cheyenne Seerden for help with the flow-cytometry experiments, Ton Schumacher and group for OT-I mice and OT-I tetramer, and Jan-Hermen Dannenberg for providing the Lck-Cre mouse model. We thank Renee X. de Menezes for help with statistical analysis and Muddassir Malik for critically reading the manuscript. This work was supported by the Dutch Cancer Society (NKI2014-7232 to F.v.L. and H.J.) and the Dutch Research Council (NWO-VICI-016.130.627 to F.v.L.; ZonMW Top 91213018 to H.J.; ZonMW Top91218022 to F.v.L. and H.J.). T.v.d.B. and E.d.W. are part of the Oncode Institute which is partly financed by the Dutch Cancer Society. The funders had no role in study design, data collection and interpretation, or in the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920372117/-/DCSupplemental.
Data availability.
Next-generation sequencing data have been deposited in Gene Expression Omnibus (GEO, National Center for Biotechnology Information) under GSE138908 and GSE138910 for ChIP-Seq and RNA-Seq data, respectively. A detailed description of all materials and methods is provided in SI Appendix, Materials and Methods.
References
- 1.Borst J., Jacobs H., Brouns G., Composition and function of T-cell receptor and B-cell receptor complexes on precursor lymphocytes. Curr. Opin. Immunol. 8, 181–190 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Hussain T., Quinn K. M., Similar but different: Virtual memory CD8 T cells as a memory-like cell population. Immunol. Cell Biol. 97, 675–684 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Jameson S. C., Lee Y. J., Hogquist K. A., Innate memory T cells. Adv. Immunol. 126, 173–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White J. T., Cross E. W., Kedl R. M., Antigen-inexperienced memory CD8+ T cells: Where they come from and why we need them. Nat. Rev. Immunol. 17, 391–400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henning A. N., Roychoudhuri R., Restifo N. P., Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18, 340–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray S. M., Kaech S. M., Staron M. M., The interface between transcriptional and epigenetic control of effector and memory CD8+ T-cell differentiation. Immunol. Rev. 261, 157–168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Omilusik K. D., Goldrath A. W., Remembering to remember: T cell memory maintenance and plasticity. Curr. Opin. Immunol. 58, 89–97 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdelsamed H. A., Zebley C. C., Youngblood B., Epigenetic maintenance of acquired gene expression programs during memory CD8 T cell homeostasis. Front. Immunol. 9, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng N. P., Araki Y., Subedi K., The molecular basis of the memory T cell response: Differential gene expression and its epigenetic regulation. Nat. Rev. Immunol. 12, 306–315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phan A. T., Goldrath A. W., Glass C. K., Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46, 714–729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlaming H., van Leeuwen F., The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma 125, 593–605 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Steger D. J. et al., DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28, 2825–2839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood K., Tellier M., Murphy S., DOT1L and H3K79 methylation in transcription and genomic stability. Biomolecules 8, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLean C. M., Karemaker I. D., van Leeuwen F., The emerging roles of DOT1L in leukemia and normal development. Leukemia 28, 2131–2138 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Onder T. T. et al., Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X., Chen C. W., Armstrong S. A., The role of DOT1L in the maintenance of leukemia gene expression. Curr. Opin. Genet. Dev. 36, 68–72 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Vlaming H. et al., Conserved crosstalk between histone deacetylation and H3K79 methylation generates DOT1L-dose dependency in HDAC1-deficient thymic lymphoma. EMBO J. 38, e101564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kagoya Y. et al., DOT1L inhibition attenuates graft-versus-host disease by allogeneic T cells in adoptive immunotherapy models. Nat. Comm. 9, 1915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheer S. et al., A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat. Comm. 10, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones B. et al., The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4, e1000190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y. et al., Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood 116, 4483–4491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chory E. J. et al., Nucleosome turnover regulates histone methylation patterns over the genome. Mol. Cell 73, 61–72.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sancho D., Gómez M., Sánchez-Madrid F., CD69 is an immunoregulatory molecule induced following activation. Trends Immunol. 26, 136–140 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Tani-ichi S. et al., Interleukin-7 receptor controls development and maturation of late stages of thymocyte subpopulations. Proc. Natl. Acad. Sci. U.S.A. 110, 612–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCaughtry T. M. et al., Conditional deletion of cytokine receptor chains reveals that IL-7 and IL-15 specify CD8 cytotoxic lineage fate in the thymus. J. Exp. Med. 209, 2263–2276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truckenbrod E. N., Jameson S. C., The virtuous self-tolerance of virtual memory T cells. EMBO J. 37, e99883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drobek A. et al., Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD8 T cells. EMBO J. 37, e98518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haluszczak C. et al., The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206, 435–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinet V. et al., Type I interferons regulate eomesodermin expression and the development of unconventional memory CD8(+) T cells. Nat. Commun. 6, 7089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Intlekofer A. M. et al., Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Lee J.-Y. Y., Hamilton S. E., Akue A. D., Hogquist K. A., Jameson S. C., Virtual memory CD8 T cells display unique functional properties. Proc. Natl. Acad. Sci. U.S.A. 110, 13498–13503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing Y., Wang X., Jameson S. C., Hogquist K. A., Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17, 565–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atherly L. O. et al., The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity 25, 79–91 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Kakaradov B. et al., Early transcriptional and epigenetic regulation of CD8+ T cell differentiation revealed by single-cell RNA sequencing. Nat. Immunol. 18, 422–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wingender G. et al., Immediate antigen-specific effector functions by TCR-transgenic CD8+ NKT cells. Eur. J. Immunol. 36, 570–582 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Fenton R. G., Marrack P., Kappler J. W., Kanagawa O., Seidman J. G., Isotypic exclusion of γ δ T cell receptors in transgenic mice bearing a rearranged β-chain gene. Science 241, 1089–1092 (1988). [DOI] [PubMed] [Google Scholar]
- 37.Goldrath A. W., Bevan M. J., Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity 11, 183–190 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst B., Lee D. S., Chang J. M., Sprent J., Surh C. D., The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity 11, 173–181 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Renkema K. R., Li G., Wu A., Smithey M. J., Nikolich-Žugich J., Two separate defects affecting true naive or virtual memory T cell precursors combine to reduce naive T cell responses with aging. J. Immunol. 192, 151–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White J. T. et al., Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat. Commun. 7, 11291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Call M. E., Wucherpfennig K. W., The T cell receptor: Critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 23, 101–125 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Ashwell J. D., Klausner R. D., Genetic and mutational analysis of the T-cell antigen receptor. Annu. Rev. Immunol. 8, 139–167 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Nayar R. et al., TCR signaling via Tec kinase ITK and interferon regulatory factor 4 (IRF4) regulates CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, E2794–E2802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogquist K. A. et al., T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Huff J. T., Plocik A. M., Guthrie C., Yamamoto K. R., Reciprocal intronic and exonic histone modification regions in humans. Nat. Struct. Mol. Biol. 17, 1495–1499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Mierlo G., Veenstra G. J. C., Vermeulen M., Marks H., The complexity of PRC2 subcomplexes. Trends Cell Biol. 29, 660–671 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Gray S. M., Amezquita R. A., Guan T., Kleinstein S. H., Kaech S. M., Polycomb repressive complex 2-mediated chromatin repression guides effector CD8+ T cell terminal differentiation and loss of multipotency. Immunity 46, 596–608 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He S. et al., Ezh2 phosphorylation state determines its capacity to maintain CD8+ T memory precursors for antitumor immunity. Nat. Comm. 8, 2125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu B. et al., Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mannering S. I., Zhong J., Cheers C., T-cell activation, proliferation and apoptosis in primary Listeria monocytogenes infection. Immunology 106, 87–95 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahrends T. et al., CD27 agonism plus PD-1 blockade recapitulates CD4+ T-cell help in therapeutic anticancer vaccination. Cancer Res. 76, 2921–2931 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Kaech S. M., Wherry E. J., Ahmed R., Effector and memory T-cell differentiation: Implications for vaccine development. Nat. Rev. Immunol. 2, 251–262 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Zhu B. et al., The protective role of DOT1L in UV-induced melanomagenesis. Nat. Commun. 9, 259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salgado C. et al., A novel germline variant in the DOT1L gene co-segregating in a Dutch family with a history of melanoma. Melanoma Res. 29, 582–589 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Zhang X. et al., Prognostic and therapeutic value of disruptor of telomeric silencing-1-like (DOT1L) expression in patients with ovarian cancer. J. Hematol. Oncol. 10, 29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong M. et al., The histone methyltransferase DOT1L promotes neuroblastoma by regulating gene transcription. Cancer Res. 77, 2522–2533 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Pace L. et al., The epigenetic control of stemness in CD8+ T cell fate commitment. Science 359, 177–186 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Chen G. et al., Ezh2 regulates activation-induced CD8+ T cell cycle progression via repressing Cdkn2a and Cdkn1c expression. Front. Immunol. 9, 549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasler H. G. et al., Histone deacetylase 7 regulates cell survival and TCR signaling in CD4/CD8 double-positive thymocytes. J. Immunol. 186, 4782–4793 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Stengel K. R. et al., Histone deacetylase 3 is required for efficient T cell development. Mol. Cell. Biol. 35, 3854–3865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akue A. D., Lee J.-Y., Jameson S. C., Derivation and maintenance of virtual memory CD8 T cells. J. Immunol. 188, 2516–2523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Kaer L., Innate and virtual memory T cells in man. Eur. J. Immunol. 45, 1916–1920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiu B.-C. C., Martin B. E., Stolberg V. R., Chensue S. W., Cutting edge: Central memory CD8 T cells in aged mice are virtual memory cells. J. Immunol. 191, 5793–5796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanzer K. G., Cookenham T., Reiley W. W., Blackman M. A., Virtual memory cells make a major contribution to the response of aged influenza-naïve mice to influenza virus infection. Immun. Ageing 15, 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinn K. M. et al., Age-related decline in primary CD8+ T cell responses is associated with the development of senescence in virtual memory CD8+ T cells. Cell Rep. 23, 3512–3524 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Felices M., Yin C. C., Kosaka Y., Kang J., Berg L. J., Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc. Natl. Acad. Sci. U.S.A. 106, 8308–8313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasanthakumar A. et al., A non-canonical function of Ezh2 preserves immune homeostasis. EMBO Rep. 18, 619–631 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobenecker M.-W. et al., Coupling of T cell receptor specificity to natural killer T cell development by bivalent histone H3 methylation. J. Exp. Med. 212, 297–306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kasler H. G., Lee I. S., Lim H. W., Verdin E., Histone Deacetylase 7 mediates tissue-specific autoimmunity via control of innate effector function in invariant Natural Killer T Cells. eLife 7, e32109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hirose S. et al., Bcl11b prevents the intrathymic development of innate CD8 T cells in a cell intrinsic manner. Int. Immunol. 27, 205–215 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Gugasyan R. et al., The NF-κB1 transcription factor prevents the intrathymic development of CD8 T cells with memory properties. EMBO J. 31, 692–706 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godfrey L. et al., DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nat. Commun. 10, 2803 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kerry J. et al., MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 18, 482–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aslam M. A., et al. , Histone methyltransferase DOT1L controls state-specific identity during B cell differentiation. bioRxiv:10.1101/826370 (25 March 2020). [DOI] [PMC free article] [PubMed]
- 76.Berdasco M., Esteller M., Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 20, 109–127 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing data have been deposited in Gene Expression Omnibus (GEO, National Center for Biotechnology Information) under GSE138908 and GSE138910 for ChIP-Seq and RNA-Seq data, respectively. A detailed description of all materials and methods is provided in SI Appendix, Materials and Methods.