Abstract
TGF-β is a master regulator of fibrosis, driving the differentiation of fibroblasts into apoptosis-resistant myofibroblasts and sustaining the production of extracellular matrix (ECM) components. Here, we identified the nuclear long noncoding RNA (lncRNA) H19X as a master regulator of TGF-β–driven tissue fibrosis. H19X was consistently upregulated in a wide variety of human fibrotic tissues and diseases and was strongly induced by TGF-β, particularly in fibroblasts and fibroblast-related cells. Functional experiments following H19X silencing revealed that H19X was an obligatory factor for TGF-β–induced ECM synthesis as well as differentiation and survival of ECM-producing myofibroblasts. We showed that H19X regulates DDIT4L gene expression, specifically interacting with a region upstream of the DDIT4L gene and changing the chromatin accessibility of a DDIT4L enhancer. These events resulted in transcriptional repression of DDIT4L and, in turn, in increased collagen expression and fibrosis. Our results shed light on key effectors of TGF-β–induced ECM remodeling and fibrosis.
Keywords: Autoimmunity, Cell Biology
Keywords: Autoimmune diseases, Epigenetics, Fibrosis
Introduction
During the development of fibrosis, the organ parenchyma is replaced with a collagen-rich, stiff connective tissue, leading to organ dysfunction and failure as seen, for example, in end-stage liver disease or idiopathic pulmonary fibrosis (IPF). Fibrosis is also a dominant pathological feature of many chronic autoimmune diseases, including systemic sclerosis (SSc), rheumatoid arthritis, Crohn’s disease, and systemic lupus erythematosus. SSc is a prototype fibrotic disease and is often used as a model disease to identify key mechanisms of tissue remodeling in fibrosis (1, 2). SSc is associated with high morbidity and the highest mortality among the rheumatic diseases; it has a standardized mortality ratio of 3.53 and average loss of life expectancy of 16 to 34 years (3). The hallmarks of SSc are abnormalities of the vasculature and immune system, eventually leading to fibrosis in multiple organs, including the skin, lungs, and heart. SSc has a chronic course, and particularly in the diffuse cutaneous subset the disease is often rapidly progressing (4). Despite advances in the identification of potential molecular targets, specific treatments for fibrotic diseases, including SSc, are limited, and there is a high clinical need for novel therapeutic concepts.
Fibroblasts from patients with fibrotic diseases display characteristic features, including overproduction of collagen and enhanced secretion of cytokines and chemokines. They also express higher levels of adhesion molecules, such as integrins, and receptors for TGF-β and PDGF (5). Moreover, they show an increased resistance to apoptosis mediated by death receptor FAS (6). A major hallmark of these cells is their differentiation into highly activated myofibroblasts. Myofibroblasts express α smooth muscle actin (αSMA) in their stress fibers and exhibit enhanced matrix adhesion and contractile properties (7). In addition, they release large amounts of extracellular matrix (ECM) components such as collagens and fibronectin (8). Resident fibroblasts transiently differentiate into myofibroblasts during the early stages of physiological wound healing and undergo apoptosis during the resolution stages. However, in scarring tissues, myofibroblast persistency contributes to fibrogenesis, a process also found in fibrotic diseases (9). Additionally, selective targeting of antiapoptotic proteins in activated myofibroblasts has been tested as a therapeutic strategy for fibrosis (10).
TGF-β is the master regulator of physiological wound healing and fibrosis by inducing the differentiation of fibroblasts into myofibroblasts (11, 12). The TGF-β canonical pathway involves phosphorylation of TGF-βR1, which in turn phosphorylates and activates SMAD2/3 that can then bind SMAD4 (13). These complexes accumulate in the nucleus where they act as transcription factors and participate in the regulation of target gene expression, leading to ECM production and myofibroblast differentiation (14).
Research in recent years has shown that noncoding RNAs play a central role in the regulation of gene expression (15, 16). Long noncoding RNAs (lncRNAs) are defined as noncoding RNA transcripts longer than 200 bp and are mostly uncharacterized and unannotated. In general, lncRNAs have more specific expression profiles than protein coding transcripts, being expressed in a manner specific to cell type, tissue, developmental stage, or disease state (17). LncRNAs exert their gene regulatory functions via a variety of mechanisms. Maternally expressed gene 3 (MEG3), for example, is a nuclear lncRNA that modulates the expression of TGF-β pathway target genes by regulating the chromatin structure of their distal enhancers (18). MEG3 has been found downregulated in the human fibrotic liver (19) and in murine cardiac fibrosis (20). Other lncRNAs regulate the behavior of other types of molecules (21), such as microRNAs. For example, myocardial infarction–associated transcript (MIAT) captures miR-24, a TGF-β regulatory miRNA, via a sponge-like activity and promotes cardiac fibrosis (22). Upregulation of the maternally imprinted lncRNA H19 has been implicated in renal fibrosis by suppressing the activity of miR-17, leading to increased fibronectin expression (23).
In this study, we identified and functionally characterized a profibrotic lncRNA H19X as a central regulator of myofibroblast differentiation and survival. We were able to show that the induction of H19X was a conditio sine qua non for the profibrotic effects of TGF-β and might be a key driver of myofibroblast persistence and fibrosis in a wide variety of TGF-β–driven fibrotic diseases.
Results
LncRNA H19X is upregulated in a variety of fibrotic diseases.
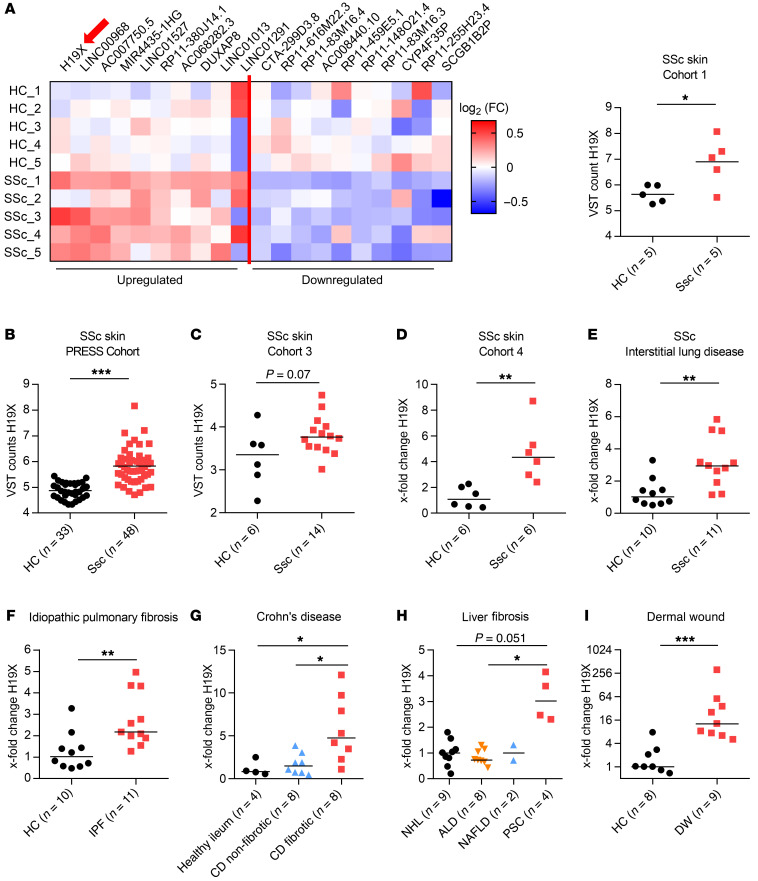
SSc is a prototype fibrotic disease. To identify lncRNAs potentially involved in the pathogenesis of fibrotic diseases, we performed whole transcriptome RNA-Seq on skin biopsies from severely affected patients with SSc with active, untreated, early diffuse cutaneous SSc and age- and sex-matched healthy controls (HCs) (n = 5 SSc vs. n = 5 HCs). In this screening experiment, H19 X-linked coexpressed lncRNA (H19X) was identified as one of the most consistently and strongly upregulated lncRNAs (P = 0.022, Figure 1A) in skin biopsies of patients with SSc. H19X, also known as MIR503HG, is an intergenic lncRNA and its gene is located on chromosome X. None of its 5 annotated transcript variants has a coding potential as demonstrated by in silico analysis using CPAT, a coding potential assessment tool (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI135439DS1).
Figure 1. H19X is upregulated in skin from patients with SSc from different cohorts in a variety of fibrotic disorders and in physiological DW healing.
Heatmap showing 20 most significantly deregulated lncRNA in skin derived from patients with SSc as revealed by RNA-Seq analysis; H19X is indicated by the arrow (cohort 1) (A). H19X differential expression in SSc versus HC skin in the PRESS cohort (B), and cohort 3 (C), as measured by RNA-Seq. H19X differential expression as measured by qPCR in cohort 4, normalized to GAPDH and RPLP0 (D). H19X differential expression in ILD-SSc versus HC lung as measured by qPCR with normalization to GAPDH and RPLP0 (E). H19X differential expression in IPF (F), Crohn’s disease (G), PSC (H), and DWs measured by qPCR with normalization to GAPDH and RPLP0 (I). Data are presented as single values and median. Differential expression analysis was carried out on variance stabilized counts using DEseq2 package 44 (A–C). Mann-Whitney test (D–F and I). Kruskal–Wallis test (G and H). *P < 0.05, **P < 0.01, ***P < 0.001.
We confirmed the upregulation of the lncRNA H19X in 3 additional independent SSc cohorts. Using RNA-Seq, H19X was found significantly upregulated (P < 0.0001, Figure 1B) in skin biopsies of patients with early diffuse cutaneous SSc who were enrolled in the large multicenter SSc cohort study PRESS (Prospective Registry of Early Systemic Sclerosis) (n = 48 SSc vs. n = 33 HCs).
We then extended our analysis to cohorts of SSc patients with more diverse disease duration and disease severity. H19X was upregulated by RNA-Seq in skin samples from patients with SSc in the third SSc cohort compared with skin from HCs (n = 14 SSc vs. n = 6 HCs, P = 0.07, Figure 1C). In the fourth cohort (n = 6 SSc vs. n = 6 HCs), we analyzed the expression of H19X with an independent technique. Using quantitative PCR (qPCR), the expression of H19X was found to be 4.3-fold higher in SSc skin samples compared with HC skin samples (P = 0.0043, Figure 1D). To assess whether H19X is also upregulated in affected internal organs of patients with SSc, we extracted total RNA from lung samples of patients with SSc-associated interstitial lung disease (SSc-ILD) and HCs. H19X was significantly upregulated in lungs of patients with end-stage SSc-ILD undergoing lung transplantation (n = 11 SSc-ILD vs. n = 11 HCs, P = 0.0181, Figure 1E). These data from multiple cohorts covering a wide range of clinical SSc phenotypes showed that H19X is upregulated in SSc, not only in the skin, but also in the lungs of affected patients.
To assess whether H19X might be upregulated in general in fibrotic processes, we analyzed its expression in additional fibrotic disorders. We measured H19X expression in total RNA isolated from the lung tissue of patients with IPF (n = 11 IPF vs. n = 11 HCs), and a significant upregulation of H19X was also confirmed in these patients compared with controls (P = 0.0256, Figure 1F). Similarly, H19X was upregulated in RNA extracts from fibrotic ileum of patients with Crohn’s disease (n = 8) versus nonfibrotic control ileum from the same subject (P = 0.0298, n = 8) and versus ileum from HCs (P = 0.0353, n = 4, Figure 1G). Furthermore, H19X expression was increased in livers of patients with primary sclerosing cholangitis (PSC) compared with normal healthy liver samples (n = 4 PSC vs. n = 9 HCs, P = 0.051, Figure 1H), while no increase was found in alcoholic liver disease with cirrhosis and nonalcoholic fatty liver disease. Pathological fibrosis is often considered as a chronic uncontrolled wound healing process since it has overlapping mechanisms with the physiological wound healing process. Therefore, we analyzed H19X expression in tissue derived from dermal wounds (DWs). Strikingly, H19X was found to be 12.77-fold higher in DWs than in normal control skin tissue (n = 9 DWs vs. n = 8 HC, P = 0.0006, Figure 1I). All together, these data suggest that H19X is upregulated in a wide variety of diseases and processes that involve physiological (wound healing) and pathological (fibrosis) tissue remodeling. When a statistically meaningful analysis was possible, we compared the expression of H19X in women and men, but we could not observe any significant difference. Therefore, we could not find evidence that H19X is involved in the increased prevalence of fibrotic diseases in females such as SSc (Supplemental Figure 1, A–C).
TGF-β is a strong inducer of H19X expression in fibroblasts.
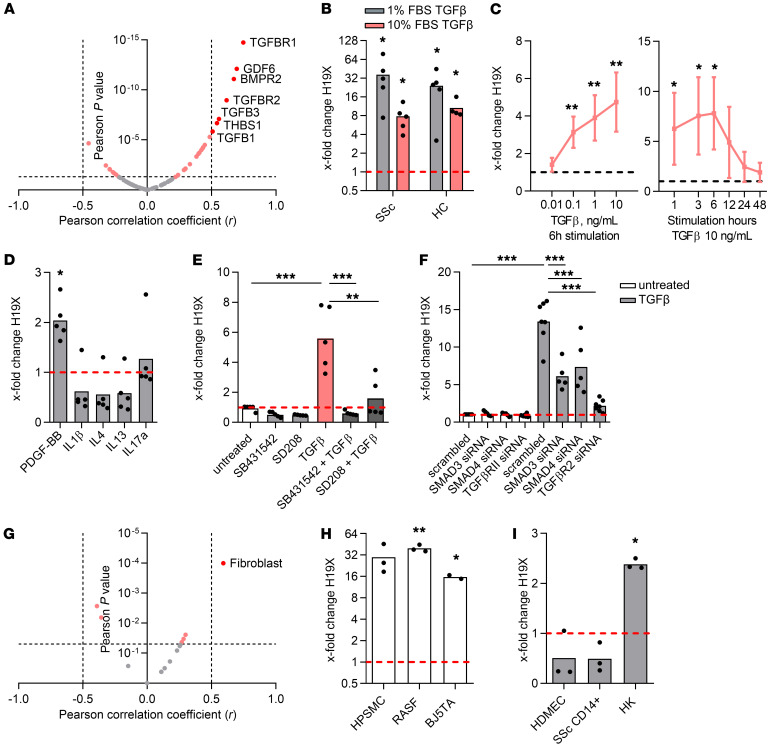
TGF-β is a key cytokine in tissue remodeling and fibrosis, which led us to analyze whether the increased expression of H19X in fibrotic tissues is associated with the expression of TGF-β pathway genes. Using the RNA-Seq data from the PRESS cohort, H19X expression significantly correlated with the expression of several TGF-β pathway mediators (Kyoto Encyclopedia of Genes and Genomes [KEGG] Pathway Database; ref. 24, Figure 2A, and Supplemental Table 2). Moreover, PAI1 mRNA, a well-characterized TGF-β marker for TGF-β pathway activation (25), was increased in most of the fibrotic tissues we analyzed, suggesting a TGF-β signaling activation parallel to the H19X upregulation (Supplemental Figure 2, A–F).
Figure 2. H19X is induced by TGF-β in a time- and dose-dependent manner in fibroblasts and related cell types.
Correlation of H19X expression with TGF-β pathway genes (KEGG Pathway Database). Significant correlations are highlighted in pink (Pearson P < 0.05). Genes with significant correlations and Pearson’s correlation coefficients r >│0.5│ are marked in red (A). H19X expression after 6 hours of 10 ng/mL TGF-β stimulation in SSc and HC dermal fibroblasts either in complete medium (10% FBS) or starvation medium (1% FBS) (B). Dose response curve of TGF-β–induced expression of H19X. SSc fibroblasts were stimulated with different concentrations of TGF-β (0.01–10 ng/mL) for 6 hours in 10% FBS. Time-dependent induction of H19X by TGF-β. SSc dermal fibroblasts were stimulated for 1–48 hours with 10 ng/mL TGF-β in 10% FBS medium (C). SSc fibroblasts were stimulated with PDGF-BB (20 ng/mL), IL-1β (10 ng/mL), IL-4 (10 ng/mL), IL-13 (10 ng/mL), or IL-17a (10 ng/mL) for 6 hours in 1% FBS medium (D). H19X expression in SSc fibroblasts after 6 hours of 10 ng/mL TGF-β stimulation and concomitant chemical inhibition of TGF-βR1 with SD208 (100 nM) or SB431542 (10 μM) in 10% FBS medium (E). Silencing of TGFBR2 and SMADs in SSc fibroblasts using 50 nM siRNA for a total of 54 hours and stimulated with TGF-β for the last 6 hours in 1% FBS medium (F). Correlation of H19X expression with different cell type–specific gene signatures. Significant correlations are highlighted in pink (Pearson P < 0.05). Fibroblast gene signature is marked in red (G). Stimulation of other types of fibroblasts or fibroblast-like cells with 10 ng/mL of TGF-β for 6 hours in 1% FBS medium (H). Stimulation of other cell types with 10 ng/mL of TGF-β for 6 hours in 10% FBS medium (I). H19X expression was measured by qPCR and normalized to GAPDH and RPLP0. Fold change was calculated respective to untreated control set as 1 (dashed line). Data are presented as single values and mean (n = 3–7) biological replicates. t test (B, C, H, and I) and 1-way ANOVA (D–F). *P < 0.05, **P < 0.01, ***P < 0.001.
Therefore, we hypothesized that TGF-β could be involved in the regulation of H19X expression. Indeed, stimulation with TGF-β resulted in a strong upregulation of H19X in primary dermal fibroblasts from patients with SSc (P = 0.0136) as well as from HCs (P = 0.0127, Figure 2B), cultured in complete medium with 10% FBS. The TGF-β–driven induction of H19X was even stronger when fibroblasts were cultured in starvation medium containing 1% FBS (P = 0.0413 SSc, P = 0.0249 HC, Figure 2B). The TGF-β–induced upregulation of H19X was dose dependent, with a steady increase over biologically relevant TGF-β concentrations (26) (0.1–10 ng/mL in complete medium with 10% FBS, Figure 2C). Time course experiments showed that the upregulation of H19X by TGF-β was strongest at 6 hours, reaching an 8-fold increase in fibroblasts cultured in complete medium with 10% FBS, indicating an early response of H19X to TGF-β. Although smaller, there was a consistent induction of H19X across all measured time points (1–48 hours, Figure 2C). Other profibrotic and proinflammatory cytokines such as IL-1β, IL-4, IL-13, and IL-17a did not induce H19X after 6 or 24 hours of stimulation. A mild absolute induction was observed after 6 hours of PDGF-BB stimulation (P = 0.0041), and a mild downregulation was recorded after 24 hours of IL-13 stimulation, but not at other time points (P = 0.0030, Figure 2D and Supplemental Figure 3A).
SMAD proteins play a major role in canonical TGF-β signaling via TGF-βR1 activation. To unravel the signaling mechanisms involved in TGF-β–dependent H19X induction, we treated fibroblasts with SB431542 and SD208, the selective inhibitors of TGF-βR1 in parallel to TGF-β stimulation. The TGF-β–driven induction of H19X significantly diminished after TGF-βR1 blockage (P < 0.001 SB431542 and P = 0.0017 SD208, Figure 2E). To analyze whether the canonical TGF-β pathway plays a role in the TGF-β–induced expression of H19X, SMAD3, SMAD4, and TGFBR2 were silenced using respective siRNAs. Indeed, the TGF-β–induced expression of H19X was reduced in silenced samples compared with scrambled controls (P = 0.0001 SMAD3 siRNA, P = 0.0006 SMAD4 siRNA, P = 0.0002 TGFBR2 siRNA, Figure 2F and Supplemental Figure 3B). These data provide further evidence that the TGF-β canonical pathway is a key regulatory pathway of H19X expression.
Moreover, in the PRESS cohort, the expression of H19X positively correlated with the fibroblast signature (27) (R = 0.59, P < 0.0001), whereas there was less or no correlation for other cell types (Figure 2G and Supplemental Table 3). To assess the cell types in which H19X can be induced by TGF-β, several cell lines were treated with TGF-β. A strong increase in H19X expression, comparable to that observed in dermal fibroblasts, was found in rheumatoid arthritis synovial fibroblasts (P = 0.0043, Figure 2H), in the foreskin fibroblast cell line BJ5TA (P = 0.040, Figure 2H), and in fibroblast-like cells, such as pulmonary artery smooth muscle cells (P = 0.0723, Figure 2H). In contrast, when we stimulated other cell types with TGF-β for 6 hours, including CD14+ cells from patients with SSc, human microvascular endothelial cells, and human keratinocytes (HKs), a decrease or a mild induction in H19X expression was observed (P = 0.0039, HK, Figure 2I). These data indicate that H19X is a TGF-β–regulated lncRNA and that its induction by TGF-β is cell type dependent. Specifically, fibroblasts of different origins and fibroblast-like cells exhibited the strongest induction by TGF-β, further supporting the hypothesis that H19X might play a role in fibrosis.
H19X mediates the profibrotic effects of TGF-β by promoting ECM production and myofibroblast differentiation.
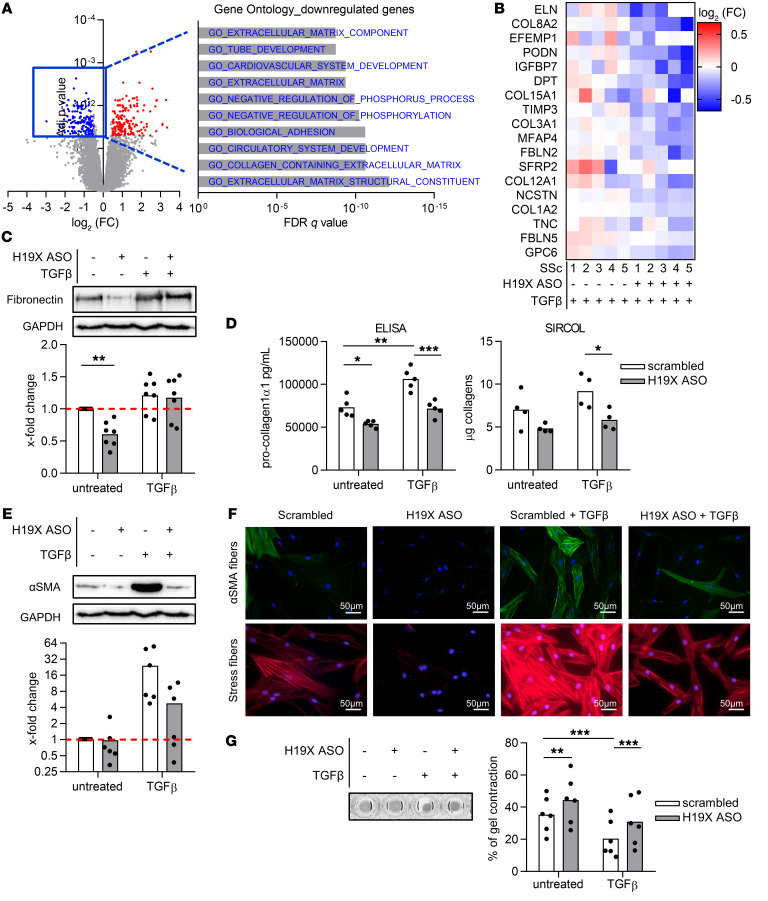
To study which genes are affected by the dysregulation of H19X in fibrotic tissues, we silenced H19X in SSc fibroblasts for 120 hours and stimulated with TGF-β for the last 72 hours (Supplemental Figure 4A). First, we used a targeted approach by analyzing a specific set of TGF-β–responsive genes known for their important roles in fibrosis, including genes encoding ECM components (collagen1α1, COL1A1 and fibronectin, FN1) and myofibroblast marker molecules (α-smooth muscle actin, ACTA2). We observed a strong and consistent reduction in the expression of these genes after H19X silencing using 2 different sets of H19X antisense oligonucleotides (ASOs, Supplemental Figure 4C). In order to have a broader overview on H19X target genes, microarray analysis was performed (Supplemental Figure 4D). At an FDR of less than 5% and absolute log2 fold change (FC) greater than 0.5, 162 genes were significantly downregulated, and 155 genes were significantly upregulated in TGF-β–stimulated cells after H19X silencing compared with TGF-β–stimulated scrambled controls. As analyzed by gene set enrichment analysis (GSEA) of 162 downregulated genes, H19X silencing had a strong effect on genes involved in ECM production. Gene ontology gene sets such as ECM structural constituents, collagen containing ECM, ECM, and ECM components were among the top 10 gene sets (Figure 3A). Moreover, different collagens (COL8A2, COL15A1, and COL3A1), other ECM genes and ECM-related genes (elastin, ELN; EGF-containing fibulin-like ECM protein, EFEMP1; podocan, PODN; dermatopontin, DPT; metalloproteinase inhibitor 3, TIMP3; microfibril-associated protein 4, MFAP4; secreted frizzled related protein 2, SFRP2; nicastrin, NCSTN; tenascin C, TNC; fibulin-5, FBLN5; glypican 6, GPC6; IGF-binding protein 3, IGFBP3; IGF-binding protein 7, IGFBP7; and fibulin-2, FBLN2) were among the top downregulated genes (log2 FC < –0.5, Figure 3B). These data suggest that the upregulation of H19X observed in fibrotic tissue might promote ECM remodeling.
Figure 3. H19X silencing reduces ECM production and myofibroblast activation.
H19X was silenced in dermal SSc fibroblasts with 25 nM ASO for a total of 120 hours and stimulated with TGF-β for the last 72 hours in 1% FBS medium. Gene ontology enrichment analysis (GSEA software) performed on significantly downregulated genes (FDR < 0.05 and absolute log2 FC < –0.5) from microarray analysis (A). Heatmap showing significant ECM-related downregulated genes as measured by microarray analysis (FDR < 0.05 and log2 FC > 0.5) (B). The protein levels of fibronectin and αSMA were analyzed by Western blot. Pictures are representative of n = 6–7 biological replicates. The protein level was semiquantified by densitometric analysis. For Western blot, semiquantification fold change was calculated compared with control set as 1 (dashed line) (C and E). The secretion of procollagen1α1 and pan-collagen into supernatants of SSc fibroblasts was quantified with ELISA and Sircol, respectively (D). αSMA and stress fiber formation were assessed by immunofluorescence staining. Scale bar: 50 μm. Pictures are representative of n = 5 biological replicates (F). Cell contraction capacity was evaluated by gel contractility assay. The picture is representative of n = 6 biological replicates (G). Data are presented as single values and mean (n = 5–7). One-way ANOVA (C–E and G). *P < 0.05, **P < 0.01, ***P < 0.001.
Given these results, we further characterized the role of H19X in the production of ECM components and in the differentiation of fibroblasts into ECM-producing myofibroblasts (Supplemental Figure 4A). Basal fibronectin expression was significantly reduced at the protein level after H19X silencing in SSc fibroblasts (P = 0.0051, Figure 3C). Moreover, protein secretion of total collagens (P = 0.0188, Figure 3D) and procollagen1α1 (P = 0.0007, Figure 3D) into the supernatant of SSc fibroblasts was impaired as revealed by Sircol assay and ELISA, respectively. Western blot analysis showed that H19X silencing in SSc fibroblasts reduced the production of αSMA at the protein level (Figure 3E). Immunofluorescence staining of αSMA fibers and phalloidin staining of stress fibers after H19X silencing in SSc fibroblasts was also strongly reduced, further confirming that H19X is a crucial factor in TGF-β–mediated actin fiber formation and myofibroblast differentiation (Figure 3F). Furthermore, SSc fibroblasts displayed a reduced capacity to contract collagen gel matrix after H19X silencing (P = 0.0026 basal condition and P = 0.0007 after TGF-β stimulation, Figure 3G). These data show that H19X expression influenced ECM gene regulation and myofibroblast differentiation. Because mitochondrial deacetylase Sirtuin-3 has recently been shown to be downregulated by TGF-β– mediating effects on collagen synthesis (28), we investigated whether H19X could be involved in the dysregulated expression of SIRT3 in fibrosis. Although we could confirm that protein levels of SIRT3 were reduced after TGF-β treatment, H19X silencing did not affect SIRT3 protein expression, indicating that in this case the TGF-β effects on SIRT3 are independent of H19X (Supplemental Figure 4E).
H19X regulates cell cycle and proliferation.
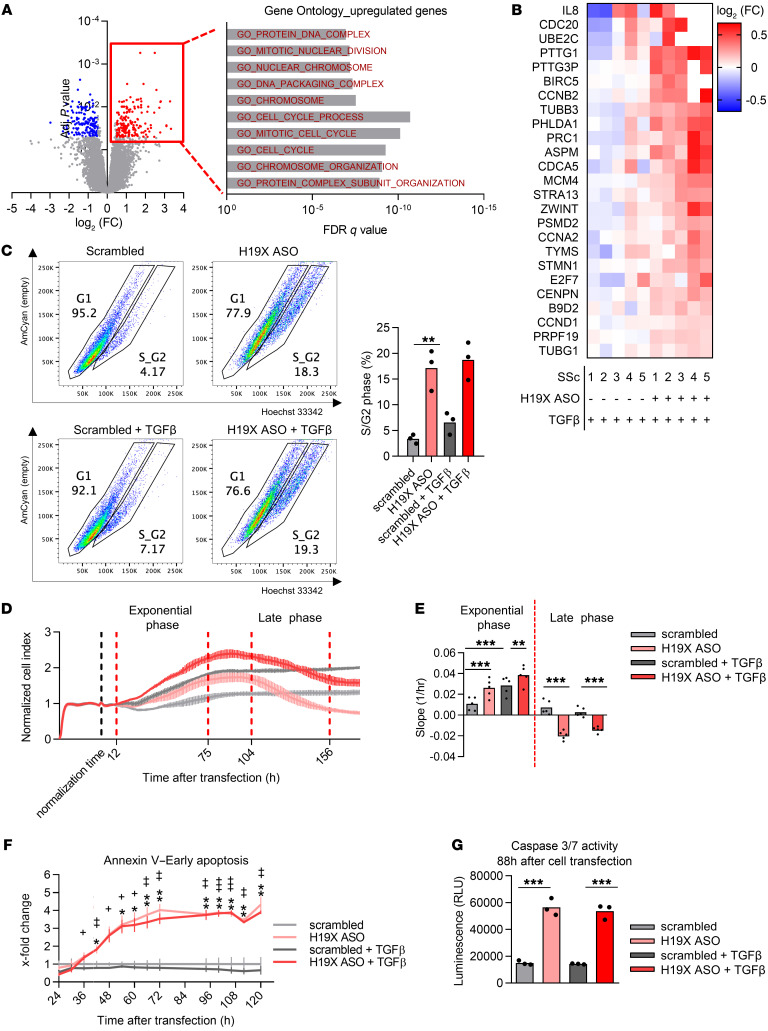
We continued with the analysis of the 155 upregulated genes after H19X silencing (FDR < 0.05 and log2 FC > 0.5, Supplemental Figure 4D). Gene ontology analysis revealed a strong involvement of H19X in cell cycle regulation (top 10 pathways shown in Figure 4A). Among the genes with the strongest upregulation (Figure 4B), there were CCND1, CCNA2, CCNB2 encoding for cyclins (cell-cycle regulators); CDC20 (regulator of the anaphase promoting complex); PTTG1 (regulatory protein of cell cycle progression); and MCM4 (involved in DNA replication). In order to confirm the role of H19X in regulating the cell cycle, we performed Hoechst 33342 staining and cell-cycle analysis by flow cytometry after H19X downregulation and TGF-β treatment. Consistent with our microarray results, H19X silencing led to a higher percentage of living fibroblasts in the S/G2 phase of the cell cycle, both at the basal level and after TGF-β stimulation (Figure 4C and Supplemental Figure 5A).
Figure 4. H19X silencing triggers fibroblast apoptosis.
H19X was silenced in dermal SSc fibroblasts with 25 nM ASO for a total of 120 hours and stimulated with TGF-β for the last 72 hours in 1% FBS medium. Gene ontology enrichment analysis performed on significantly upregulated genes (FDR <0.05 and absolute log2 FC >0.5) as revealed by microarray analysis (A). Heatmap showing significant upregulated genes that are relevant for cell cycle and proliferation as measured by microarray analysis (FDR < 0.05 and log2 FC >0.5) (B). H19X was silenced in SSc fibroblasts with 25 nM ASO for a total of 120 hours and stimulated with TGF-β for the last 48 hours in 1% FBS medium. Cells were stained with Hoechst 33342 and percentages of cells in G1 and G2/S, were acquired by flow cytometry. Dot plots are representative for n = 3 biological replicates (C). H19X was silenced in SSc fibroblasts with 25 nM ASO and in parallel stimulated with TGF-β. xCELLigence real-time measurement of cell proliferation. Measurement of the cell index normalized to the cell index value at the time of transfection (24 hours). Experiment was recorded for 180 hours after cell transfection. Representative normalized cell index curve of n = 5 biological replicates, variation is presented as SD of 4 technical replicates of the representative biological replicate (D). The slope of the cell index curve was calculated separately for the exponential growth phase and the late phase of the curve (E). Luminescence (for annexin V) as a measure of apoptosis was recorded every 6 hours starting 24 hours until 120 hours after cell transfection. Fold change was calculated respective to scrambled control. *indicates significance for untreated cells, +indicates significance for TGF-β–stimulated cells (F). Caspase-3/7 activity measured at 88 hours after cell transfection (f). Data are presented as single values and mean (n = 3–5). One-way ANOVA (C, E, and G). Two-way ANOVA (F). *P < 0.05, **P < 0.01, ***P < 0.001.
Indeed, H19X downregulation (Supplemental Figure 4B) led to changes in the proliferation of SSc fibroblasts as measured by xCELLigence real-time proliferation experiments. In the exponential phase, H19X-silenced fibroblasts showed a significant increase in cell proliferation compared with controls (P < 0.0001 basal condition and P = 0.0029 after TGF-β stimulation, Figure 4, D and E). In the late phase of the xCELLigence curve, starting on average at around 104 hours after transfection, a strong reduction in the cell index was observed for H19X-silenced cells with and without TGF-β stimulation (P = 0.0002 basal condition and P = 0.0078 in the presence of TGF-β, Figure 4, D and E). These data suggest that H19X silencing might lead to late induction of cell death. Therefore, we hypothesized that H19X could exhibit antiapoptotic effects on myofibroblasts, promoting their survival, which then leads to uncontrolled ECM deposition.
H19X promotes profibrotic effects of TGF-β by favoring myofibroblast survival.
Myofibroblasts are apoptosis-resistant cells, but they are in close proximity to the apoptotic threshold (10). As recently demonstrated, targeting apoptosis-resistant myofibroblasts might be an effective antifibrotic treatment strategy (10). Based on our results of the xCELLigence real-time proliferation experiments, we performed annexin V live measurements after H19X silencing with and without TGF-β stimulation using the same experimental design that we used for the xCELLigence analysis (Supplemental Figure 4B). Luminescence (for annexin V) and fluorescence (DNA dye) measurements were recorded between 24 and 126 hours after transfection of SSc fibroblasts. Already at 42 hours, we could record a significant difference in annexin V signals in H19X-silenced fibroblasts in comparison with scrambled transfected cells for both untreated and TGF-β–stimulated cells. This effect steadily increased until 72 hours (Figure 4F). Until about 36 hours after cell transfection, H19X-silenced cells were protected from necrosis. A delay in increased fluorescent signal was recorded starting at 54 hours after cell transfection for untreated cells and 66 hours for TGF-β–stimulated cells, marking the start of the necrotic process (Supplemental Figure 5B). Moreover, we also tested caspase-3/7 activity as a measure of activation of the apoptotic cascade after H19X silencing and parallel TGF-β stimulation. Indeed, 88 and 120 hours after cell transfection, a strong and consistent induction of caspase-3/7 activity was recorded after H19X silencing and TGF-β stimulation (P < 0.001, Figure 4G and Supplemental Figure 5C). In summary, these results suggest that H19X is a direct potent mediator of prolonged myofibroblast survival, potentiating TGF-β–driven ECM production and progressive tissue fibrosis. Inducing apoptosis of myofibroblasts by H19X silencing could be a therapeutic strategy to selectively remove ECM-overproducing cells from fibrotic tissues (10).
H19X has a human-specific function in fibrosis.
Next, we wanted to see whether H19X is conserved in mice and whether mouse models can be used to further characterize its function. A putative murine H19X transcript is annotated in the National Center for Biotechnology Information (Supplemental Figure 6A), and it was previously reported that the microRNAs associated with H19X (miR-503 and miR-424) have orthologous microRNAs in opossum, platypus, chicken, and xenopus (29). The genomic region spanning human and mouse transcripts has 60% homology. However, the homologous parts consist mainly of introns, and the annotated human and mouse transcripts profoundly differ. In addition, we analyzed the expression of the putative murine H19X (mH19X) in different well-established mouse models of fibrosis. These analyses showed that the putative mH19X was downregulated rather than upregulated in the lung tissues of mice with bleomycin-induced lung fibrosis, and its expression in skin did not differ between control mice and different fibrotic mouse models (bleomycin-induced skin fibrosis, tight skin mouse-1, injections of adenovirus overexpressing a constitutively active TGF-βR1, Supplemental Figure 6B). Moreover, the putative mH19X was not induced when murine dermal fibroblasts were stimulated with TGF-β for different time periods (Supplemental Figure 6C). These data suggest that the putative mH19X is differentially regulated and expressed in mouse fibrotic conditions and that mouse models cannot be used for functional analysis of the ortholog human gene.
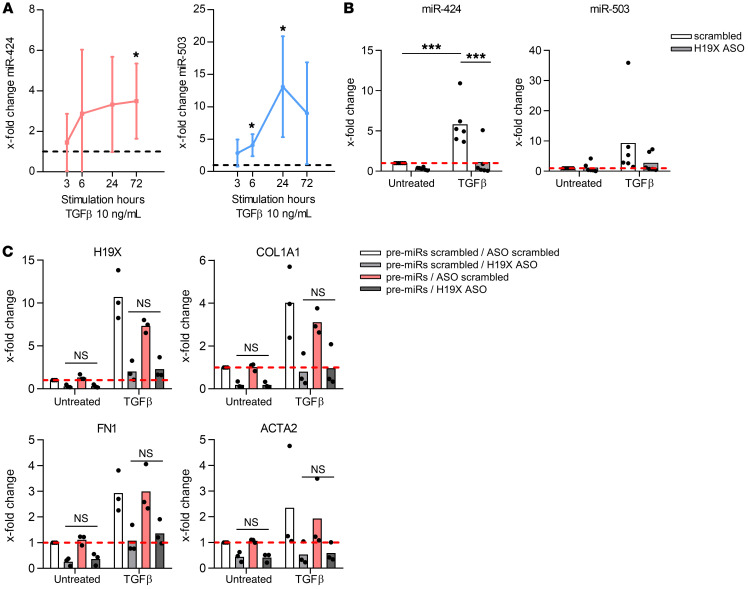
The profibrotic effects of H19X are largely independent from miR-424 and miR-503.
The H19X gene is localized on chromosome X, and the H19X gene locus encodes 2 microRNAs. The MIR424 sequence partially overlaps with the first exon of H19X, and MIR503 is embedded in the first intron (Supplemental Figure 7A). Therefore, we tested the hypothesis that H19X could exert its function, acting as a microRNA reservoir allowing a rapid release of the 2 microRNAs (30). Indeed, the expression of miR-424 and miR-503 was induced by TGF-β at 6 hours, 24 hours, and 72 hours (P < 0.05, Figure 5A). In contrast to H19X, the expression of these microRNAs was highest at 72 hours and 24 hours, respectively, and the early peak at 6 hours was not observed in SSc dermal fibroblasts. Moreover, the expression of miR-424 and miR-503 was reduced after H19X silencing (Figure 5B). These data argue that H19X could potentially function as a reservoir for these 2 microRNAs.
Figure 5. miR-424 and miR-503 are also induced by TGF-β, but do not mediate the profibrotic function of H19X.
MiR-424 and miR-503 time-dependent response to TGF-β stimulation (A) and expression after H19X silencing with 25 nM of ASO (B). Expression of miR-424 and miR-503 was analyzed by qPCR and normalized to RNU48. SSc fibroblasts were transfected with 25 nM of ASO and 100 nM of pre-miR for a total of 120 hours and stimulated with TGF-β for the last 72 hours in 1% FBS medium (C). Expression of H19X, COL1A, FN1, and ACTA2 was analyzed by qPCR and normalized to GAPDH and RPLP0. Fold change was calculated respective to untreated control set as 1 (dashed line). Data are presented as single values and mean (n = 3–6). One-way ANOVA (A–C). *P < 0.05, ***P < 0.001.
To test this hypothesis, we first silenced miR-424 (Supplemental Figure 7B) and miR-503 (Supplemental Figure 7C) in SSc fibroblasts. In contrast to the strong downregulation of COL1A1, FN1, and ACTA2 seen with H19X silencing (Supplemental Figure 4C), we could detect only a minor downregulation of FN1 after the silencing of miR-424, but not miR-503 (Supplemental Figure 7, B and C). The expression of COL1A1 and ACTA2 was not affected by the silencing of these 2 microRNAs. In the next step, we simultaneously downregulated H19X and overexpressed the 2 microRNAs to explore whether the co-upregulation of miR-424 and miR-503 could rescue the H19X-induced phenotype in H19X-silenced fibroblasts. However, the simultaneous overexpression of miR-503 and miR-424 could not restore the reduced expression of COL1A1, FN1, and ACTA2 induced by H19X silencing (Figure 5C). These results were also confirmed by the simultaneous overexpression of miR-424 and miR-503 without H19X silencing (Supplemental Figure 8). These data suggest that minor profibrotic effects of H19X might be partially due to the microRNA reservoir function of H19X as observed for FN1 expression; however, the microRNA reservoir function of H19X cannot explain the strong and widespread antifibrotic effects observed after H19X silencing. This suggests that H19X exerts most of its functions through a microRNA reservoir–independent mechanism.
H19X is a nuclear transacting lncRNA that binds to noncoding DNA regulatory elements across distinct chromosomes.
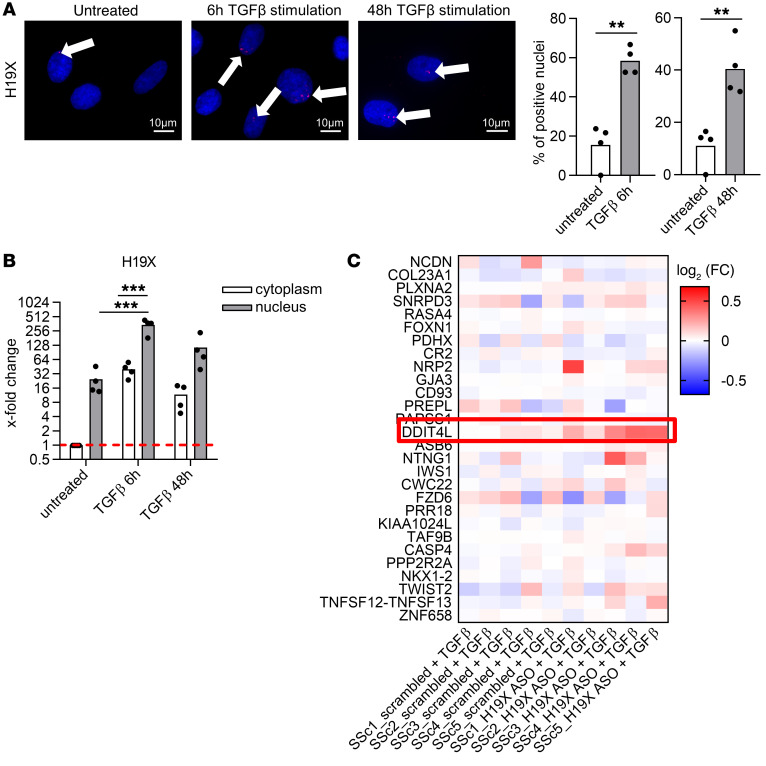
The cellular localization can give a first indication about the lncRNA mechanism of action. Therefore, we conducted H19X FISH staining of unstimulated and stimulated (TGF-β for 6 hours and 48 hours) SSc fibroblasts. We detected increased numbers of tightly localized, bright nuclear foci in TGF-β–treated cells (P = 0.0093 at 6 hours of TGF-β and P = 0.0049 at 48 hours of TGF-β, Figure 6A). This pattern resembled the type of FISH staining reported for X-inactive specific transcript (XIST), a well-characterized lncRNA, which is a part of the X chromosome inactivation center (31). The specificity of the results was confirmed by H19X silencing and by using GAPDH FISH staining as validation of the method (Supplemental Figure 9, A and B). The nuclear localization of H19X was further confirmed by cell fractionation, where nuclear H19X expression was highest after 6 hours of treatment with TGF-β (P < 0.0001, Figure 6B). XIST expression was used to confirm successful cell fractionation (Supplemental Figure 9C).
Figure 6. H19X is a nuclear lncRNA that interacts with the DNA.
SSc fibroblasts were stimulated for 6 or 48 hours with TGF-β in 1% FBS medium, and FISH staining was performed targeting H19X. Scale bar: 10 μm. Pictures are representative of n = 4 biological replicates. Percentage of nuclei positive for H19X staining (A). Cell fractions were obtained from SSc fibroblasts stimulated with TGF-β for 6 or 48 hours. H19X expression was analyzed by qPCR. Fold change was calculated respective to untreated control (B). Data are presented as single values and mean (n = 4). t test (A). One-way ANOVA (B). **P < 0.01, ***P < 0.001. Heatmap showing expression of the gene with the closest transcription start site to the putative sites of H19X interaction with the genome as measured by microarray analysis after H19X silencing; DDIT4L is highlighted in red (C).
Given the nuclear expression of H19X, we hypothesized that H19X regulates the profibrotic responses of fibroblasts by controlling gene expression at the transcriptional level. The pattern of H19X nuclear foci (Figure 6A) might indicate that H19X acts at distant genomic sites as previously described for another X-linked lncRNA (FIRRE) (32) or that it regulates the genes in its immediate genomic neighborhood, spreading over the locus from the site of its production, as observed for XIST (31). As revealed by microarray analysis of gene expression, none of the neighboring genes spanning 1 Mb from the H19X locus was affected by H19X silencing (Supplemental Figure 10A and Supplemental Table 4). This suggests that H19X might exert its profibrotic function at distant genomic regions, involving sites across different chromosomes.
To identify genomic regions that directly interact with the H19X transcript, we performed chromatin isolation by RNA purification and sequencing (ChIRP-Seq) in the fibroblast cell line BJ5TA after 6 hours of TGF-β stimulation. ChIRP-Seq is an affinity capture assay that allows the identification of genomic binding sites for a specific lncRNA. Telomerase RNA component probes were used to validate the method. LacZ probes were used as the negative control as previously described (33). After probe hybridization and pull down, RNA retrieval was assessed. On average, 15.89% and 16.77% of H19X RNA was retrieved with the even probe set and odd probe set respectively (Supplemental Figure 10B). Additionally, a specific peak at the H19X transcription start site (TSS) was identified, supporting the validity of the method (Supplemental Figure 10C). Based on ChIRP-Seq, we identified 71 peaks that were common for H19X, even and odd, corresponding to genomic regions that physically interacted with the H19X transcript (Supplemental Table 5). H19X ChIRP peaks were spread across several chromosomes, suggesting that H19X acts as a transacting lncRNA and exerts its gene regulatory function by modulating distant DNA regulatory elements.
H19X regulates DDIT4L expression via 3D genomic interactions and chromatin rearrangements.
If genes interacting with the 71 identified H19X-bound genomic regions are regulated by H19X, their transcription should be changed after H19X silencing (Supplemental Figure 4D). To address this, we first identified genes that were potentially regulated via H19X-DNA interaction by annotating the 71 regions with the closest downstream TSS. Then, we looked for gene expression changes of the aforementioned genes in our microarray data set, where we identified 28 genes with TSS potentially interacting with one of the H19X-bound genomic regions (Figure 6C). Among these genes, DNA damage–inducible transcript 4–like protein (DDIT4L, Figure 7A) had a TSS about 190 kb downstream of the H19X interaction site, and at the same time displayed increased expression after H19X silencing in all of the 5 samples paired that were analyzed. Given that the physical interaction of H19X could also influence other genes in the same genomic region as DDIT4L, we checked the expression of DDIT4L-neighboring genes (1 Mb up and downstream) in the microarray data from the H19X silencing experiment (Supplemental Figure 4D). However, none of the neighboring genes of DDIT4L were changed by H19X silencing, suggesting a specific regulation of H19X with DDIT4L (Supplemental Figure 10D). Taken together, these results identified DDIT4L as a candidate factor mediating the profibrotic effects of TGF-β–induced H19X.
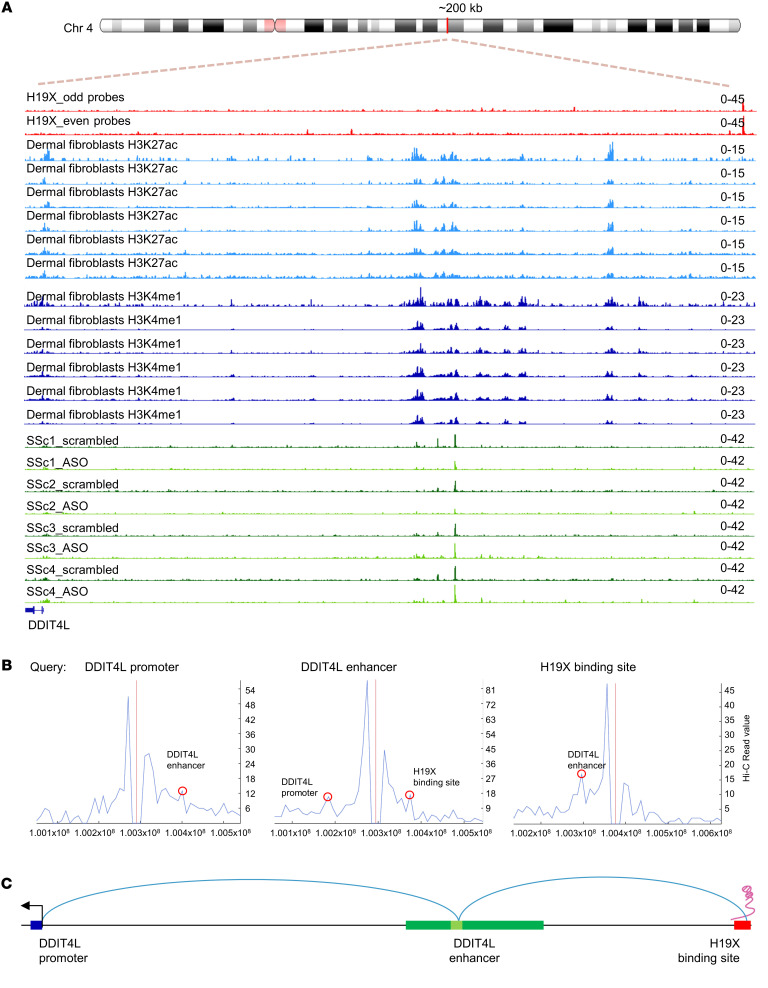
Figure 7. H19X targets DDIT4L via 3D genomic interaction and chromatin rearrangements.
BJ5TA cells were stimulated with TGF-β for 6 hours in 1% FBS medium. ChIRP peaks for H19X capture are shown as fold enrichment relative to input for 2 sets of probes (even, odd, red tracks). ChIRP tracks for H3K27ac (light blue) and H3K4me1 (dark blue) were obtained from publicly available data sets (57). ATAC-Seq tracks showing variations in chromatin accessibility after H19X silencing. H19X was silenced in dermal SSc fibroblasts with 25 nM ASO (light green) and compared with SSc fibroblasts transfected with scrambled control (dark green) (A). Hi-C data (58) obtained from publicly available data sets revealed interactions between H19X binding site, the active enhancer, and DDIT4L promoter. Query regions: DDIT4L promoter, DDIT4L enhancer (chr4:100294478), H19X binding site (chr4:100375842). Peak signals indicate interaction events and are highlighted by red circles (B). Schematic representation of DDIT4L gene (blue) and its regulatory elements: DDIT4L enhancer (green, in light green it is highlighted where H19X silencing caused variation in chromatin accessibility) and H19X binding site (red) (C).
In order to define how the interaction of H19X with the genome is influencing DDIT4L expression, we searched publicly available data sets in the WashU Epigenome Browser (34). We aimed to identify DNA regulatory elements that are in proximity with the H19X site of interaction and that might regulate DDIT4L expression. Specifically, we looked for active enhancers in primary dermal fibroblasts distal to the DDIT4L gene as defined by the presence of histone marks: histone 3 lysine 27 acetylation (H3K27ac) and histone 3 lysine 4 monomethylation (H3K4me1). With this strategy, we identified an active enhancer 95 kb upstream of the DDIT4L TSS and 56 kb downstream of the H19X binding site (Figure 7A). The same enhancer is also annotated in Ensembl (35) for the fibroblast cell line IMR90 and adult normal human dermal fibroblasts (NHDF-AD, Supplemental Figure 11). GeneHancer (36) is another database that predicts interactions between enhancers and genes, integrating data derived from different techniques such as capture Hi-C, promoter-specific genome conformation assay, and expression quantitative trait loci (eQTLs). Data for the DDIT4L promoter derived from GeneHancer predicted a likelihood interaction with the enhancer we had identified on the bases of eQTLs (37) (P = 4.6 × 10–7). This analysis supports the hypothesis that this enhancer directly regulates the expression of DDIT4L (Supplemental Figure 11).
Then, we searched for additional evidence for a role of H19X in determining the chromatin conformation of this particular enhancer by performing the assay for transposase-accessible chromatin using sequencing (ATAC-Seq) after H19X knockdown and TGF-β stimulation in SSc fibroblasts. With this technology, we observed changes in chromatin rearrangements within the DDIT4L enhancer region (chr4:100294478-100294699), indicating that the open chromatin of the enhancer closes upon H19X silencing (Figure 7A).
Therefore, we hypothesized that H19X association with the genome could influence the chromatin conformation of the enhancer, which in turn regulates the expression of DDIT4L. For this purpose, we searched publicly available data sets to identify 3D interactions using 3D Genome Browser (38) in the fibroblast cell line IMR90 with a resolution of 10 kb. Hi-C data visualized in 3D Genome Browser revealed interactions between the H19X binding site, the active enhancer, and the DDIT4L promoter. When using the DDIT4L promoter region as a query, a peak signal in the proposed DDIT4L enhancer region indicated a chromatin interaction event, further supporting the role of the enhancer in regulating DDIT4L expression. Notably, when using the enhancer region (chr4:100294478) as a query, a peak signal both in the DDIT4L promoter and at the site of H19X binding indicated a chromatin interaction event. Consistently, when using the H19X binding site (chr4:100375842) as a query, a peak signal in the enhancer region indicated a chromatin interaction event (Figure 7B). All together, these data indicate that H19X might be crucial to sustain the chromatin conformation necessary for the regulation of DDIT4L expression, promoting 3D genomic interactions between the active enhancer and DDIT4L promoter (Figure 7C).
DDIT4L mediates the collagen-inducing effects of H19X.
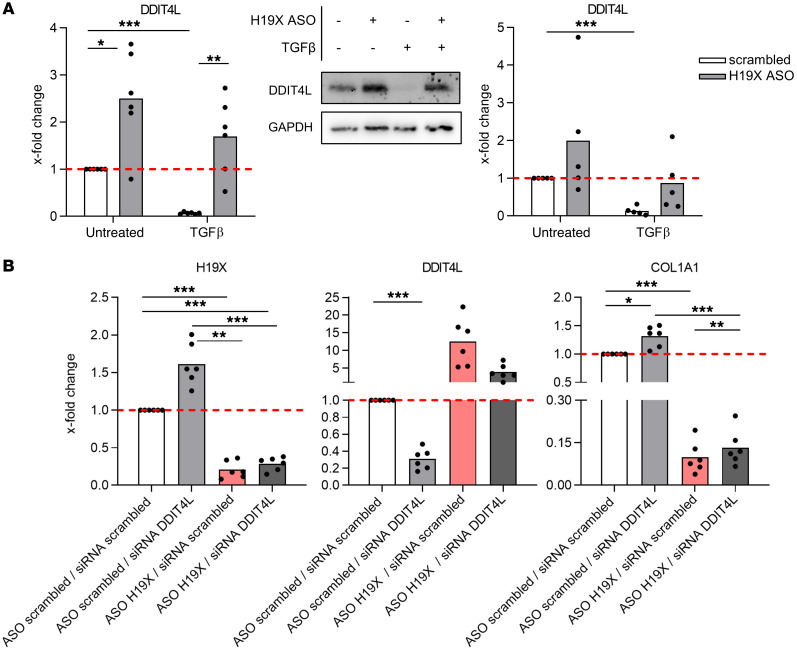
To further prove a functional role of DDIT4L, we next investigated whether its expression is regulated by TGF-β. DDIT4L expression was consistently repressed by TGF-β stimulation at the mRNA and protein level as confirmed by qPCR (P < 0.0001) and Western blot (Figure 8A), respectively. Moreover, H19X silencing resulted in DDIT4L induction as measured by qPCR (P = 0.0159 basal condition and P = 0.0085 in the presence of TGF-β, Figure 8A and Supplemental Figure 12A) and Western blot (Figure 8A), confirming the microarray results. Furthermore, DDIT4L silencing (Supplemental Figure 12B) increased the expression of COL1A1 that was reduced by H19X downregulation (Figure 8B and Supplemental Figure 12C), whereas it did not change the expression of the cell cycle genes Cyclin A2 (CCNA2) and Cyclin D1(CCND1) (Supplemental Figure 12D). These data showed that TGF-β–induced H19X mediated its profibrotic effects by disabling the transcription of DDIT4L, which acts as an inhibitor of the TGF-β pathway.
Figure 8. DDIT4L is a repressor of TGF-β–induced collagen production.
H19X was silenced in dermal SSc fibroblasts with 25 nM ASO for a total of 120 hours and stimulated with TGF-β for the last 72 hours in 1% FBS medium. The mRNA and protein levels of DDIT4L were analyzed by qPCR and Western blot respectively. Pictures are representative of n = 5 biological replicates (A). SSc fibroblasts were transfected with 25 nM of ASO and 50 nM of siRNA for 120 hours and stimulated with TGF-β for the last 72 hours in 1% FBS medium. Expression of DDIT4L, H19X, and COL1A1 was analyzed by qPCR (untreated samples) (B). qPCR data were normalized to GAPDH and RPLP0. Data are presented as single values and mean (n = 5–6). Fold change was calculated respective to untreated control set as 1 (dashed line). One-way ANOVA (A and B). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this study, we have identified lncRNA H19X as a key mediator of the TGF-β–induced effects on matrix remodeling in fibroblasts and related cell types. Matrix remodeling promoted by TGF-β is of paramount importance in physiology (e.g., wound healing) (39, 40) and fibrotic diseases (11, 41, 42). Targeting TGF-β has thus been tested as a therapeutic strategy in fibrotic diseases, but this approach was limited by its overall toxicity and the lack of appropriate drugs (43, 44). Thus, the detection of key components of TGF-β–induced fibrosis that could be therapeutically targeted has a high translational potential for the large group of fibrotic diseases that display a high unmet clinical need.
LncRNAs represent a class of molecules that can provide novel insights into the mechanistic aspects of regulation of gene expression (45). In comparison with other well-characterized lncRNAs, very little is known about the function and mechanism of H19X. H19X has been found to be downregulated in cancer. In vitro studies demonstrated that in tumor cells, H19X overexpression inhibited cell proliferation, cell invasion, and migration (46–48). H19X was also reported to be induced by hypoxia to promote angiogenesis (49). No other reports about H19X are published to date.
We propose a profibrotic mechanism according to which the presence of H19X is required for the remodeling effects of TGF-β, including ECM production and contraction, as well as myofibroblast differentiation and survival. Targeting of H19X strongly inhibited these TGF-β effects, putting H19X as a central mediator of tissue remodeling. Moreover, we demonstrated that H19X is strongly induced by TGF-β in fibroblasts and related cell types. In our experimental settings, we used SSc as a paradigm disease in order to elucidate its role in TGF-β–induced ECM remodeling and myofibroblast activation. Nevertheless, we showed that H19X was deregulated in a wide variety of tissues and fibrotic diseases, such as skin fibrosis, lung fibrosis, fibrotic ileum of patients with Crohn’s disease, the liver of patients with PSC, and tissue undergoing physiological wound healing. This suggests that targeting the H19X pathway might be beneficial in preventing fibrosis across a number of fibrotic diseases and states.
Additionally, we have identified DDIT4L as an effector of the H19X-driven effects on collagen production following a robust and stringent strategy. First, we explored the possibility that H19X exerts its function by working as a reservoir for miR-503 and miR-424. However, we could observe only minor effects after microRNA gain- and loss-of-function studies that could not explain the strong effects of H19X on ECM remodeling. LncRNA transcription can directly affect the expression of neighboring genes. In our experimental settings, the loss of profibrotic features observed after H19X silencing was not mediated by H19X neighboring genes. However, when combining publicly available histone mark data sets, ATAC-Seq experiments, and publicly available data sets for 3D interactions, we determined that H19X was necessary to maintain the open chromatin of a DDIT4L enhancer, most likely via 3D interactions.
Publications on DDIT4L are limited. Some studies have pointed to a role in cell growth by regulating the mTOR signaling pathway (50, 51). DDIT4L was also identified as potential predictor of radiation-induced fibrosis (52). Here, we demonstrated that under physiological, unstimulated conditions, the presence of DDIT4L inhibited ECM production. With activation of the TGF-β pathway, the transcription of H19X expression was induced, leading to a direct physical association of H19X RNA with a DNA regulatory element upstream to the DDIT4L gene that interacts with the DDIT4L locus, thereby suppressing DDIT4L transcription. As a result, the inhibition of the TGF-β pathway by DDIT4L was blocked, leading to TGF-β pathway activation and increased collagen production (Supplemental Figure 13). These results do not exclude the possibility that mechanisms other than via DDIT4L are contributing to the profibrotic effects of H19X.
A limitation of our study is the inapplicability of our in vitro and ex vivo human findings to an in vivo mouse model. H19X is poorly conserved in mice. We also showed that the putative mH19X regulation differed from the human ortholog and that murine fibrotic models cannot be used for translational purposes. Considering the broad biological effects of TGF-β on tissue remodeling and the key role of H19X in this process, as shown in this paper, it is also very likely that factors in addition to DDIT4L are involved in mediating its effects. For example, in our experiments, we could not find evidence that DDIT4L is responsible for the effects of H19X on the cell cycle. In this regard, appropriate annotation of the other 70 genomic regions might help to identify such additional factors. Genetic deletion of this region followed by gene expression studies could address this issue and provide a broader overview of the molecular mechanisms of H19X.
In summary, we have identified a lncRNA H19X as being consistently upregulated across different types of TGF-β–driven fibrotic diseases. Moreover, H19X was found highly upregulated in skin tissue undergoing wound healing, providing additional evidence that H19X expression might be related to a physiological and pathological mutual mechanism. TGF-β is the master regulator of both wound healing and fibrosis. H19X downregulation followed by microarray analysis and functional assays revealed that the induction of H19X was a conditio sine qua non for TGF-β–induced profibrotic effects. At last, we have identified DDIT4L as an effector of the H19X response and as a repressor of TGF-β–induced collagen synthesis. We also described a molecular mechanism whereby H19X modifies the accessibility of an enhancer distal to the DDIT4L gene, altering DDIT4L expression, which sheds light on the players in TGF-β–induced profibrotic effects.
Methods
Additional methods are detailed in the Supplemental Methods.
Patients.
Skin biopsies were obtained from the forearm of patients with SSc and HC donors. All patients fulfilled the American College of Rheumatology/European League Against Rheumatism (ACR/EULAR) 2013 classification criteria (53). Lung samples were obtained from patients with SSc-ILD and IPF undergoing lung transplantation. Patients were fulfilling the respective classification and diagnostic criteria. Normal lung tissues were obtained from organ donors whose lungs were not used for lung transplantation, as previously described (54). Small intestine samples were taken from fibrotic (stenosis) and nonfibrotic tissue (resection margin) of the terminal ileum of patients with Crohn’s disease fulfilling the Montreal classification criteria (55). Site-matched healthy colon was obtained from adenocarcinoma patients undergoing right-sided hemicolectomy. Liver samples were taken from healthy donors and from explanted livers from patients undergoing liver transplantation. Tissue from normal skin wounds was obtained from volunteers by excisional biopsies, and HC skin was taken from surgeries of patients with noninflammatory, nononcological diseases.
RNA-Seq and reverse transcription qPCR from tissue.
Skin tissue was homogenized and samples processed for RNA extraction with the QIAGEN RNeasy Fibrous Tissue kit according to the manufacturer’s protocol. Skin biopsies from 3 different cohorts were used for RNA-Seq analysis (cohort 1, PRESS and cohort 3). Poly (A) enrichment for cohort 1 or ribosomal depletion for PRESS and cohort 3 were carried out, respectively. After retrotranscription, cDNA was used to generate Illumina sequencing libraries according to the manufacturer’s protocol. Libraries were then sequenced on a HiSeq 2500. Alternatively, RNA extracted from tissue was retrotranscribed with the Transcriptor First Strand cDNA Synthesis kit (Roche), and qPCRs were performed with 2x SYBR Green Master Mix (Promega) on an Agilent Technologies Stratagene Mx3005P qPCR system.
Primary dermal fibroblast culture.
Primary dermal fibroblasts from HCs and patients with SSc were derived by outgrowth from 3 mm punch biopsy or skin resected from donors undergoing surgery. Fibroblasts were cultured in DMEM (MilliporeSigma) containing 50 U/mL penicillin and 50 μg/mL streptomycin (Gibco) and 10% FBS (Gibco) and 100 μM 2-mercaptoethanol (Gibco). Fibroblasts from passages 4–10 in monolayer culture were used for the experiments. Cells were maintained in a 5% CO2 humid 37°C incubator.
RNA interference (RNAi).
Dermal SSc fibroblasts were transfected with 25 nM ASO (Antisense LNA GapmeRs, QIAGEN), 50 nM FlexiTube siRNA (QIAGEN), 100 nM Ambion Pre-miR miRNA Precursor or 100 nM Ambion Anti-miR (Thermo Fisher Scientific) using Lipofectamine 2000 (Thermo Fisher Scientific) at a final concentration of 1.67 μL/mL. Cells were incubated with the transfection mix for 6 hours, then washed with PBS. Eventually, fresh complete medium was added to the culture. Custom-designed ASO sequences are provided in Supplemental Table 6.
Microarray and qPCR from cells.
Total RNA from cells was isolated with Zymo Quick-RNA MicroPrep RNA isolation kit. RNA was amplified and purified using a TotalPrep RNA Amplification Kit (Applied Biosystems/Ambion) and reverse transcription was performed using a T7 Oligo(dT) primer to synthesize cDNA containing a T7 promoter sequence. In vitro transcription was used to amplify and label multiple copies of biotinylated cRNA. The amplified cRNA was hybridized on Illumina HT-12 arrays. Alternatively, RNA and random hexamers were used to carry out reverse transcription using the Transcriptor First Strand cDNA Synthesis kit (Roche). Subsequent qPCRs were performed with 2x SYBR green master mix (Promega) on an Agilent Technologies Stratagene Mx3005P qPCR system.
Western blot.
Cells were lysed with RIPA buffer (MilliporeSigma) supplemented with phosphatase inhibitors (PhosphoStop, Roche) and the protease inhibitor cocktail (cOmplete ULTRA Tablets, Roche). Protein concentration was measured by the colorimetric BCA method according to the manufacturer’s protocol (Thermo Fisher Scientific). Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane overnight at 4°C. Membranes were blocked for 1 hour at room temperature in Tris-buffered saline and Tween-20 (Thermo Fisher Scientific) (TBST) containing 5% milk and probed overnight with the following antibodies: Fibronectin (1:1000, ab2413, Abcam), αSMA (1:1000, A2547, MilliporeSigma), SMAD3 (1:5000, ab40854, Abcam), SMAD4 (1:5000, ab40759, Abcam), TGF-βR2 (1:250, 701683, Thermo Fisher Scientific), DDIT4L (1:1000, AM26767PU-N, Origene), and GAPDH as loading control (1:10,000, no. 2118, Cell Signaling Technology). HRP-conjugated secondary antibodies were used for detection with ECL substrate (SuperSignal West Pico Plus, Thermo Fisher Scientific). ImageJ software was used to semiquantify the signal.
Secreted collagen analysis.
SSc dermal fibroblasts were transfected with ASO as previously described, starved for 24 hours (1% FBS medium), and treated with TGF-β in starvation medium for 72 hours. Pan-collagens or procollagen1α1 were measured in the supernatants with Sircol Assay (Biocolor Life Science Assays) or ELISA (R&D Systems) respectively per the manufacturer’s instructions. Absorbance was recorded at 555 nm for Sircol and 450 nm ELISA in Synergy HT (BioTek) plate reader. For both assays, samples were measured in triplicates. Mean absorbance was calculated and concentrations/total amount were determined using the respective standard curves.
Cell contraction assay.
ASO transfected cells were harvested and counted. After 24 hours in starvation medium, 1 × 105 cells for each condition were seeded in a collagen gel matrix in a 48-well plate at the time of TGF-β stimulation (Cell Contraction Assay, Cell Biolabs). Pictures of the contracted collagen gels were acquired 72 hours after TGF-β stimulation. ImageJ software was used to measure the gel areas. Data are reported as a percentage of gel contraction.
Immunofluorescence and phalloidin staining.
Cells were seeded in 8-chamber glass slides (Lab-Tec). Cells were transfected with 25 nM ASO 24 hours after seeding. After 24 hours of starvation and an additional 72 hours of TGF-β stimulation, cells were fixed in ice-cold methanol-acetone (7:3, both MilliporeSigma) for 10 minutes at –20°C, washed 3 times with PBS, blocked with PBS/10% FBS for 20 minutes at room temperature, and then incubated with primary anti-αSMA antibody (1:100, A2547, MilliporeSigma) for 1 hour. After 3 washes in PBS, cells were stained with the secondary antibody for 45 minutes at room temperature. For staining of stress fibers, cells were fixed in 4% paraformaldehyde (PFA) (MilliporeSigma) for 5 minutes, washed 3 times in PBS, permeabilized in PBS 0.1% Triton X-100 (MilliporeSigma), and washed again 3 times in PBS. Finally, cells were stained with 50 μg/mL of fluorescent-labeled phalloidin (MilliporeSigma) for 40 minutes at room temperature. Nuclei were counterstained with DAPI solution (1 μg/mL, Roche). Images were acquired with an Olympus BX53 microscope equipped with a DP80 camera.
Hoechst 33342 staining and flow cytometry.
Fibroblasts were transfected with 25 nM ASO as previously described for 72 hours and stimulated with TGF-β for the last 48 hours. Next, cells were harvested, washed with PBS, and stained with Hoechst 33342 at a final concentration of 5 μM for 30 minutes at 37°C. Single-cell suspensions were analyzed by flow cytometry using BD FACS Aria III. Data were analyzed with FlowJo software, version 8. The gating strategy is presented in Supplemental Figure 4A. Propidium iodide was used to exclude dead cells. Hoechst 33342 was used to assess DNA content and the percentage of cells in the different cell cycle phases G1 and S/G2.
Real-time proliferation assay.
Cellular impedance was used to assess proliferation with the xCELLigence RTCA DP instrument (ACEA Biosciences). Briefly, cells were seeded in E-Plate VIEW 16 PET (ACEA Biosciences) and transfected after 24 hours with H19X ASO or control. Fibroblasts were simultaneously treated with 10 ng/mL TGF-β. Measurements were recorded every half an hour up to 180 hours after cell transfection. RTCA software 2.0 (ACEA Biosciences) was used to analyze the data. Data were normalized at the time of transfection. The curve slope, as quantification of cell proliferation, was calculated separately for the exponential growth phase and the late phase specifically for each patient. Every condition was tested in quadruplicates.
Apoptosis assays.
Cell were seeded in a solid white 96-well plate (Nunclon Delta Surface, Thermo Fisher Scientific) transfected and stimulated using the same experimental setup described for real-time proliferation experiments. RealTime-Glo Annexin V Apoptosis and Necrosis Assay (Promega) was used to analyze apoptosis and necrosis as per the manufacturer’s instructions 24 hours after transfection. Luminescence was used to assess the exposure of phosphatidylserine on the external surface of the cell membrane during the apoptotic process. Necrosis was detected with a fluorescent DNA dye. Both signals were measured in a Synergy HT (Biotek) plate reader up to 120 hours after cell transfection. All conditions were tested in quadruplicates. The Caspase-Glo 3/7 Assay (Promega) was used as an orthogonal method to assess caspase-3 and caspase-7 enzymatic activity at 88 and 120 hours after transfection as described by the manufacturer’s protocol.
Fluorescent in situ hybridization (FISH).
Fifty-six Stellaris RNA FISH probes against H19X transcript were designed using Stellaris Probe Designer tool version 4.2 (Biosearch technologies, Supplemental Table 8). Every probe was labeled with CAL Fluor Red 610 reporter dye. Cells were grown on 18 mm round cover glasses in 12-well plates, fixed in 4% PFA, washed twice with PBS, and permeabilized in 70% ethanol for 1 hour at 4°C. Hybridization was carried out in 150 mm tissue culture plates; bottom lined with a water-saturated paper towel and a single foil of Parafilm (MilliporeSigma) placed on top of the paper towel. Hybridization buffer containing 1 μL probe stock solution was dispensed on the Parafilm and the cover glass was flipped, cell side down, on the hybridization buffer. Cells were incubated in the dark overnight at 37°C. After washing, cell nuclei were counterstained with DAPI solution (1 μg/mL). Cover glasses were then washed one more time and mounted on slides. Images were acquired with an Olympus BX53 microscope equipped with a DP80 camera. DesignReady Stellaris RNA FISH Probes, direct against human GAPDH labeled with CAL Fluor Red 610 dye (VSMF-2149-5, Biosearch Technologies) were used as positive control. Cells incubated with hybridization buffer without probes were used to assess background signal negative control.
Cell fractionation.
Cells were detached, washed with PBS, and centrifuged at 500 g at 4°C for 5 minutes. Cell pellets were resuspended in hypotonic lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.3% NP-40, and 10% glycerol) supplemented with RNase inhibitor (SUPERase-In, Thermo Fisher Scientific) and incubated in ice for 20 minutes. Next, cells were centrifuged at 1000 g at 4°C for 3 minutes. Cytoplasmic supernatant was taken and RNA precipitated in ethanol and 150 mM sodium acetate at –20°C for at least 1 hour. Nuclear pellets were washed with the hypotonic lysis buffer and centrifuged at 200 g at 4°C for 2 minutes. Precipitated cytoplasmic RNA was pelleted, washed in ice-cold 70% ethanol, and centrifuged again at 17,000 g at 4°C for 5 minutes. Finally, Trizol was added to all the fractions and RNA was extracted.
ChIRP.
ChIRP experiments were carried out as previously described (56). BJ5TA cells were starved for 24 hours and stimulated with TGF-β for 6 hours. Next, 20 million cells per ChIRP reaction were harvested, washed with PBS, and cross-linked at room temperature with 1% glutaraldehyde (glutaraldehyde solution, MilliporeSigma) for 10 minutes on a shaker. Fixation was quenched with one-tenth volume of 1.25 M glycine (Carl Roth) at room temperature for 5 minutes. Cells were washed once with ice-cold PBS and resuspended in 10 times the mass of the pellet of lysis buffer (50 mM Tris-HCl pH 7, 10 mM EDTA, 1% SDS) supplemented with protease inhibitor cocktail (cOmplete ULTRA Tablets, Roche) and RNase inhibitor (SUPERase-In, Thermo Fisher Scientific). Lysates were sonicated in Bioruptor Pico (Diagenode) with the following settings: 30 seconds on and 30 seconds off, for a total of 1 hour and 40 minutes. DNA shearing between 100 and 500 pb was checked on 1% agarose gel after reverse cross-linking with 100 μg protease K (Thermo Fisher Scientific) of 10 μL lysate for 45 minutes at 50°C and DNA fragment purification using DNA Clean & Concentrator (Zymo Research). Then, samples were centrifuged at 16,100 g at 4°C for 10 minutes. Ten microliters for INPUT were saved for later use. Two milliliters of hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-HCl pH 7, 1 mM EDTA, 15% formamide) supplemented with protease inhibitor cocktail and RNase inhibitor and 100 pmol of biotinylated probes were added to 1 mL of chromatin per ChIRP reaction. Hybridization was carried out overnight at 37°C with shaking. One hundred microliters of prewashed streptavidin magnetic beads (Dynabeads Streptavidin C1, Thermo Fisher Scientific) were added to the hybridization reaction and incubated at 37°C for 45 minutes with shaking. Beads were then washed 5 times with washing buffer (2x SSC, 0.5% SDS) using a magnetic stand. RNA extraction was performed on a small fraction in order to assess RNA retrieval. DNA was isolated by resuspending beads in 150 μL elution buffer (50 mM NaHCO3, 1% SDS) supplemented with 15 μg RNase A and 15 U of RNase H and incubated at 37°C for 30 minutes with shaking. Supernatant was separated using a magnetic stand. Similarly, DNA was also extracted from the INPUT sample. DNA was treated with protease K (Thermo Fisher Scientific) for 45 minutes at 50°C. Three hundred microliters of phenol/chloroform/isoamyl alcohol were then added and the samples spun down at 16,100 g for 5 minutes at 4°C. The aqueous phase was collected. DNA was precipitated overnight at –20°C with 30 μL NaOAc and 900 μL 100% EtOH, spun down at 16,100 g for 30 minutes at 4°C, and then washed with 1 mL of 70% EtOH. Eventually, the DNA pellets were air dried, and resuspended in 20 μL of 10 mM Tris-Cl, pH 8.5. DNA samples were than processed for library preparation following the Illumina protocol. Biotinylated probes used for ChIRP targeting H19X are listed in Supplemental Table 8. Commercially available biotinylated probes against LacZ were used as negative control (Magna ChIRP Negative Control Probe Set, Merck).
ATAC-Seq.
ATAC-Seq libraries were prepared from 50,000 transfected and TGF-β–stimulated fibroblasts. Cells were harvested, washed in PBS, and resuspended in lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40). Next, cells were centrifuged at 500 g at 4°C for 10 minutes. Transposition reaction was performed on the nuclei preparation with the NexteraTn5 Transposase (Nextera kit, Illumina) and incubated at 37°C for 30 minutes. Transposed DNA was purified with the MinElute PCR Purification Kit (QIAGEN) and amplified by PCR. The sequencing was performed on a NextSeq 500 platform.
Data availability.
PRESS RNA-Seq data accession code: GSE130955. Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130955 Enter token cfmxwywifxgrvot into the box. RNA sequencing files for cohort 3 are deposited at the European Genome-phenome Archive database under EGAO00000000316, https://www.ebi.ac.uk/ega/organisations/EGAO00000000316 RNA microarray data accession code: GSE139334. Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139334 Enter token wrmlkwgstfgrvgv into the box. ChIRP-Seq and ATAC-Seq accession code: PRJEB34589. To review ENA accession PRJEB34589, go to https://www.ebi.ac.uk/ena/data/view/PRJEB34589
Statistics.
Median and nonparametric tests were used for assessing statistical significance when the population could not be assumed to be normally distributed after Shapiro-Wilk and Kolmogorov-Smirnov normality tests. Normally distributed data were presented as mean and single values and a paired, 2-tailed parametric t test was used. When more than 2 groups were compared, a 1-way or 2-way ANOVA test was performed depending on the number of analyzed variables. The correlation coefficient between H19X expression and cell type gene expression signature and TGF-β pathway effectors was calculated using Pearson’s correlation. P values of less than 0.05 were considered significant; n refers to the number of biological replicates.
Study approval.
Approval for the use of human biological samples was obtained from the local ethics committee of the Canton of Zurich, Switzerland. All patients and controls signed written informed consent forms. All mice were bred under specific pathogen–free conditions, and all studies were approved by the local ethics committee of the veterinary office of the Canton of Zurich.
Author contributions
OD directed the project and obtained funding. OD and EP designed, analyzed, and interpreted experiments. OD, EP, and MFB wrote the manuscript. EP performed the experiments with the help of MS, FR, AW, and PB. SA and GS performed, analyzed, and helped with the interpretation of the RNA-Seq and microarray data. RL, FK, JDVB, TM, CFB, GR, WTVH, GD, and FO participated in acquisition and analysis of the data. JS, BM, and JD performed animal experiments. GK and MFB participated in the design, analysis, and interpretation of the experiments.
Supplementary Material
Acknowledgments
We thank PRESS principal investigators who collected the skin samples and gave us access to their data. This study was funded by SNF grant 310030_166259 to OD and supported by the Scleroderma Foundation PRESS (Assassi, Frech, Hinchcliff, Khanna).
Version 1. 06/30/2020
In-Press Preview
Version 2. 08/17/2020
Electronic publication
Version 3. 09/01/2020
Print issue publication
Footnotes
Conflict of interest: OD had a consultancy relationship with and/or received research funding from Actelion, Acceleron Pharma, AnaMar, Bayer, Baecon Discovery, Blade Therapeutics, Boehringer, CSL Behring, ChemomAb, Curzion Pharmaceuticals, Ergonex, Galapagos NV, GSK, Glenmark Pharmaceuticals, Inventiva, Italfarmaco, iQvia, Medac, Medscape, Mitsubishi Tanabe Pharma, Merck Sharp & Dohme (MSD), Roche, Sanofi, and Union Chimique Belge (UCB) in the area of potential treatments for scleroderma and its complications. In addition, OD has a patent on mir-29 for the treatment of systemic sclerosis issued (US8247389, EP2331143).
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4888–4905.https://doi.org/10.1172/JCI135439.
Contributor Information
Elena Pachera, Email: elena.pachera@usz.ch.
Shervin Assassi, Email: Shervin.assassi@uth.tmc.edu.
Gloria A. Salazar, Email: Gloria.SalazarCintora@uth.tmc.edu.
Mara Stellato, Email: mara.stellato@usz.ch.
Florian Renoux, Email: florian.renoux@live.fr.
Adam Wunderlin, Email: adam.wunderlin@access.uzh.ch.
Przemyslaw Blyszczuk, Email: Przemyslaw.Blyszczuk@usz.ch.
Robert Lafyatis, Email: rlafyatis@gmail.com.
Fina Kurreeman, Email: finakur@gmail.com.
Jeska de Vries-Bouwstra, Email: J.K.de_Vries-Bouwstra@lumc.nl.
Tobias Messemaker, Email: t.c.messemaker@vu.nl.
Carol A. Feghali-Bostwick, Email: feghalib@musc.edu.
Gerhard Rogler, Email: gerhard.rogler@usz.ch.
Wouter T. van Haaften, Email: w.t.van.haaften@umcg.nl.
Gerard Dijkstra, Email: gerard.dijkstra@umcg.nl.
Fiona Oakley, Email: fiona.oakley@newcastle.ac.uk.
Maurizio Calcagni, Email: Maurizio.Calcagni@usz.ch.
Janine Schniering, Email: Janine.Schniering@usz.ch.
Britta Maurer, Email: Britta.Maurer@usz.ch.
Jörg H.W. Distler, Email: joerg.distler@uk-erlangen.de.
Gabriela Kania, Email: gabriela.kania@uzh.ch.
Mojca Frank-Bertoncelj, Email: Mojca.Frank@usz.ch.
Oliver Distler, Email: Oliver.Distler@usz.ch.
References
- 1.Palumbo-Zerr K, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 2.Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest. 2007;117(3):557–567. doi: 10.1172/JCI31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikpour M, Baron M. Mortality in systemic sclerosis: lessons learned from population-based and observational cohort studies. Curr Opin Rheumatol. 2014;26(2):131–137. doi: 10.1097/BOR.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 4.Allanore Y, et al. Systemic sclerosis. Nat Rev Dis Primers. 2015;1:15002. doi: 10.1038/nrdp.2015.2. [DOI] [PubMed] [Google Scholar]
- 5.Rosenbloom J, Castro SV, Jimenez SA. Narrative review: fibrotic diseases: cellular and molecular mechanisms and novel therapies. Ann Intern Med. 2010;152(3):159–166. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]
- 6.Santiago B, Galindo M, Rivero M, Pablos JL. Decreased susceptibility to Fas-induced apoptosis of systemic sclerosis dermal fibroblasts. Arthritis Rheum. 2001;44(7):1667–1676. doi: 10.1002/1529-0131(200107)44:7<1667::AID-ART291>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Hinz B, Dugina V, Ballestrem C, Wehrle-Haller B, Chaponnier C. α-Smooth muscle actin is crucial for focal adhesion maturation in myofibroblasts. Mol Biol Cell. 2003;14(6):2508–2519. doi: 10.1091/mbc.e02-11-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jelaska A, Arakawa M, Broketa G, Korn JH. Heterogeneity of collagen synthesis in normal and systemic sclerosis skin fibroblasts. Increased proportion of high collagen-producing cells in systemic sclerosis fibroblasts. Arthritis Rheum. 1996;39(8):1338–1346. doi: 10.1002/art.1780390811. [DOI] [PubMed] [Google Scholar]
- 9.Moulin V, Larochelle S, Langlois C, Thibault I, Lopez-Vallé CA, Roy M. Normal skin wound and hypertrophic scar myofibroblasts have differential responses to apoptotic inductors. J Cell Physiol. 2004;198(3):350–358. doi: 10.1002/jcp.10415. [DOI] [PubMed] [Google Scholar]
- 10.Lagares D, et al. Targeted apoptosis of myofibroblasts with the BH3 mimetic ABT-263 reverses established fibrosis. Sci Transl Med. 2017;9(420):eaal3765. doi: 10.1126/scitranslmed.aal3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12(6):325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 12.Lafyatis R. Transforming growth factor β — at the centre of systemic sclerosis. Nat Rev Rheumatol. 2014;10(12):706–719. doi: 10.1038/nrrheum.2014.137. [DOI] [PubMed] [Google Scholar]
- 13.Goumans MJ, Mummery C. Functional analysisof the TGFbeta receptor/Smad pathway through gene ablation in mice. Int J Dev Biol. 2000;44(3):253–265. [PubMed] [Google Scholar]
- 14.Piek E, et al. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J Biol Chem. 2001;276(23):19945–19953. doi: 10.1074/jbc.M102382200. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mondal T, et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, et al. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta. 2014;1842(11):2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Piccoli MT, et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121(5):575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- 21.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13(11):971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu X, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017;7:42657. doi: 10.1038/srep42657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie H, Xue JD, Chao F, Jin YF, Fu Q. Long non-coding RNA-H19 antagonism protects against renal fibrosis. Oncotarget. 2016;7(32):51473–51481. doi: 10.18632/oncotarget.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christmann RB, et al. Association of interferon- and transforming growth factor β-regulated genes and macrophage activation with systemic sclerosis-related progressive lung fibrosis. Arthritis Rheumatol. 2014;66(3):714–725. doi: 10.1002/art.38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snowden N, Coupes B, Herrick A, Illingworth K, Jayson MI, Brenchley PE. Plasma TGF-β in systemic sclerosis: a cross-sectional study. Ann Rheum Dis. 1994;53(11):763–767. doi: 10.1136/ard.53.11.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Assassi S, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. arthritis rheumatol (Hoboken, NJ) 2015;67(11):3016–3026. doi: 10.1002/art.39289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert HF, O’Leary MH. Modification of arginine and lysine in proteins with 2,4-pentanedione. Biochemistry. 1975;14(23):5194–5199. doi: 10.1021/bi00694a027. [DOI] [PubMed] [Google Scholar]
- 29.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 30.Keniry A, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol. 2017;24(3):197–204. doi: 10.1038/nsmb.3370. [DOI] [PubMed] [Google Scholar]
- 32.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21(2):198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D, Hsu S, Purushotham D, Sears RL, Wang T. WashU Epigenome Browser update 2019. Nucleic Acids Res. 2019;47(W1):W158–W165. doi: 10.1093/nar/gkz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fishilevich S, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Rendon A, Wernisch L. Transcription factor and chromatin features predict genes associated with eQTLs. Nucleic Acids Res. 2013;41(3):1450–1463. doi: 10.1093/nar/gks1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19(1):151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lichtman MK, Otero-Vinas M, Falanga V. Transforming growth factor beta (TGF-β) isoforms in wound healing and fibrosis. Wound Repair Regen. 2016;24(2):215–222. doi: 10.1111/wrr.12398. [DOI] [PubMed] [Google Scholar]
- 40.Penn JW, Grobbelaar AO, Rolfe KJ. The role of the TGF-β family in wound healing, burns and scarring: a review. Int J Burns Trauma. 2012;2(1):18–28. [PMC free article] [PubMed] [Google Scholar]
- 41.Saito A, Horie M, Nagase T. TGF-β signaling in lung health and disease. Int J Mol Sci. 2018;19(8):E2460. doi: 10.3390/ijms19082460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabregat I, et al. TGF-β signalling and liver disease. FEBS J. 2016;283(12):2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 43.Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009;5(4):200–206. doi: 10.1038/nrrheum.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J, et al. Canonical Wnt signaling induces skin fibrosis and subcutaneous lipoatrophy: a novel mouse model for scleroderma? Arthritis Rheum. 2011;63(6):1707–1717. doi: 10.1002/art.30312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu J, et al. LncRNA MIR503HG inhibits cell migration and invasion via miR-103/OLFM4 axis in triple negative breast cancer. J Cell Mol Med. 2019;23(7):4738–4745. doi: 10.1111/jcmm.14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu F, Zhang MR, Zhou Z, Pu JX, Zhao XJ. lncRNA MIR503HG functioned as a tumor suppressor and inhibited cell proliferation, metastasis and epithelial-mesenchymal transition in bladder cancer. J Cell Biochem. 2019;120(6):10821–10829. doi: 10.1002/jcb.28373. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8(10):2814–2829. doi: 10.7150/thno.23012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiedler J, et al. Development of long noncoding RNA-based strategies to modulate tissue vascularization. J Am Coll Cardiol. 2015;66(18):2005–2015. doi: 10.1016/j.jacc.2015.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonson B, et al. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci Signal. 2017;10(468):eaaf5967. doi: 10.1126/scisignal.aaf5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morquette B, et al. REDD2-mediated inhibition of mTOR promotes dendrite retraction induced by axonal injury. Cell Death Differ. 2015;22(4):612–625. doi: 10.1038/cdd.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forrester HB, Li J, Leong T, McKay MJ, Sprung CN. Identification of a radiation sensitivity gene expression profile in primary fibroblasts derived from patients who developed radiotherapy-induced fibrosis. Radiother Oncol. 2014;111(2):186–193. doi: 10.1016/j.radonc.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 53.van den Hoogen F, et al. 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2013;72(11):1747–1755. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 54.Hsu E, Shi H, Jordan RM, Lyons-Weiler J, Pilewski JM, Feghali-Bostwick CA. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheum. 2011;63(3):783–794. doi: 10.1002/art.30159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverberg MS, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(suppl A):5A–36A. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 56.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;(61):3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PRESS RNA-Seq data accession code: GSE130955. Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130955 Enter token cfmxwywifxgrvot into the box. RNA sequencing files for cohort 3 are deposited at the European Genome-phenome Archive database under EGAO00000000316, https://www.ebi.ac.uk/ega/organisations/EGAO00000000316 RNA microarray data accession code: GSE139334. Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139334 Enter token wrmlkwgstfgrvgv into the box. ChIRP-Seq and ATAC-Seq accession code: PRJEB34589. To review ENA accession PRJEB34589, go to https://www.ebi.ac.uk/ena/data/view/PRJEB34589