Abstract
Mutation in the LMNA gene, encoding lamin A/C, causes a diverse group of diseases called laminopathies. Cardiac involvement is the major cause of death and manifests as dilated cardiomyopathy, heart failure, arrhythmias, and sudden death. There is no specific therapy for LMNA-associated cardiomyopathy. We report that deletion of Lmna in cardiomyocytes in mice leads to severe cardiac dysfunction, conduction defect, ventricular arrhythmias, fibrosis, apoptosis, and premature death within 4 weeks. The phenotype is similar to LMNA-associated cardiomyopathy in humans. RNA sequencing, performed before the onset of cardiac dysfunction, led to identification of 2338 differentially expressed genes (DEGs) in Lmna-deleted cardiomyocytes. DEGs predicted activation of bromodomain-containing protein 4 (BRD4), a regulator of chromatin-associated proteins and transcription factors, which was confirmed by complementary approaches, including chromatin immunoprecipitation sequencing. Daily injection of JQ1, a specific BET bromodomain inhibitor, partially reversed the DEGs, including those encoding secretome; improved cardiac function; abrogated cardiac arrhythmias, fibrosis, and apoptosis; and prolonged the median survival time 2-fold in the myocyte-specific Lmna-deleted mice. The findings highlight the important role of LMNA in cardiomyocytes and identify BET bromodomain inhibition as a potential therapeutic target in LMNA-associated cardiomyopathy, for which there is no specific effective therapy.
Keywords: Cardiology
Keywords: Cardiovascular disease, Fibrosis, Heart failure
Introduction
Heart failure is a major cause of morbidity and mortality in the world (1, 2). It affects an estimated 26 million people worldwide and more than 10% of the elderly in the Western world (1–3). Prevalence of heart failure continues to rise. An estimated 6.2 million Americans older than 20 years had heart failure between 2013 and 2016 (2.6% of the population) as compared with 5.7 million between 2009 and 2012 (1). Heart failure remains the number one cause of hospitalization in the United States and Europe (3).
Dilated cardiomyopathy (DCM), characterized by an increased left ventricular end-diastolic diameter (LVEDD) and a reduced left ventricular ejection fraction, is a major cause of heart failure and the most common indication for cardiac transplantation (4–6). DCM is a heterogeneous disease with several etiologies. Primary DCM is a genetic disease of the myocardium that typically exhibits an autosomal dominant pattern of inheritance. It is caused primarily by mutations in genes coding for sarcomere and cytoskeletal proteins (4, 7). Mutations in the LMNA gene, encoding the nuclear envelope protein lamin A/C (LMNA), are the second most common cause of DCM, accounting for up to 10% of familial DCM (8, 9).
Mutations in the LMNA gene cause a diverse array of diseases referred to as laminopathies, including DCM, which is a common manifestation (10–13). DCM caused by LMNA mutations (henceforth LMNA-DCM) is the major cause of premature death in laminopathies (11, 12, 14). LMNA-DCM has a phenotypic expression that is somewhat distinct from DCM caused by mutations in genes encoding sarcomere and cytoskeletal protein, because of a high prevalence of cardiac conduction defects, arrhythmias, and sudden cardiac death (8, 9, 11, 14–18). LMNA mutations are responsible for about a third of DCM cases associated with cardiac conduction defects (8). Given the poor prognosis and the high risk of sudden cardiac death, the presence of LMNA pathogenic variants typically necessitates implantation of a defibrillator (11, 12, 15, 18–20).
Despite the clinical significance of LMNA-DCM, there is no specific treatment for this potentially deadly form of heart failure, likely because of our inadequate understanding of its molecular pathogenesis. We and others have recently implicated double-stranded DNA breaks and consequent activation of the DNA damage response pathway as an upstream mechanism that instigates induction of phenotype, composed of inflammation, apoptosis, cell senescence, fibrosis, cardiac dysfunction, and premature death, in humans and mouse models of LMNA-DCM as well as in other laminopathies (21–25). Several downstream pathways, including TP53, FOXO3, PDGFRB, and MAPK, are also implicated in the pathogenesis of LMNA-DCM, illustrating the complexity of the disease (13, 26–28).
LMNA is a ubiquitously expressed protein constituent of nuclear inner membrane with low cell type or tissue specificity (13, 29). However, LMNA interacts with chromatin through lamin-associated domains (LADs) and influences gene expression in a cell type–specific manner (30–33). In human cardiomyocytes, LMNA interacts with chromatin in about 300 genomic regions, each approximately 2 million to 3 million bases in length, which collectively comprise about 20% of the genome (31). Cell type–invariant (constitutive) LADs encompass a minority of the LADs, whereas the majority are facultative and cell type–specific (30–33). Therefore, cell type–specific interventions in the heart are expected to provide insights into the specific role of each cardiac cell type in the pathogenesis of LMNA-DCM and raise the prospect of therapeutic opportunities. Accordingly, we deleted the mouse Lmna gene specifically in cardiomyocytes, the main cell type in the heart, characterized the phenotype, and identified the major dysregulated pathways. Deletion of Lmna in cardiomyocytes was associated with activation of bromodomain-containing protein 4 (BRD4). Pharmacological targeting of BET bromodomain proteins by JQ1 partially restored dysregulated gene expression, including genes encoding secretome, and improved survival, cardiac function, myocardial fibrosis, and apoptosis in the myocyte-specific Lmna-knockout mouse model of LMNA-DCM. The findings raise the prospect of interventions targeting BET bromodomain proteins for therapeutic gain in LMNA-DCM.
Results
Cardiomyocyte-specific deletion of Lmna in mice.
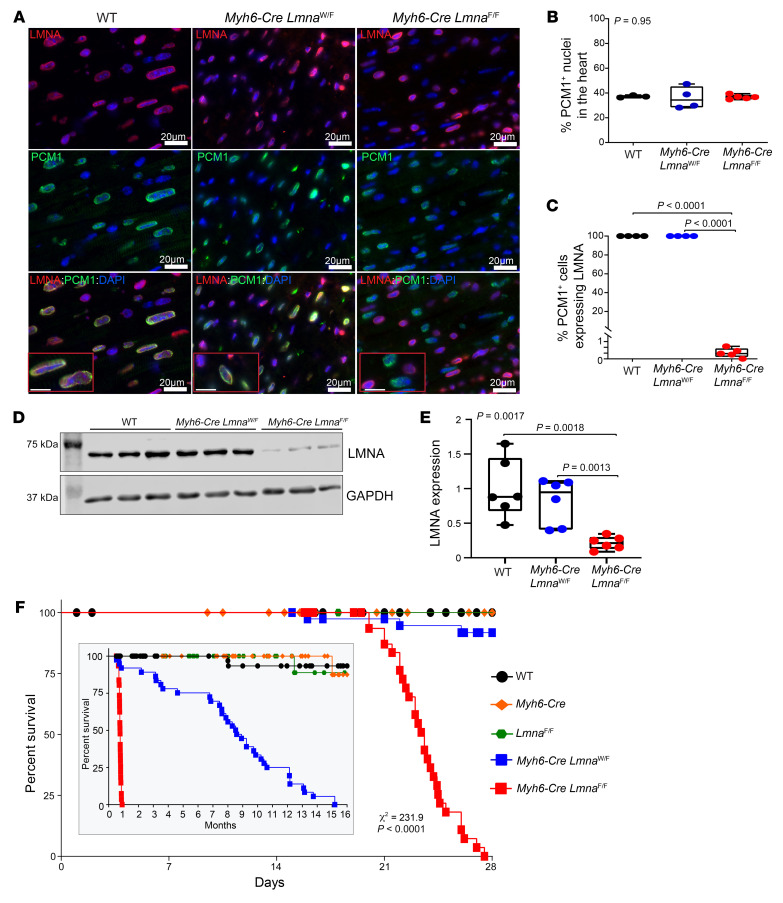
The Lmna gene was deleted in cardiomyocytes by crossing of LmnaF/F and Myh6-Cre deleter mice (34, 35). Heterozygous and homozygous Lmna-knockout mice were born per the expected Mendelian ratio and did not show discernible phenotype at birth. To determine efficiency of deletion of the Lmna gene in cardiomyocytes, thin myocardial sections from WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mouse hearts were costained with antibodies against LMNA and pericentriolar membrane-1 (PCM1), which tags cardiomyocyte nuclei in the heart (36–38). PCM1 expression was detected in approximately a third of the cells in the myocardium (Figure 1, A and B). PCM1-expressing cells also stained positive for expression of LMNA in the WT and Myh6-Cre LmnaW/F mouse hearts (Figure 1, A–C). In contrast, only 0.52% ± 0.4% of PCM1-expressing cells, i.e., myocytes, stained positive for LMNA expression in the Myh6-Cre LmnaF/F mouse hearts, indicating a near-complete absence of the LMNA protein in cardiomyocytes (Figure 1, A–C). To further validate the findings, immunoblotting, using an anti-LMNA antibody, was performed on protein extracts from cardiomyocytes isolated from WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice. The LMNA protein was reduced in Myh6-Cre LmnaW/F and was almost completely absent in Myh6-Cre LmnaF/F myocytes, as compared with WT myocytes (Figure 1, D and E). These findings supported efficient deletion of the Lmna gene in cardiomyocytes. Expression levels of the LMNA protein in cardiomyocyte extracts from mice carrying the loxP sequences flanking exon 2 of the Lmna gene (LmnaF/F) were similar to those in the WT mice (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI135922DS1). Likewise, immunoblotting of protein extracts from the noncardiomyocyte cells in the heart showed unaltered LMNA protein levels in the Myh6-Cre LmnaF/F mice (Supplemental Figure 1, C and D).
Figure 1. Conditional deletion of Lmna gene in cardiomyocytes in mice.
(A) Representative immunofluorescence staining of thin myocardial section from 3-week-old WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice showing localization of LMNA (red) at the nuclear membrane in PCM1-labeled (green) cardiomyocytes. Nuclei were counterstained with DAPI (blue). Scale bars: 20 μm, 10 μm (insets). (B) Quantitative data of PCM1-labeled nuclei in WT (n = 3), Myh6-Cre LmnaW/F (n = 4), and Myh6-Cre LmnaF/F (n = 5, P = 0.95). (C) Quantitative data of PCM1-labeled nuclei showing expression of LMNA in WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F (n = 5, P < 0.0001). (D) Western blots showing expression of LMNA in isolated cardiomyocyte cell lysates in 2-week-old WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F and the corresponding GAPDH as a loading control. (E) Quantitative data on LMNA expression levels in cardiomyocytes in WT (n = 6), Myh6-Cre LmnaW/F (n = 6), and Myh6-Cre LmnaF/F mice (n = 6, P = 0.0017). (F) Kaplan-Meier curve showing the survival of WT (n = 52), Myh6-Cre (n = 38), LmnaF/F (n = 50), Myh6-Cre LmnaW/F (n = 40), and Myh6-Cre LmnaF/F (n = 37) mice during the first 4 weeks (χ2 = 231, P < 0.0001) and 16 months (inset; χ2 = 344, P < 0.0001) after birth. P values shown in B, C, and E were calculated by 1-way ANOVA followed by Tukey’s post hoc pairwise comparison test.
Gross morphology.
Myh6-CreLmnaW/F and Myh6-Cre LmnaF/F mice had normal growth rates for the first 3 weeks of life, but the Myh6-Cre LmnaF/F mice showed stunted growth after 3 weeks of life, as indicated by failure to gain weight (Supplemental Figure 2). At 4 weeks of age, the Myh6-Cre LmnaF/F mice had a 40.2% ± 2.1% lower body weight as compared with the WT mice. The LmnaF/F, Myh6-Cre, and Myh6-Cre LmnaW/F mice had a normal growth curve during this period (Supplemental Figure 2).
Survival.
The Myh6-Cre LmnaF/F mice showed increased mortality, notably starting at 3 weeks of age and rapidly progressing to 100% mortality by 4 weeks (Figure 1F). The Myh6-Cre LmnaW/F mice also exhibited an increased mortality albeit at an older age, reaching 100% mortality by 15 months of age (Figure 1F and inset). The median survival time in Myh6-Cre LmnaF/F was 22 days, whereas it was about 8 months in the Myh6-Cre LmnaW/F mice (Figure 1F and inset). WT, LmnaF/F, and Myh6-Cre had normal survival (Figure 1F). The survival rates between male and female Myh6-Cre LmnaF/F were similar (Supplemental Figure 3).
Cardiac function.
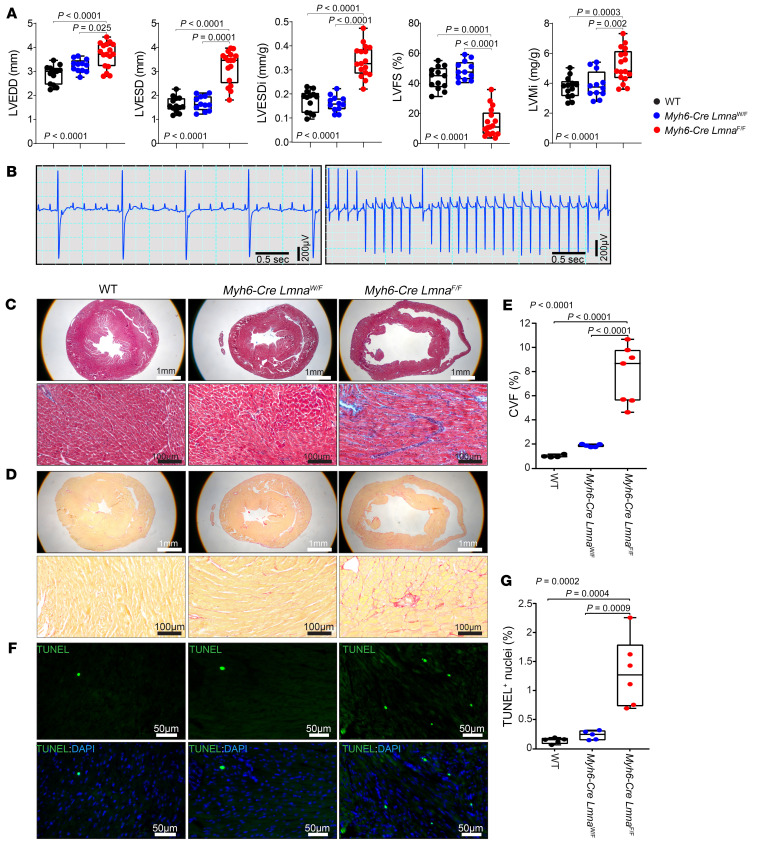
The Myh6-Cre LmnaF/F mice exhibited cardiac dilatation and systolic dysfunction, as indicated by a markedly increased LVEDD and left ventricular end-systolic diameter (LVESD), and reduced left ventricular fractional shortening (LVFS) at 3 weeks of age compared with WT mice (Figure 2A and Supplemental Table 1). The heterozygous Lmna-deficient mice had normal cardiac function at 3 weeks of age (Supplemental Table 1). However, these mice developed cardiac dilatation and dysfunction at 10 months of age, the last time point analyzed (Supplemental Table 2). The Myh6-Cre and LmnaF/F mice, which were included as control, did not show evidence of cardiac dysfunction compared with WT mice (Supplemental Tables 1 and 2). There was no sex-dependent effect on indices of cardiac function in Myh6-Cre LmnaF/F mice, as both sexes showed significant cardiac dysfunction at 3 weeks of age (Supplemental Figure 4).
Figure 2. Cardiac phenotype in 3-week-old WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice.
(A) Selected echocardiographic parameters showing left ventricular end-diastolic diameter (LVEDD), left ventricular end-systolic diameter (LVESD), left ventricular end-systolic diameter indexed to body weight (LVESDi), left ventricular fractional shortening (LVFS), and left ventricular mass indexed to body weight (LVMi) in 3-week-old WT (n = 13), Myh6-Cre LmnaW/F (n = 12), and Myh6-Cre LmnaF/F (n = 17) mice. P values shown were obtained using ordinary 1-way ANOVA and Bonferroni’s post hoc test for comparisons of the LVEDD, LVESDi, and LVMi, and Kruskal-Wallis and Dunn’s post hoc test for comparisons for LVESD and LVFS. (B) Selected representative surface ECG recordings showing third-degree atrioventricular block (no association between P and QRS waves) and ventricular tachycardia observed in the Myh6-Cre LmnaW/F (n = 19) and Myh6-Cre LmnaF/F mice (n = 19). (C and D) Masson’s trichrome–stained (top panel) and Picrosirius red–stained (bottom panel) representative myocardial sections in 3-week-old WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice. (E) Corresponding quantitative data on percentage collagen volume fraction (CVF) in myocardial sections in the WT (n = 4), Myh6-Cre LmnaW/F (n = 5), and Myh6-Cre LmnaF/F (n = 7) mice. (F) TUNEL-stained thin myocardial sections in 3-week-old WT, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice. TUNEL-positive cells are shown in green and nuclei, counterstained with DAPI, in blue. (G) Quantitative data showing percentage of TUNEL-positive nuclei in the WT (n = 5), Myh6-Cre LmnaW/F (n = 5), and Myh6-Cre LmnaF/F (n = 6) mice. P values shown in E and G were calculated using ordinary 1-way ANOVA and Tukey’s post hoc test.
Cardiac arrhythmias.
Myh6-Cre LmnaW/F and Myh6-Cre LmnaF/F mice showed sinus bradyarrhythmias; atrial flutter; atrioventricular blocks, including complete heart block; ectopic ventricular beats; and ventricular tachycardia, all occurring within the first 3 weeks in the Myh6-Cre LmnaF/F mice and at an older age in the Myh6-Cre LmnaW/F mice (Figure 2B and Supplemental Tables 3 and 4). Advanced atrioventricular block (second and third degree) was common and was observed in approximately two-thirds of the 3-week-old Myh6-Cre LmnaF/F mice. Likewise, ventricular tachycardia (≥3 ventricular ectopic beats in a row with a rate of >300 bpm) was recorded in about 20% of the Myh6-Cre LmnaF/F mice (Supplemental Table 3). WT, LmnaF/F, and Myh6-Cre mice did not exhibit notable cardiac arrhythmias, except for premature atrial and ventricular contractions (Supplemental Tables 3 and 4).
Myocardial fibrosis and apoptosis.
Collagen volume fraction (CVF), a measure of myocardial fibrosis, was increased about 8-fold in 3-week-old Myh6-Cre LmnaF/F as compared with WT mice (Figure 2, C–E). Likewise, the number of TUNEL-positive cells was increased about 10-fold (Figure 2, F and G).
The heterozygous Lmna-knockout mice did not exhibit significant myocardial fibrosis or apoptosis at 3 weeks of age but showed 4-fold increased myocardial fibrosis and approximately 7-fold increased apoptosis as compared with corresponding WT mice at 10 months of age (Supplemental Figure 5). The LmnaF/F mice did not show evidence of myocardial fibrosis, as percentage CVF was similar to that in the WT mice (Supplemental Figure 6).
Differentially expressed genes in cardiomyocytes isolated from Myh6-Cre LmnaF/F mouse hearts.
To identify transcriptomic changes in cardiomyocytes that are consequent to deletion of the Lmna gene but precede the onset of cardiac dysfunction, cardiac phenotype was assessed in 2-week-old mice. There was no discernible evidence of cardiac dysfunction on echocardiogram at 2 weeks of age (Supplemental Table 5). Likewise, CVF was not significantly different between the WT and Myh6-Cre LmnaF/F mice, and the number of TUNEL-positive cells was also similar between the 2 genotypes (Supplemental Figure 7). Therefore, cardiomyocytes were isolated from 2-week-old WT and Myh6-Cre LmnaF/F mice to extract RNA for RNA-Seq.
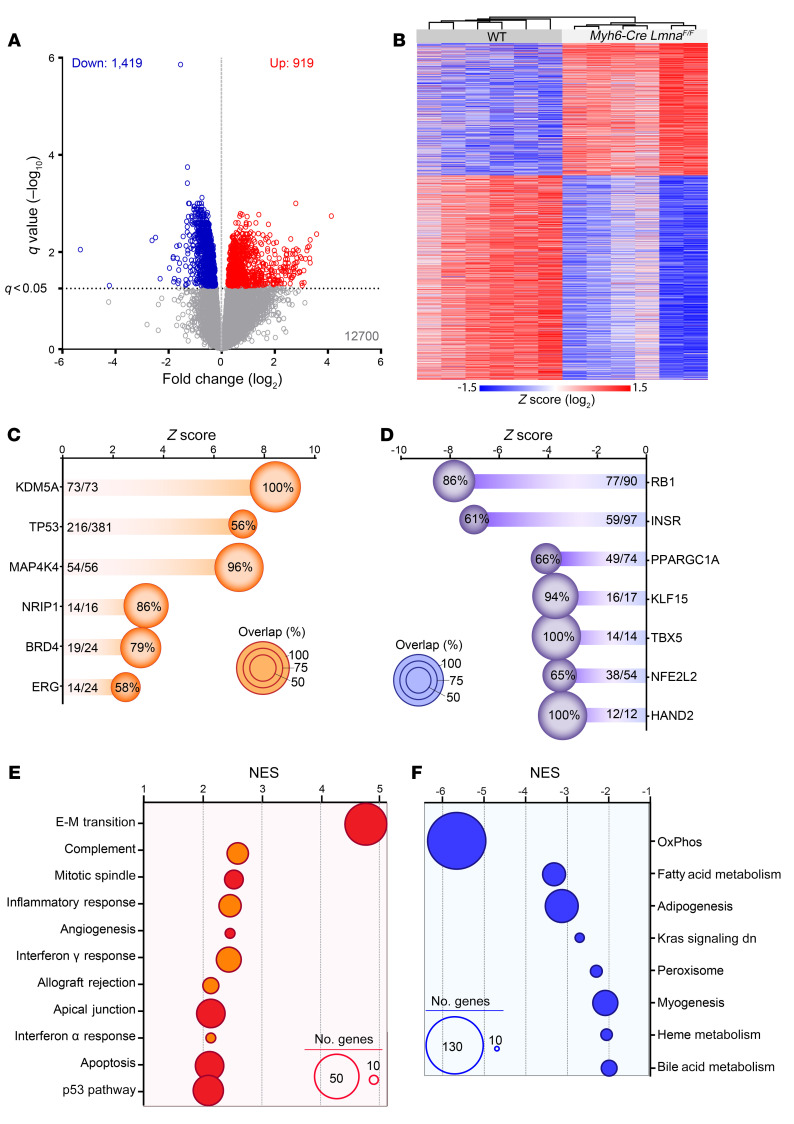
Principal component analysis of the RNA-Seq data showed segregation of the samples according to the mouse genotype (Supplemental Figure 8). A total of 2338 genes were differentially expressed in the Myh6-Cre LmnaF/F as compared with WT cardiomyocytes, of which 1419 genes were downregulated, whereas transcript levels of 919 genes were increased (Figure 3A). Unsupervised hierarchical clustering showed distinct grouping of the samples per genotypes (Figure 3B).
Figure 3. Differentially expressed genes in 2-week-old WT and Myh6-Cre LmnaF/F mouse cardiomyocytes.
(A) Volcano plot depicting differentially expressed genes (DEGs) showing downregulation of 1419 and upregulation of 919 genes in Myh6-Cre LmnaF/F (n = 6) compared with WT mouse cardiomyocytes (n = 6). (B) Heatmap and unsupervised hierarchical clustering of the DEGs in WT and Myh6-Cre LmnaF/F mouse cardiomyocytes, showing clustering according to the genotype. (C and D) Ingenuity Pathway Analysis (IPA) for inferred activated (C) and suppressed (D) upstream regulators of the DEGs in Myh6-Cre LmnaF/F mouse cardiomyocytes. Z score and percentage of overlap of the DEGs with the IPA database are depicted on the graphs. The size of each circle is proportional to the percentage of genes considered as downstream targets of each regulator in the RNA-Seq data set. (E and F) Top significantly enriched hallmark pathways activated (E) or inhibited (F), inferred using Gene Set Enrichment Analysis (GSEA). Normalized enrichment score (NES) and number of genes involved (size of the circle) are depicted for each pathway.
Ingenuity Pathway Analysis (IPA) of differentially expressed genes (DEGs) predicted activation of the transcriptional regulators lysine demethylase 5A (KDM5A), TP53, mitogen-activated protein kinase 4 (MAP4K4), nuclear receptor–interacting protein 1 (NRIP1), BRD4, and ERG1 in the Myh6-Cre LmnaF/F mouse cardiomyocytes (Figure 3C). In contrast, retinoblastoma 1 (RB1), insulin receptor (INSR), and peroxisome proliferator–activated receptor-γ coactivator 1α (PPARGC1α) were predicted to be suppressed (Figure 3D).
Gene set enrichment analysis (GSEA) of DEGs against Molecular Signatures Database hallmark gene sets identified enrichment for genes involved in epithelial-mesenchymal transition (EMT), inflammation, and apoptosis (Figure 3E), whereas genes involved in oxidative phosphorylation, metabolism, and myogenesis were underrepresented (Figure 3F).
The DEGs were analyzed to identify genes whose protein products are known to be or expected to be secreted, i.e., the secretome. A total of 168 of 2338 DEGs (7.2%) coded for secretome, which are depicted in a heatmap in Supplemental Figure 9A. To validate the RNA-Seq data, quantitative reverse transcriptase PCR (RT-qPCR) was performed, which showed changes concordant with the RNA-Seq findings for 17 of 18 genes tested (13 increased and 4 decreased), and 1 gene was unchanged (Supplemental Figure 9, B and C). DEGs coding for secretome predicted TGF-β1 as the most activated and α-catenin as the most suppressed regulators of gene expression (Supplemental Figure 9D). A GSEA plot showing enrichment of genes in the TGF-β1 pathway is illustrated in Supplemental Figure 9E. Immunoblotting of cardiomyocyte extracts suggested increased levels of the active TGF-β1 protein in the Myh6-Cre LmnaF/F cardiomyocytes (Supplemental Figure 9, F and G). In accord with activation of TGF-β1, the DEGs encoding secretome were involved in EMT, angiogenesis, STAT signaling, hypoxia, and apoptosis (Supplemental Figure 9H).
Activation of BRD4 in Myh6-Cre LmnaF/F cardiomyocytes.
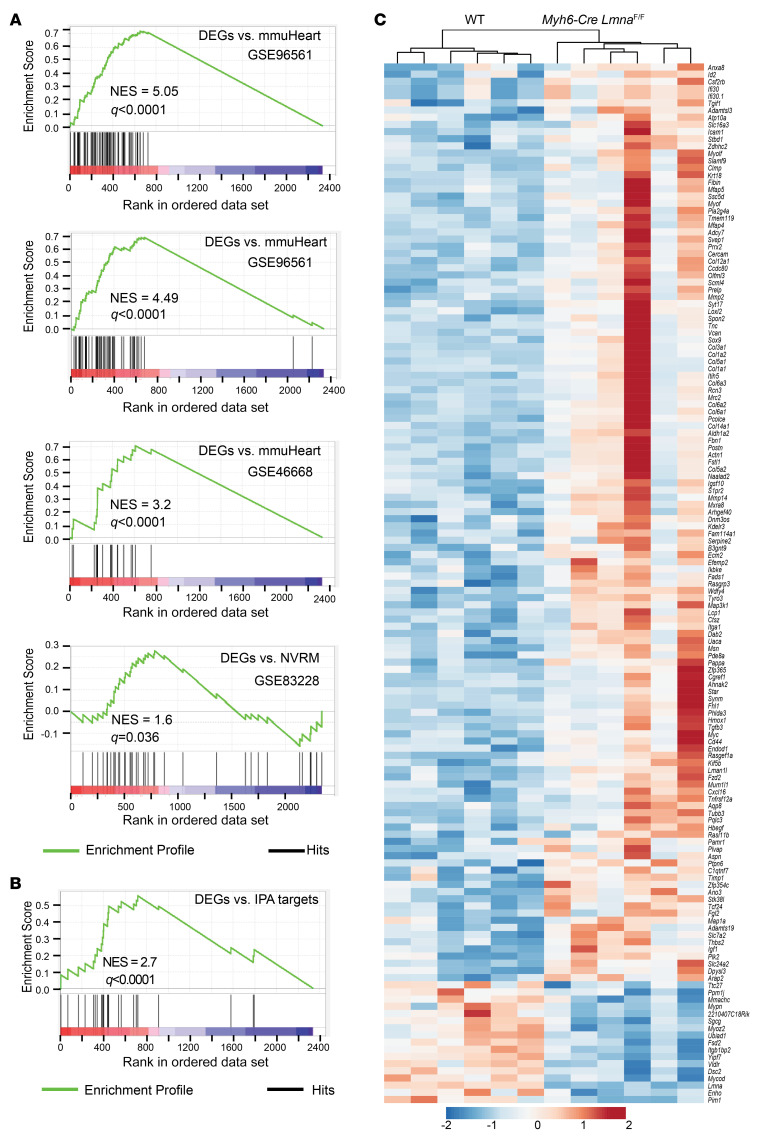
Predicted activation of BRD4 based on the DEGs was in accord with activation of EMT in the heart and the prominent expression of myocardial fibrosis in Myh6-Cre LmnaF/F hearts, rendering BRD4 a potential therapeutic target. To strengthen the evidence for activation of BRD4, DEGs in the Myh6-Cre LmnaF/F cardiomyocytes were analyzed against 3 publicly available mouse heart and 1 rat cardiomyocyte RNA-Seq data sets with defined BRD4 targets (39–41). BRD4 targets in each data set were defined as genes whose transcript levels were increased in the pathological state (including intervention) but were reduced subsequently upon inhibition of BRDs or BRD4. As shown in the GSEA plots in Figure 4A, BRD4 target genes, identified in 4 publicly available data sets, were enriched in Myh6-Cre LmnaF/F cardiomyocytes. Similarly, DEGs in Myh6-Cre LmnaF/F cardiomyocytes were also enriched for the BRD4 target genes gathered from the IPA data sets (Figure 4B). Transcript levels of the BRD4 target genes, identified in all 4 data sets and from the IPA database, in the Myh6-Cre LmnaF/F cardiomyocytes are depicted in a heatmap (Figure 4C). As shown, transcript levels of 123 BRD4 target genes were upregulated in the Myh6-Cre LmnaF/F cardiomyocytes as compared with WT myocytes, whereas expression of 17 genes was suppressed (P < 0.0001; Figure 4C).
Figure 4. Evidence of BET bromodomain activation in Myh6-Cre LmnaF/F mouse cardiomyocytes.
(A) GSEA plot of DEGs in Myh6-Cre LmnaF/F mouse cardiomyocytes against predicted BRD target genes gathered from whole mouse heart after transaortic constriction or myocardial infarction (GSE96561, GSE96561, and GSE48110) and in rat neonatal ventricular cardiomyocytes (NVRM; GSE83228). (B) GSEA of DEGs in Myh6-Cre LmnaF/F mouse cardiomyocytes against predicted BRD target genes obtained from IPA. (C) Heatmap and unsupervised hierarchical clustering of 145 DEGs in Myh6-Cre LmnaF/F mouse cardiomyocytes obtained after overlapping with the GSEA data shown in A and B.
To determine whether increased transcript levels of BRD4 were because of increased BRD4 protein levels, immunoblotting was performed on cardiomyocyte protein extracts from the WT and Myh6-Cre LmnaF/F mice. There was no difference in the BRD4 protein levels between the 2 groups (Supplemental Figure 10). Likewise, BRD2 and BRD3 protein levels were unchanged (Supplemental Figure 10).
BRD4 ChIP-Seq.
To determine whether increased transcript levels of BRD4 target genes in the Myh6-Cre LmnaF/F cardiomyocytes were because of increased recruitment of BRD4 to chromatin, BRD4 ChIP-Seq was performed on myocyte chromatin extracts using an anti-BRD4 antibody. Representative Integrative Genomics Viewer (IGV) tracks showing BRD4 enrichment in the corresponding regions in the Myh6-Cre LmnaF/F myocytes are shown (Figure 5A). The total number of peaks in each ChIP-Seq experiment and the intersects showing shared peaks between the samples in each genotype are presented in the Venn diagrams in Supplemental Figure 11A. BRD4 ChIP peaks were redistributed in the Myh6-Cre LmnaF/F as compared with WT cardiomyocyte genomes, as only about 10,776 of 44,947 (24%) of the peaks were shared between the 2 genotypes (Supplemental Figure 11B). Approximately 6000 BRD4-enriched peaks were overlapped in at least 2 genomes in the WT or Myh6-Cre LmnaF/F myocytes, of which 758 (12.6%) were exclusive to WT, and 1946 (32.4%) to Myh6-Cre LmnaF/F genomes (Supplemental Figure 11C). Distributions of BRD4 ChIP-Seq peaks differed in their localizations to genomic partitions between the WT and Myh6-Cre LmnaF/F myocytes, as more peaks were located in the upstream (P = 0.0007) and downstream (P < 0.0001) genomic regions in the Myh6-Cre LmnaF/F genomes (Supplemental Figure 11D).
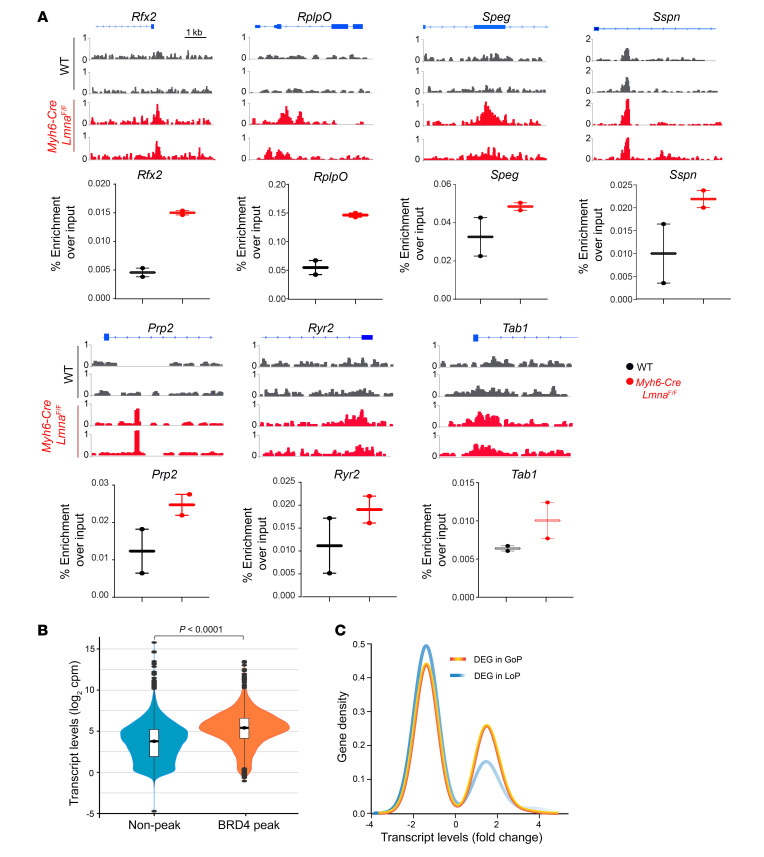
Figure 5. BRD4 activation in Myh6-Cre LmnaF/F mouse cardiomyocytes.
(A) ChIP-qPCR in WT and Myh6-Cre LmnaF/F mouse cardiomyocyte chromatin extracts (n = 2 for each genotype) and IGV tracks from ChIP-Seq showing BRD4 enrichment in the corresponding regions assessed. (B) Violin plots depicting transcript levels of all genes at nonpeak and BRD4 peak regions in Myh6-Cre LmnaF/F mouse cardiomyocytes (P < 0.0001 by Kruskal-Wallis). (C) Transcript levels of DEGs plotted against gene density at the GoP and LoP genomic regions, showing a higher density of genes with increased transcript levels in the GoP regions.
To validate the BRD4 ChIP-Seq findings, ChIP was performed on an independent set of chromatin extracts from the WT and Myh6-Cre LmnaF/F myocytes. The chromatin precipitates were analyzed for the enrichment of the genomic DNA mapped to the peak regions in the discovery ChIP-Seq experiments. As shown in Figure 5, A and B, genomic DNA located at the BRD4 ChIP-Seq peaks were enriched in both WT and Myh6-Cre LmnaF/F myocytes in the independent validation ChIP experiments, albeit the enrichment was greater in the Myh6-Cre LmnaF/F than in the WT myocytes.
To determine whether recruitment of BRD4 to chromatin affected gene expression in the Myh6-Cre LmnaF/F myocytes, transcript levels of the annotated genes located in the BRD4 peak and outside of the peak regions were compared. Globally, transcript levels of genes located at the peak regions were higher as compared with those located outside of the peak regions (Figure 5C). Representative genome browser tracks of the transcript levels at the peak regions as compared with regions outside of peaks are shown in Supplemental Figure 12A. Likewise, gain of peak (GoP), defined as peak present in the Myh6-Cre LmnaF/F and absent in the WT, was associated with a greater number of genes (a higher gene density) that were upregulated, as compared with loss of peak (LoP), defined as peaks absent in the Myh6-Cre LmnaF/F but present in the WT myocytes (Supplemental Figure 12B). Likewise, a greater number of DEGs located at the GoP regions were upregulated as compared with DEGs located at the LoP regions (Figure 5C and Supplemental Figure 12B). The difference in the gene density was also remarkable for the upregulated DEGs located at the upstream genomic regions (Supplemental Figure 12C).
Given the differences in genomic partition of BRD4 peaks, distribution of the annotated DEGs in each genomic segment was compared between the WT and Myh6-Cre LmnaF/F myocytes (Supplemental Figure 12D). Only distribution of DEGs located in the upstream genomic regions differed significantly between the WT and Myh6-Cre LmnaF/F myocytes (Supplemental Figure 12D). Finally, LoP in the Myh6-Cre LmnaF/F genome was associated with a larger number of DEGs located in the upstream region (39/54; 72%) whose expression levels were suppressed (Supplemental Figure 12E).
Dose titration studies with JQ1.
Given the data indicating activation of BDR4 in the Myh6-Cre LmnaF/F myocytes, phenotypic effects of pharmacological BET bromodomain inhibition were analyzed in the Myh6-Cre LmnaF/F mice. The primary endpoint was survival, and the secondary endpoints were myocardial function and fibrosis. The latter was selected based on evidence of enrichment of the DEGs for EMT, presence of myocardial fibrosis in Myh6-Cre LmnaF/F hearts, and the prominent antifibrotic effects of BRD4 (41–43).
JQ1, a well-established specific paninhibitor of BET bromodomain proteins, was selected to treat the Myh6-Cre LmnaF/F mice (44). The 3- to 4-week life span of the Myh6-Cre LmnaF/F mice necessitated administration of JQ1 at a very young age, which was counterbalanced by potential fortuitous effects arising because of the prominent role of epigenetic regulation during cardiac development and myocyte maturation/proliferation in the early postnatal stage (45–47). Consequently, a series of time course studies were performed in the WT mice to determine the effects of injection of JQ1 at the commonly used dose of 50 mg/kg/d of JQ1 at postnatal day 4 (P4), P7, P10, and P14 on survival and body growth (42, 44). JQ1 administration at 50 mg/kg/d was associated with increased mortality, growth retardation, and alopecia at the injection site, which were more pronounced when JQ1 was injected at earlier rather than later time points (Supplemental Figure 13). Injection of JQ1 at a later time point (after P14) was not considered because of the potential confounding effects of early mortality of the Myh6-Cre LmnaF/F mice. Therefore, the JQ1 dose was reduced to 20 mg/kg/d, which has been shown to effectively inhibit BET bromodomain protein activity (48, 49). JQ1 administered at 20 mg/kg/d did not affect survival or gain in weight in the WT mice (Supplemental Figure 14). Consequently, the subsequent experiments in WT and Myh6-Cre LmnaF/F mice were performed with 20 mg/kg/d injected subcutaneously beginning at P14.
Effects of JQ1 on cardiomyocyte transcripts and regulatory pathway.
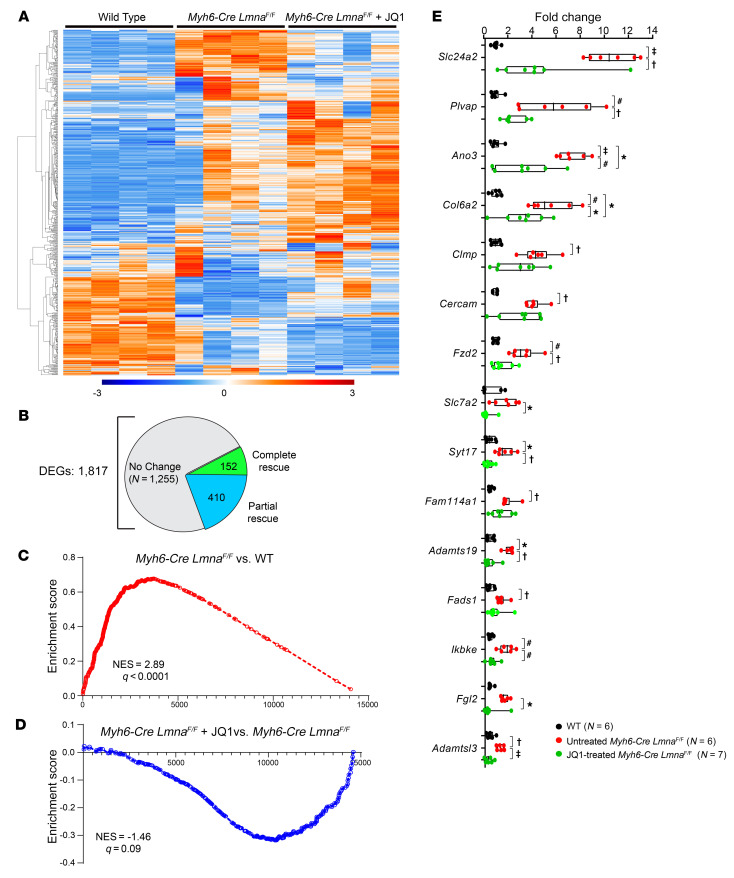
RNA was extracted from cardiomyocytes isolated from 3-week-old WT mice, Myh6-Cre LmnaF/F mice treated with a vehicle, and Myh6-Cre LmnaF/F mice treated with JQ1 (20 mg/kg/d) for 1 week (P14 to P21). Gene expression was analyzed by RNA-Seq. Treatment with JQ1 shifted the transcriptomic profile of the cardiomyocytes toward that of the WT, as illustrated in the heatmap (Figure 6A). Overall, a total of 1817 genes were differentially expressed between the WT and Myh6-Cre LmnaF/F myocytes. Treatment with JQ1 normalized transcript levels of 152 genes (no difference between JQ1-treated Myh6-Cre LmnaF/F and WT myocytes) and significantly lowered transcript levels of another 410 genes (Figure 6B). Overall, transcript levels of 562 DEGs (31%) had reversed toward normal (Figure 6B). To determine efficacy of treatment with JQ1 in suppressing BRD4 activity, transcript levels of the BRD4 target genes in the untreated and JQ1-treated Myh6-Cre LmnaF/F myocytes were compared. GSEA showed suppression of the BRD4 target gene transcript levels in the JQ1-treated myocytes as compared with untreated Myh6-Cre LmnaF/F myocytes (Figure 6, C and D).
Figure 6. Effect of BET bromodomain inhibition on the transcriptome of Myh6-Cre LmnaF/F mouse cardiomyocytes.
(A) Heatmap of the DEGs in WT, Myh6-Cre LmnaF/F, and JQ1-treated Myh6-Cre LmnaF/F mouse cardiomyocytes (n = 4 for each). (B) Pie chart depicting the number of genes that were rescued and partially rescued upon JQ1 treatment in Myh6-Cre LmnaF/F mouse cardiomyocytes from the differentially expressed genes between WT and Myh6-Cre LmnaF/F myocytes. (C and D) GSEA plots of the BRD4 target genes showing induction in Myh6-Cre LmnaF/F compared with WT cardiomyocytes (NES = 2.89, q < 0.0001) (C), and partial suppression in JQ1-treated Myh6-Cre LmnaF/F compared with Myh6-Cre LmnaF/F (NES = –1.46, q = 0.09) (D). (E) RT-qPCR data showing transcript levels of selected BRD4 target genes in WT (n = 6), untreated (n = 6), and JQ1-treated Myh6-Cre LmnaF/F mouse cardiomyocytes (n = 7). P values were obtained with ordinary 1-way ANOVA or Kruskal-Wallis; *P < 0.05, †P < 0.01, #P < 0.001, ‡P < 0.0001.
To validate the RNA-Seq data, transcript levels of 32 BRD4 target genes were quantified by RT-qPCR in cardiomyocyte RNA extracts from the WT mice, Myh6-Cre LmnaF/F mice treated with a vehicle (10% DMSO and 10% 2-hydroxypropyl-β-cyclodextrin), and Myh6-Cre LmnaF/F mice treated with JQ1 (20 mg/kg/d for 7 days, starting at P14). Treatment with JQ1 normalized elevated transcript levels of 15 of 30 selected BRD4 target genes and significantly reduced, but did not normalize, levels of another 7 target genes (Figure 6E and Supplemental Figure 15A). JQ1 treatment had no effect on transcript levels of 9 selected DEGs, whereas the transcript level of 1 candidate target gene was similar among the 3 groups, i.e., not differentially expressed (Supplemental Figure 15B).
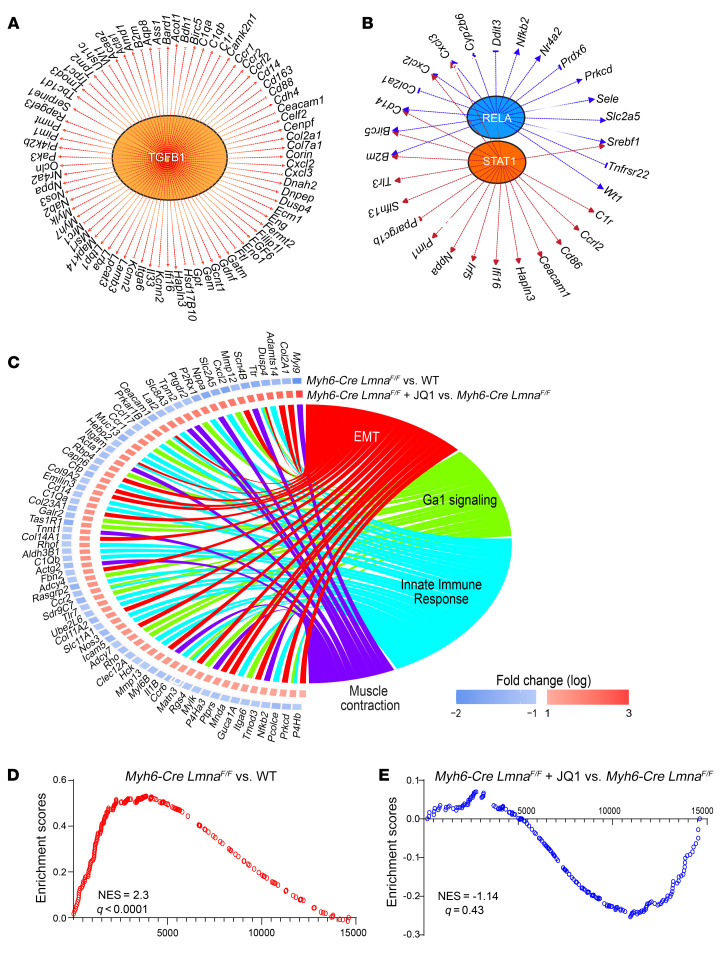
Genes whose transcript levels were significantly affected by JQ1 treatment were analyzed by IPA to infer the transcriptional regulators. The approach predicted activation of over a dozen regulators of gene expression, including TGF-β1, STAT1, and RELA (Figure 7, A and B). A complete list of dysregulated growth/paracrine factors and transcriptional regulators is provided in Supplemental Table 6. Biologically, genes regulated by JQ1 were involved in EMT and innate immune response, among other functions (Figure 7C). Concordant with this finding, GSEA showed activation of the EMT pathway in Myh6-Cre LmnaF/F mice and its reversal upon treatment with JQ1 (Figure 7, D and E).
Figure 7. Upstream regulators and biological pathways altered after BET bromodomain inhibition in Myh6-Cre LmnaF/F mouse cardiomyocytes.
(A and B) Major putative inferred upstream regulators and their targets from IPA overlap analysis of the completely and partially rescued genes upon JQ1 treatment in Myh6-Cre LmnaF/F. (C) Circos plot showing the top significantly enriched hallmark pathways inferred from the completely and partially rescued genes upon JQ1 treatment in Myh6-Cre LmnaF/F using GSEA, with the log2 fold change depicting the relative expression of the genes involved in these pathways: (I) Myh6-Cre LmnaF/F compared with WT cardiomyocytes; (II) JQ1-treated Myh6-Cre LmnaF/F compared with Myh6-Cre LmnaF/F. (D and E) GSEA plots of EMT showing induction in Myh6-Cre LmnaF/F compared with WT cardiomyocytes (NES = 2.3, q < 0.0001) (D), and partial suppression in JQ1-treated Myh6-Cre LmnaF/F compared with Myh6-Cre LmnaF/F (NES = –1.14, q = 0.43) (E).
The RNA-Seq data were also analyzed for the effects of treatment with JQ1 on expression levels of genes encoding secreted proteins (secretome). Treatment with JQ1 shifted the secretome profile of the Myh6-Cre LmnaF/F toward that of the WT myocytes, involving 195 genes, as depicted in the heatmap shown in Supplemental Figure 16A. Treatment with JQ1 normalized transcript levels of 21 genes encoding secretome and significantly reduced transcript levels of another 50 genes, albeit the latter group was not normalized (Supplemental Figure 16B). The findings were tested for validation by RT-qPCR, which was in agreement with the RNA-Seq data, as transcript levels of 11 of 12 upregulated genes (92%) were either normalized or partially reduced (Supplemental Figure 16C). Analysis of the DEGs encoding secretome predicted activation of several growth and paracrine factors, including GDF15, TGF-β1, and FGF21 among others, which might contribute to the pathogenesis of the phenotype (Supplemental Figure 16D). The findings of a similar analysis to predict transcriptional regulators of secretome are presented in Supplemental Figure 16E. Finally, genes encoding secretome were mostly involved in extracellular matrix formation and deposit (Supplemental Figure 16F).
Effects of JQ1 on survival.
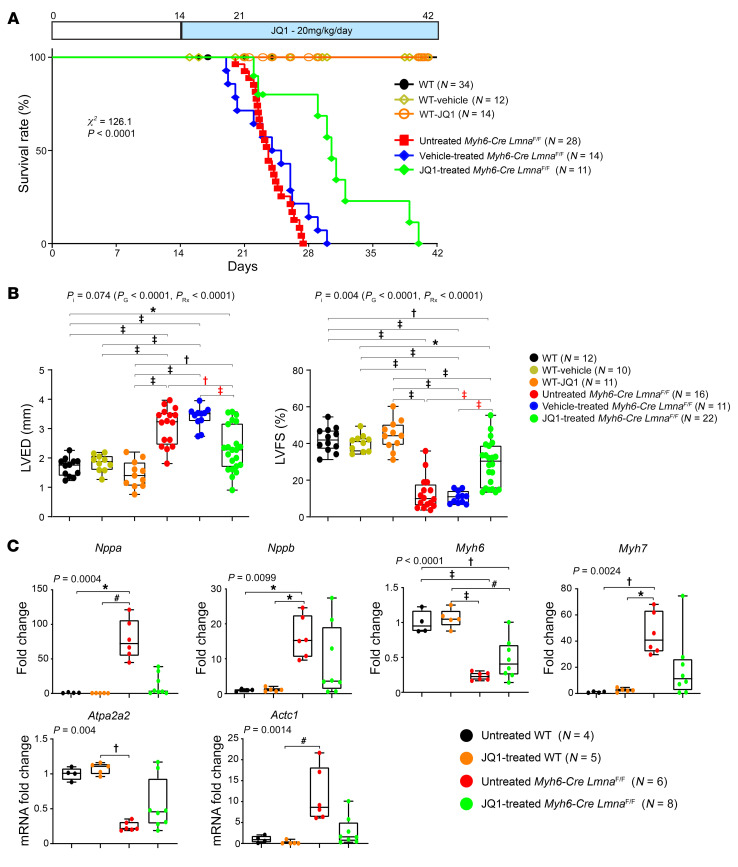
To determine effects of treatment with JQ1 on survival, WT and Myh6-Cre LmnaF/F mice were treated with daily subcutaneous injection of either a vehicle (10% DMSO and 10% 2-hydroxypropyl-β-cyclodextrin) or JQ1 (20 mg/kg/d) starting at P14. A group of untreated littermate mice were included as a control. Treatment with JQ1 prolonged median survival of the Myh6-Cre LmnaF/F mice from 23 days to 32 days and the maximum life span from 28 to 42 days (χ2 = 126.1, P < 0.0001; Figure 8A). There were no differences in the survival rates between untreated mice and Myh6-Cre LmnaF/F mice treated with the vehicle (χ2 = 1.69, P = 0.19). Likewise, treatment with JQ1 had no effect on the survival rate of the WT mice.
Figure 8. Phenotypic effects of BET bromodomain protein inhibition in Myh6-Cre LmnaF/F mice.
(A) Kaplan-Meier survival curves in the untreated (black line; n = 34), vehicle-treated (yellow line; n = 12), and JQ1-treated WT (orange line; n = 14), as well as untreated (red line; n = 28), vehicle-treated (blue line; n = 14), and JQ1-treated Myh6-Cre LmnaF/F (green line; n = 11), mice. (B) Selected echocardiographic parameters after 1 week of treatment in 3-week-old untreated (black dots; n = 12), vehicle-treated (yellow dots; n = 10), and JQ1-treated WT (orange dots; n = 11), as well as untreated (red dots; n = 16), vehicle-treated (blue dots; n = 11), and JQ1-treated (green dots; n = 22) Myh6-Cre LmnaF/F, mice. P values shown were obtained using 2-way ANOVA and Bonferroni’s post hoc test for comparisons; *P < 0.05, †P < 0.01, #P < 0.001, ‡P < 0.0001. PI, P value for interaction; PG, P value for genotype, PRx, P value for treatment effect. (C) Expression levels of known markers of cardiac dysfunction, quantified by RT-qPCR, after 1 week of treatment in 3-week-old untreated (black dots; n = 4) and JQ1-treated WT (orange dots; n = 5) mice, as well as untreated (red dots; n = 6) and JQ1-treated (green dots; n = 8) Myh6-Cre LmnaF/F mice. Because untreated and vehicle-treated mice were indistinguishable, only untreated and JQ1-treated mice were analyzed. P values shown were obtained with ordinary 1-way ANOVA or Kruskal-Wallis; *P < 0.05, †P < 0.01, #P < 0.001, ‡P < 0.0001.
Effects of JQ1 on body weight.
Treatment with JQ1 had no discernible effect on body weight in the Myh6-Cre LmnaF/F or WT mice (Supplemental Figure 17). Myh6-Cre LmnaF/F mice, regardless of the treatment groups, had a smaller body weight after 3 weeks of age as compared with the WT mice.
Effects of JQ1 on cardiac function.
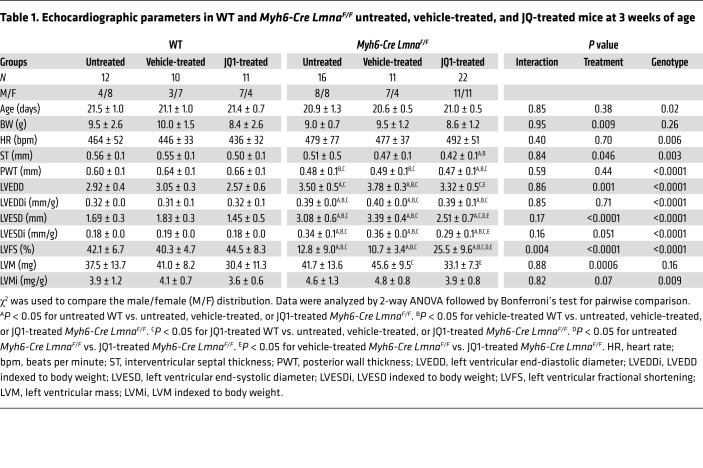
WT and Myh6-Cre LmnaF/F mice were treated with a vehicle or JQ1 at 20 mg/kg/d starting at P14, and cardiac size and function were assessed by echocardiography at 3 weeks of age. Treatment with JQ1 improved left ventricular size and function in the Myh6-Cre LmnaF/F mice, as indicated by a smaller LVEDD and LVESD, improved LVFS, and reduced left ventricular mass (Table 1 and Figure 8B). Genotype-by-treatment interaction analysis showed significant effects of the genotype alone, treatment alone, and genotype-by-treatment interactions on the LVFS (Table 1). Treatment with JQ1 did not have a discernible effect on echocardiographic indices of cardiac size and function in the WT mice (Table 1).
Table 1. Echocardiographic parameters in WT and Myh6-Cre LmnaF/F untreated, vehicle-treated, and JQ-treated mice at 3 weeks of age.
Effects of JQ1 on molecular markers of cardiac dysfunction.
Cardiac hypertrophy and failure are associated with changes in expression of a number of genes referred to as markers of cardiac hypertrophy and dysfunction, including Nppa, Nppb, Myh6, Myh7, Atp2a2, and Actc1 (50–52). To corroborate the beneficial effects of JQ1 on survival and cardiac function, transcript levels of the molecular markers of cardiac dysfunction were quantified by RT-qPCR on cardiac RNA extracts. Transcript levels of Nppa, Nppb, Myh7, and Actc1 were increased and those of Atp2a2 and Myh6 were reduced in the Myh6-Cre LmnaF/F hearts, which was in accord with the anticipated changes in cardiac hypertrophy and failure (50–52). Treatment with JQ1 completely or partially rescued transcript levels of the molecular markers of cardiac hypertrophy and dysfunction (Figure 8C).
Effects of JQ1 on cardiac arrhythmias.
Phenotypic characteristics of human LMNA-DCM include conduction defects and ventricular arrhythmias, which were also observed in the Myh6-Cre LmnaF/F mice (Figure 2B and Supplemental Table 3). Treatment with JQ1 significantly reduced the frequency of supraventricular arrhythmias, atrioventricular blocks, and ventricular arrhythmias, which was in accord with improved survival in the JQ1-treated Myh6-Cre LmnaF/F mice (Supplemental Table 7).
Effects of JQ1 on myocardial fibrosis.
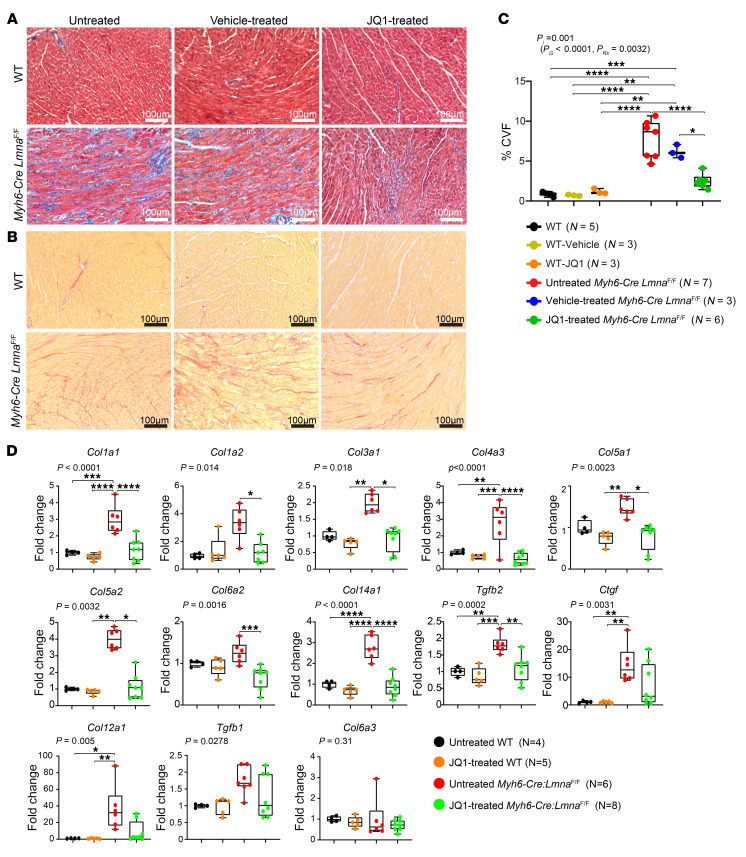
Given the marked dysregulation of EMT signature in the Myh6-Cre LmnaF/F cardiomyocytes and in view of the well-established effects of BRD4 in mediating fibrosis in various organs, effects of JQ1 administration on myocardial fibrosis were assessed by determination of CVF and transcript levels of genes involved in fibrosis (40, 42). CVF comprised 8.3% ± 2.6% and 6.6% ± 0.9% of the myocardial area in the untreated and vehicle-treated Myh6-Cre LmnaF/F mice, respectively (P < 0.01 compared with WT; Figure 9, A–C). Treatment with JQ1 reduced CVF by about 68% (P < 0.0001) in comparison with WT and by approximately 60% (P = 0.013) in comparison with vehicle-treated Myh6-Cre LmnaF/F mice (Figure 9, A and B). Consequently, CVF in JQ1-treated Myh6-Cre LmnaF/F mice was similar to that in the WT mice.
Figure 9. Effect of BET bromodomain protein inhibition on myocardial fibrosis in the Myh6-Cre LmnaF/F mice.
(A and B) Representative Masson’s trichrome–stained (top panels) and Picrosirius red–stained (bottom panels) myocardial sections after 1 week of treatment in 3-week-old untreated, vehicle, and JQ1-treated WT and Myh6-Cre LmnaF/F mice. (C) Corresponding quantitative data showing the percentage of CVF in myocardial sections in untreated (black dots; n = 5), vehicle-treated (yellow dots; n = 3), and JQ1-treated WT (orange dots; n = 3), as well as untreated (red dots; n = 7), vehicle-treated (blue dots; n = 3), and JQ1-treated (green dots; n = 6) Myh6-Cre LmnaF/F, mice. P values were calculated using 2-way ANOVA and Tukey’s post hoc test for comparisons; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (D) Transcript levels of selected BRD4 target genes involved in fibrosis in the heart as quantified by RT-qPCR in untreated (black dots; n = 4) and JQ1-treated (orange dots; n = 5) WT mice, as well as untreated (red dots; n = 6) and JQ1-treated (green; n = 8) Myh6-Cre LmnaF/F mice. P values shown were obtained with ordinary 1-way ANOVA or Kruskal-Wallis; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To corroborate the findings, transcript levels of 13 genes involved in myocardial fibrosis were quantified by RT-qPCR on whole-heart RNA extracts. As shown in Figure 9D, treatment with JQ1 normalized or reduced transcript levels of 12 of 13 (92%) genes in the Myh6-Cre LmnaF/F mouse hearts (Figure 9D). Among genes analyzed, only transcript levels of Col6a3 were not different among 4 experimental groups (Figure 9D).
Effects of JQ1 on myocardial apoptosis.
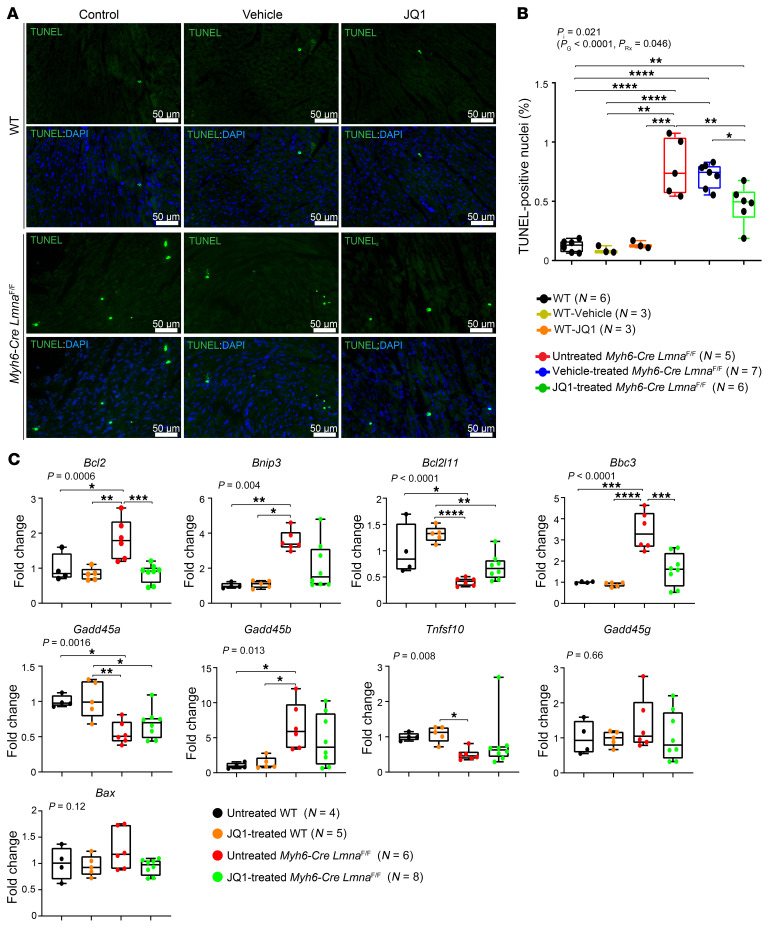
Given increased apoptosis in the myocardium of Myh6-Cre LmnaF/F mice, the effect of treatment with JQ1 on myocardial apoptosis was assessed by TUNEL assay and transcript levels of the molecular markers of apoptosis. The number of TUNEL-positive cells was increased markedly (about 7-fold) in the myocardium of Myh6-Cre LmnaF/F mice at 3 weeks of age (Figure 10, A and B). Treatment with JQ1 was associated with a 50% reduction in the number of TUNEL-positive cells in the myocardium of Myh6-Cre LmnaF/F mice as compared with untreated or vehicle-treated mice (P < 0.009 and P < 0.002, respectively).
Figure 10. Effect of BET bromodomain protein inhibition on myocardial apoptosis in the Myh6-Cre LmnaF/F mice.
(A) Representative TUNEL-stained thin myocardial sections in 3-week-old untreated, vehicle-treated, and JQ1-treated WT mice along with untreated, vehicle-treated, and JQ1-treated Myh6-Cre LmnaF/F mouse hearts. Nuclei were counterstained with DAPI. (B) Corresponding quantitative data showing percentage of TUNEL-labeled nuclei in myocardial sections in untreated (black dots; n = 6), vehicle-treated (yellow dots; n = 3), and JQ1-treated (orange dots; n = 3) WT and untreated (red dots; n = 5), vehicle-treated (blue dots; n = 7), and JQ1-treated (green dots; n = 6) Myh6-Cre LmnaF/F mouse hearts. P values were calculated using 2-way ANOVA and Tukey’s post hoc test for comparisons; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) Transcript levels of selected BRD4 target genes involved in apoptosis, as quantified by RT-qPCR, are depicted in 3-week-old untreated (black dots; n = 4) and JQ1-treated (orange dots; n = 5) WT mice, and in untreated (red dots; n = 6) and JQ1-treated (green dots; n = 8) Myh6-Cre LmnaF/F mouse hearts. P values shown were obtained with ordinary 1-way ANOVA or Kruskal-Wallis; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To corroborate the findings, transcript levels of 9 markers of apoptosis were quantified by RT-qPCR on whole-heart RNA extracts in the experimental groups. Transcript levels of Bcl2, Bnip3, Bbc2, and Bcl2l1 were fully rescued (not different from the WT), whereas that of Gadd45b remained increased in the myocardium of Myh6-Cre LmnaF/F (Figure 10C). Transcript levels of Gadd45a and Tnfsf10, which were reduced in the Myh6-Cre LmnaF/F myocardium, did not change upon treatment with JQ1 (Figure 10C). Finally, treatment with JQ1 had no significant effect on transcript levels of Gadd45g and Bax, which were unaltered in the myocardium of Myh6-Cre LmnaF/F mice (Figure 10C).
Discussion
We report that genetic deletion of the Lmna gene exclusively in cardiomyocytes led to a dose-dependent cardiac dilatation and dysfunction; cardiac arrhythmias, including conduction defects and ventricular arrhythmias; myocardial fibrosis; and apoptosis; and resulted in total mortality within 4 weeks in the homozygous and 15 months in the heterozygous mice. Transcriptomic analysis identified dysregulation of a dozen transcriptional regulators, including the epigenetic reader BRD4, which was activated in the Lmna-null cardiomyocytes. Treatment of Lmna-deficient mice with JQ1, a well-characterized and specific inhibitor of the BET bromodomain proteins, prolonged survival and improved cardiac function, cardiac arrhythmias, myocardial fibrosis, and apoptosis (44). These findings in a genetically defined mouse model of LMNA-DCM set the stage for advancing the findings on beneficial effects of inhibition of BRD4 toward treatment of human patients with DCM in laminopathies.
Because LMNA is a ubiquitously expressed protein, mutations in the LMNA gene are expected to affect multiple cell types (13, 29). The ensuing clinical phenotype associated with the LMNA mutations is the consequence of changes in multiple cell types. Deletion of the Lmna gene exclusively in cardiomyocytes recapitulated several components of LMNA-associated DCM in humans, including cardiac dysfunction, arrhythmias, fibrosis, and apoptosis. The cardiac phenotype in the Myh6-Cre LmnaF/F mice is not distinct from that observed in mice with systemic deletion of Lmna or expressing LMNA with missense mutations but perhaps is more pronounced (28, 53, 54). The similarities in the findings between myocyte-specific and systemic deletion of the Lmna gene signify the crucial role of cardiomyocytes in cardiomyopathies resulting from mutations in the LMNA gene. However, the beneficial effects of treatment with JQ1 might be in part mediated through the effects on systemic delivery of JQ1 on other cardiac cell types, in addition to its effects on cardiomyocytes.
BRD4 was among the top dysregulated epigenetic factors in Myh6-Cre LmnaF/F myocytes. The major function of BRD4 is to promote transcription by binding to acetylated histones, transcription factors, and other chromatin-associated proteins, resulting in recruitment of positive regulators of gene expression and displacement of the negative regulators (55). Consistent with this knowledge, transcript levels of 123 of 145 BRD4 target genes (85%), collected from 4 public data sets and IPA, were increased in Myh6-Cre LmnaF/F myocytes. Furthermore, ChIP-Seq data showed enriched recruitment of BRD4 and overall an increased expression of genes associated with these BRD4 peaks in the Myh6-Cre LmnaF/F myocytes, which provided further evidence in support of activation of BRD4-mediated transcription. Consequently, multiple lines of evidence of activation of BRD4 in cardiomyocytes set the rationale for pharmacological inhibition of BRD4 in Myh6-Cre LmnaF/F myocytes. The approach was also in accord with the prominent phenotype of myocardial fibrosis in the Myh6-Cre LmnaF/F mice and the well-established role of BRD4 in EMT, as well as the emerging role of inhibitors of BRD4 in reversing fibrosis and attenuating myocardial dysfunction (39–42).
The beneficial effects of treatment with JQ1 were concordant across multiple phenotypes, including prolonged survival, reduced cardiac conduction defects and arrhythmias, and improved cardiac function, fibrosis, and apoptosis, which were corroborated by complementary methods whenever applicable. Likewise, administration of JQ1 partially restored dysregulated cardiomyocyte transcriptome, particularly transcripts of the genes involved in inflammation and those encoding cardiac secretome and their upstream regulators, such as the TGF-β1 pathway. Partial restoration of secretome was associated with attenuation of EMT and myocardial fibrosis. Similarly, administration of JQ1 attenuated myocardial apoptosis, which is likely a phenotype effect of the secretome; however, the identity of myocardial cells undergoing apoptosis in the Myh6-Cre LmnaF/F hearts could not be ascertained by the TUNEL assay. Improvement of cardiac function was likely the consequence of effects of JQ1 on myocytes, including secretome, as well as its effects on nonmyocyte cells. The rescue, while remarkable for the relatively large effect sizes on all assessed phenotypes, was incomplete. Treatment with JQ1 delayed premature death, improved cardiac function, and reduced arrhythmias but did not fully abrogate the LMNA-DCM phenotype. The incompleteness of the rescue likely had multiple reasons, including the multifarious nature of the pathways involved in the pathogenesis of LMNA-DCM, such as activation of KDM5A, MAP4K4, and TP53, as well as suppression of RB1 and INSR transcriptional regulators, among others, as well as the study design pertaining to dose, timing, and route of administration of JQ1. Several of the upstream regulatory pathways predicted to be dysregulated in the Myh6-Cre LmnaF/F myocytes, including TP53, have been previously implicated in the pathogenesis of LMNA-associated DCM in humans and mouse models (21, 28). However, administration of JQ1 rescues only a fraction of the dysregulated pathways and a few of them only partially, which in part may explain incomplete phenotypic rescue. The complexity of the LMNA-DCM phenotype would necessitate targeting of multiple pathogenic pathways for a superior therapeutic gain. This notion is in accord with the current therapeutic practice in treatment of complex phenotypes, such as heart failure, wherein the effect size of each therapeutic modality, while significant, is relatively modest, necessitating polypharmacy.
The findings in LMNA-DCM mice are in agreement with previous data on the profibrotic effects of BRD4 and the beneficial effects of JQ1, a first-in-class paninhibitor of BET bromodomain proteins, on attenuation of myocardial fibrosis and cardiac dysfunction in pressure overload and myocardial infarction mouse models (39, 41). Suppression of cardiac arrhythmias upon treatment with JQ1 is remarkable and, to our knowledge, a novel finding. Several mechanisms are likely to be involved in the beneficial effects of JQ1 in heart failure and profibrotic conditions. The core mechanism likely pertains to competitive displacement of BRD proteins by JQ1 from acetyl-lysine recognition motif on histones, and the consequent suppression of aberrant gene expression (44).
The molecular mechanism(s) responsible for activation of BRD4 in the Myh6-Cre LmnaF/F mouse heart is unknown. Coimmunoprecipitation studies do not show an interaction between the LMNA and BRD4 proteins. In addition, transcript levels of Brd4 were not significantly changed in Myh6-Cre LmnaF/F myocytes, suggesting a post-transcriptional mechanism(s) for activation of BRD4, such as hyperphosphorylation, which is typically mediated by CDK9 (56). Previous data suggest that activation of BRD4 in cardiac fibroblasts is mediated, in part, through p38 MAP kinase (MAPK14) and TGF-β1 pathways (42). The activation leads to recruitment of BRD4 to enhancers provoking expression of proinflammatory and profibrotic genes through a yet-to-be-defined mechanism(s) (42). It is noteworthy that TGF-β1 and MAP4K4, the latter known to regulate p38, were among the top activated transcriptional coregulators in Myh6-Cre LmnaF/F myocytes (Figure 3C and refs. 57, 58). Several alternative mechanisms might also be involved, such as chromatin conformational and epigenetic changes in the absence of LMNA, which might lead to activation of BRD4 in cardiomyocytes.
The findings showing beneficial effects of treatment with JQ1 in Myh6-Cre LmnaF/F mice serve as proof-of-principle data that set the stage for additional preclinical studies in heterozygous Lmna-knockout mice and pilot clinical studies in humans to advance the utility of targeting BRD4 in LMNA-DCM. Given the paucity of specific therapies for LMNA-DCM, inhibition of BET bromodomain, with specific and relatively safe drugs, could emerge as an effective therapy that targets a specific pathogenic pathway in LMNA-DCM.
Methods
Detailed methods are provided in Supplemental Methods.
Mouse models.
Myh6-Cre, Myh6-Cre LmnaW/F, and Myh6-Cre LmnaF/F mice were generated per conventional methods (34, 35). Oligonucleotide primers used in PCR reactions are listed in Supplemental Table 8.
Survival.
Survival was analyzed by construction of Kaplan-Meier survival plots.
Echocardiography and electrocardiography.
Cardiac size and function were assessed in age- and sex-matched mice at 2 weeks, 3 weeks, and 10 months by echocardiography, as published (21, 28, 59). Indices of cardiac size and function were measured in 5–6 cardiac cycles. Left ventricular fractional shortening and mass were calculated from the measured indices.
Needle electrodes were placed subcutaneously and connected to an ECG recording unit to obtain surface ECG.
Isolation of cardiomyocytes.
Cardiomyocytes were isolated by collagenase perfusion of the myocardium, as published (28).
Histology.
Myocardial fibrosis was quantified by determination of CVF in myocardial sections as published (28, 59, 60).
Immunoblotting and immunofluorescence.
Expression levels of the proteins of interest were detected and quantified by immunoblotting, as published (21, 59). Immunofluorescence was performed to detect expression and localization of LMNA and PCM1 proteins, using specific antibodies, as published (21, 59, 60). A detailed list of antibodies used is provided in Supplemental Table 8.
Detection of apoptosis.
Apoptosis was detected by nick-end labeling of DNA with the TUNEL assay, as previously described (21, 59, 60). Percentage of TUNEL-positive nuclei was determined in 6400 to 16,500 DAPI-positive nuclei per mouse.
Quantitative real-time PCR.
Transcript levels of selected genes were determined by RT-qPCR, using specific TaqMan gene expression assays or SYBR Green specific primers, and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA levels, as published (21, 59). TaqMan probes and SYBR Green primers are detailed in Supplemental Table 8.
RNA-Seq.
RNA-Seq was performed on ribosome-depleted RNA extracted from the hearts of 2-week-old WT and Myh6-Cre LmnaF/F as well as 3-week-old WT, vehicle-treated, and JQ1-treated mice, as published (21, 59, 60). Samples with an RNA integrity number (RIN) readout of more than 8 were used for library preparation and sequencing to generate 75 or 100 bp runs. DEGs were identified using edgeR, voom, and DESeq2 following RUV (remove unwanted variation) adjustment and annotated using the GENCODE gene model (https://www.gencodegenes.org/mouse) (61–64). Pathway analysis was performed by Gene Set Enrichment Analysis (GSEA, version 2.2.3, http://software.broadinstitute.org/gsea) using the ranked gene list (65). Activation of BRD4 was further validated against BRD4 target genes reported in 4 independent public data sets (39, 41). DEGs encoding the secreted proteins were obtained as published (59).
ChIP-Seq.
Myocytes were isolated, chromatin was cross-linked with 1% formaldehyde, nuclei were extracted and homogenized in a nuclear lysis buffer, and chromatin was sheared by sonication, as published (31). After overnight immunoprecipitation with an anti-BRD4 antibody, the complexes were immunoprecipitated. The chromatin elution was treated with RNase A and proteinase K, and the DNA was extracted using phenol/chloroform and ethanol precipitation. The protocol was also followed for the DNA sample used for sequencing as well as ChIP-PCR. Libraries were prepared, and single reads of 50 bp length were sequenced.
Sequencing of BRD4 ChIP-Seq libraries yielded 36 to 65 million single-end reads. Reads were aligned to the mouse genome (mm10 release) by Bowtie version 2.3.4.3, and uniquely mapping reads were analyzed. ChIP-Seq peaks were called using MACS2 from Galaxy (version 2.1.1) (66). Enriched regions were intersected using Multiple Intersect tools (Galaxy version 2.27.1), and regions present in at least 2 samples per genotype were considered BRD4-bound regions (q < 0.05). BRD4-bound regions that were present in the Myh6-Cre LmnaF/F but absent in the WT samples were considered as gain of peak (GoP) and those that were unique to WT as loss of peak (LoP). Plots showing gene density versus expression were generated using the density plots and the sort map function in R (https://cran.r-project.org/) and EaSeq (http://easeq.net) (67). Data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE142129). ChIP-Seq signal and genomic localization were visualized using Integrative Genomics Viewer (IGV). Genomic locations showing BRD4 enrichment at its putative targets were obtained from IGV.
ChIP-qPCR.
The ChIP-Seq findings were tested for validation in an independent set of ChIP experiment using an anti-BRD4 antibody on cardiomyocyte chromatin extracted from the WT and Mhy6-Cre LmnaF/F mice, as described above. Genomic regions showing enrichment for the BRD4 recruitment were visualized using IGV (68). The sequences flanking the enriched regions were used to design oligonucleotide primers, and qPCR was performed to determine enrichment of the BRD4 recruitment in the WT and Mhy6-Cre LmnaF/F samples.
Statistics.
Normality of data distribution was assessed by Shapiro-Wilk test. Data that followed a Gaussian distribution pattern were presented as mean ± SD and compared using unpaired t test, ordinary 1-way ANOVA, or 2-way ANOVA, followed by Tukey’s or Bonferroni’s multiple-comparisons test, as appropriate. Otherwise, data were presented as the median values and compared by Kolmogorov-Smirnov, Kruskal-Wallis, or Mann-Whitney test followed by Dunn’s test correction for multiple comparison. Survival rates were analyzed by construction of Kaplan-Meier survival plots and comparison of the survival rate by log-rank (Mantel-Cox) test. A q value of less than 0.05 and a P value of less than 0.05 were considered significant.
Study approval.
The animal care and use committee of the University of Texas Health Science Center approved the mouse studies.
Author contributions
GA performed most of the molecular biology and genomic experiments, including preparation of samples for RNA-Seq and ChIP-Seq; performed echocardiography and electrocardiographic rhythm monitoring; analyzed the data; and drafted as well as edited the manuscript. LR performed part of the RT-qPCR and ChIP-qPCR experiments, and immunoblotting. SJM performed bioinformatics analysis of RNA-Seq data. CC performed bioinformatics analysis of RNA-Seq and ChIP-Seq data. MJR performed bioinformatics analysis of RNA-Seq data. GC assisted in mouse genotyping. PG analyzed the RNA-Seq and ChIP-Seq data and edited the manuscript. AJM developed the concept, supervised the experiments, interpreted the findings, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NIH, National Heart, Lung, and Blood Institute (NHLBI R01-HL088498, 1R01-HL132401, R01-HL151737, and S10-OD018135); the Leducq Foundation (14 CVD 03); the National Institute on Aging, NIH (R21-AG060413-01); the American Heart Association (19POST34410032); the Ewing Halsell Foundation; the George and Mary Josephine Hamman Foundation; and the TexGen Fund from the Greater Houston Community Foundation.
Version 1. 06/02/2020
In-Press Preview
Version 2. 08/10/2020
Electronic publication
Version 3. 09/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4740–4758.https://doi.org/10.1172/JCI135922.
See the related Commentary at BETs that cover the spread from acquired to heritable heart failure.
Contributor Information
Gaelle Auguste, Email: Gaelle.Auguste@uth.tmc.edu.
Leila Rouhi, Email: Leila.Rouhigharabaei@uth.tmc.edu.
Scot J. Matkovich, Email: sjmatkovich@lilly.com.
Cristian Coarfa, Email: coarfa@bcm.edu.
Matthew J. Robertson, Email: matthew.robertson@bcm.edu.
Grazyna Czernuszewicz, Email: Grazyna.Z.Czernuszewicz@uth.tmc.edu.
Priyatansh Gurha, Email: Priyatansh.Gurha@uth.tmc.edu.
Ali J. Marian, Email: Ali.J.Marian@uth.tmc.edu.
References
- 1.Benjamin EJ, et al. Heart disease and stroke statistics — 2019 update: a report from the American Heart Association. Circulation. 2019;139(10):e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 2.Savarese G, Lund LH. Global public health burden of heart failure. Card Fail Rev. 2017;3(1):7–11. doi: 10.15420/cfr.2016:25:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrosy AP, et al. The global health and economic burden of hospitalizations for heart failure: lessons learned from hospitalized heart failure registries. J Am Coll Cardiol. 2014;63(12):1123–1133. doi: 10.1016/j.jacc.2013.11.053. [DOI] [PubMed] [Google Scholar]
- 4.McNally EM, Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circ Res. 2017;121(7):731–748. doi: 10.1161/CIRCRESAHA.116.309396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metra M, et al. Advanced chronic heart failure: a position statement from the Study Group on Advanced Heart Failure of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail. 2007;9(6–7):684–694. doi: 10.1016/j.ejheart.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Alraies MC, Eckman P. Adult heart transplant: indications and outcomes. J Thorac Dis. 2014;6(8):1120–1128. doi: 10.3978/j.issn.2072-1439.2014.06.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366(7):619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arbustini E, et al. Autosomal dominant dilated cardiomyopathy with atrioventricular block: a lamin A/C defect-related disease. J Am Coll Cardiol. 2002;39(6):981–990. doi: 10.1016/S0735-1097(02)01724-2. [DOI] [PubMed] [Google Scholar]
- 9.Parks SB, et al. Lamin A/C mutation analysis in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy. Am Heart J. 2008;156(1):161–169. doi: 10.1016/j.ahj.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrzynska A, Gonzalo S, Shanahan C, Askjaer P. The nuclear lamina in health and disease. Nucleus. 2016;7(3):233–248. doi: 10.1080/19491034.2016.1183848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatkin D, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341(23):1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 12.Taylor MR, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41(5):771–780. doi: 10.1016/S0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152(6):1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quarta G, et al. Mutations in the lamin A/C gene mimic arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2012;33(9):1128–1136. doi: 10.1093/eurheartj/ehr451. [DOI] [PubMed] [Google Scholar]
- 15.Anselme F, et al. Implantable cardioverter-defibrillators in lamin A/C mutation carriers with cardiac conduction disorders. Heart Rhythm. 2013;10(10):1492–1498. doi: 10.1016/j.hrthm.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21(3):285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 17.Brodsky GL, Muntoni F, Miocic S, Sinagra G, Sewry C, Mestroni L. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation. 2000;101(5):473–476. doi: 10.1161/01.CIR.101.5.473. [DOI] [PubMed] [Google Scholar]
- 18.van Berlo JH, et al. Meta-analysis of clinical characteristics of 299 carriers of LMNA gene mutations: do lamin A/C mutations portend a high risk of sudden death? J Mol Med. 2005;83(1):79–83. doi: 10.1007/s00109-004-0589-1. [DOI] [PubMed] [Google Scholar]
- 19.van Berlo JH, Duboc D, Pinto YM. Often seen but rarely recognised: cardiac complications of lamin A/C mutations. Eur Heart J. 2004;25(10):812–814. doi: 10.1016/j.ehj.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 20.van Tintelen JP, et al. High yield of LMNA mutations in patients with dilated cardiomyopathy and/or conduction disease referred to cardiogenetics outpatient clinics. Am Heart J. 2007;154(6):1130–1139. doi: 10.1016/j.ahj.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 21.Chen SN, et al. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with LMNA (Lamin A/C) mutations. Circ Res. 2019;124(6):856–873. doi: 10.1161/CIRCRESAHA.118.314238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redwood AB, et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011;10(15):2549–2560. doi: 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graziano S, Kreienkamp R, Coll-Bonfill N, Gonzalo S. Causes and consequences of genomic instability in laminopathies: Replication stress and interferon response. Nucleus. 2018;9(1):258–275. doi: 10.1080/19491034.2018.1454168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbs-Seymour I, Markiewicz E, Bekker-Jensen S, Mailand N, Hutchison CJ. Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell. 2015;14(2):162–169. doi: 10.1111/acel.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho S, et al. Mechanosensing by the lamina protects against nuclear rupture, DNA damage, and cell-cycle arrest. Dev Cell. 2019;49(6):920–935.e5. doi: 10.1016/j.devcel.2019.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroud MJ, Banerjee I, Veevers J, Chen J. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ Res. 2014;114(3):538–548. doi: 10.1161/CIRCRESAHA.114.301236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J, et al. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572(7769):335–340. doi: 10.1038/s41586-019-1406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auguste G, Gurha P, Lombardi R, Coarfa C, Willerson JT, Marian AJ. Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res. 2018;122(5):678–692. doi: 10.1161/CIRCRESAHA.117.312052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Röber RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105(2):365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 30.van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169(5):780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheedipudi SM, et al. Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ Res. 2019;124(8):1198–1213. doi: 10.1161/CIRCRESAHA.118.314177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23(2):270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38(4):603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y, Zheng Y. Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun. 2013;440(1):8–13. doi: 10.1016/j.bbrc.2013.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100(1):169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, Frisén J. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res. 2011;317(2):188–194. doi: 10.1016/j.yexcr.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Bergmann O, Jovinge S. Isolation of cardiomyocyte nuclei from post-mortem tissue. J Vis Exp. 2012;(65):4205. doi: 10.3791/4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preissl S, et al. Deciphering the epigenetic code of cardiac myocyte transcription. Circ Res. 2015;117(5):413–423. doi: 10.1161/CIRCRESAHA.115.306337. [DOI] [PubMed] [Google Scholar]
- 39.Anand P, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154(3):569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stratton MS, Haldar SM, McKinsey TA. BRD4 inhibition for the treatment of pathological organ fibrosis. F1000Res. 2017;6:F1000 Faculty Rev-1015. doi: 10.12688/f1000research.11339.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan Q, et al. BET bromodomain inhibition suppresses innate inflammatory and profibrotic transcriptional networks in heart failure. Sci Transl Med. 2017;9(390):eaah5084. doi: 10.1126/scitranslmed.aah5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stratton MS, et al. Dynamic chromatin targeting of BRD4 stimulates cardiac fibroblast activation. Circ Res. 2019;125(7):662–677. doi: 10.1161/CIRCRESAHA.119.315125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song S, et al. Inhibition of BRD4 attenuates transverse aortic constriction- and TGF-β-induced endothelial-mesenchymal transition and cardiac fibrosis. J Mol Cell Cardiol. 2019;127:83–96. doi: 10.1016/j.yjmcc.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senyo SE, Lee RT, Kühn B. Cardiac regeneration based on mechanisms of cardiomyocyte proliferation and differentiation. Stem Cell Res. 2014;13(3 Pt B):532–541. doi: 10.1016/j.scr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497(7448):249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segatto M, et al. Epigenetic targeting of bromodomain protein BRD4 counteracts cancer cachexia and prolongs survival. Nat Commun. 2017;8(1):1707. doi: 10.1038/s41467-017-01645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter GE, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell. 2017;67(1):5–18.e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman MS, Nguyen T, Watson MJ, Hull RW, Yu HG. Transcriptome profiling reveals novel BMI- and sex-specific gene expression signatures for human cardiac hypertrophy. Physiol Genomics. 2017;49(7):355–367. doi: 10.1152/physiolgenomics.00122.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsybouleva N, et al. Aldosterone, through novel signaling proteins, is a fundamental molecular bridge between the genetic defect and the cardiac phenotype of hypertrophic cardiomyopathy. Circulation. 2004;109(10):1284–1291. doi: 10.1161/01.CIR.0000121426.43044.2B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lutucuta S, et al. Induction and reversal of cardiac phenotype of human hypertrophic cardiomyopathy mutation cardiac troponin T-Q92 in switch on-switch off bigenic mice. J Am Coll Cardiol. 2004;44(11):2221–2230. doi: 10.1016/j.jacc.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arimura T, et al. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum Mol Genet. 2005;14(1):155–169. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 54.Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum Mol Genet. 2005;14(15):2167–2180. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- 55.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 56.Wang R, et al. Uncovering BRD4 hyperphosphorylation associated with cellular transformation in NUT midline carcinoma. Proc Natl Acad Sci U S A. 2017;114(27):E5352–E5361. doi: 10.1073/pnas.1703071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, et al. MAP4K4 deletion inhibits proliferation and activation of CD4(+) T cell and promotes T regulatory cell generation in vitro. Cell Immunol. 2014;289(1–2):15–20. doi: 10.1016/j.cellimm.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, Niswander L. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 2006;125(5):957–969. doi: 10.1016/j.cell.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 59.Cheedipudi SM, et al. Exercise restores dysregulated gene expression in a mouse model of arrhythmogenic cardiomyopathy. Cardiovasc Res. 2020;116(6):1199–1213. doi: 10.1093/cvr/cvz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karmouch J, et al. Distinct cellular basis for early cardiac arrhythmias, the cardinal manifestation of arrhythmogenic cardiomyopathy, and the skin phenotype of cardiocutaneous syndromes. Circ Res. 2017;121(12):1346–1359. doi: 10.1161/CIRCRESAHA.117.311876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32(9):896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23(4):349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 68.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.