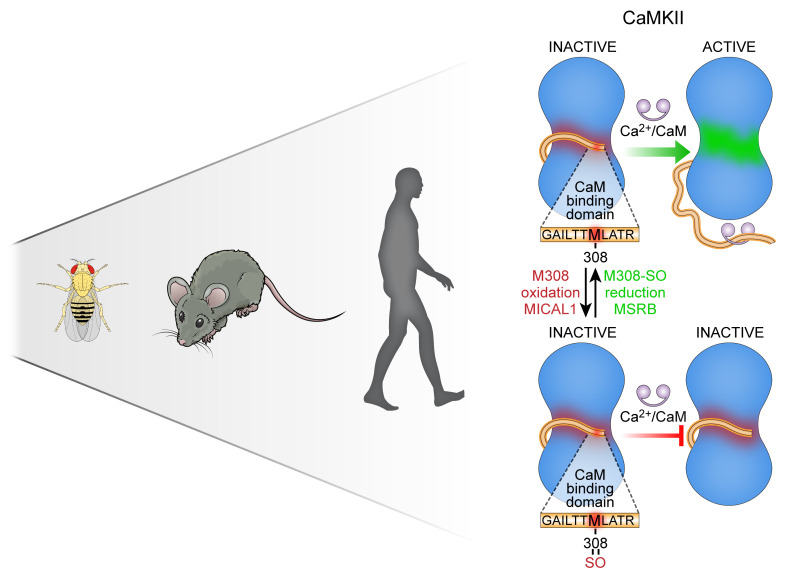
Abstract
Oxidant stress can contribute to health and disease. Here we show that invertebrates and vertebrates share a common stereospecific redox pathway that protects against pathological responses to stress, at the cost of reduced physiological performance, by constraining Ca2+/calmodulin-dependent protein kinase II (CaMKII) activity. MICAL1, a methionine monooxygenase thought to exclusively target actin, and MSRB, a methionine reductase, control the stereospecific redox status of M308, a highly conserved residue in the calmodulin-binding (CaM-binding) domain of CaMKII. Oxidized or mutant M308 (M308V) decreased CaM binding and CaMKII activity, while absence of MICAL1 in mice caused cardiac arrhythmias and premature death due to CaMKII hyperactivation. Mimicking the effects of M308 oxidation decreased fight-or-flight responses in mice, strikingly impaired heart function in Drosophila melanogaster, and caused disease protection in human induced pluripotent stem cell–derived cardiomyocytes with catecholaminergic polymorphic ventricular tachycardia, a CaMKII-sensitive genetic arrhythmia syndrome. Our studies identify a stereospecific redox pathway that regulates cardiac physiological and pathological responses to stress across species.
Keywords: Cardiology, Cell Biology
Keywords: Molecular biology, Protein kinases
Introduction
Ca2+/calmodulin-dependent protein kinase II (CaMKII) is a serine/threonine kinase with important roles in cardiovascular physiology and disease (1–4). Despite the critical role of CaMKII in fight-or-flight responses and human diseases, common pathways shared between vertebrates and invertebrates that regulate CaMKII-mediated cardiac stress responses remain unknown. CaMKII is initially activated after binding calcified calmodulin (Ca2+/CaM) (5). The CaM-binding domain is a discrete region of amino acids that is conserved in all CaMKII isoforms (α, β, γ, and δ) across species (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI133181DS1), and is nearly invariant in more than 120,000 humans in the genome aggregation database (gnomAD). Here we show that MICAL1 (molecule interacting with Cas-L 1), a methionine monooxygenase thought to exclusively target actin (6–8), selectively catalyzes oxidation of M308 to reduce Ca2+/CaM binding and prevent pathological CaMKII activation. MICAL1 together with methionine sulfoxide reductase B (MSRB) (8, 9), a methionine reductase, set the stereospecific redox status of M308 to regulate physiological and pathological cardiac responses to stress. We used in silico modeling, chemical and cellular studies, and developed in vivo models to demonstrate that the redox status of M308 determines physiological and pathological cardiac responses to stress by regulating Ca2+/CaM binding and CaMKII activity in mice and flies. Our models revealed unanticipated evidence that the redox status of M308 is a critical regulator of fight-or-flight responses across species. As a therapeutic proof of concept, we demonstrate that this pathway can be targeted to rescue the arrhythmic phenotype of human induced pluripotent stem cell–derived (hiPSC-derived) cardiomyocytes with catecholaminergic polymorphic ventricular tachycardia (CPVT), a genetic arrhythmia syndrome linked to CaMKII hyperactivation (10–13). Our findings provide evidence for an essential and expanded role of MICAL1 in Ca2+-sensing biology beyond the cytoskeleton, and suggest, based on the conserved nature of the CaMKII Ca2+/CaM-binding domain in all CaMKII isoforms, that our findings will have broad implications for CaMKII signaling in diverse cell types governing multiple biological systems.
Results
The redox state of CaMKII M308 is set by MICAL1 and MSRB to control CaM binding.
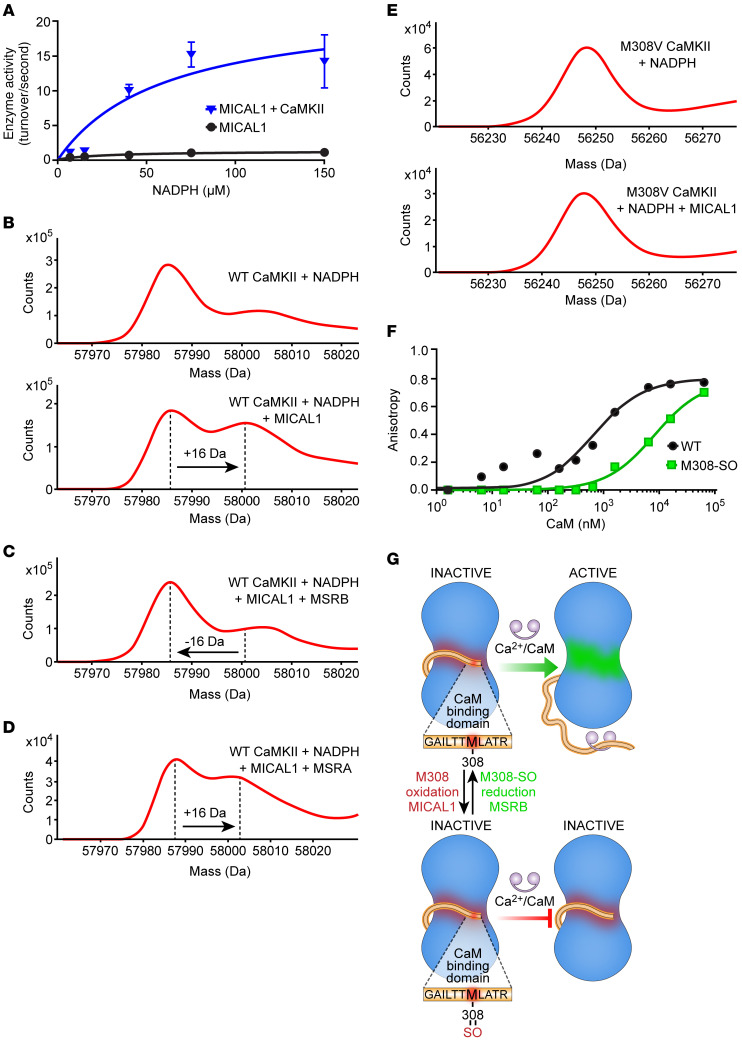
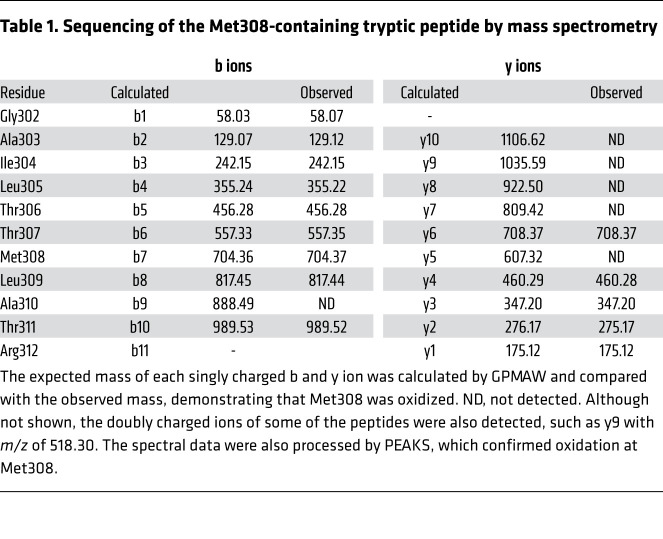
CaMKII function is known to be enhanced by oxidative modifications of regulatory domain methionines (MM281/282) where oxidation prevents CaMKII autoinhibition, causing excessive CaMKII activity (14). However, it is unknown whether CaMKII methionine oxidation is mediated by enzymes. We began our studies with an aim to identify an oxidase capable of activating CaMKII. The family of MICAL proteins represents the only known example of enzymes that catalyze methionine oxidation. MICAL1 is the best-characterized MICAL protein, based on available structure-activity studies (15–17), and it has 10- to 100-fold higher activity at baseline compared with MICAL2 and MICAL3 (18). We incubated full-length CaMKIIδ in the presence of MICAL1 and NADPH (17, 19), and observed a significant increase in NADPH consumption in the presence of CaMKIIδ (Figure 1A), suggesting that CaMKII is either a MICAL1 activator or a substrate. In the presence of MICAL1 and NADPH, mass spectrometry (MS) revealed a +16-Da shift in the mass of CaMKIIδ, consistent with the addition of a single oxygen (Figure 1B), demonstrating that CaMKII is a MICAL1 substrate. In contrast, there was no mass increase for CaMKIIδ induced by NADPH in the absence of MICAL1 (Figure 1B). Methionine oxidation by MICAL1 is stereospecific (8, 9, 20) and generates the R-stereospecific sulfoxide, whereas methionine oxidation by hydrogen peroxide is not stereospecific and generates an equal amount of R- and S-sulfoxide epimers (20). To confirm that MICAL1 oxidation of CaMKII was direct and stereospecific we incubated the MICAL1-oxidized CaMKII with MSRA and MSRB, the S- and R-stereospecific reductases, respectively. We found that MSRB, the R-stereospecific partner of MICAL1 (8, 9), reversed the +16-Da shift (Figure 1C). However, MSRA, the S-stereospecific reductase, with activity toward diverse oxidized proteins, including oxidized CaMKIIδ MM281/282 (14, 21), was ineffective (Figure 1D). These findings indicate that MICAL1 generated only the R-stereospecific CaMKII sulfoxide through a catalytic reaction and not indirectly through hydrogen peroxide formation. CaMKII MS/MS peptide sequencing identified methionine 308 (M308) as the residue oxidized to M308-sulfoxide (M308-SO) by MICAL1 (Figure 1B, Supplemental Figure 1B, and Table 1). Incubation of mutant CaMKIIδ M308V with MICAL1 and NADPH did not produce a mass shift (Figure 1E), confirming the identity of this residue as the only MICAL1 target under these assay conditions. These assays were performed in the absence of Ca2+/CaM, suggesting that CaMKII activation is not required for M308 oxidation by MICAL1.
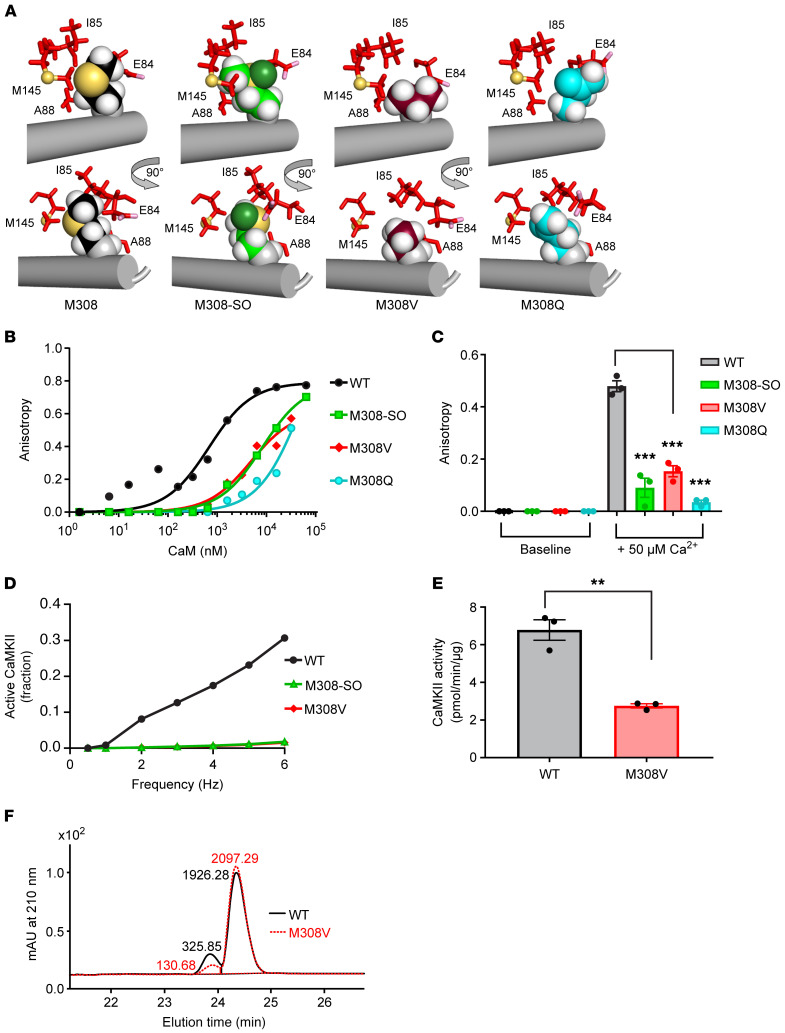
Figure 1. The redox state of CaMKII M308 is set by MICAL1 and MSRB to control CaM binding.
(A) Enzymatic activity of MICAL1 as measured by NADPH absorbance at 340 nm in the absence and presence of CaMKII with increasing concentrations of NADPH. The curves represent nonlinear regression fit derived from experimental data (curves are derived from n = 14 or 15 experimental measurements in each group; each data point represents the average of n = 2–3 measurements at different NADPH concentrations. (B) Mass spectrometry showing a +16-Da shift in the mass of WT CaMKII after incubation with MICAL1 and NADPH (bottom panel) but not with NADPH only (top panel). (C) Methionine sulfoxide reductase B (MSRB) reverses the +16-Da shift in the mass of WT CaMKII treated with MICAL1 and NADPH. (D) Methionine sulfoxide reductase A (MSRA) does not reverse the +16-Da shift in the mass of WT CaMKII treated with MICAL1 and NADPH. (E) Mass spectrometry of CaMKII M308V mutant incubated with NADPH only (top panel). CaMKII M308V mutant does not exhibit a +16-Da shift in mass after incubation with MICAL1 and NADPH (bottom panel). Findings in B–E are representative of at least 2 independent experiments. (F) Fluorescence anisotropy measurements show binding of FITC-labeled M308-sulfoxide (M308-SO) and WT peptides from CaMKII CaM-binding domain at various (n = 11) CaM concentrations. The curves represent nonlinear regression fit derived from the experimental data. Each data point represents the average of n = 2 measurements at each CaM concentration. (G) A schematic model in which the redox status of M308, set by MICAL1 and MSRB, regulates CaM binding, the requisite initial step for CaMKII activation.
Table 1. Sequencing of the Met308-containing tryptic peptide by mass spectrometry.
Analysis of crystallographic structures of fully calcified CaM [(Ca2+)4-CaM] bound to WT CaMKII peptides showed that CaM side chains E84, I85, A88, and M145 make close contacts (<4.5 Å) with reduced M308, with the terminal methyl group of M145 close to the sulfur moiety of M308 (22). Our structural modeling of the M308-SO (R-stereoisomer) predicted an approximately 180° rotation of the M308-SO side chain compared with the side chain of the reduced M308 (Supplemental Figure 1C). The rotation of the M308-SO side chain caused a loss of energetically favorable hydrophobic-hydrophobic interactions and increased destabilizing hydrophobic-hydrophilic interactions with the surrounding CaM amino acids. The M308-SO oxygen is predicted to be solvent exposed. The network of interactions between CaM M145 and Met-SO at position 308 was significantly different from the interface with reduced M308. The sulfur atom of M308-SO interacted more closely with E84 than M145, causing reorientation of both E84 and I85. As CaM M145 is known to interact with multiple amino acids in the CaM-binding domain of CaMKII (22), our findings suggest that the perturbations caused by M308-SO could have secondary effects on the interactions between CaM and other amino acids in the CaM-binding domain of CaMKII. Given our structural analysis and modeling prediction, we designed FITC-labeled peptides of the CaMKII CaM-binding domain to measure CaM binding to M308 in reduced and sulfoxide forms, using fluorescence anisotropy (22). Compared with the peptide synthesized with methionine in the 308 position, the peptide synthesized with M308-SO showed significantly decreased binding to CaM (M308 peptide Kd 0.684 μM, M308-SO peptide Kd 8.2 μM) (Figure 1F). Taken together, these findings established M308 on CaMKII as a potentially novel target for MICAL1-catalyzed oxidation and MSRB-catalyzed reduction in vitro. The known connection between Ca2+/CaM binding and CaMKII activation suggests a model where redox modulation of M308 tunes CaMKII activity (Figure 1G).
CaMKII hyperactivation and increased mortality in MICAL1-knockout mice after cardiac stress.
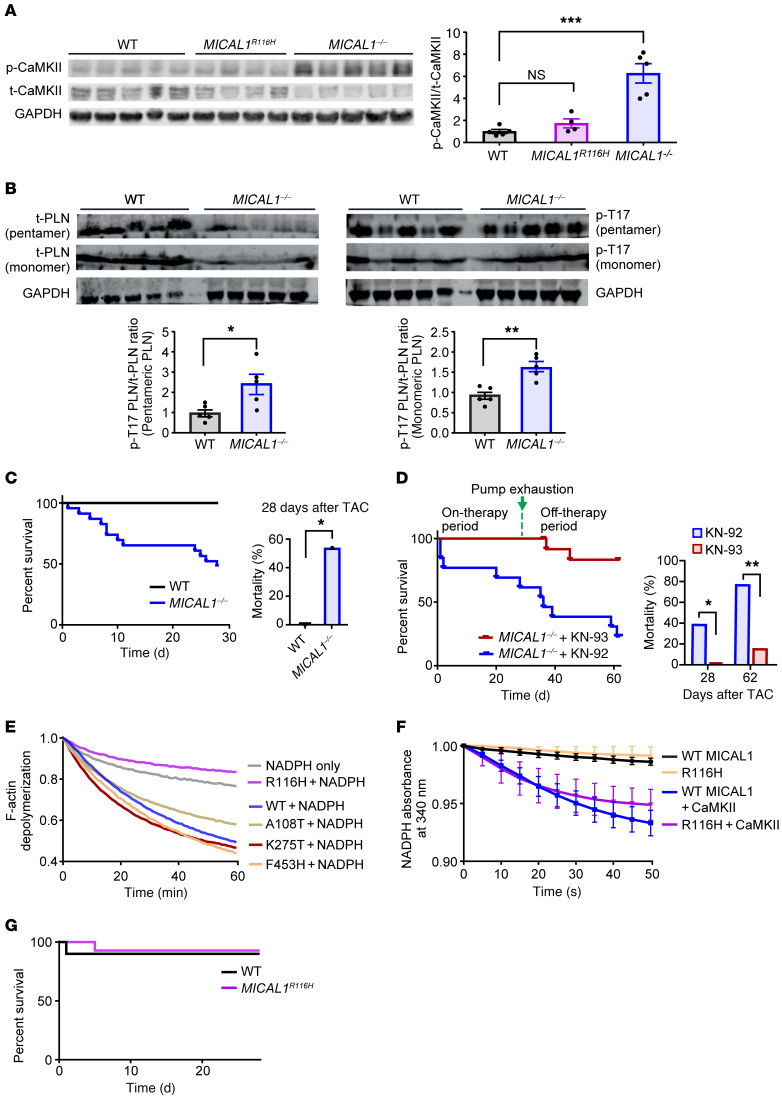
Based on our finding that CaMKIIδ is a stereospecific MICAL1 substrate at M308 in vitro, and that Met308-SO significantly decreases binding to Ca2+/CaM, we next asked if CaMKII activity was increased in hearts from MICAL1-knockout (MICAL1–/–)mice. We found that MICAL1–/– heart lysates exhibited increased CaMKII activation, compared with hearts from WT littermate controls, based on increased levels of autophosphorylated-T287 CaMKII (ref. 23 and Figure 2A; see complete unedited blots in the supplemental material) and phosphorylated phospholamban at T17 (p-T17 PLN), a known downstream target of active CaMKII (ref. 24 and Figure 2B). To assess whether the absence of MICAL1 led to other changes in the sarcoplasmic reticulum (SR) we measured the levels of 2 other SR proteins, ryanodine receptor type 2 (RyR2) and the sarcoplasmic endoplasmic reticulum Ca2+ ATPase (SERCA), but found no difference between WT and MICAL1–/– mice (Supplemental Figure 2, A and B). Moreover, we isolated adult cardiomyocytes from WT and MICAL1–/– mouse hearts and measured intracellular Ca2+ (Supplemental Figure 2C). We did not find any differences at baseline between WT and MICAL1–/– adult mouse cardiomyocytes in cytoplasmic Ca2+ decay slope, diastolic Ca2+ concentration, Ca2+ peak amplitude, or SR Ca2+ content (Supplemental Figure 2C). Excessive CaMKII activation predisposes to adverse responses to pathological stress, including transaortic banding surgery, a model of chronic left ventricular pressure overload (3, 4, 25). Within 4 weeks of transaortic banding surgery, 54% of MICAL1–/– mice had died compared with no deaths in their WT littermates (see Kaplan-Meier survival curve in Figure 2C). To assess the mode of death in MICAL1–/– mice after transaortic banding, we performed serial echocardiograms to monitor cardiac function and implanted subcutaneous telemeters to assess for cardiac arrhythmias. There was no difference in left ventricular function between WT and MICAL1–/– mice at baseline, 3 days, 7 days, and 14 days after transaortic banding (Supplemental Figure 2D). Two MICAL1–/– mice died during ECG monitoring. Both mice had significant arrhythmias preceding their death — the first mouse had severe bradycardia and the second mouse exhibited episodes of a polymorphic agonal rhythm (Supplemental Figure 2E) — suggesting that excess mortality was linked to cardiac arrhythmias, a known consequences of excessive CaMKII activity (26, 27). In order to determine whether CaMKII hyperactivation was sufficient to explain the increased mortality in MICAL1–/– mice, we repeated the aortic banding studies in MICAL1–/– mice infused with either KN-93, a small-molecule CaM and CaMKII inhibitor, or with an inactive KN-93 congener, KN-92, using osmotic minipumps that can deliver these drugs for 4 weeks (Supplemental Methods and refs. 25, 28). KN-93, but not KN-92, infusion rescued the mortality in MICAL1–/– mice after aortic banding surgery during the drug infusion period (see Kaplan-Meier survival curve in Figure 2D). Interestingly, mortality in the MICAL1–/– KN-93–treated group was evident after pump exhaustion, between 4 and 8 weeks (see Kaplan-Meier survival curve in Figure 2D). These findings suggest that CaMKII is excessively activated in the hearts of mice lacking MICAL1, and that hyperactivation of CaMKII contributes to excess mortality after pathological myocardial stress.
Figure 2. MICAL1–/– mice show hyperactivation of CaMKII at baseline and increased mortality with pathological cardiac stress.
(A and B) Hearts of MICAL1–/– mice exhibit higher levels of (A) active, autophosphorylated-T287 CaMKII (p-CaMKII) and (B) phosphorylated phospholamban at T17 (p-T17 PLN) at baseline compared with WT littermate mice. MICAL1R116H mice do not exhibit increased p-CaMKII at baseline. Quantification of the Western blot data is shown (right panel in A and bottom panels in B) (WT n = 5 mice, MICAL1R116H n = 4 mice, MICAL1–/– n = 5 mice). t-CaMKII, total CaMKII; t-PLN, total PLN. (C) Survival curve of WT littermate and MICAL1–/– mice within 4 weeks of transaortic constriction (TAC). WT n = 8, MICAL1–/– n = 24; P = 0.01. (D) Mortality of the MICAL1–/– mice at 28 days is reversed by administration of KN-93 (n = 13) via osmotic minipumps (drug delivery for 28 days), but not its inactive congener KN-92 (n = 13). Mortality reappears in the MICAL1–/– KN-93–treated group after pump exhaustion (P < 0.05 at 4 weeks, P < 0.01 at 8 weeks). Quantification of the mortality data (left panel) is shown (right panels). (E) Screening of various MICAL1 mutants identifies R116H that does not depolymerize F-actin. Depolymerization of pyrene F-actin is measured as a reduction in the fluorescence signal of pyrene F-actin (excitation wavelength at 350 nm and emission wavelength at 407 nm). Data represented as the mean of n = 2–3 replicates. (F) R116H accelerates NADPH consumption at the same rate as the WT MICAL1 after addition of CaMKII (measured as absorbance at 340 nm). Data represented as the mean of n = 3 replicates. (G) Survival curve of WT and MICAL1R116H mice within 4 weeks of transaortic constriction (WT n = 10, MICAL1R116H n = 14) shows no difference between WT littermate and MICAL1R116H mice (P = 1). *P < 0.05; **P < 0.01; ***P < 0.001 by 1-way ANOVA with Tukey’s multiple-comparisons test (A), 2-tailed Student’s t test (B), or 2-tailed Fischer’s exact test (C and D).
MICAL1 R116H shows selective loss of actin targeting and retains protection against CaMKII hyperactivation.
F-actin has been the only known substrate for MICAL1 (6, 7). Given the important role of actin in cardiac function, the known effects of MICAL1 on actin dynamics (6, 7, 29, 30), and the potential contribution of defective actin dynamics to increased mortality in MICAL1–/– mice, we screened MICAL1 mutants with the goal to identify a mutant protein that could distinguish F-actin and CaMKII (Figure 2E). We found that MICAL1 R116H was unable to support F-actin depolymerization (Figure 2E), but sustained its ability to accelerate NADPH oxidation in the presence of CaMKII at the same level as WT MICAL1 (Figure 2F).
We used computational modeling to suggest a potential structural basis for the differences in activities seen between WT MICAL1 and R116H for CaMKII and actin. To catalyze the stereospecific oxidation of M308 by O2, MICAL1 requires the binding of CaMKII with its M308 sulfur group in closer proximity to the isoalloxazine of the reduced FAD (Supplemental Figure 2F). Our structural analysis revealed that M308 is well positioned for oxidation by the hydroxyperoxide group of FAD (Supplemental Figure 2F). The MICAL1 monooxygenase domain opens its catalytic site after cofactor reduction (16), facilitating the protein-substrate binding to catalyze its oxidation (Supplemental Figure 2, F and G). Modeling showed that R116 is located in a critical site of the MICAL1 monooxygenase domain (15, 16), interacts with the ribose moiety of FAD, and participates in the FAD adenosine-binding pocket (Supplemental Figure 2, F and G). This pocket anchors the FAD tail, allowing the isoalloxazine head to bend and swing into the catalytic site upon FAD reduction. Compared with WT, the R116H mutation weakened the MICAL1–FAD adenosine interaction by widening the MICAL1-binding pocket, and removing a polar interaction with its ribose moiety (Supplemental Figure 2G). In addition, this mutation removed a positive charge stabilizing the FAD phosphates. These perturbations may impair positioning of the isoalloxazine head and weaken the coupling between FAD reduction and MICAL1 conformational changes. We postulate that, in contrast to the flexible linker between the CaMKII association and regulatory domains, F‑actin is a less adaptable substrate for MICAL1 R116H. Taken together, our experimental findings and enzymatic and structural modeling suggest that R116H MICAL1 has differential effects on CaMKII and F-actin compared with WT MICAL1.
In order to separate the effects of MICAL1 on actin and CaMKII in vivo, we developed a MICAL1 R116H–knockin (MICAL1R116H) mouse using CRISPR/Cas9 technology (Supplemental Figure 2H and Methods). MICAL1R116H mice were born at Mendelian ratios with no obvious morphometric differences compared with their WT littermates. At baseline, MICAL1R116H mice, unlike MICAL1–/– mice, did not exhibit elevated levels of myocardial autophosphorylated-T287 CaMKII (Figure 2A), indicating that R116H sustains its ability to oxidize CaMKII in vivo and that oxidation of CaMKII M308 by MICAL1 prevents excessive CaMKII activity. The MICAL1R116H mice were protected from the excessive mortality seen in MICAL1–/– mice following transaortic banding surgery (Figure 2G). F-actin staining with phalloidin did not reveal evidence of disordered actin at baseline in MICAL1–/– hearts, nor in MICAL1R116H hearts, compared with WT littermate controls (Supplemental Figure 2I), suggesting redundancy in actin regulation among the MICAL isoforms in the heart. We interpret our data to suggest that MICAL1 constrains CaMKII activity in vivo, that loss of MICAL1 activity is sufficient to promote CaMKII-triggered heart disease, and that these observations are not due to loss of MICAL1 functions related to actin oxidation.
M308 is a redox switch that determines CaMKII activity.
Based on the apparent importance of MICAL1 for constraining CaMKII activity and the identification of M308 in the CaM-binding domain of CaMKII as a critical MICAL1 target, we returned to structural modeling to interrogate the relationship between oxidized and reduced CaMKII M308, and M308 mutants (M308V and M308Q), to Ca2+/CaM binding (Figure 3A). Computational modeling of M308V suggested that M308V reduces affinity for (Ca2+)4-CaM, as the contact surface area between valine and (Ca2+)4-CaM would be about half of that observed for reduced Met, with a complete loss of close contacts (≤4.5 Å) with CaM I85 and M145 (Figure 3A), leaving a gap between M308V and CaM. M308Q was predicted to be the worst perturbation. Similar to M308V, it reduced the contact surface between CaM and CaMKII to approximately 75% of that of WT. Reorientation of M308Q eliminated close contacts with the atoms of M145. Similar to the M308-SO side chain, M308Q moved away from the CaM pocket toward the solvent. There were additional destabilizing interactions in the M308Q mutant relative to M308-SO. Thus, our structural models suggested that the reduced form of methionine would be optimal for binding (Ca2+)4-CaM, and predicted that oxidation of methionine to M308-SO, or replacement of methionine with valine or glutamine, would reduce affinity for (Ca2+)4-CaM. To test this hypothesis, we designed additional FITC-labeled peptides modeled after the CaMKII CaM-binding domain to measure Ca2+/CaM binding using fluorescence anisotropy. Compared with the peptide synthesized with M308, peptides synthesized with M308-SO or mutated to M308V or M308Q showed significantly decreased binding to CaM in titrations over a wide range of CaM concentrations and after the addition of Ca2+ (Figure 3, B and C). Because MICAL1 oxidation produces the R-stereospecific sulfoxide (8, 9), we next asked if the R-stereospecific M308-SO peptide would lead to decreased Ca2+/CaM binding. The process for synthesizing the M308-SO–containing peptide produced a racemic mixture of both R and S epimers (Supplemental Figure 3A). We treated the racemic peptide mixture with MSRA to reduce the S-stereospecific M308-SO and then purified the R-stereospecific M308-SO–containing peptide (Supplemental Figure 3A). The R-stereospecific M308-SO peptide showed decreased binding to Ca2+/CaM compared with the peptide with reduced methionine (Supplemental Figure 3B), indicating that R-stereospecific oxidation of M308 was sufficient to reduce Ca2+/CaM binding.
Figure 3. Met308 is critical for CaM binding and CaMKII activity.
(A) Model interfaces between (Ca2+)4-CaM residues E84, I85, A88, and M145 and CaMKII residues M308 (WT), M308-SO, M308V, or M308Q shown from 2 viewpoints (rotated by 90° about the vertical y axis). CaM side chains are red sticks, with E84 carboxyl oxygens (OE1, OE2) highlighted in pink, M145 sulfur (SD) atom shown as a yellow-orange sphere, CaMKII WT M308 (black), mutant M308-SO (green), M308V (burgundy), and M308Q (cyan) heteroatoms are shown as spheres; C-α carbon atoms (gray), hydrogen atoms (white), M308 and M308-SO, sulfur (yellow-orange), side chain oxygen of M308-SO (forest green). Models were aligned based on the backbone of CaMKII residues 294–311 (gray cylinder). (B) Fluorescence anisotropy measurements show binding of WT CaMKII CaM-binding-domain peptide compared with peptides with M308-SO, M308V, or M308Q at various (n = 9–11) CaM concentrations (WT and M308-SO data are the same as shown in Figure 1F). The curves represent nonlinear regression fit derived from the experimental data. Each data point represents the average of n = 2 measurements at each CaM concentration. (C) Fluorescence anisotropy measurements of the same peptides as B at baseline and after the addition of 50 μM Ca2+. Data represented as the mean of n = 3 replicates ± SEM. (D) Modeling of WT CaMKII holoenzyme activity compared with M308-SO CaMKII and M308V CaMKII with increasing frequency stimulation. Note that M308-SO and M308V curves are superimposed. (E) CaMKII kinase activity for WT and M308V recombinant proteins using syntide as a substrate with radioactive 32P-ATP. Data represented as the mean of n = 3 replicates. Bars denote mean ± SEM. (F) CaMKII kinase activity for WT and M308V recombinant proteins using syntide as a substrate with nonradioactive ATP using HPLC (representative image of n = 2 replicates). mAU, milliabsorbance units. **P < 0.01; ***P < 0.001 by 1-way ANOVA with Tukey’s multiple-comparisons test (C) or 2-tailed Student’s t test (E).
We next used a computational model of CaMKII activation within the holoenzyme structure (see Supplemental Methods) to determine the consequences of diminished Ca2+/CaM binding affinity in oxidized compared with reduced M308. Ca2+/CaM binding initially disrupts autoinhibition of the catalytic subunit, permitting T287 autophosphorylation between neighboring subunits (5). T287 autophosphorylation sustains CaMKII activity in the event of Ca2+/CaM dissociation, but it also simultaneously causes Ca2+/CaM trapping that biases against inactivation and autoinhibition. CaMKII activation responds to the frequency and duration of cytoplasmic Ca2+ transients (31). Using computational modeling (see Supplemental Methods), we compared CaMKII activity between WT, M308-SO, and M308V across a range of CaM and Ca2+ concentrations at various Ca2+ concentration pulse frequencies and durations (Figure 3D and Supplemental Figure 3, C and D). Our modeling data predicted that M308 oxidation prevents a relentless, feed-forward CaMKII activation process, providing a critical checking mechanism to constrain excessive CaMKII activity (Figure 3D and Supplemental Figure 3, C and D). Based on these results, we directly measured the consequences of CaMKII activity using an artificial substrate to assess the role of M308. Because the M308-SO and M308V peptides showed similarly reduced Ca2+/CaM binding affinity (Figure 3, B and C), and were predicted by our modeling to have similar effects on CaMKII activity (Figure 3D and Supplemental Figure 3, C and D), we used full-length M308V CaMKIIδ to quantify the impact of Met308 oxidation on Ca2+/CaM-dependent activation. Using 2 independent CaMKII activity assays (see Supplemental Methods), we verified decreased activity of M308V compared with WT CaMKIIδ (Figure 3, E and F), suggesting that M308V is hypomorphic but not inactive. Taken together, these studies show that M308-SO and M308V profoundly reduce Ca2+/CaM binding, and subsequent CaMKII activity.
M308 constrains CaMKII responses in cardiac myocytes and in vivo in mice.
CaMKII is activated by Ca2+/CaM and, in turn, catalyzes the phosphorylation of Ca2+ homeostatic proteins in myocardium (2). Phospholamban is a CaMKII substrate (32) and an inhibitor of SERCA2a that operates to sequester cytoplasmic Ca2+ to the SR, allowing myocardial relaxation. Based on our finding that M308 oxidation could reduce Ca2+/CaM binding and CaMKII activation, we next used computational modeling of myocardial Ca2+-homeostatic proteins in the presence and absence of M308 oxidation (Supplemental Figure 4, A and B). This modeling predicted a slowing of Ca2+ transient decay in the M308V model compared with WT under conditions of elevated CaMKII activity (Supplemental Figure 4B, left panel). Furthermore, the difference between WT and M308V Ca2+ dynamics in the model was dependent on CaMKII targeting of phospholamban and SR Ca2+ uptake (Supplemental Figure 4B, right panel). We next developed a CaMKIIδ M308V–knockin (CaMKIIδM308V) mouse (Supplemental Figure 4C and Methods) to test the consequences of reduced Ca2+/CaM binding and CaMKII activation in cardiac myocytes and in vivo. Ventricular myocytes isolated from CaMKIIδM308V mice showed a significant decrease in the cytoplasmic Ca2+ concentration decay rate during diastole compared with ventricular myocytes isolated from WT littermates (Figure 4A). These data suggested that the consequences of M308 oxidation, mimicked in M308V ventricular myocytes, affect intracellular Ca2+ fluxes, at least in part, by slowing uptake of Ca2+ by SERCA2a.
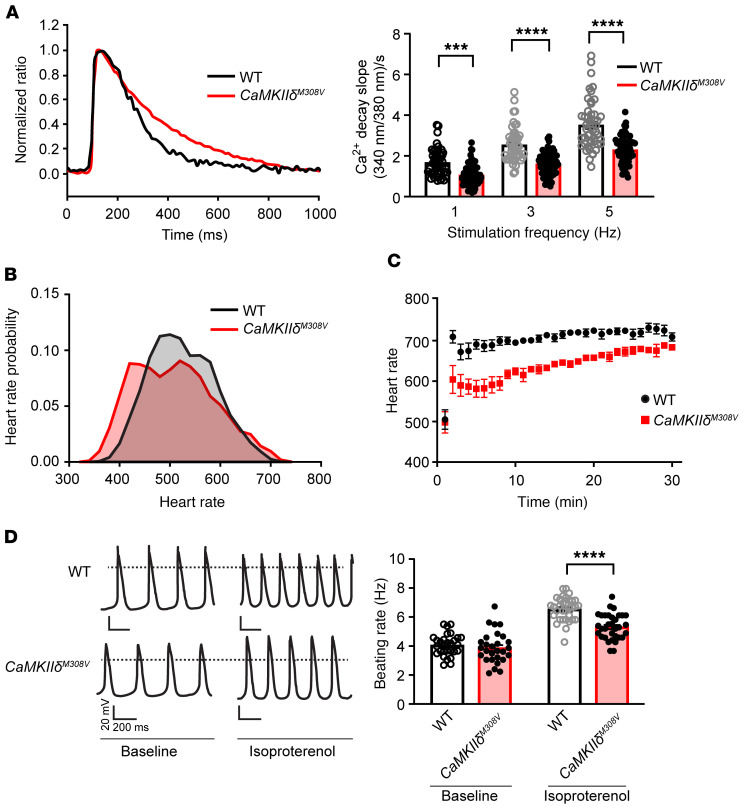
Figure 4. M308 determines fight-or-flight responses in mouse heart.
(A) Left: Representative Ca2+ transients from ventricular myocytes isolated from WT and CaMKIIδM308V mice. Right: Diastolic Ca2+ decay rates in WT and CaMKIIδM308V ventricular myocytes at various pacing frequency stimuli (WT n = 4 mice, n = 49 cells; CaMKIIδM308V n = 4 mice, n = 66–72 cells). (B) Heart rate at baseline averaged over a 24-hour period recorded by implanted telemeters in WT and CaMKIIδM308V mice (WT n = 5, CaMKIIδM308V n = 7). WT median heart rate = 520 bpm, CaMKIIδM308V median heart rate = 505 bpm (P < 0.0001). (C) Heart rate responses to isoproterenol injection (0.4 mg/kg/mouse, intraperitoneal) in WT compared with CaMKIIδM308V mice (WT n = 5, CaMKIIδM308V n = 7). (D) Left: Representative tracings of beating rate of isolated sinoatrial cells from WT and CaMKIIδM308V mice at baseline and after isoproterenol treatment (right panel). Right: Quantitative analysis of beating rates (WT n = 6 mice, n = 28–33 cells; CaMKIIδM308V n = 7 mice, n = 28–33 cells). ***P < 0.001, ****P < 0.0001 by 1-way ANOVA with Tukey’s multiple-comparisons test (A and D).
CaMKII inhibition slows heart rate in mice by reducing SR Ca2+ release (1, 33). Based on our findings of slowed cytosolic Ca2+ uptake in cardiac myocytes, we asked if CaMKIIδM308V mice would exhibit reduced heart rates. We found that unrestrained CaMKIIδM308V mice, implanted with heart rate and activity monitors, showed significantly lower heart rates compared with WT littermate controls averaged over a 24-hour period (Figure 4B). Furthermore, CaMKIIδM308V mice had diminished heart rate acceleration (Figure 4C) with less autophosphorylated T287 CaMKII compared with WT mice following isoproterenol injection (Supplemental Figure 4D and Supplemental Methods). Heart rate is a fundamental physiological response in which cardiac pacemaker cells play a predominant role, integrating autonomic nervous system inputs (34). In order to determine whether the heart rate slowing in CaMKIIδM308V mice was due to innate characteristics of sinoatrial nodal pacemaker cells, we measured spontaneous and isoproterenol-stimulated rates in isolated sinoatrial nodal pacemaker cells. Consistent with our in vivo findings, sinoatrial cells from CaMKIIδM308V mice exhibited significantly slower responses to isoproterenol treatment compared with sinoatrial cells isolated from WT littermates (Figure 4D). Taken together, these findings show that M308V impairs fight-or-flight physiological cardiac responses in mice.
Loss of normal heart tube physiology and altered circadian rhythms in M308V Drosophila mutants.
In contrast to Homo sapiens and Mus musculus that express 4 CaMKII isoforms, Drosophila melanogaster has a single CaMKII, and like Homo sapiens and Mus musculus, M308 is conserved in the CaM-binding domain of Drosophila (Supplemental Figure 1A). We next sought to investigate whether the redox status of M308 is shared among vertebrates and invertebrates to regulate physiological and pathological cardiac function. Based on our Ca2+/CaM-binding assay results (Figure 3, B and C) and modeling of CaMKII activity (Figure 3D and Supplemental Figure 3, C and D), we developed CaMKII M308V–knockin (CaMKIIM308V) Drosophila melanogaster using CRISPR/Cas9 technology (Supplemental Figure 5A and Methods) to mimic the loss of Ca2+/CaM binding associated with M308 oxidation by MICAL1. CaMKII has been shown to regulate the core circadian oscillator, across multiple species (35). CaMKII activation accelerates the circadian rhythm cycle through phosphorylation of core clock proteins such as CLOCK and loss-of-function manipulations in Drosophila melanogaster and mice result in increased period length (35, 36). In addition, hypoactive CaMKII variants are known to prolong circadian rhythm in Clunio marinus midges (37). To assess whether the M308V mutation results in a hypoactive CaMKII variant in flies, we monitored locomotor activity of WT and CaMKIIM308V flies in constant darkness (see Supplemental Methods). Consistent with a hypoactive CaMKII variant, CaMKIIM308V flies had a longer circadian rhythm period compared with WT flies (Figure 5, A and B). In addition, CaMKIIM308V flies had decreased average locomotion (Figure 5C) and sustained wake-sleep rhythm strength (Figure 5D) under constant darkness compared with WT flies.
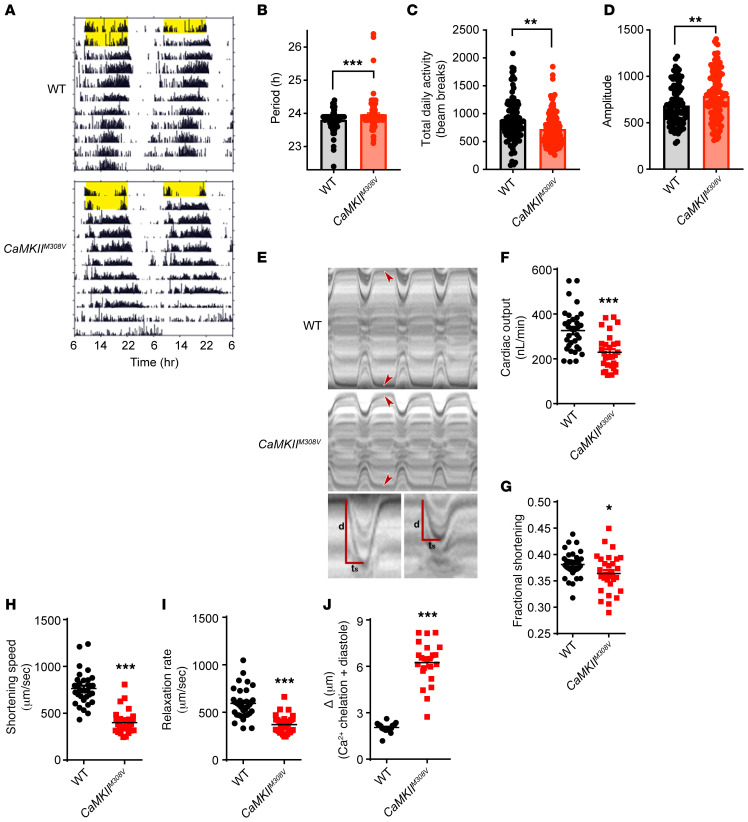
Figure 5. M308 determines circadian rhythms and heart tube physiology in Drosophila melanogaster.
(A) Representative actograms of WT and CaMKIIM308V flies. Actograms are double plotted with yellow bars representing light exposure. (B) CaMKIIM308V flies have a significantly longer free-running circadian period, as estimated by χ2 periodogram (WT n = 99, CaMKIIM308V n = 120). Bars denote mean ± SEM. (C) CaMKIIM308V flies exhibit significantly decreased average daily locomotion during constant darkness, as measured by beam-break counts per day (WT n = 99, CaMKIIM308V n = 120). Bars denote mean ± SEM. (D) Circadian χ2 amplitude indicates increased rhythm strength of CaMKIIM308V flies under constant darkness (WT n = 99, CaMKIIM308V n = 120). Bars denote mean ± SEM. (E) Representative M-mode kymograms generated from high-speed videos of beating WT and CaMKIIM308V fly heart tubes. Top panel shows cardiac cycle dynamics and heart wall contraction over time. Bottom panel shows individual systolic intervals of WT and CaMKIIM308V flies. CaMKIIM308V flies exhibit reduced wall movement distance (d) during systole and a prolonged time of shortening (ts) resulting in significantly diminished shortening speed. (F–I) CaMKIIM308V flies have markedly impaired heart function compared with WT flies. CaMKIIM308V flies at baseline exhibit (F) decreased cardiac output, (G) decreased fractional shortening, (H) decreased shortening speed, and (I) decreased relaxation rate (WT n = 33, CaMKIIM308V n = 30). (J) CaMKIIM308V heart tube diastolic diameter is markedly increased after intracellular Ca2+ chelation with EGTA-AM compared with WT heart tubes (WT n = 20, CaMKIIM308V n = 23). *P < 0.05, **P < 0.01, ***P < 0.001 by 2-tailed Student’s t test (B–D and F–I) or 2-tailed Mann-Whitney test (J).
CaMKII activation is critical for mouse cardiac fight-or-flight physiology (1, 38); however, the role of CaMKII in Drosophila heart tube function is not known. Heart contraction and relaxation are regulated by Ca2+ dynamics and Ca2+-dependent steric modulation of myosin cross-bridge cycling on actin (39). To test for potential effects of CaMKII M308 on heart tube physiology we assessed beating hearts using high-speed video microscopy and motion analysis programs (Figure 5E, Supplemental Methods, and ref. 40). Compared with the WT flies, CaMKIIM308V flies had strikingly impaired cardiac function. CaMKIIM308V flies had significantly lower cardiac output (Figure 5F), lower fractional shortening (Figure 5G), markedly decreased shortening speed (Figure 5H) and relaxation rate (Figure 5I), significantly smaller diastolic and systolic diameters (Supplemental Figure 5, B and C), and shorter diastolic intervals (Supplemental Figure 5D). Our findings in Drosophila melanogaster hearts suggested that oxidized M308 diminishes fight-or-flight responses in flies. Given the known effects of CaMKII on Ca2+ dynamics, and our modeling and experimental findings showing slowed intracellular Ca2+dynamics in CaMKIIδM308V mouse cardiomyocytes (Figure 4A and Supplemental Figure 4B), we hypothesized that CaMKIIM308V flies have impaired heart tube physiology due, at least in part, to altered Ca2+ dynamics. To test this hypothesis, we treated WT and CaMKIIM308V flies with extracellular and intracellular Ca2+ chelators (EGTA and EGTA-AM, see Supplemental Methods). After Ca2+ chelation, CaMKIIM308V flies showed a significantly greater increase in heart tube lumen diameter during diastole compared with WT (Figure 5J and Supplemental Figure 5E), suggesting impaired Ca2+ dynamics in CaMKIIM308V heart tubes. Our findings in Drosophila melanogaster demonstrate the critical role of M308 redox status in invertebrate cardiac function regulation.
M308V protects against a genetic human arrhythmia.
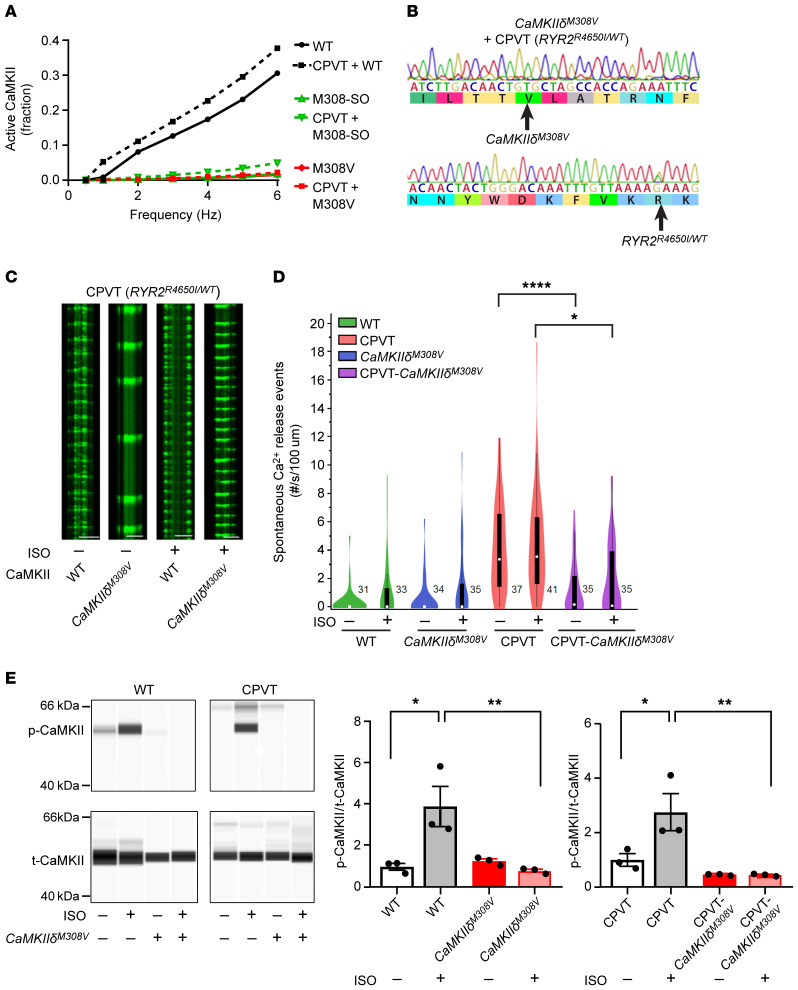
CPVT is a dominant genetic arrhythmia syndrome due to mutations that destabilize the closed state of the cardiac ryanodine receptor (RyR2) intracellular Ca2+ channel (41). CPVT heart cells have increased RyR2 Ca2+ leak, commonly measured as spontaneous Ca2+ release events (42), and cell membrane hyperexcitability due to loss of intracellular Ca2+ homeostasis (13, 41). CaMKII hyperactivation contributes to CPVT because it is bound to RyR2 and catalyzes phosphorylation of serine 2814 that further enhances RyR2 Ca2+ release from intracellular stores (43). Importantly, CaMKII inhibition can prevent CPVT in a humanized knockin mouse model (11), suggesting that identification of new pathways for inhibiting CaMKII could have therapeutic benefits in CPVT. In CPVT the genetic instability of RyR2 induces Ca2+ leak that activates CaMKII, and activated CaMKII amplifies the RyR2 Ca2+ leak, creating a feed forward, proarrhythmic phenotype. Based on mathematical modeling, we predicted that modification of CaMKII M308 by oxidation or mutation to M308V could significantly inhibit CaMKII activity in CPVT (Figure 6A). To directly study the potential role of M308 oxidation in CPVT, we introduced M308V in hiPSCs with CPVT (RYR2R4650I/WT) using CRISPR/Cas9 technology. Using Sanger sequencing, we confirmed the presence of M308V and CPVT mutations in the hiPSCs (Figure 6B), and did not find any of the potential CRISPR-mediated off-target effects on genes predicted by computational analysis as the top potential off-targets (see Methods). We confirmed that hiPSCs met a cardiomyocyte differentiation profile using cTnT staining measured by flow cytometry (Supplemental Figure 6). The hiPSC-derived cardiomyocytes harboring the CPVT mutation exhibited increased spontaneous Ca2+ release event frequency, both at baseline and after isoproterenol (Figure 6, C and D), consistent with the reported CPVT cellular arrhythmia phenotype (11, 41). In contrast, CPVT hiPSC–derived cardiomyocytes harboring CaMKIIδM308V were resistant to increased spontaneous Ca2+ release events (Figure 6, C and D). As expected, the CaMKIIδM308V mutant WT and CPVT hiPSC–derived cardiomyocytes showed significantly lower T287 autophosphorylated CaMKII following isoproterenol challenge compared with isogenic hiPSC-derived cardiomyocytes with WT CaMKIIδ (Figure 6E). Taken together, these in vivo and in vitro findings confirm a role for M308 in determining fundamental physiological and disease responses to CaMKII.
Figure 6. Prevention of CaMKII hyperactivation by M308V rescues catecholaminergic polymorphic ventricular tachycardia (CPVT).
(A) Modeling comparing WT, M308-SO, and M308V CaMKII holoenzyme activity under normal and CPVT conditions. Note that M308-SO and M308V curves under normal and CPVT conditions are superimposed. (B) Successful introduction of M308V (CaMKIIδM308V) in iPSCs from patients with CPVT (RyR2R4650I/WT) was confirmed with Sanger sequencing. (C) Representative confocal line scans of hiPSC-derived cardiomyocytes expressing GCaMP6f-Junctin nanosensor at baseline and after isoproterenol (ISO) treatment. Horizontal scale bars: 20 μm. (D) Spontaneous Ca2+ release events measured with the GCaMP6f-Junctin nanosensor in hiPSC-derived cardiomyocytes from controls (WT), patients with CPVT (CPVT), and their isogenic lines with homozygous CaMKIIδM308V. Recordings at baseline and after isoproterenol treatment (WT n = 31–33 cells, WT-CaMKIIδM308V n = 34–35 cells, CPVT n = 37–41 cells, CPVT-CaMKIIδM308V n = 35 cells). (E) Left: Western blots showing autophosphorylated-T287 CaMKII (p-CaMKII) and total CaMKII (t-CaMKII) in WT and CPVT hiPSC–derived cardiomyocytes with WT or CaMKIIδM308V at baseline and after isoproterenol treatment. Quantification is shown in right panels (n = 3 in each group). *P < 0.05; **P < 0.01; ****P < 0.0001 by 1-way ANOVA with Tukey’s multiple-comparisons test (D and E).
Discussion
CaMKII is a multifunctional serine/threonine protein kinase that enhances myocardial physiological responses to stress (1, 38, 44). However, excessive CaMKII activity is now recognized to contribute to heart failure and arrhythmias, common forms of heart disease (2–4, 25, 45). Our findings reveal that invertebrates and vertebrates rely on an ancient CaM-binding domain redox switch to regulate physiological and pathological cardiac responses to stress. Our data support a new model of CaMKII regulation in which M308 oxidation by MICAL1 is a molecular brake constraining excessive CaMKII activity by inhibiting Ca2+/CaM binding. M308 and the CaM-binding domain are highly conserved among all species and all CaMKII isoforms. M308 first appeared in free-living unicellular eukaryotes with origins more than 600 million years ago (46). Our finding that MICAL1-catalyzed oxidation of M308 holds CaMKII activity in check by diminishing Ca2+/CaM binding provides critical missing information required for understanding why oxidant stress does not excessively bias CaMKII toward hyperactivation under normal, physiological conditions, as was predicted by computational modeling (Figure 3D and Supplemental Figure 3, C and D). High intensity physiological stimulation in excitable cells, such as nerve and muscle, including myocardium, promotes T287 autophosphorylation in CaMKII (31, 47). Autophosphorylation prevents autoinhibition by a mechanism that is similar to oxidation of MM281/282, but is a more potent CaMKII-activating posttranslational modification because it induces a 1000-fold increase in Ca2+/CaM binding to CaMKII, so-called CaM trapping (48). CaMKII basal autophosphorylation at T306/T307 can also prevent or reduce Ca2+/CaM binding in vitro (49–51); however, to our knowledge these sites have not been shown to be dynamically regulated, nor to affect physiological/pathological processes in vivo. MM281/282 oxidation activates CaMKII without attendant CaM trapping, presumably due to oxidation of M308 (14). In contrast to the MM281/282 module that appears to arise in vertebrate evolution (52), M308 is present in primordial CaMKII, suggesting that a check against excessive CaMKII activation during oxidized conditions has always been fundamental to CaMKII molecular physiology. Thus, M308 is an oxidant sensor that constrains and prevents excessive CaMKII activity by shifting the affinity of Ca2+/CaM binding to reduce initial activation and to counterbalance activity-sustaining CaM trapping. This dual role of oxidation to activate and check CaMKII likely provides balance for multilevel control preventing CaMKII from causing injury or dysfunction under physiological redox stress.
Our studies demonstrate that the CaMKII M308 redox status is determined by MICAL1 and its stereospecific partner MSRB (8, 9). Our findings show that MICALs are multifunctional, directing cellular processes more broadly than initially recognized, since their discovery almost 20 years ago. This is also consistent with the different multidomain structures of MICAL proteins, suggesting additional levels of regulation. Our findings indicate that regulation of protein function by methionine R-sulfoxidation is a general and conserved mechanism to control protein function, rather than an actin-specific regulatory mechanism. The R116H mutation in MICAL1 demonstrates that the dual proclivities of MICAL1 for F-actin and CaMKII can be disarticulated. We found that MICAL1 absence led to CaMKII hyperactivation at baseline, and increased mortality with pathological stress. These findings suggest that the MICAL isoforms (MICAL1, MICAL2, and MICAL3) in the heart are unlikely to all similarly target CaMKII, and therefore probably perform, at least partially, nonredundant tasks. In contrast, MICAL1 knockout did not cause obvious baseline cardiac actin defects, suggesting redundancy in actin regulation among the MICAL isoforms in the heart. MICAL1 mutations are now recognized to cause human diseases (53), suggesting that CaMKII dysregulation could be a downstream consequence. Relatively little is known about MICAL regulation (54), but it is possible that upstream signals could also modify MICAL substrate specificity. We speculate that MICAL may regulate other, yet unknown, protein targets, including enzymes, thereby enabling fine-tuned redox control of cellular signaling. Our findings uncover an unanticipated biological function for MICALs beyond the cytoskeleton, in cellular Ca2+-sensing physiology and signaling across species.
Methods
Mouse experiments
MICAL1–/–, MICAL1R116H, and CaMKIIδM308V mice.
All mouse experiments were carried out under protocols approved by the Johns Hopkins University animal care and use committee. MICAL1–/– mice were generated as described previously (55). MICAL1R116H- and CaMKIIδM308V-knockin mice were generated using CRISPR technology at the Johns Hopkins University Transgenic Mouse Core. Pronuclear injection of 1-cell C57BL/6J embryos (Jackson Laboratory) was performed with standard microinjection techniques (56) using a mix of Cas9 protein (30 ng/μL, PNABio), tracrRNA (0.6 μM, Dharmacon), crRNA (0.6 μM, IDT), and ssDNA oligo (10 ng/μL, IDT) diluted in RNase-free injection buffer (10 mM Tris-HCl, pH 7.4, 0.25 mM EDTA). Injected embryos were transferred into the oviducts of pseudopregnant ICR females (Envigo) as described previously (56). The following gRNA and ssODN oligonucleotides were used for the generation of the knockin mice using CRISPR: MICAL1R116H mouse guide RNA, AAAAGCGTATCAAGTTCTCTAGG; MICAL1R116H mouse ssODN, TGTCTCGTGGTAGGTGCCGGGCCTTGCGGACTTCGGGCTGCTGTGGAGTTGGCACTGTTAGGTGCCCGAGTGGTACTTGTGGAAAAGCATATCAAGTTCTCTAGACACAATGTGCTCC ATCTCTGGCCCTTCACCATCCATGACCTTCGGGCACTTGGGGCCAAGAAGTTCTACGGGCGCTTCTGTAC; CaMKIIδM308V mouse guide RNA, CGCCATCTTGACAACTATGCTGG; and CaMKIIδM308V mouse ssODN, GCTACCTGATTTTAGGTCAGAGGTTTGTTAGGGCCAACTATAATCCATTTCTCTTATTTCTTTGATAGGGCGCaataTTGACCACCGTGCTGGCTACGAGAAATTTTTCAGGTATACATGTTTGAACTCTGTTCCTTGTCCTCTCACCTCT.
Successful introduction of the mutations was confirmed in both lines with Sanger sequencing. Before any experiments, founder mice were backcrossed with WT C57BL/6J mice obtained from Jackson Laboratory. The mice used in our experiments were littermates to minimize the impact of potential off-target effects of CRISPR. We used online algorithms (CRISPR RGEN Tools) to search for potential off-target effects. We performed this analysis for the MICAL1R116H and CaMKIIδM308V mice and CaMKIIM308V flies and found no potential off-target regions in the targeted gene–containing chromosomes, allowing up to 3 mismatches between the gRNA and DNA region. Previous studies have shown that gRNA with mismatches greater or equal to 3 have minimal or no effect on gene function (57), suggesting that our findings are extremely unlikely due to off-target effects.
Drosophila melanogaster experiments
CaMKIIM308V Drosophila melanogaster.
CaMKIIM308V flies were generated by Rainbow Transgenic Flies, Inc. on the w118 background. Successful introduction of the desired mutation was confirmed by Sanger sequencing and the flies were shipped to Johns Hopkins University. To minimize potential off-target effects, the CaMKIIM308V flies were backcrossed 5 times with WT iso31 flies before experiments. During the backcrossing process, the presence of the CaMKIIM308V-carrying allele was confirmed by Sanger sequencing. To generate the CaMKIIM308V flies we used the gRNA AGCCATACTTACGACAATGTTGG and the ssODN GTAGACTGTCTCAAGAAATTTAATGCGCGGCGCAAGCTAAAGGGAGCCATACTTACGACAGTGTTGGCGACCAGAAATTTTTCGAGTATGTAACTTAACTTAATTAATCTTGACCACATCAAT.
Recombinant protein experiments
Recombinant proteins and mutagenesis.
WT CaMKII and M308V CaMKIIδ recombinant proteins were generated using the Bac-to-Bac baculovirus system (Invitrogen) and purified on a CaM-agarose column. Sf9 cells infected with recombinant CaMKII baculovirus were harvested 48 hours after infection by centrifugation. Cells were resuspended in homogenization buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 5% betaine, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and protease inhibitors [P8340, MilliporeSigma]), and sonicated to release cell contents. Cell lysate was then centrifuged at 15,000 g and 4°C for 20 minutes. The supernatant was removed and added to 3 volumes of equilibration buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM CaCl2, 4 mM magnesium acetate, 1 mM DTT, protease inhibitors) and CaM-agarose, and incubated for 1 hour at 4°C. Resin was washed 3 times with wash buffer (50 mM Tris-HCl pH 7.5, 1 mM CaCl2, 1 mM magnesium acetate, 1 mM DTT, protease inhibitors), followed by 2 washes with 1 M NaCl wash buffer (50 mM HEPES pH 7.5, 1 M NaCl, 1 mM CaCl2, 1 mM magnesium acetate, 1 mM DTT, protease inhibitors), and 2 washes with 1 M NaCl wash buffer without protease inhibitors. CaMKII was eluted from the resin in 5 fractions of elution buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 2.5 mM EDTA, 1 mM DTT). Protein fractions were dialyzed overnight at 4°C into storage buffer (50 mM HEPES pH 7.5, 1 mM EDTA, 50% glycerol, 10% ethylene glycol). The fractions containing CaMKII protein were determined by SDS-PAGE. The experiments shown in Figure 1 were done using C-terminally 6×His-tagged WT CaMKII. WT MICAL1 gene was subcloned into the pET-28a vector (Novagen; kanamycin resistance with N-terminal 6×His tag). WT MICAL1 full-length recombinant proteins and mutant MICAL1 proteins were generated using an E. coli bacterial expression system, as described previously (8, 17). MSRA and MSRB proteins were generated by Geumsoo Kim (National Heart, Lung, and Blood Institute), as described previously (58). Mutagenesis of MICAL1 and CaMKIIδ cDNA was performed using the QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies. Introduction of the various mutations was confirmed using Sanger sequencing at the Johns Hopkins University Genetics Resources Core Facility. Recombinant WT CaM was generated as described previously (22).
MICAL1 activity assessed by NADPH absorbance at 340 nm.
The enzymatic activity of MICAL1 recombinant proteins (100 nM) was monitored by the rate of NADPH consumption, measured as decreasing light absorbance at 340 nm, as described previously (17, 19). All reactions were done in 50 mM HEPES buffer, pH 7.4. To obtain MICAL1 kinetics with CaMKII, reactions were performed at various NADPH concentrations (50, 100, and 150 μM). Kinetic values were derived using the initial slope of NADPH consumption. WT and MICAL1 R116H (100 nM) activities were measured with CaMKII (100 nM) using NADPH (200 μM).
hiPSC studies
hiPSC maintenance and genome editing.
All the iPSC lines in this study were maintained in mTeSR1 medium (STEMCELL Technologies) and passaged in Versene Solution (15040066, Thermo Fisher Scientific) every 5 days. Culture dishes were precoated with Geltrex (Thermo Fisher Scientific) diluted 1:100. The procedures for CRISPR/Cas9 genome editing were described in detail in a previous publication (59). To introduce the M308V mutation in CaMKIIδ, we first synthesized the single-stranded gRNA (EnGen RNA synthesis kit, New England BioLabs): 5′-CTGGCTACAAGGAATTTCTC-3′. We then transfected the gRNA along with a targeted donor template (5′-CACTGACTTTTGTGTTCTAATTTTTTTCACAGGGTGCCATCTTGACAACTGTGCTAGCCACCAGAAATTTCTCAGGTACATGCATTGGGAACTCTGCTTC-3′) into both WT hiPSCs containing a doxycycline-inducible Cas9 allele and an isogenic iPSC line harboring the RYR2 mutation R4651I (RYR2R4651I/WT). After exposure to doxycycline (5 μM) for 18 hours, cells were plated at low density to isolate single cells. A 300-bp region surrounding the target site was amplified by PCR and positive colonies were identified by restriction digestion with NheI. Introduction of the homozygous M308V mutation was confirmed by Sanger sequencing. We predicted Cas9-gRNA off-target sites using http://crispr.mit.edu/ Amplicons containing the top predicted off-target genes (CaMKIIα, CaMKIIβ, CaMKIIγ, cTnT5, PLEC, WDR33, STMP1, SLC6A19, and LOC105376183) were amplified from all 4 iPSC lines and Sanger sequenced.
Statistics.
Statistical analysis between 2 groups was calculated by 2-tailed Student’s t test and 1-way ANOVA with Tukey’s multiple-comparisons test for more than 2-group comparisons unless otherwise specified. Statistical analysis of differences between categorical data was done using Fisher’s exact test. The Kds for the CaMKII CaM-binding peptides were calculated using GraphPad Prism 7 software nonlinear regression 1-site-specific binding analysis. The D’Agostino-Pearson test was performed to test for normality. For non-normally distributed data, the Mann-Whitney test was used to test for statistical significance. Statistical values are NS > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001, unless otherwise specified. Bars denote mean ± SEM. All statistical analyses were done using GraphPad Prism software.
Author contributions
KK and MEA designed the project. KK, VJB, LL, HMI, YW, MCV, IDB, JMG, DZ, DHW, EDL, KRM, FL, BM, Qiang Wang, and RLL performed experiments and analyzed data. ACW performed CaMKII activity simulations. HMI and MAS performed structural modeling of CaMKII and CaM interactions. DHG and TJH performed intracellular Ca2+ simulations. ALK provided critical advice. VNG, WTP, MNW, AC, MAB, MAS, and RLL supervised experiments and assisted with data interpretation. Qinchuan Wang assisted with CRISPR construct design. KK and MEA wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Chip Hawkins and the Transgenic Mouse Core at Johns Hopkins University for generating our transgenic mice using CRISPR technology. We are grateful to the Johns Hopkins Genetics Resources Core Facility for their assistance with Sanger sequencing. We thank Djahida Bedja, Nadan Wang, Michelle Leppo, Christian Oeing, and the Cardiovascular Physiology and Surgery Core at Johns Hopkins University for technical assistance. We are grateful to Geumsoo Kim for providing MSRA and MSRB recombinant proteins. We thank Jinying Yang for animal model maintenance. We are grateful to Heping Cheng for providing the adenoviral construct for the GCaMP6f-Junctin nanosensor. We acknowledge Charles Steenbergen for his advice on actin fluorescence imaging analysis. We thank Harry C. Dietz and Gregg L. Semenza for helpful comments. We are grateful to Shawn Roach and Teresa Ruggle for their help with graphic art and figure preparation. This work was supported by NIH grant R35 HL140034 (to MEA). KK was supported by NIH grant T32HL007227 and AHA postdoctoral fellowship 18POST34030257. LL and RLL were supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, grant ZIA HL000225. ACW was supported by MOST-107-2636-B-002- 001. MNW was supported by NIH grant NS079584. VNG was supported by NIH grants AG021518 and GM065204. DHG and TJH were supported by NIH grants HL134824 and HL135096. AC was supported by NIH grant R01HL124091. MAS was supported by NIH grant R01GM57001. MAB was supported by NIH grant R21NS108842 and the Mirowsky award at Johns Hopkins University.
Version 1. 08/04/2020
Electronic publication
Version 2. 09/01/2020
Print issue publication
Footnotes
Conflict of interest: MEA is a named inventor on patents claiming to treat heart disease by CaMKII inhibition. No income derives from these activities.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4663–4678.https://doi.org/10.1172/JCI133181.
Contributor Information
Klitos Konstantinidis, Email: kkonsta4@jhmi.edu.
Vassilios J. Bezzerides, Email: vbezzerides@gmail.com.
Lo Lai, Email: lucy.lai1983@gmail.com.
An-Chi Wei, Email: awei2jhu@gmail.com.
Yuejin Wu, Email: ywu95@jhmi.edu.
Jonathan M. Granger, Email: jgrange1@jhu.edu.
Donghui Zhang, Email: dongh.zhang@hubu.edu.cn.
Elizabeth D. Luczak, Email: Betsy.Luczak@jhmi.edu.
Fujian Lu, Email: Fujian.Lu@childrens.harvard.edu.
Bruno Manta, Email: bruno.manta@gmail.com.
Qiang Wang, Email: wangqiang@fudan.edu.cn.
Qinchuan Wang, Email: Qinchuan.Wang@jhmi.edu.
Alex L. Kolodkin, Email: kolodkin@jhmi.edu.
Vadim N. Gladyshev, Email: vgladyshev@rics.bwh.harvard.edu.
Thomas J. Hund, Email: thomas.hund@osumc.edu.
Anthony Cammarato, Email: acammar3@jhmi.edu.
Mario A. Bianchet, Email: bianchet@jhmi.edu.
Rodney L. Levine, Email: rlevine@nih.gov.
Mark E. Anderson, Email: Mark.Anderson@jhmi.edu.
References
- 1.Wu Y, et al. Calmodulin kinase II is required for fight or flight sinoatrial node physiology. Proc Natl Acad Sci U S A. 2009;106(14):5972–5977. doi: 10.1073/pnas.0806422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swaminathan PD, Purohit A, Hund TJ, Anderson ME. Calmodulin-dependent protein kinase II: linking heart failure and arrhythmias. Circ Res. 2012;110(12):1661–1677. doi: 10.1161/CIRCRESAHA.111.243956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backs J, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106(7):2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ling H, et al. Requirement for Ca2+/calmodulin-dependent kinase II in the transition from pressure overload-induced cardiac hypertrophy to heart failure in mice. J Clin Invest. 2009;119(5):1230–1240. doi: 10.1172/JCI38022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J. 2002;364(pt 3):593–611. doi: 10.1042/BJ20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334(6063):1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung RJ, et al. Mical links semaphorins to F-actin disassembly. Nature. 2010;463(7282):823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee BC, et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell. 2013;51(3):397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15(12):1445–1454. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Pasquale E, et al. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis. 2013;4:e843. doi: 10.1038/cddis.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N, et al. Calmodulin kinase II inhibition prevents arrhythmias in RyR2(R4496C+/-) mice with catecholaminergic polymorphic ventricular tachycardia. J Mol Cell Cardiol. 2011;50(1):214–222. doi: 10.1016/j.yjmcc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Bezzerides VJ, et al. Gene therapy for catecholaminergic polymorphic ventricular tachycardia by inhibition of Ca2+/calmodulin-dependent kinase II. Circulation. 2019;140(5):405–419. doi: 10.1161/CIRCULATIONAHA.118.038514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, et al. Insights into the pathogenesis of catecholaminergic polymorphic ventricular tachycardia from engineered human heart tissue. Circulation. 2019;140(5):390–404. doi: 10.1161/CIRCULATIONAHA.119.039711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson JR, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadella M, Bianchet MA, Gabelli SB, Barrila J, Amzel LM. Structure and activity of the axon guidance protein MICAL. Proc Natl Acad Sci U S A. 2005;102(46):16830–16835. doi: 10.1073/pnas.0504838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siebold C, et al. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc Natl Acad Sci U S A. 2005;102(46):16836–16841. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alqassim SS, Urquiza M, Borgnia E, Nagib M, Amzel LM, Bianchet MA. Modulation of MICAL monooxygenase activity by its calponin homology domain: structural and mechanistic insights. Sci Rep. 2016;6:22176. doi: 10.1038/srep22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Yesilyurt HG, Yoon J, Terman JR. The MICALs are a family of F-actin dismantling oxidoreductases conserved from Drosophila to humans. Sci Rep. 2018;8(1):937. doi: 10.1038/s41598-017-17943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zucchini D, Caprini G, Pasterkamp RJ, Tedeschi G, Vanoni MA. Kinetic and spectroscopic characterization of the putative monooxygenase domain of human MICAL-1. Arch Biochem Biophys. 2011;515(1–2):1–13. doi: 10.1016/j.abb.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Manta B, Gladyshev VN. Regulated methionine oxidation by monooxygenases. Free Radic Biol Med. 2017;109:141–155. doi: 10.1016/j.freeradbiomed.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He BJ, et al. Oxidation of CaMKII determines the cardiotoxic effects of aldosterone. Nat Med. 2011;17(12):1610–1618. doi: 10.1038/nm.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans TI, Shea MA. Energetics of calmodulin domain interactions with the calmodulin binding domain of CaMKII. Proteins. 2009;76(1):47–61. doi: 10.1002/prot.22317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou LL, Lloyd SJ, Schulman H. Activation of the multifunctional Ca2+/calmodulin-dependent protein kinase by autophosphorylation: ATP modulates production of an autonomous enzyme. Proc Natl Acad Sci U S A. 1986;83(24):9497–9501. doi: 10.1073/pnas.83.24.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattiazzi A, Kranias EG. The role of CaMKII regulation of phospholamban activity in heart disease. Front Pharmacol. 2014;5:5. doi: 10.3389/fphar.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R, et al. Calmodulin kinase II inhibition protects against structural heart disease. Nat Med. 2005;11(4):409–417. doi: 10.1038/nm1215. [DOI] [PubMed] [Google Scholar]
- 26.Luo M, et al. Diabetes increases mortality after myocardial infarction by oxidizing CaMKII. J Clin Invest. 2013;123(3):1262–1274. doi: 10.1172/JCI65268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan PD, et al. Oxidized CaMKII causes cardiac sinus node dysfunction in mice. J Clin Invest. 2011;121(8):3277–3288. doi: 10.1172/JCI57833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong MH, et al. The KN-93 molecule inhibits calcium/calmodulin-dependent protein kinase II (CaMKII) activity by binding to Ca2+/CaM. J Mol Biol. 2019;431(7):1440–1459. doi: 10.1016/j.jmb.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Grintsevich EE, Yesilyurt HG, Rich SK, Hung RJ, Terman JR, Reisler E. F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat Cell Biol. 2016;18(8):876–885. doi: 10.1038/ncb3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frémont S, et al. Oxidation of F-actin controls the terminal steps of cytokinesis. Nat Commun. 2017;8:14528. doi: 10.1038/ncomms14528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279(5348):227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 32.Gupta RC, Kranias EG. Purification and characterization of a calcium-calmodulin-dependent phospholamban kinase from canine myocardium. Biochemistry. 1989;28(14):5909–5916. doi: 10.1021/bi00440a030. [DOI] [PubMed] [Google Scholar]
- 33.Gao Z, et al. Catecholamine-independent heart rate increases require Ca2+/calmodulin-dependent protein kinase II. Circ Arrhythm Electrophysiol. 2011;4(3):379–387. doi: 10.1161/CIRCEP.110.961771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monfredi O, Maltsev VA, Lakatta EG. Modern concepts concerning the origin of the heartbeat. Physiology (Bethesda) 2013;28(2):74–92. doi: 10.1152/physiol.00054.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci. 2007;27(46):12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kon N, et al. CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes Dev. 2014;28(10):1101–1110. doi: 10.1101/gad.237511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaiser TS, et al. The genomic basis of circadian and circalunar timing adaptations in a midge. Nature. 2016;540(7631):69–73. doi: 10.1038/nature20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckendorf J, van den Hoogenhof MMG, Backs J. Physiological and unappreciated roles of CaMKII in the heart. Basic Res Cardiol. 2018;113(4):29. doi: 10.1007/s00395-018-0688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisner DA, Caldwell JL, Kistamás K, Trafford AW. Calcium and excitation-contraction coupling in the heart. Circ Res. 2017;121(2):181–195. doi: 10.1161/CIRCRESAHA.117.310230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viswanathan MC, et al. Distortion of the actin A-triad results in contractile disinhibition and cardiomyopathy. Cell Rep. 2017;20(11):2612–2625. doi: 10.1016/j.celrep.2017.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Priori SG, Chen SR. Inherited dysfunction of sarcoplasmic reticulum Ca2+ handling and arrhythmogenesis. Circ Res. 2011;108(7):871–883. doi: 10.1161/CIRCRESAHA.110.226845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262(5134):740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 43.van Oort RJ, et al. Ryanodine receptor phosphorylation by calcium/calmodulin-dependent protein kinase II promotes life-threatening ventricular arrhythmias in mice with heart failure. Circulation. 2010;122(25):2669–2679. doi: 10.1161/CIRCULATIONAHA.110.982298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan W, Bers DM. Ca-dependent facilitation of cardiac Ca current is due to Ca-calmodulin-dependent protein kinase. Am J Physiol. 1994;267(3 pt 2):H982–H993. doi: 10.1152/ajpheart.1994.267.3.H982. [DOI] [PubMed] [Google Scholar]
- 45.Wagner S, et al. Ca2+/calmodulin-dependent protein kinase II regulates cardiac Na+ channels. J Clin Invest. 2006;116(12):3127–3138. doi: 10.1172/JCI26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451(7180):783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279(5352):870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 48.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256(5060):1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 49.Colbran RJ. Inactivation of Ca2+/calmodulin-dependent protein kinase II by basal autophosphorylation. J Biol Chem. 1993;268(10):7163–7170. [PubMed] [Google Scholar]
- 50.Hanson PI, Schulman H. Inhibitory autophosphorylation of multifunctional Ca2+/calmodulin-dependent protein kinase analyzed by site-directed mutagenesis. J Biol Chem. 1992;267(24):17216–17224. [PubMed] [Google Scholar]
- 51.Lu CS, Hodge JJ, Mehren J, Sun XX, Griffith LC. Regulation of the Ca2+/CaM-responsive pool of CaMKII by scaffold-dependent autophosphorylation. Neuron. 2003;40(6):1185–1197. doi: 10.1016/S0896-6273(03)00786-4. [DOI] [PubMed] [Google Scholar]
- 52.Anderson ME. Oxidant stress promotes disease by activating CaMKII. J Mol Cell Cardiol. 2015;89(pt B):160–167. doi: 10.1016/j.yjmcc.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dazzo E, et al. Mutations in MICAL-1cause autosomal-dominant lateral temporal epilepsy. Ann Neurol. 2018;83(3):483–493. doi: 10.1002/ana.25167. [DOI] [PubMed] [Google Scholar]
- 54.Yoon J, Kim SB, Ahmed G, Shay JW, Terman JR. Amplification of F-actin disassembly and cellular repulsion by growth factor signaling. Dev Cell. 2017;42(2):117–129.e8. doi: 10.1016/j.devcel.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Battum EY, et al. The intracellular redox protein MICAL-1 regulates the development of hippocampal mossy fibre connections. Nat Commun. 2014;5:4317. doi: 10.1038/ncomms5317. [DOI] [PubMed] [Google Scholar]
- 56. Behringer R, Gertsenstein M, Vintersten Nagy K, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 4th Ed. Cold Spring Harbor Press; 2003. [Google Scholar]
- 57.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim JC, You Z, Kim G, Levine RL. Methionine sulfoxide reductase A is a stereospecific methionine oxidase. Proc Natl Acad Sci U S A. 2011;108(26):10472–10477. doi: 10.1073/pnas.1101275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang G, et al. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat Protoc. 2017;12(1):88–103. doi: 10.1038/nprot.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.