Abstract
BACKGROUND
The effects of the novel coronavirus disease 2019 (COVID-19) in pregnancy remain relatively unknown. We present a case of second trimester pregnancy with symptomatic COVID-19 complicated by severe preeclampsia and placental abruption.
METHODS
We analyzed the placenta for the presence of severe acute respiratory syndrome coronavirus 2 (SARS–CoV-2) through molecular and immunohistochemical assays and by and electron microscopy and measured the maternal antibody response in the blood to this infection.
RESULTS
SARS–CoV-2 localized predominantly to syncytiotrophoblast cells at the materno-fetal interface of the placenta. Histological examination of the placenta revealed a dense macrophage infiltrate, but no evidence for the vasculopathy typically associated with preeclampsia.
CONCLUSION
This case demonstrates SARS–CoV-2 invasion of the placenta, highlighting the potential for severe morbidity among pregnant women with COVID-19.
FUNDING
Beatrice Kleinberg Neuwirth Fund and Fast Grant Emergent Ventures funding from the Mercatus Center at George Mason University. The funding bodies did not have roles in the design of the study or data collection, analysis, and interpretation and played no role in writing the manuscript.
Keywords: COVID-19, Reproductive Biology
Keywords: Molecular pathology
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS–CoV-2) is a novel betacoronavirus causing the deadly pandemic of coronavirus disease 2019 (COVID-19). The risks and specific effects of SARS–CoV-2 in pregnant women remain unknown. No adverse outcomes were reported among a small cohort of pregnant women who presented with COVID-19 in the late third trimester of pregnancy in Wuhan, China (1). However, little is known about maternal and neonatal outcomes as a result of infection in the first and second trimesters of pregnancy.
Hypertensive disorders of pregnancy (HDP) complicate 2%–8% of pregnancies and rarely occur in the second trimester (2). HDP in women with COVID-19 have been noted in limited case reports from China and New York City (3, 4). At the same time, a subset of nonpregnant patients with COVID-19 have demonstrated significant liver enzyme abnormalities as well as coagulopathy (5, 6). These laboratory abnormalities significantly overlap with the findings in severe preeclampsia, a subset of HDP, defined by associated hemolysis, elevated liver enzymes, and low platelet counts as well as proteinuria and elevated blood pressure. This overlap leads to a diagnostic dilemma when faced with pregnant patients with COVID-19, hypertension, and coagulopathy.
We present the case of a woman with COVID-19 in the second trimester of pregnancy, who had severe hypertension, coagulopathy, and preeclampsia. This case highlights the association between COVID-19 and HDP and demonstrates clear SARS–CoV-2 invasion of the placenta, with associated placental inflammation distinct from typical preeclampsia.
Case report
In mid-March 2020, a previously healthy 35-year-old non-Hispanic Asian American woman, gravida 3, para 1011 (G3P1011), presented at 22 weeks’ gestation with symptoms of COVID-19 infection. Her husband had exposure to an individual with laboratory-confirmed COVID-19 and had symptoms of the disease. Ten days before admission, the patient developed fever and cough, with acute worsening of symptoms in the 4 days before admission including fever, malaise, nonproductive cough, diffuse myalgias, anorexia, nausea, and diarrhea. On the morning of presentation, the patient awoke with vaginal bleeding and abdominal pain. Her initial vital signs showed that she was afebrile with a pulse of 110, a respiratory rate of 22 breaths per minute, and an oxygen saturation of 99% on room air. Her blood pressure was elevated at 150/100 mmHg. A physical examination was notable for dark blood in the vaginal vault without cervical dilation. SARS–CoV-2 RNA was detected by reverse transcription PCR (RT-PCR) in a nasopharyngeal swab obtained from the patient on admission.
Her past medical history was significant for psoriasis, without current symptoms. The patient had a prior pregnancy that was complicated by term gestational hypertension that resolved with delivery. Her current antepartum course was notable for normal blood pressure and normal baseline preeclampsia evaluation.
The patient was admitted to the labor and delivery department. Her chest x-ray was significant for a hazy opacity throughout the left lung. A transabdominal ultrasound revealed an active fetus, normal amniotic fluid volume, an estimated fetal weight of 485 grams, which measured within 1 week of gestational age, and a posterior fundal placenta with a retroplacental clot that was concerning for placental abruption. Laboratory studies revealed elevated liver transaminases, profound thrombocytopenia, and increased urine protein consistent with preeclampsia, as well as a prolonged partial thromboplastin time and decreased fibrinogen consistent with disseminated intravascular coagulation (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI139569DS1). A blood smear revealed normochromic, normocytic red cells with unremarkable morphology, atypical lymphocytes, suggestive of viral infection, and severe thrombocytopenia (Supplemental Figure 1). Rotational thromboelastometry (ROTEM) showed severe deficiencies in clot formation. The patient was resuscitated with 4 units of cryoprecipitate, 4 pools of packed platelets, 2 grams of tranexamic acid (TXA), 5 grams of fibrinogen concentrate, and 2 units of fresh-frozen plasma. This resulted in an improvement of the coagulopathy, but the thrombocytopenia and elevated blood pressure persisted.
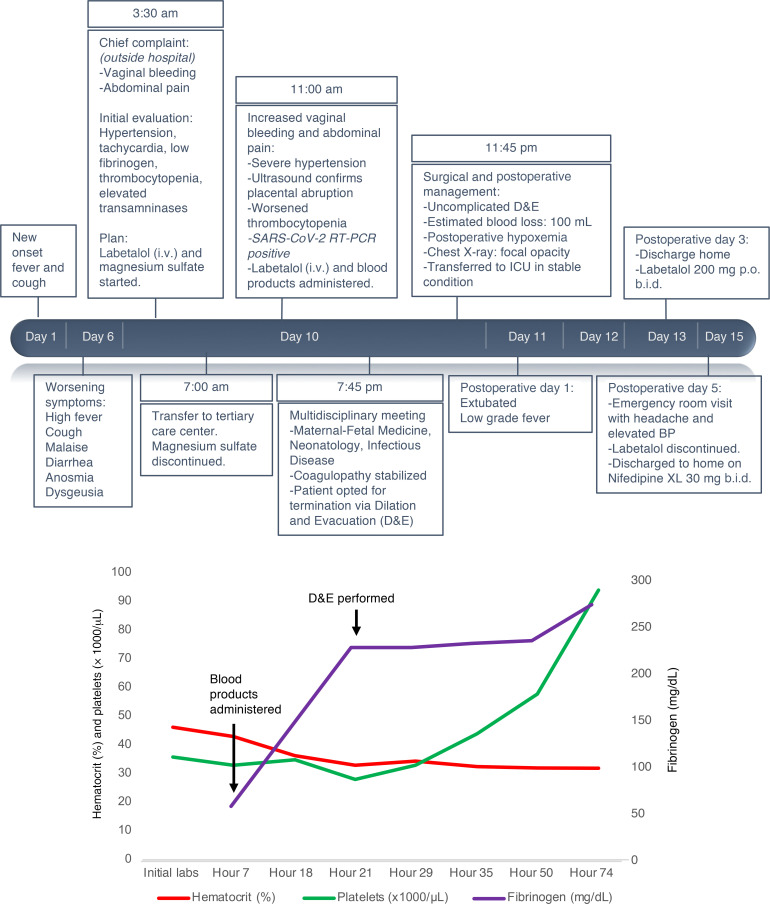
The combination of hypertension, proteinuria, elevated transaminases, and low platelet counts supported a diagnosis of severe preeclampsia, for which delivery is the definitive treatment. Multidisciplinary consultations with materno-fetal medicine, neonatology, and infectious disease departments were conducted via telemedicine. The patient opted for termination of the pre-viable pregnancy to reduce the risk of serious maternal morbidity or death. Termination of the pregnancy was performed via dilation and evacuation (D&E) under general endotracheal anesthesia. Intraoperative findings included a retroplacental clot and were otherwise unremarkable. On postoperative day 1, she was extubated and weaned to room air; however, lymphopenia developed. Her coagulation markers improved, and she was discharged to self-isolation on postoperative day 3 (Figure 1). Home blood pressure monitoring was initiated with close provider telemedicine support. An emergency room visit on postoperative day 4 was required to titrate the patient’s antihypertensive medications. She consented to a pathology examination and to the release of tissue for research-related testing according to our IRB-regulated protocol.
Figure 1. Case timeline.
(A) Clinical course of a 35-year-old G3P1011 at 22 weeks’ gestation with COVID-19 associated with preeclampsia and placental abruption. Timeline from the onset of symptoms to the immediate postpartum period including resuscitation products, hypertension management, and surgical preparation for D&E. BP, blood pressure. (B) Patient’s platelet count (green line), hematocrit (red line), and serum fibrinogen (purple line) throughout hospitalization. Major clinical events are highlighted with arrows.
Results
Microbiologic investigation.
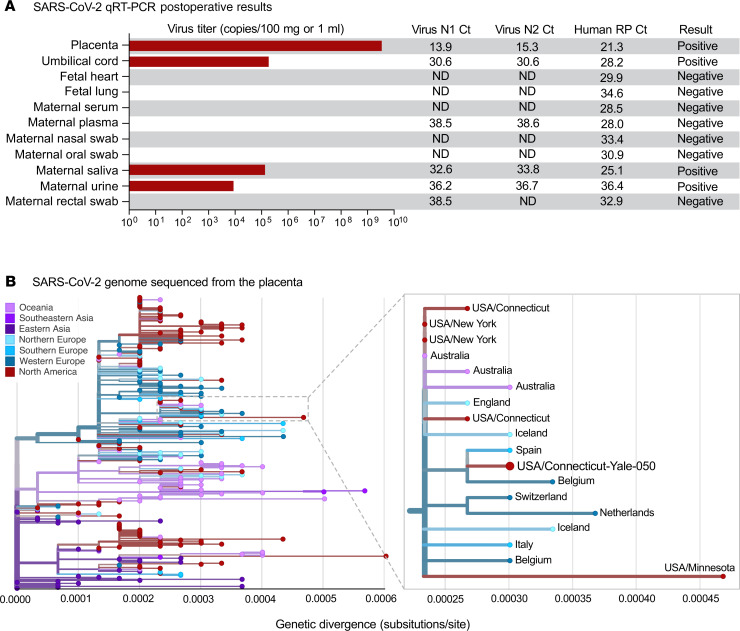
We performed the US CDC quantitative RT-PCR (qRT-PCR) assay, which showed that the placenta (3 × 107 virus copies/mg) and umbilical cord (2 × 103 virus copies/mg) were positive for SARS–CoV-2 RNA (Figure 2A). Fetal heart and lung tissues were also tested and met the human RNA control (RNase P) standards and were negative for viral RNA (Figure 2). Maternal samples were also collected postoperatively, and although the oral and nasal swabs were negative, the saliva and urine were still positive for SARS–CoV-2 (Figure 2A). Virus from the placenta was whole-genome sequenced (Yale-050) and was phylogenetically similar to other SARS–CoV-2 detected locally (Connecticut, USA) and abroad (Europe and Australia) (Figure 2B). In addition, the SARS–CoV-2 genome from the placenta did not contain any unique amino acid substitutions compared with other sequenced SARS–CoV-2.
Figure 2. Examination of SARS–CoV-2 RNA in maternal and fetal tissue.
(A) SARS–CoV-2 qRT-PCR results of fetal and maternal samples using the CDC assay which consists of the N1 and N2 primers and probes targeting the coronavirus nucleocapsid and the RP primers and probe targeting human RNase P as an internal control. Ct values from both N1 and N2 must be below 38 for the result to be positive, as internally validated. Virus titers are shown as the average calculation from N1 and N2 Ct values. For the tissues, 80–160 mg was used for extractions, and for the swabs in viral transport media and other liquid samples, 0.25–0.4 mL was used. ND, not detected. (B) The SARS–CoV-2 genome sequenced from the infected placenta was combined with 289 other genomes available from GISAID from around the world. The phylogenetic tree was constructed using IQ-TREE (http://www.iqtree.org/) within the Nextstrain Augur pipeline, and the results were visualized using Auspice (28). Genetic divergence is shown as substitutions per site from the root. An enlarged view of the 18 genomes in the clade that contains the SARS–CoV-2 genome sequenced from the placenta (USA/Connecticut-Yale-050) is shown. The clade is defined by 3 nucleotide substitutions, A28881A, G28882A, and G28883C, providing the equivalent of approximately 95% branch support. The consensus SARS–CoV-2 genome from the placenta (Yale-050) can be found using NCBI’s BioProject PRJNA614976 and the phylogenetic data can be visualized at Nexstrain (https://nextstrain.org/community/grubaughlab/CT-SARS-CoV-2/paper2). The Acknowledgments for the sequences obtained from GISAID can be found at Nexstrain (https://github.com/grubaughlab/CT-SARS-CoV-2/tree/master/paper2).
Serologic testing.
Levels of anti–SARS–CoV-2 IgG and IgM antibodies in this patient were among the highest observed in 56 COVID-19+ patients admitted to Yale New Haven Hospital. Further quantification revealed endpoint dilution titers of 1:1600 for IgM and 1:25,600 for IgG (Supplemental Figure 2).
Pathology.
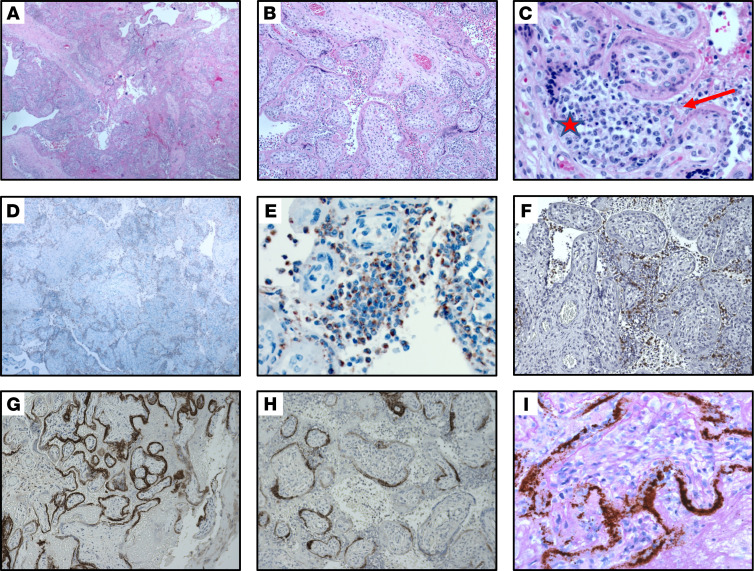
On gross examination, the placenta showed a marginal adherent blood clot associated with a focal placental infarct supportive of the clinical diagnosis of abruption and was otherwise unremarkable. On histological examination, the placenta was remarkable for the presence of diffuse perivillous fibrin and an inflammatory infiltrate composed of macrophages as well as T lymphocytes, as demonstrated by IHC for CD68 and CD3, consistent with histiocytic intervillositis (Figure 3, A–F). No dark pigment was noted in the intervillous space. Maternal vessels did not show features of decidual vasculopathy. The fetal organs were grossly and microscopically unremarkable (Supplemental Figure 4). SARS–CoV-2 localized predominantly to the syncytiotrophoblast cells of the placenta, as demonstrated by IHC for the SARS–CoV-2 spike protein (Figure 3, G and H, and Supplemental Figure 3A, negative control), and by SARS–CoV-2 RNA in ISH (Figure 3I and Supplemental Figure 3B, negative control).
Figure 3. Histopathology of placenta.
(A–C). Section of placenta stained with H&E showing histiocytic intervillositis. Original magnification, ×40 (A), ×100 (B), ×400 (C). (C) Star indicates intervillous space infiltrated by immune cells. Arrow indicates perivillous fibrin. (D and E). Immunohistochemical stain for CD68 showing the majority of intervillous inflammatory infiltrate positive (brown stain) for this macrophage marker. Original magnification, ×40 (D), ×400 (E). (F) Staining for CD3, a marker of T lymphocytes. Original magnification, ×100. (G and H). Immunohistochemical staining for SARS–CoV-2 spike protein, demonstrating virus localization predominantly in syncytiotrophoblast cells. Original magnification, ×50 (G), ×400 (H). (I) In situ analysis for the presence of SARS–CoV-2 RNA shows strong positive staining within the placenta . Original magnification, ×400.
Electron microscopy.
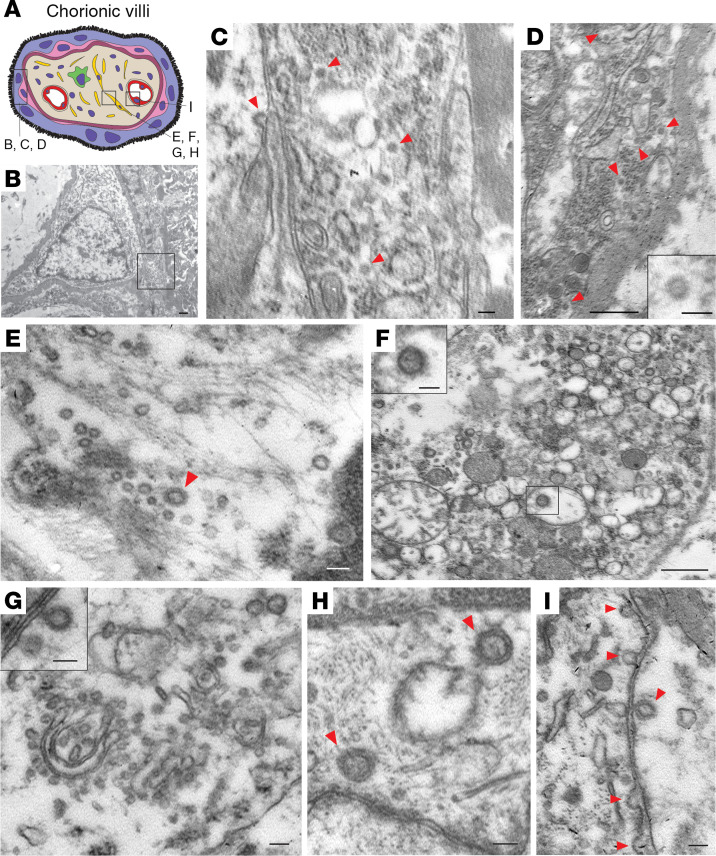
Electron microscopic analysis of immersion-fixed placental tissue showed a relatively well-preserved ultrastructure of the placenta (Figure 4). Analysis of the placental region adjacent to the umbilical cord identified virus particles within the cytosol of placental cells. The size of these virus particles was 75–100 nm in diameter, which is consistent with the size and appearance of SARS–CoV-2 (7).
Figure 4. Electron microscopic images of coronavirus particles in different placental cell types.
(A) Schematic illustration of chorionic villi. Cytotrophoblasts, syncytiotrophoblasts, fibroblasts, and endothelial cells are indicated in pink, purple, yellow, and red, respectively. (B) Cytotrophoblast (on the left with the nucleus) and syncytiotrophoblast (on the right). Scale bar: 500 nm. (C) Enlarged image of boxed area in B. Cytoplasm of a syncytiotrophoblast cell with virus particles (red arrowheads). Scale bar: 100 nm. (D) Cytoplasm of a cytotrophoblast cell with virus particles indicated by red arrowheads. Scale bar: 500 nm and 100 nm (insert). (E–H) Fibroblast with virus particles indicated by red arrowheads. Scale bars: 100 nm (E), 500 nm (F) (insert, 100 nm), 100 nm (G) (insert, 100 nm), 100 nm (H). (I) Endothelial cell within chorionic villus with budding viral particles indicated by red arrowheads. Scale bar: 100 nm.
Discussion
This report describes a case of second trimester COVID-19 associated with preeclampsia and SARS–CoV-2 infection of the placenta. The distinction between preeclampsia and COVID-19 is important, as it may have implications for the patient’s future pregnancy outcomes. This patient previously had gestational hypertension, which increased her risk in this pregnancy for preeclampsia. Although preeclampsia, placental abruption, and disseminated intravascular coagulopathy (DIC) — which are commonly seen together in obstetric practice — accounted for many of the clinical and laboratory findings in this patient, the histopathological features and viral infection of the placenta suggest a prominent role for COVID-19 in this patient’s presentation. This is highlighted by the presence of high levels of SARS–CoV-2 and the invasion of intervillous macrophages (intervillositis) within the placenta. These findings suggest that COVID-19 may have contributed to the placental inflammation that ultimately resulted in early-onset preeclampsia and a worsening maternal condition.
Intervillositis, as seen in this case, describes a placental pathology consisting of fibrin deposits and mononuclear cell infiltration of the intervillous spaces. It is associated with high rates of miscarriage, fetal growth restriction, and severe early preeclampsia (8, 9). This entity is typically idiopathic or autoimmune in nature, but can also be seen in association with infections such as CMV (10) as well as malaria (11). Massive fibrin deposition was also noted as a feature of placental pathology in pregnant women with SARS (12) as well as in those with malaria (13), and recently in a pregnant woman with SARS–CoV-2 infection without preeclampsia with intervillous fibrin deposition, who experienced a second trimester miscarriage (14). In the latter case, it remains unknown whether COVID-19 precipitated the intervillositis. However, massive macrophage infiltration alongside fibrin deposition has recently been observed in lung tissue examined at autopsy from patients with severe COVID-19, raising the possibility of a common immunopathology leading to macrophage recruitment and activation causing tissue damage (15). Further studies of placenta from women with COVID-19 may help address whether this is a histological feature associated with placental SARS–CoV-2 infection.
The pathophysiology of coagulopathy during COVID-19 remains unknown. A DIC-like coagulopathy has been reported in patients with COVID-19 and is associated with poor outcomes (16). However, our patient’s thrombocytopenia and hypofibrinogenemia were more severe than what would have been expected with COVID-19 alone (16). Both SARS–CoV-2 and HDP are reported to reduce angiotensin-converting enzyme 2 (ACE2) activity, leading to increased tissue levels of angiotensin II (17–19). The imbalance of the RAS system seen in COVID-19 patients may therefore contribute to hypertensive complications including preeclampsia in pregnant patients with COVID-19. We hypothesize that SARS–CoV2’s use of the ACE2 receptor could unmask HDP earlier in women who are predisposed to HDP and that COVID-19 causes significant placental pathology leading to an exacerbation of the severity of the patient’s condition.
Importantly, we found no amino acid differences in the SARS–CoV-2 genome sequenced from the placenta compared with others sequenced from around the world (Figure 2), suggesting that placental invasion is not a unique feature of this virus and does not require adaptation. Rather, given the patient’s high serum titers of anti–SARS–CoV-2 spike protein antibodies, a potential mechanism to explain placental invasion in this case would be antibody-dependent transcytosis mediated by the neonatal Fc receptor (FcRn), which has previously been observed with other viruses including CMV, HIV, and Zika (20–22). Future studies are warranted to investigate protective versus pathological roles of humoral immunity in SARS–CoV-2 infection during pregnancy and to assess for possible virus transmission to the fetus.
SARS–CoV-2 localized to placental syncytiotrophoblast cells, the outer layer of multinucleated cells that cover the chorionic villi and that are in contact with maternal blood in the intravillous space. Syncytiotrophoblasts form a cellular layer between maternal and fetal circulation, contribute to transplacental transfer of protective antibodies, and, in some cases, are permissive to viral infection and to subsequent transmission to fetal cells. In this case, we did not find definitive evidence for fetal infection. However, future studies should further characterize SARS–CoV-2 presence in the placenta and fetus through cellular colocalization studies and antibody-based electron microscopy analysis.
This report highlights a case of acute placental infection with SARS–CoV-2 that may have potentiated severe, early-onset preeclampsia. Future efforts to correctly diagnose and identify the underlying processes of COVID-19–associated HDP are critical for directing patient care and counseling pregnant women during the pandemic.
Methods
Microbiological investigation.
RNA was postoperatively extracted from homogenized placenta, umbilical cord, fetal lungs, heart kidney tissues (27–160 mg; stored in formalin), and maternal oral, nasal, and rectal swabs, saliva, urine, plasma, and serum and tested for the presence of SARS–CoV-2 and human RNase P using the US CDC qRT-PCR assay as described previously (23). The qRT-PCR results were confirmed via independent replicates. SARS–CoV-2 RNA from the placenta was sequenced using the portable MinION platform (Oxford Nanopore). The sequenced data were processed and used for phylogenetic analysis as described previously (24).
SARS–CoV-2 s1 spike protein IgM and IgG serology testing.
ELISAs for IgG and IgM antibodies against SARS–CoV-2 were performed on maternal plasma as described by Amanat et al. (25). Initial screening of plasma samples from 56 COVID-19+ patients and 24 uninfected health care workers from the Yale New Haven Hospital was performed with a 1:50 dilution, and endpoint titers were further tested for the case study patient using serial 4-fold dilutions beginning at 1:100. The cutoff was set with a confidence level of 99%, as described by Frey et al. (26). The endpoint titer is defined as the reciprocal of the highest dilution that gives a reading above the cutoff.
Placenta and fetal organ examination.
The placenta and fetal parts were immediately fixed in 10% buffered formalin. After 3 days of formalin fixation, placenta and fetal lung, heart, liver, and kidney were sampled and embedded in paraffin. The placenta was sampled extensively, with 2 sections of the cord and peripheral membranes and 10 full-thickness sections of the placental parenchyma, which included both the maternal and fetal sides. The sections obtained were stained with H&E. Immunohistochemical stainings were performed on selected slides using antibodies against CD68 and CD3 standard techniques and against SARS–CoV-2 using the monoclonal SARS–CoV/SARS–CoV-2 (COVID-19) spike antibody, clone 1A9 (Genetex; dilution 1:200). ISH for SARS–CoV-2 was performed with RNAScope (Advanced Cell Diagnostics [ACD]) using probes directed against SARS–CoV-2 on formalin-fixed, paraffin-embedded tissue cut at a thickness of 3 μm (27). Negative control probes (bacterial gene dapB) were included to assess background signals, as were the positive control probes for the housekeeping gene peptidylprolyl isomerase B (PPIB). The ISH sections were counterstained using periodic acid–Schiff (PAS).
Electron microscopy.
Tissues were fixed in 10% formaldehyde for 10 days. This solution was replaced by 4% paraformaldehyde and 0.3% glutaraldehyde for 20 hours. Tissue blocks were then processed for electron microscopy. Ultrathin sections were cut on a Leica Ultra-Microtome, collected on Formvar-coated single-slot grids, and analyzed with a Tecnai 12 BioTwin electron microscope (FEI).
Study approval.
This study was approved by the Yale University IRB (HIC no. 2000027690). Written informed consent was obtained from the patient before inclusion in the study.
Author contributions
HH provided primary clinical care to the patient described in this case report and contributed to the study’s conception and design, data collection and interpretation, literature searches, and writing of the original draft of the manuscript. SFF obtained consent from the patient for research, designed and oversaw all laboratory studies and interpretations, and aided in drafting the manuscript. HSL designed and supervised all clinical care and the study design, provided data interpretation, and contributed to both the drafting and clinical revisions of the manuscript. TLH, YY, KSB, and SP contributed to electron microscopy analyses. CBFV, JRF, AFB, TA, ALW, and NDG contributed to virus detection, sequencing, and phylogenetics. ALC, RAM, RT, MH, and CL contributed to histological studies. FL and AMR contributed to antibody detection. CDO, BG, PV, ACM, JF, and CDC contributed to patient enrollment, specimen procurement, and tissue processing. AI, AIK, TLH, UD, KHC, AMG, GA, KLF, HST, CMP, and UMR aided with study design, provided data interpretation, and contributed critical revisions of the manuscript. All authors reviewed and approved the final version of the manuscript. HH and SFF share first authorship. HH appears first in the author list because she was primarily involved in the preparation and revisions of the manuscript until publication.
Supplementary Material
Acknowledgments
We thank the patient in this case for her willingness to provide detailed medical data, and Drs. Baxi, August, and Webster for participating in the patient’s care. This study was supported by the Beatrice Kleinberg Neuwirth Fund and by Fast Grant Emergent Ventures funding from the Mercatus Center at George Mason University. The funding bodies did not have roles in the design of the study or data collection, analysis, and interpretation and played no role in writing the manuscript.
Version 1. 06/23/2020
In-Press Preview
Version 2. 08/17/2020
Electronic publication
Version 3. 09/01/2020
Print issue publication
Funding Statement
This study was in part supported by Fast Grant funding support from the Emergent Ventures at the Mercatus Center, George Mason University.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(9):4947–4953.https://doi.org/10.1172/JCI139569.
Contributor Information
Hillary Hosier, Email: hillary.hosier@yale.edu.
Shelli F. Farhadian, Email: shelli.farhadian@yale.edu.
Raffaella A. Morotti, Email: raffaella.morotti@yale.edu.
Uma Deshmukh, Email: uma.deshmukh@yale.edu.
Alice Lu-Culligan, Email: alice.lu@yale.edu.
Katherine H. Campbell, Email: katherine.campbell@yale.edu.
Yuki Yasumoto, Email: yuki.yasumoto@yale.edu.
Chantal B.F. Vogels, Email: chantal.vogels@yale.edu.
Arnau Casanovas-Massana, Email: arnau.casanovas@yale.edu.
Pavithra Vijayakumar, Email: pavithra.vijayakumar@yale.edu.
Bertie Geng, Email: bertie.geng@yale.edu.
Camila D. Odio, Email: camiodio@gmail.com.
John Fournier, Email: john.fournier@yale.edu.
Anderson F. Brito, Email: anderson.brito@yale.edu.
Joseph R. Fauver, Email: joseph.fauver@yale.edu.
Feimei Liu, Email: feimei.liu@yale.edu.
Tara Alpert, Email: tara.alpert@yale.edu.
Reshef Tal, Email: reshef.tal@yale.edu.
Klara Szigeti-Buck, Email: klara.szigeti@yale.edu.
Sudhir Perincheri, Email: sudhir.perincheri@yale.edu.
Aileen M. Gariepy, Email: Aileen.Gariepy@yale.edu.
Gabriela Aguilar, Email: gabriela.aguilar@yale.edu.
Kristen L. Fardelmann, Email: kristen.fardelmann@yale.edu.
Malini Harigopal, Email: malini.harigopal@yale.edu.
Hugh S. Taylor, Email: hugh.taylor@yale.edu.
Christian M. Pettker, Email: christian.pettker@yale.edu.
Anne L. Wyllie, Email: anne.wyllie@yale.edu.
Aaron M. Ring, Email: aaron.ring@yale.edu.
Nathan D. Grubaugh, Email: nathan.grubaugh@yale.edu.
Albert I. Ko, Email: albert.ko@yale.edu.
Tamas L. Horvath, Email: tamas.horvath@yale.edu.
Akiko Iwasaki, Email: akiko.iwasaki@yale.edu.
Uma M. Reddy, Email: uma.reddy@yale.edu.
Heather S. Lipkind, Email: heather.lipkind@yale.edu.
References
- 1.Chen H, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steegers EA, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-eclampsia. Lancet. 2010;376(9741):631–644. doi: 10.1016/S0140-6736(10)60279-6. [DOI] [PubMed] [Google Scholar]
- 3. doi: 10.5858/arpa.2020-0901-sa. Schwartz DA. An analysis of 38 pregnant women with COVID-19, their newborn infants, and maternal-fetal transmission of SARS-CoV-2: maternal coronavirus infections and pregnancy outcomes [published online March 17, 2020]. Arch Pathol Lab Med . [DOI] [PubMed]
- 4.Breslin N, et al. COVID-19 infection among asymptomatic and symptomatic pregnant women: Two weeks of confirmed presentations to an affiliated pair of New York City hospitals. Am J Obstet Gynecol MFM. 2020;2(2):100118. doi: 10.1016/j.ajogmf.2020.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, et al. Coagulopathy and antiphospholipid antibodies in patients with COVID-19. N Engl J Med. 2020;382(17):e38. doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goldsmith CS, Tamin A. Transmission electron microscopic image of an isolate from the first U.S. case of COVID-19, formerly known as 2019-nCoV. https://phil.cdc.gov/Details.aspx?pid=23336 Accessed July 2, 2020.
- 8.Bos M, et al. Clinical outcomes in chronic intervillositis of unknown etiology. Placenta. 2020;91:19–23. doi: 10.1016/j.placenta.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Nowak C, et al. Perinatal prognosis of pregnancies complicated by placental chronic villitis or intervillositis of unknown etiology and combined lesions: about a series of 178 cases. Placenta. 2016;44:104–108. doi: 10.1016/j.placenta.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Freitag L, von Kaisenberg C, Kreipe H, Hussein K. Expression analysis of leukocytes attracting cytokines in chronic histiocytic intervillositis of the placenta. Int J Clin Exp Pathol. 2013;6(6):1103–1111. [PMC free article] [PubMed] [Google Scholar]
- 11.Walter PR, Garin Y, Blot P. Placental pathologic changes in malaria. A histologic and ultrastructural study. Am J Pathol. 1982;109(3):330–342. [PMC free article] [PubMed] [Google Scholar]
- 12.Ng WF, et al. The placentas of patients with severe acute respiratory syndrome: a pathophysiological evaluation. Pathology. 2006;38(3):210–218. doi: 10.1080/00313020600696280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordi J, et al. Massive chronic intervillositis of the placenta associated with malaria infection. Am J Surg Pathol. 1998;22(8):1006–1011. doi: 10.1097/00000478-199808000-00011. [DOI] [PubMed] [Google Scholar]
- 14.Baud D, et al. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. JAMA. 2020;323(21):2198–2200. doi: 10.1001/jama.2020.7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaleyeva LM, et al. Downregulation of apelin in the human placental chorionic villi from preeclamptic pregnancies. Am J Physiol Endocrinol Metab. 2015;309(10):E852–E860. doi: 10.1152/ajpendo.00272.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anton L, et al. Activation of local chorionic villi angiotensin II levels but not angiotensin (1-7) in preeclampsia. Hypertension. 2008;51(4):1066–1072. doi: 10.1161/HYPERTENSIONAHA.107.103861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci. 2020;63(3):364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, et al. The Neonatal Fc receptor (FcRn) enhances human immunodeficiency virus type 1 (HIV-1) transcytosis across epithelial cells. PLoS Pathog. 2013;9(11):e1003776. doi: 10.1371/journal.ppat.1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidji E, McDonagh S, Genbacev O, Tabata T, Pereira L. Maternal antibodies enhance or prevent cytomegalovirus infection in the placenta by neonatal Fc receptor-mediated transcytosis. Am J Pathol. 2006;168(4):1210–1226. doi: 10.2353/ajpath.2006.050482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerman MG, et al. Cross-reactive Dengue virus antibodies augment Zika virus infection of human placental macrophages. Cell Host Microbe. 2018;24(5):731–742.e6. doi: 10.1016/j.chom.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. doi: 10.1101/2020.03.30.20048108. Vogels CBF, et al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays. Posted on medRxiv April 26, 2020. [DOI]
- 24. doi: 10.1101/2020.03.25.20043828. Fauver JR, et al. Coast-to-coast spread of SARS-CoV-2 in the United States revealed by genomic epidemiology. Posted on medRxiv March 26, 2020. [DOI]
- 25. doi: 10.1101/2020.03.17.20037713. Amanat F, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans [preprint]. Posted on medRxiv April 16, 2020. [DOI] [PMC free article] [PubMed]
- 26.Frey A, Di Canzio J, Zurakowski D. A statistically defined endpoint titer determination method for immunoassays. J Immunol Methods. 1998;221(1-2):35–41. doi: 10.1016/S0022-1759(98)00170-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadfield J, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.