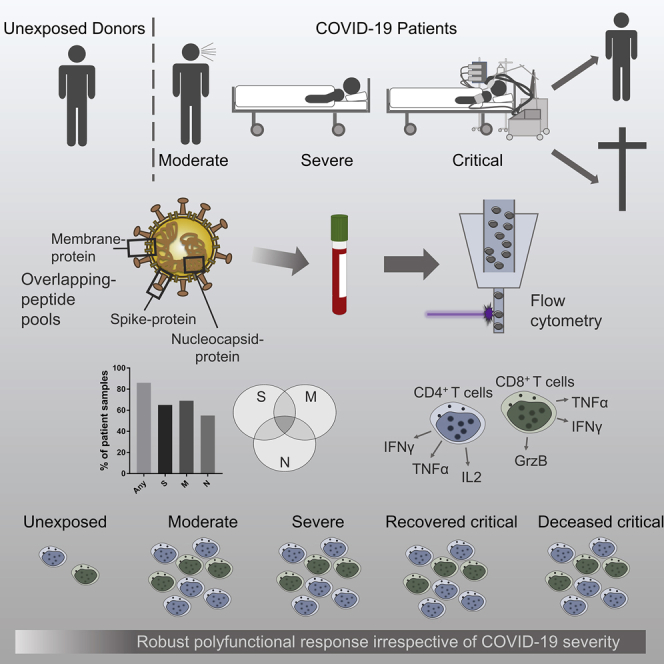
Summary
T cell immunity toward SARS-CoV-2 spike (S-), membrane (M-), and nucleocapsid (N-) proteins may define COVID-19 severity. Therefore, we compare the SARS-CoV-2-reactive T cell responses in moderate, severe, and critical COVID-19 patients and unexposed donors. Overlapping peptide pools of all three proteins induce SARS-CoV-2-reactive T cell response with dominance of CD4+ over CD8+ T cells and demonstrate interindividual immunity against the three proteins. M-protein induces the highest frequencies of CD4+ T cells, suggesting its relevance for diagnosis and vaccination. The T cell response of critical COVID-19 patients is robust and comparable or even superior to non-critical patients. Virus clearance and COVID-19 survival are not associated with either SARS-CoV-2 T cell kinetics or magnitude of T cell responses, respectively. Thus, our data do not support the hypothesis of insufficient SARS-CoV-2-reactive immunity in critical COVID-19. Conversely, it indicates that activation of differentiated memory effector T cells could cause hyperreactivity and immunopathogenesis in critical patients.
Keywords: COVID-19, SARS-CoV-2, immunity, spike, nucleocapsid, S/M/N protein-reactive T cells, immunopathology, critical COVID-19
Graphical Abstract
Highlights
M-, N-, and S-proteins induce T cell reactivity with differing immune dominance
M-protein triggers a strong CD4+ T cell response
ICU and non-ICU COVID-19 patients have comparable SARS-CoV-2 T cell responses
Viral clearance and COVID-19 survival are not associated with SARS-CoV-2 immunity
Thieme et al. demonstrate the T cell response against M-, N-, and S-proteins of SARS-CoV-2, with M-protein triggering a stronger CD4+ T cell response. ICU COVID-19 patients are capable of generating functional SARS-CoV-2 immunity non-inferior to non-ICU COVID-19 patients. SARS-CoV-2 clearance and COVID-19 survival are not associated with the magnitude of T cell response.
Introduction
The clearance of viral pathogens requires an effective T cell response directed against protein antigens expressed by the virus.1 The T cell response against the severe acute respiratory syndrome-related coronavirus (SARS-CoV)-2 virus, which caused the ongoing pandemic, is presumably initiated by respiratory professional antigen-presenting cells (APCs) that can engulf viral antigens, as shown for the 2002–2003 SARS-CoV infection.2 As the production of type I interferons (IFN-Is), which can directly and indirectly induce and enhance antiviral effector T cell responses,3 is suppressed in SARS-CoV-2 infection, especially in severe cases,4,5 one may hypothesize that the reduced IFN-I response results in an impaired T cell immunity in severe SARS-CoV-2 infection, as observed in SARS-CoV-1 infection.6 Activated T cells can migrate to the site of infection, where they facilitate viral clearance. However, antigen-specific T cells can contribute to immune pathogenesis. There is mounting evidence that the latter is the major reason for critical coronavirus disease 2019 (COVID-19) disease manifestations,7,8 as evidenced by the recent findings of the successful treatment of COVID-19 patients with immunosuppressive dexamethasone.9
The SARS-CoV-2 contains 4 structural proteins: the spike glycoprotein (S), the envelope (E) protein, the membrane (M) protein and the nucleocapsid (N) protein.10 The S-protein mediates host cell entry by binding to the angiotensin-converting enzyme 2 (ACE2).11 Due to its surface exposure and crucial role in infecting host cells, the S-protein is an attractive therapeutic target—for instance, for antibodies that block the S-ACE2 interaction. In fact, it was shown that patients who recovered from COVID-19 developed virus-neutralizing anti-S immunoglobulin (Ig) titers.12 Given the requirement for T cell help in the generation of high-affinity IgG antibodies, this finding indicates that S-protein-reactive T cell immunity was formed in those patients.13,14 Accordingly, very recent studies identified SARS-CoV-2 S-protein-reactive T cell responses in patients suffering from moderate, severe, and critical COVID-19.8,15 Furthermore, it was shown that the amount of SARS-CoV-2 S-protein-reactive CD4+ T cells increased with disease progression.16 Besides the S-protein, N- and M-proteins were also suggested as potential targets for diagnostic and prophylactic approaches.7,10 In fact, B cell responses against the N-protein seemed to be the first to arise 4–8 days after symptom onset for the 2002–2003 SARS-CoV infection,17,18 which indicates that the N-reactive T cell response also is prevalent during this time frame. The M-protein is the most abundant surface protein of SARS-CoV virions, suggesting a high antigenicity.19 Grifoni et al.20 provided data on T cell responses toward a diverse set of SARS-CoV-2 epitopes in recovered COVID-19 patients. However, no analysis of the T cell reactivity toward these antigens in hospitalized patients and no association with disease severity are available. These data are important to understand possible immunopathogenic effects provided by T cells reactive to certain SARS-CoV-2 proteins. A dominant T cell response toward viral antigens has been associated with immunopathogenesis, for example, in patients with hemorrhagic fever following dengue virus infection.21 The observation of similar mechanisms in COVID-19 could influence vaccination and treatment strategies profoundly. It has been shown that studying T cell responses may be more reliable than antibody testing in Swedish and French cohorts.22,23 Information on early T cell responses in critical COVID-19 patients in comparison to non-critical is lacking, but it is important to define strategies addressing epidemiological questions. Another unresolved question is the influence of the magnitude of the T cell response for the clinical disease course.
For these reasons, we identified, characterized, and compared S-, M-, and N-reactive T cell responses in COVID-19 patients with different clinical manifestation during the acute disease phase and at recovery.
Results
Study Design
We analyzed 65 blood samples drawn at different time points after hospital admission of a cohort of 28 COVID-19 patients with moderate (32 samples), severe (16 samples), and critical (17 samples) disease manifestation (Tables 1 and S1). The patients were grouped into the respective category according to their worst manifestation using the German Robert Koch Institute symptom classification, as previously described.8 This way, 7 patients with a total of 16 samples were categorized as moderate, 9 patients with 27 samples as severe, and 12 patients with 22 samples as critical. In agreement with other studies,24 we observed significantly more males within the group of critical COVID-19 patients compared to the moderate and severe cases. We further analyzed 10 samples from SARS-CoV-2 unexposed individuals that were collected and stored before the pandemic. Unexposed donors were significantly younger than COVID-19 patients (median age [range] 55 [42–64] versus 69 [26–91] years) (Table S2).
Table 1.
Patient Characteristics
| Characteristics | Moderate | Severe | Critical | P = |
|---|---|---|---|---|
| No. patients (%) | 7 (25) | 9 (32.1) | 12 (42.9) | |
| Age, y, median (range) | 68 (58.5–82.5) | 81 (69–83) | 58 (54.75–70.5) | ns |
| Gender, male/female (%) | 4/3 (57.1/42.9) | 3/6 (33.3/66.6) | 11/1 (91.7/8.3) | 0.022 |
| Chest CT abnormalities | ||||
| Bilateral ground-glass opacity (%) | 5 (71.4) | 7 (77.8) | 11 (91.7) | ns |
| Pleural effusion (%) | 0 (0) | 1 (11.1) | 1 (8.3) | ns |
| Acute respiratory distress syndrome (%) | 0 (0) | 1 (11.1) | 5 (41.7) | ns |
| Treatments | ||||
| Oxygen therapy (%) | 2 (28.6) | 7 (77.8) | 11 (91.7) | 0.017 |
| Intravenous antibiotics (%) | 5 (71.4) | 8 (88.9) | 11 (91.7) | ns |
| Admission to intensive care unit (%) | 0 (0) | 0 (0) | 10 (83.3) | <0.001 |
| Mechanical ventilation (%) | 0 (0) | 0 (0) | 10 (83.3) | <0.001 |
| No. samples | 16 | 27 | 22 | – |
| No. samples/patient, median (range) | 2 (1–3) | 3 (2–4) | 2 (1–4) | – |
Patients were grouped according to the most severe COVID-19 category as per Robert Koch Institute classification.8 Comparison between patient ages was made with the Kruskal-Wallis test; comparisons of patient gender, chest CT abnormalities, and treatments were made with Fisher’s exact test. p < 0.05 was considered significant.
Simultaneous Analysis of S-, M-, and N-Proteins Is Required to Avoid Underestimation of T Cell Immunity
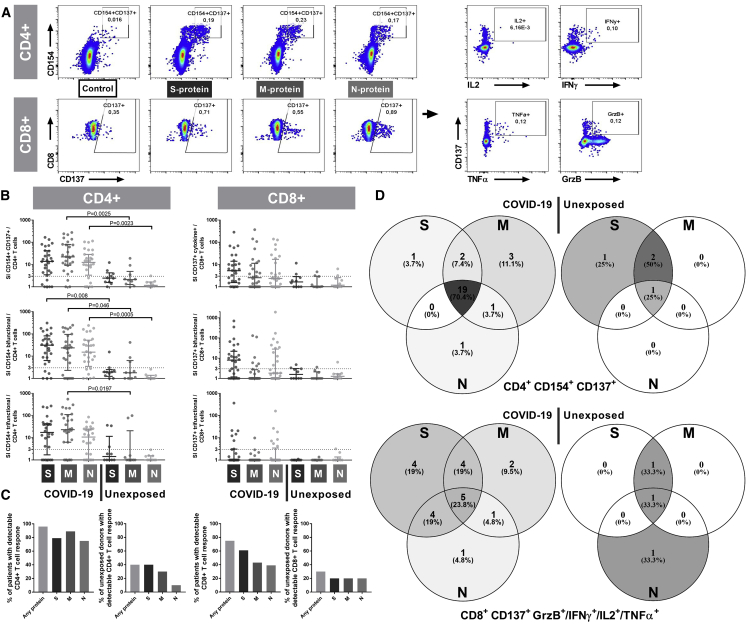
By stimulation with S-, M-, or N-protein overlapping peptide pools (OPPs), we could show that all 3 proteins have the capacity to induce SARS-CoV-2-reactive CD4+ and CD8+ T cells (Figure 1A). To this end, we used pools of mainly 15-mer peptide with 11 amino acid overlaps spanning the whole N- and M-protein or in silico predicted immunodominant sequence domains of S-protein (Figure S1). Large OPPs have been shown to allow monitoring of antigen-specific T cell responses independent of human leukocyte antigen (HLA) type.25 This approach is therefore time and cost-efficient and allows the monitoring of T cell reactivity in larger cohorts. After 16 h of stimulation, antigen-reactive T cell responses were detected by intracellular staining using flow cytometry. The gating strategy is presented in Figure S2. Activation markers CD154 and CD137 in CD4+ T cells and CD137 in combination of production of any of interleukin (IL)-2, IFN-γ, tumor necrosis factor α (TNF-α), and/or granzyme B (GrzB) in CD8+ T cells (CD137+ cytokine+ CD8+ T cells) were used to define SARS-CoV-2-reactive T cells. We regarded responses as detectable if the frequency in the specifically stimulated sample exceeded the unstimulated DMSO control 3 times (stimulation index > 3). The presented frequencies show values in the stimulated samples after subtraction of the unstimulated control (Figures 1 and S3).
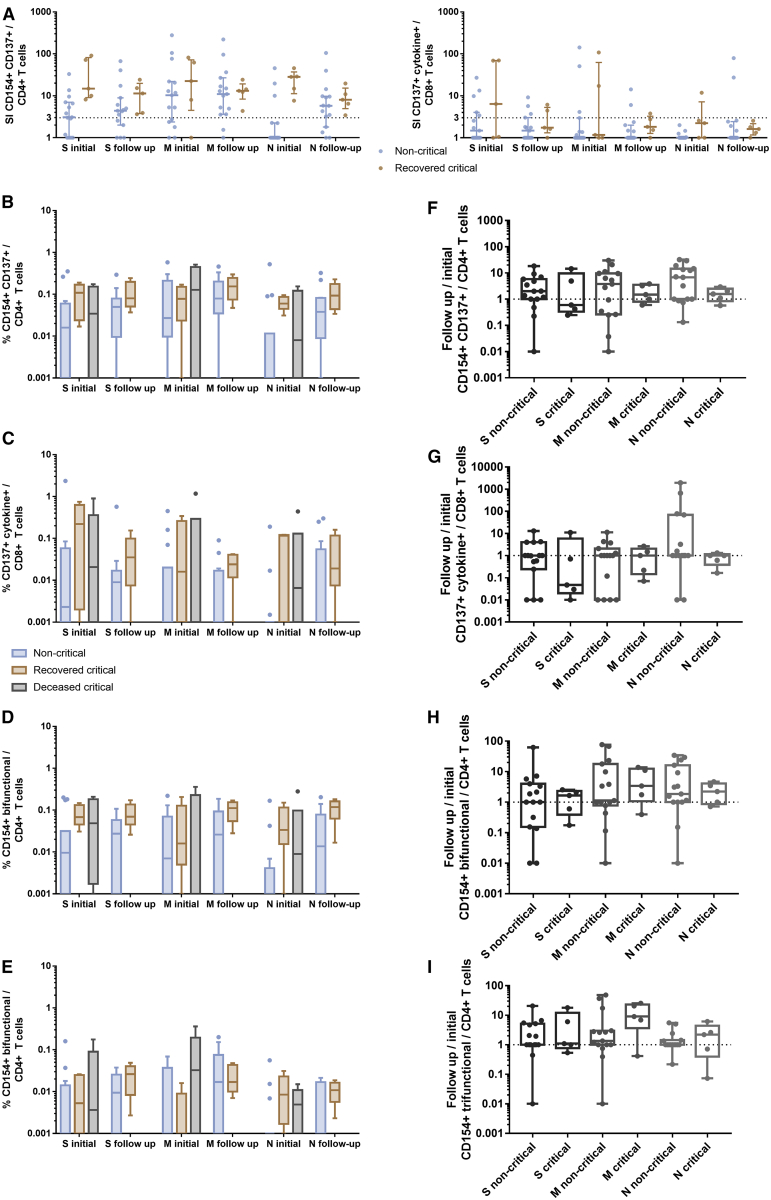
Figure 1.
SARS-CoV-2-Reactive T Cells Are Induced by the S-, M- and N-Proteins with Interindividual Patterns
Peripheral blood mononuclear cells (PBMCs) isolated from 65 blood samples collected from 28 COVID-19 patients with moderate, severe, or critical disease and blood samples of 10 unexposed donors collected and cryopreserved before the COVID-19 pandemic were stimulated for 16 h with S-, M-, or N-protein OPPs. Antigen-reactive T cells were determined by flow cytometry and identified according to the gating strategy presented in Figure S2. Maximum values of each COVID-19 patient were compared to unexposed donors.
(A) Representative plots of CD4+ T cells and CD8+ T cells after stimulation with S-, M-, and N-protein OPPs. Antigen-reactive CD4+ T cells were identified by CD154 and CD137 expression and antigen-reactive CD8+ T cells by CD137 expression and production of any cytokines out of IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+).
(B) Stimulation index (SI) of CD154+ CD137+ CD4+ T cells (SARS-COV-2-specific CD4+ T cells), CD137+ cytokine+ CD8+ T cells (SARS-COV-2-specific CD8+ T cells) and bifunctional and trifunctional CD154+ CD4+ and CD137+ CD8+ T cells. Bi- and trifunctional T cells were calculated by Boolean gating of IL-2, IFN-γ, TNF-α, IL-4, and GrzB production. SI was calculated by dividing the measured T cell subset response by the respective response in the DMSO control. Values >3 were considered detectable in the following analyses. The maximum value of each COVID-19 patient is depicted. Scatterplots show line at median; error bars represent the interquartile ranges. The statistical comparison was done with the Kruskal-Wallis test and the Dunn’s multiple comparisons test. p < 0.05 was considered significant.
(C) Frequency of patient samples with detectable (SI > 3) CD4+ (left) and CD8+ (right) T cell responses in at least 1 sample after stimulation with S-, M-, or N-protein (total of 65 samples of 28 COVID-19 patients and 10 samples of 10 unexposed donors).
(D) Venn diagrams of 28 COVID-19 patients and 10 unexposed donors with detectable (SI > 3) SARS-Cov-2-reactive CD4+ or CD8+ T cells after stimulation with S-, M-, or N-protein in at least 1 sample. A total of 27 COVID-19 patients and 4 unexposed donors showed CD4+ T cell reactivity and 21 COVID-19 patients and 3 unexposed donors showed CD8+ T cell reactivity toward at least 1 of the tested SARS-CoV-2-S-, M-, and N-proteins.
See also Figures S1, S2, and S3 and Table S2.
Considering the response rate per patient population, CD4+ T cell and CD8+ T cell responses were detectable in at least 1 sample in 27 (96.4%) and 21 (75%) COVID-19 patients, respectively (Figures 1B–1D). Considering the response rate per sample, we detected a CD4+ T cell response in 56 and a CD8+ T cell response in 33 of 65 patient samples against at least one of the SARS-CoV-2 proteins (Figures S3A–S3C). However, none of the proteins induced CD4+ or CD8+ T cell responses in all 56 and 33 positive samples, respectively. Within the 56 responding samples, M-protein OPPs induced a detectable CD4+ T cell response in the highest number of samples (M = 45, N = 36, S = 42), whereas for the 33 responding samples within CD8+ subsets, the S-protein OPP was dominant (M = 13, N = 14, S = 26) (Figures S3A–S3C). Therefore, testing a single protein OPP would underestimate T cell immunity in at least 20% of patients. Although the immunogenicity of the three proteins and its role in the clinical course require follow-up investigations, our data advocate that measuring T cell responses induced by all three proteins is of high relevance when studying SARS-CoV-2-reactive T cells, especially in the early phase of the disease. In accordance with other studies,15,20 SARS-Cov-2-reactive T cells were detectable also in samples of unexposed donors that were frozen before the outbreak of COVID-19. We measured detectable CD4+ T cells in 4 and CD8+ T cells in 3 of 10 unexposed donors (Figures 1B–1D). Overall, the level of T cell responses was lower in the unexposed donors, confirming the specificity of SARS-CoV-2 peptide stimulation in COVID-19 patients.
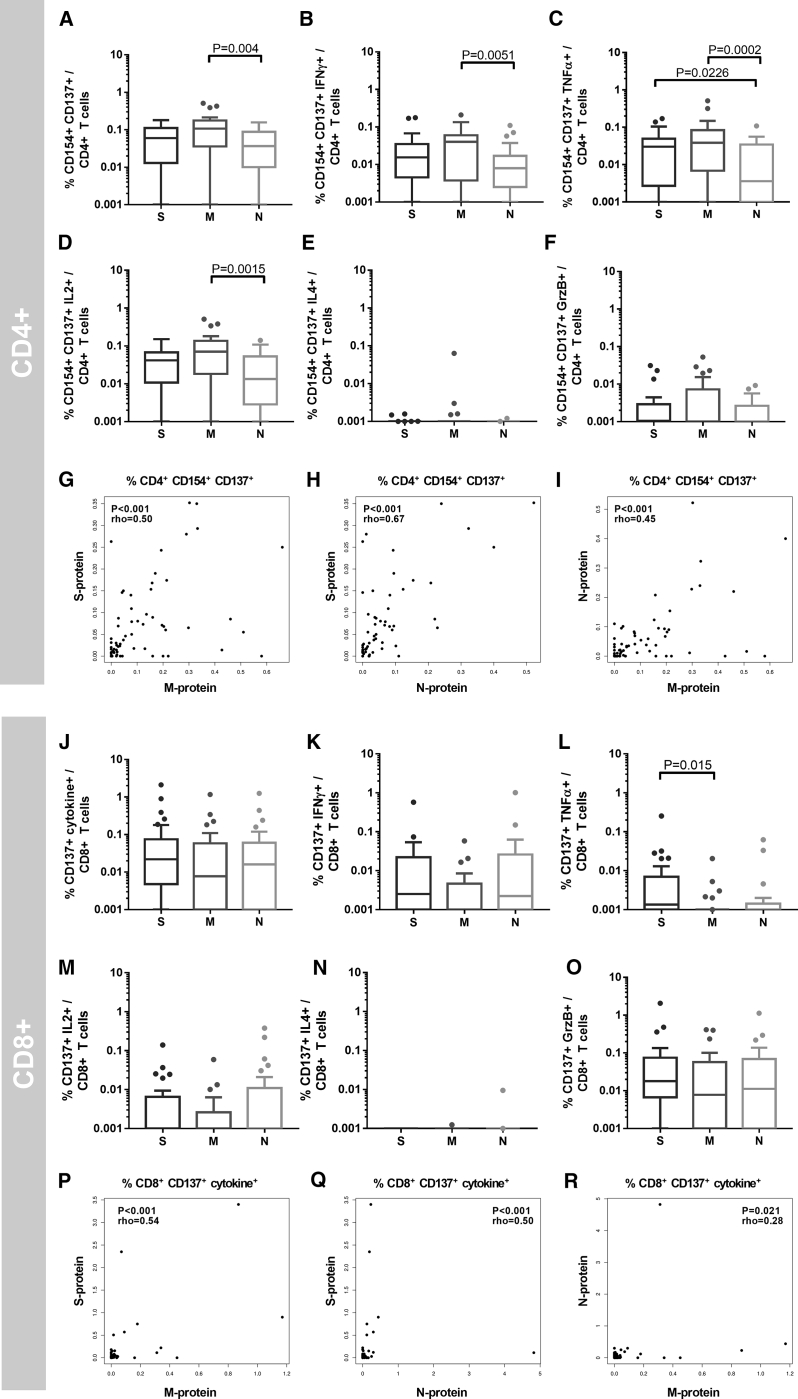
Magnitude and Functionality of T Cells Directed against S-, M-, and N-Proteins Show Different Patterns for CD4+ and CD8+ T Cells
Comparing the magnitude of response against the three proteins, we found that the M-protein OPP induced the highest frequencies of reactive CD4+ T cells, similar to the number of detectable CD4+ T cell responses described above (Figure 2A). Compared to the S- and N-reactive CD4+ T cells, we found a consistent trend of higher frequencies of M-reactive CD4+ T cells expressing cytokines and effector molecules such as IL-2, IFN-γ, TNF-α, and GrzB. N-protein induced the lowest responses in comparison to the other proteins (Figures 2B–2F).
Figure 2.
SARS-CoV-2 Reactive T Cells Display a Higher M-Protein Reactivity in CD4+ T Cells and S- and N-Protein Reactivity in CD8+ T Cells
From a total of 28 COVID-19 patients, 65 blood samples were drawn at 1 or at multiple time points after SARS-CoV-2 positive PCR tests. PBMCs were stimulated for 16 h with S-, M-, or N-protein OPPs. The gating strategy is presented in Figure S2.
(A–F) Mean frequencies of samples of individual COVID-19 patients (n = 28).
(A) CD154+ CD137+ CD4+ T cells (antigen-specific CD4+ T cells), and (B) IFN-γ-, (C) TNF-α-, (D) IL-2-, (E) IL-4-, and (G) GrzB-producing antigen-specific CD4+ T cells. The statistical analysis was performed with the Friedman test for non-parametric data and with Dunn’s multiple comparisons test. p < 0.05 was considered significant. Boxplots show medians and interquartile ranges. Whiskers and outliers were calculated with the Tukey method.
(G–I) Correlation of M-, N-, and S-protein OPP-reactive (CD154+ CD137+) CD4+ T cells. The calculation was performed with Spearman’s rank correlation coefficient.
(J–O) Mean frequencies per COVID-19 patient (n = 28). (J) CD137+ IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+) CD8+ T cells (antigen-specific CD8+ T cells), and (K) IFN-γ-, (L) TNF-α-, (M) IL-2-, (N) IL-4-, and (O) GrzB-producing CD137+ CD8+ T cells. The statistical analysis was performed with the Friedman test for non-parametric data and with Dunn’s multiple comparisons test. p < 0.05 was considered significant. Boxplots show medians and interquartile ranges. Whiskers and outliers were calculated with the Tukey method.
(P–R) Correlation analysis of M-, N-, and S-protein-reactive (CD137+ cytokine+) CD8+ T cells. The calculation was performed with Spearman’s rank correlation coefficient.
See also Figures S1 and S2.
The patterns observed for S-, N-, and M-protein reactivity of CD4+ T cells were not found in CD8+ T cells (Figures 2J–2O). In fact, a tendency of the higher frequencies of S- or N-protein reactive CD8+ T cells compared to M-protein was observed, but without reaching statistical significance after correction for multiple testing. Analyzing a possible correlation in frequencies of T cells reactive to S-, N-, and M-proteins, we did not observe a strong correlation in CD4+ and CD8+ T cell-restricted immunity (Figures 2G–2I and 2P–2R).
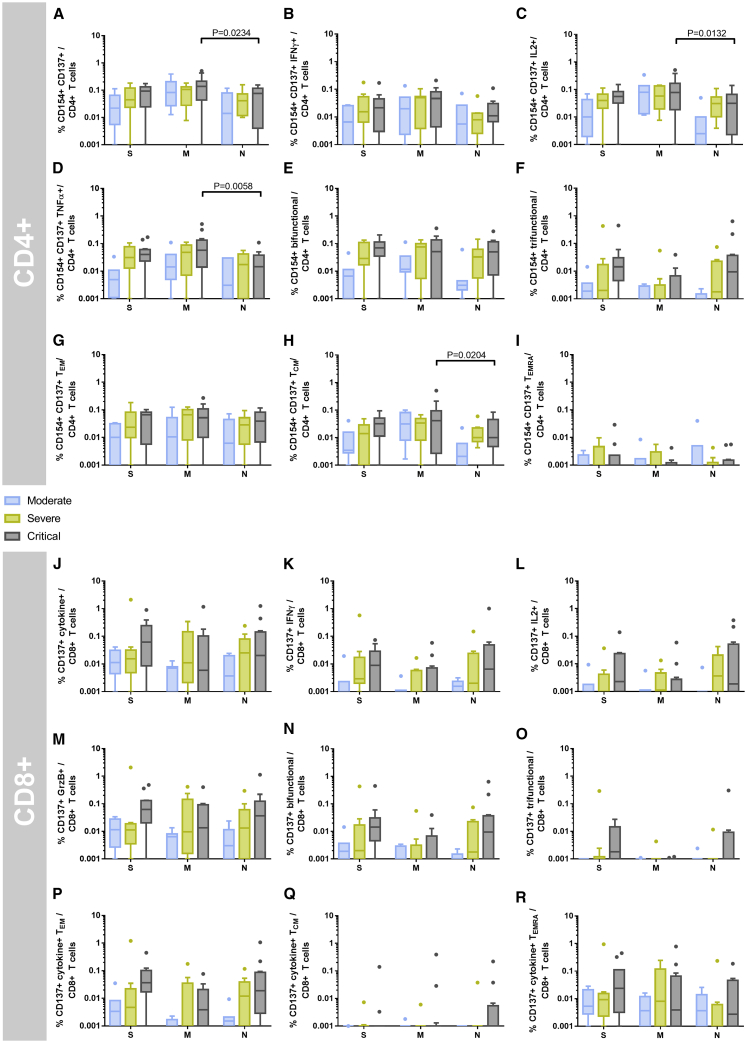
SARS-CoV-2-Reactive T Cell Immunity in Critical Patients Is Robust and Non-inferior Compared to Moderate COVID-19 Cases
The exact role of SARS-CoV-2-reactive T cell immunity for COVID-19 progression is unknown at present. We therefore investigated differences in the T cell immunity between moderate, severe, and critical COVID-19 patients. A defective switch between innate and adaptive immunity has been described in previous studies to differentiate patients with favorable and unfavorable outcomes after SARS-CoV infection.26 In contrast to the endemic SARS-CoV infection, we detected a comparable or even slightly higher magnitude of CD4+ and CD8+ T cells reactive to S-, M-, and N-proteins in critical COVID-19 as compared to moderate and severe cases (Figure 3).
Figure 3.
Critical COVID-19 Patients Maintain a Strong SARS-CoV-2 S-, M-, and N-Protein-Reactive CD4+ and CD8+ T Cell Response
From a total of 28 COVID-19 patients, 65 blood samples were drawn at 1 or at multiple time points after COVID-19 diagnosis. The mean frequency was calculated for each individual COVID-19 patient. COVID-19 severity was assessed at the time of sampling as per the guidelines of the German Robert Koch Institute and the patients grouped according to their worst disease category (n = 7 moderate, n = 9 severe, and n = 12 critical samples). PBMCs were stimulated for 16 h with S-, M-, or N-protein OPPs and analyzed by flow cytometry. The gating strategy to identify SARS-CoV-2 S-, M-, or N-protein-reactive T cells is presented in Figure S2.
(A–D) Frequencies of (A) CD154+ CD137+ CD4+ T cells (antigen-specific CD4+ T cells), and (B) IFN-γ-, (C) IL-2-, and (D) TNF-α-producing antigen-specific CD4+ T cells.
(E and F) Frequencies of polyfunctional CD154+ CD4+ T cells. (E) Bifunctional and (F) trifunctional CD154+ CD4+ T cells were analyzed by Boolean gating of IL-2, IFN-γ, TNF-α, IL-4, and GrzB production. A detailed composition of bi- and trifunctional cells is presented in Figure S4.
(G–I) Frequencies of antigen-specific CD4+ (G) TEM, (H) TCM, and (I) TEMRA cells.
(J–M) Frequencies of (J) CD137+ IL-2, IFN-γ, TNF-α and/or GrzB (CD137+ cytokine+) CD8+ T cells (antigen-specific CD8+ T cells), and (K) IFN-γ-, (L) IL-2-, and (M) GrzB-producing CD137+ CD8+ T cells.
(N and O) Frequencies of polyfunctional CD137+ CD8+ T cells. (N) Bifunctional and (O) trifunctional CD137+ CD8+ T cells were analyzed by Boolean gating of IL-2, IFN-γ, TNF-α, IL-4, and GrzB production. Composition of bi- and trifunctional cells is presented in Figure S4.
(P–R) Frequencies of antigen-specific CD8+ (P) TEM, (Q) TCM, and (R) TEMRA cells.
Statistical comparisons were done with 2-way repeated-measures ANOVA and with Tukey’s multiple comparison test. p < 0.05 was considered significant. Boxplots show medians and interquartile ranges. Whiskers and outliers were calculated with the Tukey method.
See also Figures S1, S2, and S4– S6 and Table S1.
Examining a limited number of subjectively selected functions of virus-reactive T cells may generate distorted and incomplete interpretations of the function and phenotype of these cells.27 Polyfunctional T cells, which express more than one cytokine or effector molecule, have been described as a hallmark of protective immunity in viral infections.27, 28, 29, 30 Addressing this point, we analyzed the IFN-γ, TNF-α, IL-2, and IL-4 cytokines, as well as effector molecule GrzB expression in parallel to differentiation stage phenotyping. Not only the quantity but also the functionality of T cell immunity were similar or even higher in patients with critical COVID-19 severity compared to moderate and severe cases (Figures 3E, 3F, 3N, and 3O). The cytokine and effector molecule expression of bifunctional CD4+ T cells was dominated by IFN-γ, TNF-α, IL-2 (Figure S4B), and antigen-reactive IFN-γ-, IL-2-, and TNF-α-producing CD4+ T cells constituted over 70% of trifunctional CD4+ T cells (Figure S4C). As expected, the majority of polyfunctional CD8+ T cells produced the cytotoxic effector molecule GrzB, most commonly in combination with IFN-γ and TNF-α (Figures S4B and S4C). A T helper 2 (TH2) cell-dominated response was associated with immunopathology and eosinophil infiltration after vaccination with N-protein-expressing vaccinia virus in a mouse SARS-CoV model.31,32 However, we have no indication of ongoing TH2 responses in critical patients since only very few IL-4-producing T cells were observed in all of the samples (Figures 2E and 2N).
An immunodominance of T cell responses toward certain peptides has been associated with immunopathology in flaviviridae.21,33, 34, 35 However, the relative composition of T cell responses against S-, M- and N-proteins in COVID-19 patient samples was mostly uniform across the different disease severities (Figure S5).
In line with data showing an association between polyfunctionality and the stage of phenotypic differentiation,27 we observed similar frequencies of CD4+ and CD8+ T cells with effector memory (TEM)/TEMRA phenotypes in critical COVID-19 samples compared to moderate and severe samples (Figures S6A and S6B). The presence of S-, N-, and M-reactive T cells with an advanced differentiation phenotype early after diagnosis in our study indicates preexisting cellular immunity as demonstrated by the detection of SARS-CoV-2-cross-reactive T cells in unexposed donors in our study and other studies.15,20 Of note, few unexposed donors also showed detectable polyfunctional T cells (Figure 1B). The memory composition of SARS-CoV-2-reactive T cells in unexposed donors largely resembled the COVID-19 patients, with the exception of fewer CD4+ and CD8+ TEM and more central memory (TCM) and naive (TNAIVE) cells (Figures S6A and S6B).
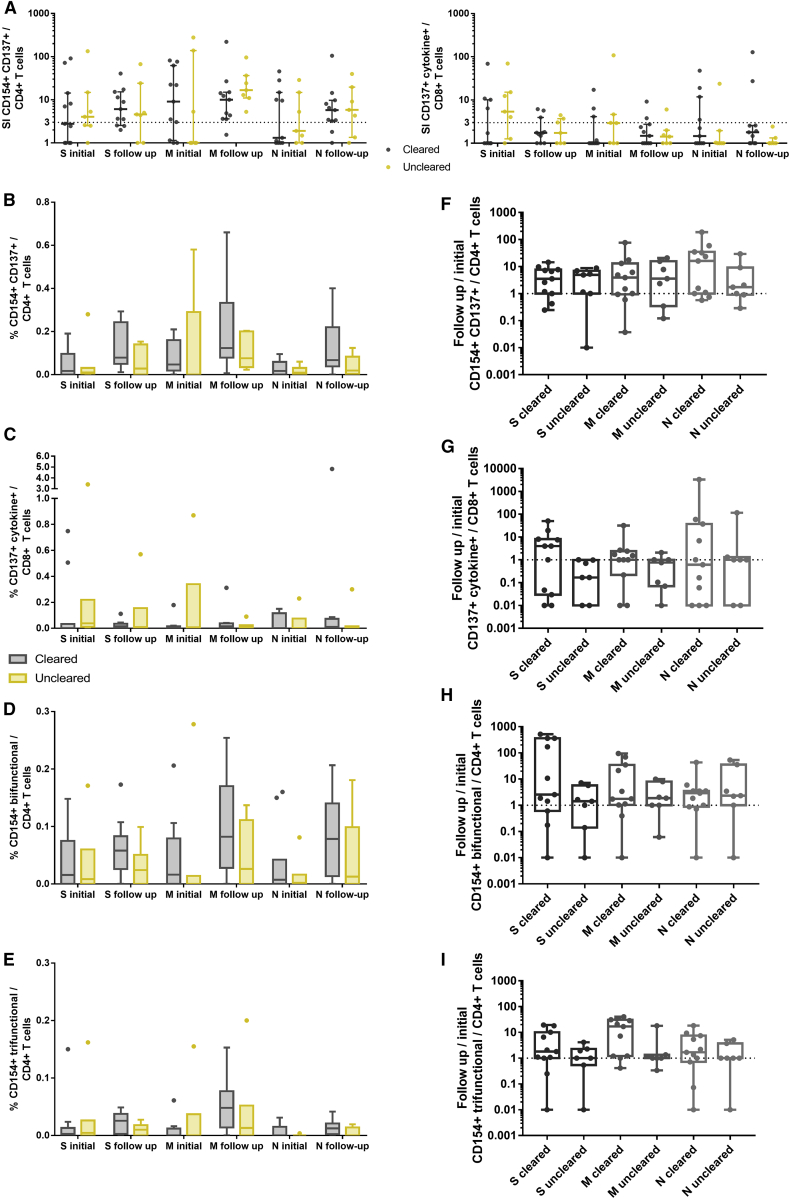
Viral Clearance and COVID-19 Recovery Are Not Associated with Changes in SARS-CoV-2-Reactive T Cell Responses
It is still an open question as to which extent the T cell immunity contributes to viral clearance and what the basis for a critical disease manifestation in certain individuals is. To investigate the possible involvement of impaired/insufficient adaptive immune responses, we stratified our patients according to their virus clearance status. Patients who were discharged from the hospital or had a minimum of 2 negative SARS-CoV-2 RNA PCR samples without a positive sample thereafter were categorized as cleared. Patients who were not discharged and had repetitive positive PCR results were grouped as uncleared. All of the patients from whom this information or follow-up samples were not available were excluded from this analysis. Consequently, we could include 11 patients in the cleared cohort and 7 patients in the uncleared cohort. Their clinical characteristics are described in Table S3. In the cleared group, we analyzed an initial blood sample before viral clearance and a follow-up sample after clearance. In the uncleared group, the first and the last available samples were obtained. There were no significant differences in sampling time related to the date of the first positive PCR and to the hospital admission, as well as in the time between the initial sample and the follow-up sample between the cleared and uncleared cohort (Table S3). We observed no significant differences regarding detectable T cell responses between the two cohorts (Figure 4A). In addition, the frequencies of SARS-CoV-2 protein-reactive CD4+ and CD8+ T cells, the frequencies of polyfunctional CD4+ T cells, and the ratio of initial and follow-up samples did not differ between the groups (Figures 4B–4I). Patients who did not clear the virus achieved a comparable magnitude of SARS-CoV-2-reactive T cell immunity as compared to the clearing patients already at the initial time point. Furthermore, we measured higher titers of neutralizing antibodies in patients who did not successfully clear the virus compared to patients who cleared the virus (Figure S7).
Figure 4.
T Cell Reactivity Does Not Differ between Viral PCR-Positive and -Negative Patients
Analysis of T cell responses of patients who cleared SARS-CoV-2 (n = 11) and patients who did not clear SARS-CoV-2 according to RT-PCR and were not discharged from the hospital during the observation period (uncleared, n = 7). In the cleared group, a sample before and after viral clearance was analyzed (initial and follow-up sample, respectively). In the uncleared group, the first (initial) and the last (follow-up) available sample were analyzed. PBMCs were stimulated for 16 h with S-, M-, or N-protein OPPs and analyzed by flow cytometry. The gating strategy to identify SARS-CoV-2 S-, M-, or N-protein-reactive T cells is presented in Figure S2.
(A) SI of CD154+ CD137+ CD4+ T cells (antigen-specific CD4+ T cells), and CD137+ IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+) CD8+ T cells (antigen-specific CD8+ T cells). SI was calculated by dividing the measured T cell subset response by the respective response in the DMSO control. Scatterplots show line at median; error bars represent interquartile ranges. The statistical comparison was done with 2-way repeated-measures ANOVA and with Sidak’s multiple comparisons test. p < 0.05 was considered significant.
(B–E) Comparison of the frequencies of CD154+ CD137+ CD4+ T cells, CD137+ IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+) CD8+ T cells, and bifunctional and trifunctional CD154+ CD4+ T cells. Bi- and trifunctional CD154+ CD4+ T cells were calculated by Boolean gating of IL-2, IFN-γ, TNF-α, IL-4, and GrzB production. The statistical comparisons were done with 2-way repeated-measures ANOVA and with Sidak’s multiple comparison test. p < 0.05 was considered significant. Boxplots show medians and interquartile ranges. Whiskers and outliers were calculated with the Tukey method.
(F–I) Relative changes of the follow-up sample compared to the initial sample. A value of 1 indicates no change as compared to the initial sample, lower values indicate a reduction, and values >1 indicate an increase in the frequency of the subset; base 10 logarithmic scale. The statistical comparison was done with the Kruskal-Wallis test and Dunn’s multiple comparisons test. p < 0.05 was considered significant. Boxplots show medians and minimum and maximum values.
See also Figures S1, S2, and S7 and Table S3.
We did not find any changes in SARS-CoV-2-reactive T cells associated with recovery from COVID-19, which is in line with the viral clearance data. Comparisons of clinical characteristics and sample timing are summarized in Table S4. The magnitude of S-, M-, and N-reactive T cells measured at the time of critical COVID-19 did not show significant differences in comparison to the corresponding measurements performed directly after patient recovery (Figure 5). The T cell data of recovering critical patients were comparable with the magnitude and functionality of S-, M-, and N-reactive T cell immunity measured at the initial visit in critical patients with lethal outcomes (Figures 5B–5I), suggesting that other cell subsets or cell subsets located at another site (e.g., the infected organ) are responsible for antiviral control and disease manifestation. Critical patients appear to have a generally higher level of SARS-CoV-2-reactive T cells as compared to the non-critical controls, but it must be kept in mind that the critical patients are probably later in their infectious course (Table S4), with more time for T cell proliferation. Accordingly, the differences between critical and moderate patients decrease in follow-up (Figure 5).
Figure 5.
T Cell Reactivity Does Not Differ between Patients with Non-critical Disease, Patients Who Recovered from COVID-19, and Patients Who Died
Analysis of T cell responses in samples of patients who recovered from critical COVID-19 (n = 5) during critical disease (initial) and shortly before discharge from the intensive care unit (follow-up), compared to samples of critical COVID-19 patients who died (n = 6) and to samples of non-critical COVID-19 patients (n = 15). PBMCs were stimulated for 16 h with S-, M-, or N-protein OPPs and analyzed by flow cytometry. The gating strategy to identify SARS-CoV-2 S-, M-, or N-protein-reactive T cells is presented in Figure S2.
(A) SI of CD154+ CD137+ CD4+ T cells (antigen-specific CD4+ T cells) and CD137+ IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+) CD8+ T cells (antigen-specific CD8+ T cells). The SI was calculated by dividing the measured T cell subset response by the respective response in the DMSO control. Scatterplots show line at median; error bars represent interquartile ranges. The statistical comparison was done with 2-way repeated-measures ANOVA and with Sidak’s multiple comparisons test. p < 0.05 was considered significant.
(B–E) Comparison of the frequencies of CD154+ CD137+ CD4+ T cells; CD137+ IL-2, IFN-γ, TNF-α, and/or GrzB (CD137+ cytokine+) CD8+ T cells; and bifunctional and trifunctional CD154+ CD4+ T cells. Bi- and trifunctional CD154+ CD4+ T cells were calculated by Boolean gating of IL-2, IFN-γ, TNF-α, IL-4, and GrzB production. The statistical comparisons between non-critical and recovered critical were done with 2-way repeated-measures ANOVA and with Sidak’s multiple comparison test. p < 0.05 was considered significant. Boxplots show medians and interquartile ranges. Whiskers and outliers were calculated with the Tukey method.
(F–I) Relative changes of the follow-up sample compared to the initial sample. A value of 1 indicates no change as compared to the initial sample, lower values indicate a reduction, and values >1 indicate an increase in the frequency of the subset; base 10 logarithmic scale. The statistical comparison was done with the Kruskal-Wallis test and with Dunn’s multiple comparisons test. p < 0.05 was considered significant. Boxplots show medians and minimum and maximum values.
See also Figures S1 and S2 and Table S4.
Discussion
Our data provide a comprehensive characterization of the T cell response against S-, M- and N-SARS-CoV-2 proteins in patients with different COVID-19 severity and in unexposed donors. The reactivity demonstrates individual patterns, indicating that all three proteins should be considered in cellular monitoring to avoid underestimation, especially in the early phase of the disease. Moreover, our findings suggest potential targets of humoral immunity. Considering the role of CD4+ TH cells for antibody generation and the higher CD4+ T cell response against M-protein, our results highlight the M-protein as an additional target for antibody monitoring and vaccine development. Here, our data are in line with the article by Grifoni et al.,20 demonstrating the immunogenic properties of certain M- and N-related immunodominant peptides in COVID-19 convalescent patients. Similar to the data by Grifoni et al.20 and Weiskopf et al.,16 we showed dominance of CD4+ T cell responses over CD8+ T cell responses. However, in comparison to these two studies, CD8+ T cell frequencies were substantially lower in our study. We could detect a CD8+ T cell response in 75% of the COVID-19 patients and in 30% of the unexposed donors. The CD8+ T cell response appeared to be functional, since a substantial number of the cells produced GrzB and IFN-γ. Discrepancies between our findings and the results of two other studies can be explained by different methodological protocols and different populations analyzed in the studies. Thus, our study analyzed patients with differing COVID-19 severity, while convalescent patients or patients with acute respiratory distress syndrome were analyzed in studies by Grifoni et al.20 and Weiskopf et al.,16 respectively. Furthermore, instead of using the two activation markers CD137 and CD69 for the definition of antigen-specific CD8+ T cells, we used activation marker CD137 together with at least 1 cytokine. In addition, OPPs containing 15-mers were applied in our study for T cell stimulation, whereas Weiskopf et al.16 and Grifoni et al.20 used PPs consisting mainly of 9- to 11-amino acid-long peptides for the optimal stimulation of CD8+ T cells, as identified by a bioinformatics epitope prediction approach.36 In fact, 9- to 11-mers are described to be optimal for being presented via major histocompatibility complex (MHC) class I molecules for CD8+ T cell activation, and there are some concerns that CD8+ T cell responses can be underestimated using 15-mer peptides. However, previous studies by other groups and our group37, 38, 39 demonstrated 15-mer as a non-inferior T cell stimulation source, and appropriate experimental setup can avoid underestimation.37 The induction of CD4+ and CD8+ T cell responses in unexposed donors in our cohort is comparable to that of previous investigations and can probably be attributed to cross-reactivity toward common cold coronaviruses.15,40
A prolonged positivity of SARS-CoV-2 RT-PCR has been observed for up to several weeks after infection.41 Factors that influence this finding in certain individuals and the infectiousness of the detected virus have yet to be investigated. Seroconversion, as a sign of the development of adaptive immunity, does not implicate PCR negativity.42 Similarly, we did not find differences in the T cell responses of patients who cleared the virus to patients who did not clear it. Neutralizing antibodies were higher in the serum of patients who did not clear the virus, implicating that the prevention of virus entry in the host cells leads to a prolonged residence of the virus in the extracellular space and stronger activation of adaptive immunity. However, this finding needs to be confirmed in a larger cohort.
Patients with critical COVID-19 demonstrated equal, or even slightly higher, frequencies of CD4+ and CD8+ T cells reactive to S-, M-, and N-OPPs, demonstrating the ability of critical COVID-19 patients to maintain a substantial cellular immunity that was induced following infection. The exact time point of infection in our cohort, as in most other investigations studying natural infections, is not known. The slightly higher response of critical patients may therefore reflect a longer disease course in the critical cohort. It may also be that the higher magnitude and functionality of the T cell response observed in critical COVID-19 cases simply reflects a more severe infection course with a stronger immunogenic environment, provided by a higher viral burden and inflammatory bystander activation. Yet another possible explanation lies in the observation that SARS-CoV-2 triggers the production of high amounts of circulating chemokines, which may prevent T cells from reaching the infected tissue.43
Although the direct antiviral capacity must be proven in future studies, polyfunctional T cells are commonly regarded as a parameter of protective immunity.27,29,30,44 To this end, the presence of IFN-γ- and TNF-α-coproducing CD4+ and CD8+ T cells indicating an effector/memory phenotype and long-term protection was also shown for the 2002–2003 SARS-CoV infection.45,46 However, they can also provide immune damage contributing to immunopathogenesis,47 and the correlation of IFN-γ-induced protein-10 (IP-10), T cell proliferation, and COVID-19 progression observed in a recent study may point toward this hypothesis.43 In this context, our finding of an advanced differentiation stage of SARS-CoV-2-reactive T cells found at early time points and in unexposed donors raises the question about the beneficial effect of preexisting immunity for the course of infection. One could speculate that even though it appears to be generally protective, preexisting SARS-CoV-2 reactive T cells with effector phenotype, which are cross-reactive with common cold coronaviruses,15 can lead to hyperactive response and immunopathogenesis in severe infection. It has been observed that preformed antigen-specific T cells skewed toward certain peptides due to previous infection with a different serotype can cause immunopathology in flaviviridae infection.21,33, 34, 35 It is therefore important to note that we did not find this skewed response in our critical patients as compared to the moderate cases; however, a more detailed approach using individual peptides may be beneficial to study this question more closely in the future.
Another important finding is the lack of association between COVID-19 recovery and SARS-CoV-2-reactive T cell immunity in critical COVID-19. Thus, the magnitude and functionality of M-, N-, and S-reactive T cells were comparable before and after recovery from COVID-19 in critical cases. More important, there were no significant differences in SARS-CoV-2 reactive T cell immunity between deceased and recovered critical COVID-19 patients at any analyzed time point.
Although further studies are required, our data may suggest that other cell subsets or other locations should be targeted to elucidate the exact role of T cells in COVID-19 progression. Our study highlights that all three main SARS-CoV-2 structural proteins should be evaluated for diagnostics and therapeutic strategies to avoid the underestimation of cellular immunity, especially in early COVID-19. Critical patients mount a robust SARS-CoV-2-specific T cell response that could be involved in immunopathogenesis, but certainly disfavors the hypothesis of an impaired T cell response as a reason for life-threatening COVID-19.
Limitations of Study
Our study has some limitations. Thus, the exact time point of infection in our cohort, as described above, is not known, and the slightly higher T cell response detected in critical patients may reflect a longer disease course in the critical cohort. Therefore, further prospective studies considering a longer follow up in non-critical patients enabling a better sampling time match between the groups would be required. Furthermore, only three SARS-CoV-2 OPPs were available at the time point of the study initiation (M-, S-, and N-proteins), while 26 further proteins remained unconsidered but could theoretically elicit immune responses.48 Our analysis was limited to TH1 cytokines and CD8+ T cell effector molecules. Unbiased or broader analysis may reveal protective or detrimental T cell subtypes in non-critical or critical COVID-19 patients, respectively. Due to the relatively short history of SARS-CoV-2 infection in humans, the longevity of T cell memory remains uncertain.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti CCR7 conjugated to PerCP-Cy5.5 (clone G043H7) | BioLegend | Cat#: 353220, RRID: AB_10916121 |
| anti CD4 conjugated to A700 (clone OKT4) | BioLegend | Cat#: 317426, RRID: AB_571943 |
| anti CD8 conjugated to V500 (clone RPA-T8) | BD Biosciences | Cat#: 560775, RRID: n/a |
| anti CD45RA conjugated to BV605 (clone HI100) | BioLegend | Cat#: 304134, RRID: AB_2563814 |
| anti Granzyme B conjugated to FITC (clone GB11) | BioLegend | Cat#: 515403, RRID: AB_2114575 |
| anti IL2 conjugated to PE (clone MQ1-17H12) | BioLegend | Cat#: 500307, RRID: AB_315094 |
| anti IL4 conjugated to PE-Dazzle594 (clone MP4-25D2) | BioLegend | Cat#: 500832, RRID: AB_2564036 |
| anti CD137 (4-1BB) conjugated to PE-Cy7 (clone 4B4-1) | BioLegend | Cat#: 309818, RRID: AB_2207741 |
| anti CD154 (CD40L) conjugated to A647 (clone 24-31) | BioLegend | Cat#: 310818, RRID: AB_492970 |
| anti TNFα conjugated to eFluor450 (clone MAb11) | eBioscience | Cat#: 48-7349-42, RRID: AB_2043889 |
| anti IFNγ conjugated to BV650 (clone 4S.B3) | BioLegend | Cat#: 502538, RRID: AB_2563608 |
| anti CD3 conjugated to BV785 (clone OKT3) | BioLegend | Cat#: 317330, RRID: AB_2563507 |
| Biological Samples | ||
| Blood and serum from SARS-CoV2 infected patients | Marien Hospital Herne, University Hospital of the Ruhr-University Bochum, Germany | n/a |
| Blood and serum from SARS-CoV2 infected patients | University Hospital Essen, University Duisburg-Essen, Germany | n/a |
| Blood from patients not infected with SARS-CoV2 (collected and biobanked prior SARS-CoV2 pandemic) | University Hospital Essen, University Duisburg-Essen, Germany | n/a |
| Blood from patients not infected with SARS-CoV2 (collected and biobanked prior SARS-CoV2 pandemic) | Knappschaftskrankenhaus Bochum, University Hospital of the Ruhr-University Bochum, Germany | n/a |
| Chemicals, Peptides, and Recombinant Proteins | ||
| PepTivator SARS-CoV-2 Protein S (covering the immunodominant sequence domains of the surface (or spike) glycoprotein (“S”) of SARS-Coronavirus 2 (GenBank MN908947.3, Protein QHD43416.1)) | Miltenyi Biotec | Cat#: 130-126-701 |
| PepTivator SARS-CoV-2 Protein N (covering the complete sequence of the nucleocapsid phosphoprotein (“N”) of SARS-Coronavirus 2 (GenBank MN908947.3, Protein QHD43423.2)) | Miltenyi Biotec | Cat#: 130-126-699 |
| PepTivator SARS-CoV-2 Protein M (covering the complete sequence of the membrane glycoprotein (“M”) of SARS-Coronavirus 2 (GenBank MN908947.3, Protein QHD43419.1)) | Miltenyi Biotec | Cat#: 130-126-703 |
| RPMI 1640 | Life Technologies | Cat#: 21875-034 |
| Penicillin-Streptomycin-Glutamin (L-glutamine 200 mM, penicillin 10,000 units, streptomycin 10 mg/mL, sterile-filtered) | Sigma Aldrich | Cat#: G6784-10X5ML |
| FBS Good, EU approved regions, filtrated bovine serum, 0.2 μm sterile filtered | PAN-Biotech | Cat#: P40-37500 |
| Brefeldin A from Penicillium brefeldianum, ≥ 99% (HPLC and TLC) | Sigma Aldrich | Cat#: B7651-5MG |
| Fixable Viability Dye eFluor 780 | eBioscience | Cat#: 65-0865-14 |
| DMEM | Thermo Fisher | Cat#: 11965092 |
| GMEM | Thermo Fisher | Cat#: 21710025 |
| Poly-L-Lysin | Sigma-Aldrich | Cat#: P4832 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher | Cat#: 15140122 |
| Lipofectamin 2000 | Thermo Fisher | Cat#: 11668027 |
| L-Glutamine (200 mM) | Thermo Fisher | Cat#: 25030024 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher | Cat#: 11140035 |
| Mifepristone | Thermo Fisher | Cat#: H11001 |
| Critical Commercial Assays | ||
| Intracellular Fixation & Permeabilization Buffer Set | Thermo Fisher Scientific | Cat#: 88-8824-00 |
| Luciferase Assay System | Promega | Cat#: E1501 |
| Deposited Data | ||
| Raw and analyzed data - Stimulation of PBMCs from COVID19 patients with S Protein OPPs. | 8 | http://medxriv.org/lookup/doi/10.1101/2020.04.28.20083089 |
| Experimental Models: Cell Lines | ||
| Human embryo kidney (HEK) 293T cells | ATCC | ATCC CRL-3216 |
| I1-Hybridoma | ATCC | ATCC CRL-2700 |
| Vero E6 cells | Christian Drosten/Marcel Müller (Charite, Berlin, Germany) | n/a |
| BHK-G43 | BHK-21 cells co-transfected with the plasmids pSwitch (Invitrogen) and pGeneC-VSVG | n/a |
| Recombinant DNA | ||
| pCAGGS-SARS-S | 11 | n/a |
| VSV∗ΔG(FLuc) | Berger Rentsch and Zimmer, 2011 | n/a |
| Software and Algorithms | ||
| FlowJo version 10.6.2 | BD Biosciences | https://www.flowjo.com/ |
| R, version 3.6.2 | 51 | https://www.r-project.org/ |
| GraphPad Prism version 7 | GraphPad | https://www.graphpad.com/ |
| Venny version 2.1 | 52 | https://bioinfogp.cnb.csic.es/tools/venny/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Nina Babel (nina.babel@charite.de)
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Human Samples
28 patients with moderate (n = 7), severe (n = 9) and critical (n = 12) COVID-19, categorized by their most severe diagnosis were recruited. The degree of COVID-19 severity was evaluated according to the guidelines of the Robert Koch Institute, Germany, as previously described8. In addition, we used samples of 10 donors collected before the COVID-19 pandemic and frozen at −80°C as an unexposed control group. The study was approved by the ethical committee of the Ruhr-University Bochum (20-6886) and University Hospital Essen (20-9214-BO), and written informed consent was obtained from all participants. The clinical and demographic patient parameters are shown in Table 1. Patients with moderate and severe COVID-19 were recruited after the first symptoms were reported and a positive SARS-CoV-2 PCR confirmed the disease (in median 4 days after the diagnostic test). The sampling time is presented in Table S2. For the Figure 1, the maximum value of all samples of an individual patient was included. In Figures 2 and 3, the mean frequency was calculated of all measured samples of an individual patient. For Figures 4 and S7, we included patients that were discharged from the hospital or had a minimum of 2 negative SARS-CoV-2 RNA PCR samples without a positive sample thereafter in the cleared group. The uncleared group consisted of patients that were not discharged and had repetitive positive PCR results. All patients from whom this information or follow-up samples were not available were excluded from this analysis. Figure 5 shows a more detailed analysis of critical patients (n = 11). A first sample was collected during critical COVID-19 on ICU in all presented patients. Of patients with critical disease who survived, we collected a second sample at the day of the planned discharge from the intensive care unit (recovered critical group) (n = 5). Samples of critically ill patients who deceased later on from the disease were categorized as deceased critical (n = 6). One patient categorized as critical only had a short stay on ICU and was not included in this analysis. For moderate and severely ill patients, the initial and the last available samples were used for the non- critical control group. Figures S3, S4, S5, and S6 include all acquired and pooled patient samples. Samples were grouped into COVID-19 severity groups according to the symptom presentation at time of sampling.
Primary Cell Culture and Cell Line Cultivation
Primary cells were incubated for OPP stimulation in a humidified incubator at 37°C and 5% CO2. Cells were maintained in RPMI (Life Technologies) medium supplemented with 1% Penicillin-Streptomycin-Glutamin and 10% FCS. Cells were slowly frozen in freeze-medium (FCS + 10% DMSO, Life Technologies) and stored at −80°. Vero E6 cells were maintained in Dulbecco’s minimal essential medium (DMEM; Life Technologies, Zug, Switzerland) supplemented with 10% fetal bovine serum (FBS) and non-essential amino acids (Life Technologies). Baby hamster kidney (BHK-21) cells were maintained in Glasgow’s minimal essential medium (GMEM, Life Technologies) supplemented with 10% FBS. BHK-G43 was maintained in GMEM containing 5% FBS. I1-Hybridoma maintained in minimal essential medium (MEM, Life Technologies) supplemented with 15% FBS. Human embryo kidney (HEK) 293T cells were cultured using DMEM with 10% FBS.
Method Details
Preparation of PBMCs
PBMC were prepared from EDTA collection tubes (S-Monovette K3, Sarstedt) by gradient centrifugation: 15 mL blood was diluted with PBS/BSA (GIBCO) at a 1:1 ratio, underlaid with 15 mL Ficoll-Paque Plus (GE Healthcare) and centrifuged for 20 min at 800 rpm. The mononuclear cell layer was isolated and cells were washed with PBS. Afterward, PBMCs were frozen in FCS + 10% DMSO (Sigma Aldrich) and stored at −80°C until further usage.
Stimulation with OPPs
SARS-CoV-2 PepTivator peptide pools (Miltenyi Biotec), consisting mainly of 15-mer sequences with 11 amino acids (aa) overlap containing overlapping peptide pools (OPP) spanning in silico predicted immunodominant50 parts of the S-protein, or, covering the complete sequence of the N- and M-protein, were used for peripheral blood mononuclear cells (PBMC) stimulation. The purity of each OPP was > 70%. The peptides were dissolved per manufacturer’s directions in sterile water and used at a concentration of 1 μg/ml. After thawing, PBMCs were rested overnight before being plated in a 96-UWell plate (Sarstedt) in RPMI media (Life Technologies) supplemented with 1% Penicillin-Streptomycin-Glutamine (Sigma Aldrich), and 10% FCS (PAN-Biotech). The cells were stimulated for 16 h at 37°C and after 2 h, Brefeldin A (1 μg/ml, Sigma Aldrich) was added. As a positive control, cells were stimulated with SEB (1 μg/ml, Sigma Aldrich) and as a negative control, cells were left untreated.
Flow cytometry
After stimulation with SARS-CoV-2 OPP, PBMCs were stained with fluorophore conjugated antibodies for flow cytometry analysis. Mastermixes, containing all antibodies for intra or extracellular staining in the optimal concentrations were prepared directly before staining. At first, cells were surface stained with CCR7-PerCP-Cy5.5 (clone G043H7) (BioLegend), CD4-A700 (clone OKT4) (BioLegend), Fixable Viability Dye eFluor780 (eBioscience), CD8-V500 (clone RPA-T8) (BD Biosciences) and CD45RA-BV605 (clone HI100) (BioLegend) for 10 minutes at room temperature in the dark. After thoroughly washing with PBS/BSA, cells were fixed and permeabilized using Intracellular Fixation & Permeabilization Buffer Set according to the manufacturer’s instructions (Thermo Fisher Scientific). Then, the cells were stained with Granzyme B-FITC (clone GB11) (BioLegend), IL2-PE (clone MQ1-17H12) (BioLegend), IL4-PE-Dazzle594 (clone MP4-25D2) (BioLegend), CD137 (4-1BB)-PE-Cy7 (clone 4B4-1) (BioLegend), CD154 (CD40L)-A647; (clone 24-31) (BioLegend), TNFa-eFluor450 (clone MAb11) (eBioscience), IFNg-BV650 (clone 4S.B3) (BioLegend), CD3-BV785 (clone OKT3) (BioLegend) for 30 minutes at room temperature in the dark. All samples were washed thoroughly with PBS and acquired on a CytoFlex flow cytometer (Beckman Coulter).
Measurement of SARS-CoV-2 neutralizing antibodies
For the virus neutralization assay, sera were incubated for 30 min at 56°C in order to inactivate complement factors. A propagation-incompetent VSV∗ΔG(FLuc) pseudovirus system bearing the SARS-CoV-2 spike protein in the envelope was incubated with quadruplicates of twofold serial dilutions of immune sera in 96-well plate prior to infections of Vero E6 cells (1x104 cells / well) in DMEM + 10% FBS (Life Technologies). At 18 hours post infection, firefly luciferase (FLuc) reporter activity was determined after addition of 25 μL of firefly luciferase ONE-Glo™ substrate (Promega) using a GloMax® plate reader (Promega) and the reciprocal antibody dilution causing 50% inhibition of the luciferase reporter calculated (PVND50).
Quantification and Statistical Analysis
Data analysis and graphical representation
Flow cytometry data were analyzed using FlowJo version 10.6.2 (BD Biosciences). Gating was performed using single stains and fluorescence minus one controls and adjusted according to the DMSO controls for each individual sample. Gating strategies are presented in Figure S2. Unspecific activation in unstimulated controls was subtracted from stimulated samples to account for specific activation in the presented frequencies. Negative values were set to zero. Stimulation index (SI) was calculated by dividing the measured T cell subset response by the respective response in the DMSO control. If the DMSO control was negative, the minimum value across that subset was used for calculation. SI below 1 was set to 1. SI above 3 was considered detectable response. Relative changes of the follow up sample compared to the initial samples were calculated by dividing the frequency of the respective subset in the follow up sample by the frequency in the initial sample. If the initial sample was 0, the minimum value across that subset was used for calculation. If the value was below 0.01, it was set to 0.01. Multifunctional T cells were analyzed using Boolean gating of IL2, IL4, IFNγ, TNFα, and GrzB producing T cells in combination with CD154 for CD4+ and CD137 for CD8+ T cells. Statistical analysis was performed using R, version 3.6.251 and GraphPad Prism v7, which was also used for graphical representation. Venn diagrams were prepared using Venny v2.152.
Statistical analysis
Non-parametric statistical tests were used where applicable. Patient age and time point of sampling were compared with Kruskal-Wallis-Test. Patient gender, chest CT abnormalities and treatments were compared with Fisher’s exact test. Comparisons of SI between COVID-19 patient samples and unexposed donors were done with Kruskal-Wallis test and Dunn’s multiple comparisons test. Differences in T cells responses of mean patient frequencies and of all patient samples together were analyzed with Friedman test and Dunn’s multiple comparison test. As sphericity was not assumed, Geisser-Greenhouse correction was applied. Correlation between the T cell responses toward the different peptides was analyzed by Spearman’s rank correlation coefficient. Differences in T cell responses of patient samples grouped according to COVID-19 severity were analyzed by repeated-measurement two-way ANOVA and Tukey’s multiple comparison test. Comparison of SI and frequencies of T cell subsets in patients with different SARS-CoV-2 clearance and of patients with different clinical course in follow up was performed using Two-way repeated-measurement ANOVA and Sidak’s multiple comparisons test. Comparisons of relative changes of T cell subset frequencies and of neutralizing antibodies in follow up in these patients were done with Kruskal-Wallis test and Dunn’s multiple comparisons. P values below 0.05 were considered significant; only significant P values are reported.
Acknowledgments
We feel deep gratitude to the patients who donated their blood samples and clinical data for this project. We would like to acknowledge the excellent technical assistance as well as the expertise of the immune diagnostic laboratory (Sarah Skrzypczyk, Eva Kohut, Julia Kurek, and Jan Zapka) of the Center for Translational Medicine at Marien Hospital Herne. This work was supported by grants from the Stiftung Mercator, the Bundesministerium für Bildung und Forschung (BMBF) e:KID (01ZX1612A), and the BMBF NoChro (FKZ 13GW0338B).
Author Contributions
T.H.W., U.S., T.B., and N.B. conceived and designed the study. M.A., K.P., T.L.M., S.P., and E.S. performed the experiments. M.A., K.P., A.D., F.S.S., S.D., M.J.K., M.M.B., T.B., O.W., C.T., and B.H. organized patient recruitment, data collection, and sampling. C.J.T., M.A., A.B.-N., U.S., T.R., and N.B. analyzed the data. C.J.T., M.A., U.S., T.R., C.W., U.D., and N.B. wrote the paper. All of the authors reviewed and edited the paper.
Declaration of Interests
The authors declare no competing interests.
Published: August 29, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xcrm.2020.100092.
Supplemental Information
References
- 1.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vijay R., Perlman S. Middle East respiratory syndrome and severe acute respiratory syndrome. Curr. Opin. Virol. 2016;16:70–76. doi: 10.1016/j.coviro.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 4.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anft M., Paniskaki K., Blazquez-Navarro A., Doevelaar A., Seibert F.S., Hoelzer B., Skrzypczyk S., Kohut E., Kurek J., Zapka J. A possible role of immunopathogenesis in COVID-19 progression. medRxiv. 2020 doi: 10.1101/2020.04.28.20083089. [DOI] [Google Scholar]
- 9.Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E. Effect of Dexamethasone in Hospitalized Patients with COVID-19: Preliminary Report. medRxiv. 2020 doi: 10.1101/2020.06.22.20137273. [DOI] [Google Scholar]
- 10.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nisreen M.A.O., Marcel A.M., Wentao L. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. J. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sette A., Moutaftsi M., Moyron-Quiroz J., McCausland M.M., Davies D.H., Johnston R.J., Peters B., Rafii-El-Idrissi Benhnia M., Hoffmann J., Su H.P. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann M.F., Zinkernagel R.M. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997;15:235–270. doi: 10.1146/annurev.immunol.15.1.235. [DOI] [PubMed] [Google Scholar]
- 15.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 16.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H.-S., Hsieh Y.-C., Su I.-J., Lin T.H., Chiu S.C., Hsu Y.F., Lin J.H., Wang M.C., Chen J.Y., Hsiao P.W. Early detection of antibodies against various structural proteins of the SARS-associated coronavirus in SARS patients. J. Biomed. Sci. 2004;11:117–126. doi: 10.1007/BF02256554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Y.-J., Goh P.-Y., Fielding B.C., Shen S., Chou C.F., Fu J.L., Leong H.N., Leo Y.S., Ooi E.E., Ling A.E. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin. Diagn. Lab. Immunol. 2004;11:362–371. doi: 10.1128/CDLI.11.2.362-371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y., Wen J., Tang L., Zhang H., Zhang X., Li Y., Wang J., Han Y., Li G., Shi J. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinformatics. 2003;1:118–130. doi: 10.1016/S1672-0229(03)01016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duangchinda T., Dejnirattisai W., Vasanawathana S., Limpitikul W., Tangthawornchaikul N., Malasit P., Mongkolsapaya J., Screaton G. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc. Natl. Acad. Sci. USA. 2010;107:16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020 doi: 10.1116/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallais F., Velay A., Wendling M.-J., Nazon C., Partisani M., Sibilia J., Candon S., Fafi-Kremer S. Intrafamilial Exposure to SARS-CoV-2 Induces Cellular Immune Response without Seroconversion. medRxiv. 2020 doi: 10.1101/2020.06.21.20132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kern F., Surel I.P., Brock C., Freistedt B., Radtke H., Scheffold A., Blasczyk R., Reinke P., Schneider-Mergener J., Radbruch A. T-cell epitope mapping by flow cytometry. Nat. Med. 1998;4:975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 26.Cameron M.J., Ran L., Xu L., Danesh A., Bermejo-Martin J.F., Cameron C.M., Muller M.P., Gold W.L., Richardson S.E., Poutanen S.M., Canadian SARS Research Network Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 2007;81:8692–8706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duvall M.G., Precopio M.L., Ambrozak D.A., Jaye A., McMichael A.J., Whittle H.C., Roederer M., Rowland-Jones S.L., Koup R.A. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur. J. Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Subbarao K. The Immunobiology of SARS. Annu. Rev. Immunol. 2007;25:443–472. doi: 10.1146/annurev.immunol.25.022106.141706. [DOI] [PubMed] [Google Scholar]
- 29.Harari A., Vallelian F., Meylan P.R., Pantaleo G. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J.I Immunol. 2005;174:1037–1045. doi: 10.4049/jimmunol.174.2.1037. [DOI] [PubMed] [Google Scholar]
- 30.Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A., Suthar M., Harkema J., Whitmore A., Pickles R. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLOS Med. 2006;3:e525. doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181:6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 33.Screaton G., Mongkolsapaya J., Yacoub S., Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 2015;15:745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds C.J., Suleyman O.M., Ortega-Prieto A.M., Skelton J.K., Bonnesoeur P., Blohm A., Carregaro V., Silva J.S., James E.A., Maillère B. T cell immunity to Zika virus targets immunodominant epitopes that show cross-reactivity with other Flaviviruses. Sci. Rep. 2018;8:672. doi: 10.1038/s41598-017-18781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mongkolsapaya J., Duangchinda T., Dejnirattisai W., Vasanawathana S., Avirutnan P., Jairungsri A., Khemnu N., Tangthawornchaikul N., Chotiyarnwong P., Sae-Jang K. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 36.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiecker F., Streitz M., Ay B., Cherepnev G., Volk H.D., Volkmer-Engert R., Kern F. Analysis of antigen-specific T-cell responses with synthetic peptides--what kind of peptide for which purpose? Hum. Immunol. 2004;65:523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmeister B., Bunde T., Rudawsky I.M., Volk H.D., Kern F. Detection of antigen-specific T cells by cytokine flow cytometry: the use of whole blood may underestimate frequencies. Eur. J. Immunol. 2003;33:3484–3492. doi: 10.1002/eji.200324223. [DOI] [PubMed] [Google Scholar]
- 39.Schmueck M., Fischer A.M., Hammoud B., Brestrich G., Fuehrer H., Luu S.-H., Mueller K., Babel N., Volk H.-D., Reinke P. Preferential expansion of human virus-specific multifunctional central memory T cells by partial targeting of the IL-2 receptor signaling pathway: the key role of CD4+ T cells. J.I Immunol. 2012;188:5189–5198. doi: 10.4049/jimmunol.1103763. [DOI] [PubMed] [Google Scholar]
- 40.Stervbo U., Rahmann S., Roch T., Westhof T.H., Babel N. SARS-CoV-2 reactive T cells in uninfected individuals are likely expanded by beta-coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.07.01.182741. [DOI] [Google Scholar]
- 41.Xiao A.T., Tong Y.X., Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa460. ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wajnberg A., Mansour M., Leven E., Bouvier N.M., Patel G., Firpo A., Mendu R., Jhang J., Arinsburg S., Gitman M. Humoral immune response and prolonged PCR positivity in a cohort of 1343 SARS-CoV 2 patients in the New York City region. medRxiv. 2020 doi: 10.1101/2020.04.30.20085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Munoz-Ruiz M., Mckenzie D., Hayday T., Francos Quijorna I. A consensus Covid-19 immune signature combines immuno-protection with discrete sepsis-like traits associated with poor prognosis. medRxiv. 2020 doi: 10.1101/2020.06.08.20125112. [DOI] [Google Scholar]
- 44.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babel N., Volk H.-D., Reinke P. BK polyomavirus infection and nephropathy: the virus-immune system interplay. Nat. Rev. Nephrol. 2011;7:399–406. doi: 10.1038/nrneph.2011.59. [DOI] [PubMed] [Google Scholar]
- 48.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger Rentsch M., Zimmer G. A Vesicular Stomatitis Virus Replicon-Based Bioassay for the Rapid and Sensitive Determination of Multi-Species Type. I Interferon. Plos One. 2011;6 doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Development Core Team R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2020 https://www.r-project.org/ [Google Scholar]
- 52.Oliveros J.C. Venny 2.1. An interactive tool for comparing lists with Venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.