Abstract
Automated assays for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies in coronavirus disease 2019 (COVID-19) diagnostics have recently come available. We compared the performance of the Elecsys® Anti–SARS-CoV-2 and LIAISON® SARS-CoV-2 S1/S2 IgG tests. The seroconversion panel comprised of 120 samples from 13 hospitalized COVID-19 patients. For the sensitivity and specificity testing, samples from COVID-19 outpatients >15 days after positive nucleic acid amplification test (NAAT) result (n = 35) and serum control samples collected before the COVID-19 era (n = 161) were included in the material. Samples for the detection of possible cross-reactions were also tested. Based on our results, the SARS-CoV-2 antibodies can be quite reliably detected 2 weeks after NAAT positivity and 3 weeks after the symptom onset with both tests. However, since some COVID-19 patients were positive only with Elecsys®, the antibodies should be screened against N-antigen (Elecsys®) and reactive samples confirmed with S antigen (LIAISON®), but both results should be reported. In some COVID-19 patients, the serology can remain negative.
Keywords: Antibody, COVID-19, Elecsys, LIAISON, SARS-CoV-2, Serology
Highlights
-
•
Elecsys® and LIAISON® SARS-CoV-2 antibody tests performed reliably.
-
•
SARS-CoV-2 antibodies should be confirmed with different antigens to increase accuracy.
-
•
In some patients, the COVID-19 serology may remain completely negative.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), which was for the first time met in China in the end of the year 2019 (Zhu et al., 2020). After that, the virus has caused a severe pandemic (Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, 2020). The acute COVID-19 is diagnosed by nucleic acid amplification test (NAAT). SARS-CoV-2 antibodies are formed in the blood usually within 2–3 weeks after infection (Okba et al., 2020), and their determination can be used in epidemiological surveys and as a support in the diagnostics of prolonged and obscure cases. However, CE-marked, in vitro diagnostics suitable and US Food and Drug Administration Emergency Use Authorized SARS-CoV-2 antibody tests have not come on the market until recently, and there exist a few articles on the performance of fully automated test platforms (Egger et al., 2020; Kohmer et al., 2020; Merrill et al., 2020; Montesinos et al., 2020; Plebani et al., 2020a; Tang et al., 2020a, Tang et al., 2020b; Tré-Hardy et al., 2020). In this paper, we compared the performance of the fully automated Elecsys® Anti–SARS-CoV-2 test detecting antibodies against nucleocapsid N protein and LIAISON® SARS-CoV-2 S1/S2 IgG test detecting antibodies against spike protein S1 and S2 antigens.
2. Materials and methods
2.1. Evaluation samples
The seroconversion panel part of the study comprised of 120 samples from 13 patients [age 55 years (median), range 20–79; 8 males] of whom the seroconversion time was sought. The patients had been admitted to Tampere University Hospital or other communal hospitals in Fimlab Laboratories operation region due to aggravated COVID-19 respiratory tract symptoms, i.e., difficulty breathing with positive NAAT result. During hospitalization, blood cell count from EDTA blood was analyzed from the patients almost daily. After this routine analysis, the residual samples were collected from these patients, and the EDTA plasma was separated and stored −20 °C until the evaluation.
The other part of the study concerning sensitivity and specificity of the tests was partly based on the seroconversion panel [n = 5, age 55 years (median), range 34–79; 2 males], but also residual plasma/serum samples from the COVID-19 NAAT positive outpatients were traced and collected for evaluation [n = 35, age 47 years (median), range 11–95; 12 males]. All these patients had had respiratory tract symptoms including rhinitis, cough, sore throat, chest pain, and/or difficulty breathing, with or without fever. In this part, the follow-up time after positive NAAT result was at least 16 days. The control material comprised 161 serum samples from apparently healthy adults [age 45 years (mean), range 32–65; 72 males] with mildly to moderately increased total cholesterol who were part of the chitosan study before the COVID-19 era (Lehtimäki et al., 2005). These samples had been stored −20 °C before the comparison. The use of these samples for control purposes had an approval from The Ethics Committee of the Tampere University Hospital District, and all participants had given their written informed consent.
For the detection of possible cross-reactions in the tests, follow-up plasma/serum samples from other coronavirus and influenza A/B polymerase chain reaction (PCR)–positive patients and serum/plasma samples from acute Epstein-Barr virus (EBV: IgG VCA and IgM antibodies positive, and IgG EBNA antibodies negative)–, hepatitis B core antibody (HBcAb)–, antinuclear antibody (ANA)–, and rheumatoid factor (RF)–positive patients were included in the study material (n = 43). EBV-, HBcAb-, and ANA-positive samples had been collected in year 2019 and RF-positive samples in year 2017 before the COVID-19 pandemic. The samples from other coronavirus and influenza A/B patients had been collected in April–May 2020.
2.2. Methods
SARS-CoV-2 antibodies were analyzed using Elecsys® Anti–SARS-CoV-2 test (Roche Diagnostics GmbH, Mannheim, Germany) detecting the antibodies against nucleocapsid N protein and LIAISON® SARS-CoV-2 S1/S2 IgG test (DiaSorin S.p.A., Saluggia, Italy) detecting the antibodies against spike (S) protein S1 and S2 antigens. Primary COVID-19 diagnosis had been based on in-house real-time reverse-transcription (RT)-PCR test detecting E-gene target sequence according to Corman et al. (2020); Allplex™ 2019-nCoV Assay (Seegene Inc., Seoul, South Korea) detecting target sequences E, N, and RdRp; or Abbott RealTime SARS-CoV-2 Assay (Abbott Laboratories, Abbott Park, IL) detecting target sequences N and RdRp. The used RT-PCR method had been chosen based on the availability. The primary COVID-19 diagnosis was based on 1 RT-PCR result. All the serologically discrepant cases had been tested with at least 1 of the available commercial RT-PCR tests detecting several viral domains.
3. Results and discussion
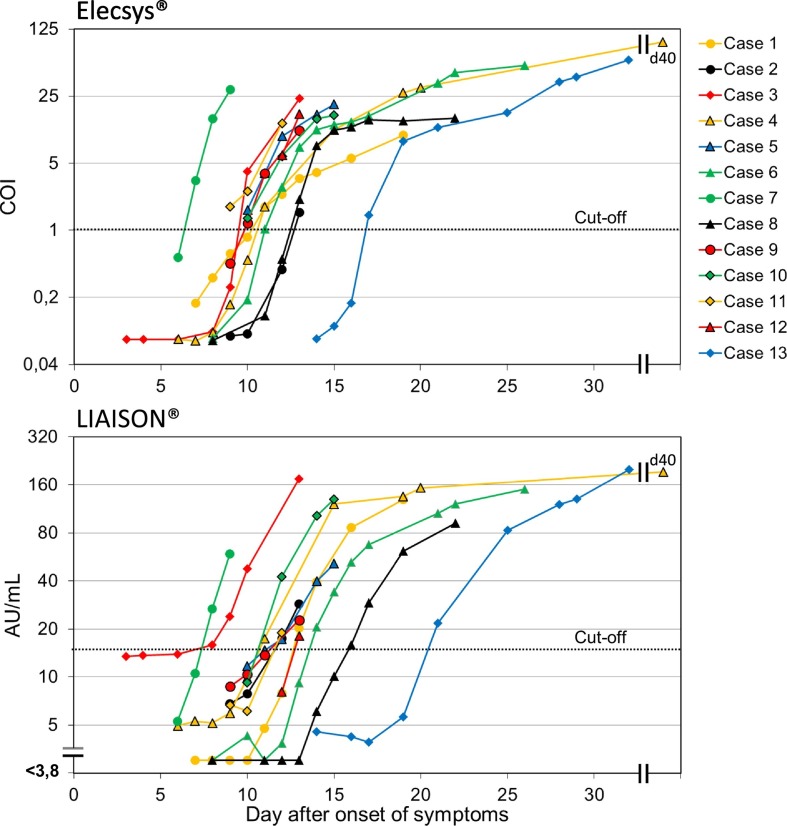
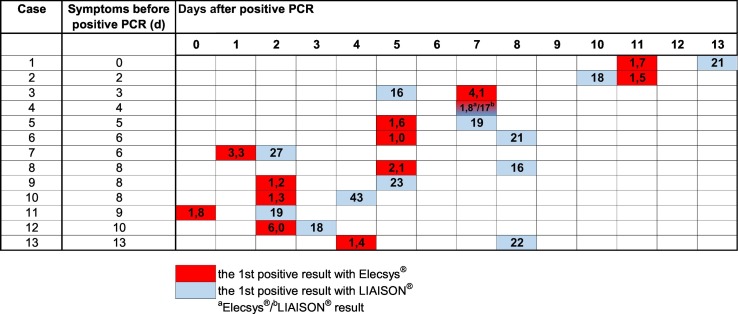
The antibody kinetics of the patients in the seroconversion panel is illustrated in Fig. 1 and the seroconversion times in Fig. 2 . The time interval from the positive NAAT result to seroconversion was 5 days (median, range 0–11 days) with Elecsys® and 7 days (median, range 2–13 days) with LIAISON®. The seroconversion was first detected with Elecsys® in 10 and with LIAISON® in 2 patients, and at the same time in 1 patient. In both methods, the seroconversion had happened in 85% (11/13) of the patients within 8 days and in all within 13 days after the positive NAAT result. The time interval from the onset of symptoms to seroconversion was 11 days with Elecsys® (median, range 7–17 days) and 12 days (median, range 8–21) with LIAISON®, respectively. Although in some studies a decline in SARS-CoV-2 antibodies has been detected a few weeks after seroconversion (Favresse et al., 2020a: Seow et al., 2020), in our seroconversion panel, the antibody trend was rising in all patients with both methods. However, the follow-up time was quite short.
Fig. 1.
The SARS-CoV-2 antibody kinetics in COVID-19 patients determined by Elecsys® Anti–SARS-CoV-2 and LIAISON® SARS-CoV-2 S1/S2 IgG tests. The analyses were carried out of 83 samples in the seroconversion panel.
Fig. 2.
SARS-CoV-2 antibody seroconversion time in COVID-19 patients determined by Elecsys® Anti–SARS-CoV-2 and LIAISON® SARS-CoV-2 S1/S2 IgG tests. Antibody level of the first positive sample (COI for Elecsys® and AU/mL for LIAISON®) is shown in the bar. The cutoff values for the positive result are ≥1 COI and ≥15 AU/mL.
Elecsys® detects total antibodies against N protein, while LIAISON® detects only IgG antibodies against S1 and S2 antigens. The S protein is an immunogenic surface structure of the SARS-CoV-2 involved in the virus attachment to the host cells, and its functional subunits (S1 and S2) are used in the immunoassays. N protein is, in turn, the major structural protein of the SARS-CoV-2 involved in the replication processes of the virus (Infantino et al., 2020). The positive antibody test result means that the person has had COVID-19, but it does not definitely tell about protective immune response. N protein–based tests may be more sensitive to detect past COVID-19, but S protein may be a possible target for neutralizing antibodies, and SARS-CoV-2 antibodies against that antigen may better predict the protective immunity (Walls et al., 2020). It has been suggested that the immune response against S antigens might come earlier than that against N antigen (Liu et al., 2020). However, in some clinical test comparisons, the observed seroconversion time has been shorter with the tests detecting total antibodies to N antigen compared to those detecting IgG antibodies to S antigens, although the studies using systematic seroconversion panels with follow-up samples are sparse (Montesinos et al., 2020; Tang et al., 2020a). In our seroconversion panel, the first positive result was detected in most cases earlier with Elecsys® using N antigen than with LIAISON® using S1/S2 antigens. Since it has been shown that SARS-CoV-2 IgG and IgM antibodies can occur simultaneously or sequentially, either IgG or IgM first (Long et al., 2020), it can be speculated whether the IgM in the Elecsys® total antibody composition causes the earlier positive reaction in some cases compared to LIAISON®. However, it is also possible that it is a result of differences in the test chemistry or the antibody response to different antigens (Long et al., 2020; Tang et al., 2020b). The seroconversion times with Elecsys® and LIAISON® in our study were well in line with the other studies (Egger et al., 2020; Okba et al., 2020).
Sensitivity and specificity of the tests are shown in Table 1 . The sensitivity and specificity of the Elecsys® were 92.5% and 98.8% and of LIAISON® 87.5% and 97.5%, respectively. The positive (PPV) and negative (NPV) predictive values with different assumed COVID-19 prevalence in the population are shown in the table. Elecsys® seemed to be more sensitive and specific than LIAISON®, but the differences were minor and the number of tested samples was limited. In the study by Egger et al. (2020), the sensitivity and specificity of the Elecsys® were 100% and 99.8 in the samples taken over 15 days after symptom onset, respectively, but the number of COVID-19 positive cases in that time point was only 18. In the study by Favresse et al. (2020b), the specificity of the Elecsys® was 100% and the sensitivity was over 90% in the samples taken 14 days after positive NAAT or symptom onset, and the results were also in line with Tang et al. (2020b). According to the study by Tré-Hardy et al. (2020), the sensitivity and specificity of the LIAISON® test were 91% and 100%, respectively, evaluated from the samples taken 2 weeks after the positive NAAT result. The results were quite similar in the study by Plebani et al. (2020a), in which the samples were collected over 12 days after the onset of symptoms. In one comparison of several tests, the Elecsys® and LIAISON® performed quite equally, the sensitivities being 75.6% in both and specificities being 97% and 100%, respectively. However, in that study, the time interval after positive NAAT to serological test varied from 2 to 49 days, so some seroconversions had probably not happened yet, and also, the control group was quite small, consisting of 19 persons (Kohmer et al., 2020). In the study by Merrill et al. (2020), Elecsys® seemed to be somewhat more sensitive and specific than LIAISON®, but the sensitivity remained quite low in both methods since most test samples were collected within 1 week after positive NAAT.
Table 1.
Sensitivity and specificity of the Elecsys® Anti–SARS-CoV-2 and LIAISON® SARS-CoV-2 S1/S2 IgG tests.
| Test and result | COVID-19 NAAT test result |
Sensitivity (%) | Specificity (%) | PPV (%) (COVID-19 prevalence 1/5/10%) |
NPV (%) (COVID-19 prevalence 1/5/10%) |
|
|---|---|---|---|---|---|---|
| Positive (n = 40) | Negative (n = 161) | |||||
| Elecsys® Anti–SARS-CoV-2 | ||||||
| Positive | 37 | 2b | 92.5 (CI: 79.6–98.4) |
98.8 (CI: 95.6–99.9) |
42.9/79.7/89.2 | 99.9/99.6/99.2 |
| Negative | 3a | 159 | ||||
| LIAISON® SARS-CoV-2 S1/S2 IgG | ||||||
| Positive | 35 | 4b | 87.5 (CI: 73.2–95.8) |
97.5 (CI: 93.8–99.3) |
26.2/65.0/79.7 | 99.9/99.3/98.6 |
| Negative | 5 | 157 | ||||
The PPV and NPV with different assumed COVID-19 prevalence in the population are shown in the table. NAAT = nucleic acid amplification test; CI = 95% confidence interval.
These samples were also negative with LIAISON®.
All false-positive antibody results were obtained from different control samples.
Some research groups have intended to optimize the cutoffs of Elecsys® and LIAISON® tests and ended up to the levels of 0.165 COI (cut-off index) regarding Elecsys® and 6.1–6.2 AU/mL regarding LIAISON® (Favresse et al., 2020b, Favresse et al., 2020c; Plebani et al., 2020a, Plebani et al., 2020b; Tré-Hardy et al., 2020). We applied also a receiver operating characteristic curve performance analysis and determined the optimal cutoff for the tests in our material using Youden index. The optimal cutoff for Elecsys® was 0.137 COI with sensitivity and specificity of 97.5% and 96.9%, and for LIAISON®, the optimal cutoff was 11.9 AU/mL with sensitivity and specificity of 90.0% and 96.9%, respectively. According to our results, the optimized cutoff for Elecsys® was well in line with the other studies, but for LIAISON®, it was somewhat higher. These optimized cutoffs need to be validated more and taken into consideration when the serological methods are applied into the clinical use.
SARS-CoV-2 antibodies remained negative in 3 NAAT-positive COVID-19 patients in both Elecsys® and LIAISON® tests. The follow-up times after positive NAAT were 28, 45, and 53 days, respectively. Plebani et al. (2020a) have reported COVID-19 cases that are not able to produce detectable antibodies to N or S antigens. Thus, the negative serology does not exclude the possibility of previous COVID-19 infection. Two NAAT-positive COVID-19 patients were positive with Elecsys® but remained negative with LIAISON®. The follow-up times were 34 and 77 days, respectively. From the control sample material collected before COVID-19 era, 4 samples were positive with LIAISON® and 2 with Elecsys®. All of the false-positive results were from different control samples. Based on our results, the overall specificity of the antibody testing increases, reaching 100% if the positive antibody results are confirmed with another method. However, if the positive Elecsys® result is confirmed with LIAISON®, the sensitivity decreases. Thus, in this sense, to increase the accuracy of the overall testing results and taking also the financial impact into account, the SARS-CoV-2 antibodies should be screened against N antigen (Elecsys®) and reactive samples confirmed with S antigen test (LIAISON®), but both SARS-CoV-2 antibody test results should be reported parallel for clinical evaluation and related to the patient's clinical picture. A paired sample should be taken into consideration if there is a discrepancy within the 2 results and the time from the onset of symptoms is inadequate.
Cross-reactivities were tested for several conditions as shown in Table 2 . Two human coronavirus (HCoV)-OC43–positive patients had also a positive LIAISON® test result 60 and 70 days after HCoV-OC43 diagnosis (levels 19 and 21 AU/mL, respectively). However, antibodies against SARS-CoV-2 N protein by Elecsys® were totally negative (0.076 and 0.084 COI, respectively). Since the samples had been collected in April and May 2020, it is possible that these patients may also have had a nondiagnosed COVID-19 and thus specific SARS-CoV-2 antibodies, but it is unlikely since the COVID-19 morbidity rates in Finland have so far remained considerably low (134 diagnosed cases/100,000 persons until July 31, 2020), and the seroprevalence has been below 0.5% in general population determined by a gold standard method, i.e., virus microneutralization (Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, 2020; Serological population study of the coronavirus epidemic by Finnish Institute for Health and Welfare, 2020). Furthermore, S-protein antibody positivity (LIAISON®) without detectable N protein antibodies (Elecsys®) among the true-positive COVID-19 patients seemed to be atypical according to our results. Thus, the LIAISON® results were estimated to be false positive. Otherwise, all the cross-reactivity results were negative. SARS-CoV-2 belongs to betacoronaviruses like HCoV-OC43 and HCoV-HKU1. HCoV-229E and HCoV-NL63 belong to alphacoronaviruses. The cross-reactions are usually seen within Alpha- and Betacoronavirus genera but not between them (Huang et al., 2020).
Table 2.
Cross-reactivity testing for Elecsys® Anti–SARS-CoV-2 and LIAISON® SARS-CoV-2 S1/S2 IgG tests against several conditions to evaluate potential interference.
| Possible cross-reactive samples | n | Days after positive NAAT to sample collection, median (range) | Elecsys® Anti–SARS-CoV-2 positive result | LIAISON® SARS-CoV-2 S1/S2 IgG positive result |
|---|---|---|---|---|
| Human coronavirus OC43a | 13 | 35 (4–100) | 0 | 2d |
| Human coronavirus NL63a | 2 | 40 (27–52) | 0 | 0 |
| Human coronavirus 229Ea | 1 | 82 | 0 | 0 |
| Human coronavirus OC43 and human bocavirusa | 1 | 35 | 0 | 0 |
| Influenza A virusa | 5 | 58 (41–85) | 0 | 0 |
| Influenza A and B virusa | 1 | 54 | 0 | 0 |
| Acute EBV (IgG VCA and EBNA, and IgM antibodies positive)b | 5 | - | 0 | 0 |
| HBcAb positiveb | 5 | - | 0 | 0 |
| ANA positiveb | 5 | - | 0 | 0 |
| RF positivec | 5 | - | 0 | 0 |
NAAT = nucleic acid amplification test, Allplex Respiratory Panel 1 & 3, Seegene; EBV = Epstein–Barr virus; HBcAb = hepatitis B core antibody; ANA = antinuclear antibody; RF = rheumatoid factor.
Samples collected in April–May, 2020.
Samples collected in year 2019.
Samples collected in year 2017.
Time after diagnosis to sample collection 60 and 70 days, respectively.
In this study, we compared the performance of the fully automated Elecsys® Anti–SARS-CoV-2 test detecting antibodies against nucleocapsid N protein and LIAISON® SARS-CoV-2 S1/S2 IgG test detecting antibodies against spike protein S1 and S2 antigens. The seroconversion was detected in most cases earlier with Elecsys® than with LIAISON®, but the antibodies could be quite reliably detected 2 weeks after NAAT positivity and 3 weeks after the symptom onset with both methods. Elecsys® was somewhat more sensitive and specific than LIAISON®, but the differences were minor. However, since some patients were seropositive only with Elecsys®, we conclude that the SARS-CoV-2 antibodies should be screened against N antigen (Elecsys®) and reactive samples confirmed with S antigen test (LIAISON®), but both test results should be reported parallel for clinical evaluation and related to the patient's clinical picture. Furthermore, in some patients, the COVID-19 serology may remain completely negative. Because clear guidelines for the use of SARS-CoV-2 serology are lacking at the moment, these aspects should be taken into consideration when these processes are assessed.
Acknowledgments
Acknowledgments
We thank Nina Peltonen for delivering the chitosan study control samples, Auni Collings for checking the written language, and all others who have enabled this study and taken part in developing COVID-19 diagnostics in Fimlab Laboratories operation region.
Funding
The study was supported by Tampere Tuberculosis Foundation and Competitive State Research Financing of Expert Responsibility area of Tampere.
Conflict of interest
No conflict of interest.
Authors' contributions
HF has participated in planning the study, collecting the serological samples, setting up the serological methods, analyzing the samples and the results, and writing the manuscript.
AR has participated in setting up the serological methods and analyzing the samples.
BL has set up and evaluated the in-house and commercial NAAT methods.
TL has delivered the chitosan study control samples and participated in writing the manuscript.
AMH has participated in writing the manuscript.
TS has participated in writing the manuscript.
JA has participated in planning the study, collecting the serological samples, analyzing the results, and writing the manuscript.
References
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Bundschuh C., Wiesinger K., Gabriel C., Clodi M., Mueller T. Comparison of the Elecsys® Anti–SARS-CoV-2 immunoassay with the EDI™ enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta. 2020;509:18–21. doi: 10.1016/j.cca.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favresse J., Eucher C., Elsen M., Graux C., Goebels P., Laffineur K. Unexpected kinetics of anti–SARS-CoV-2 total antibodies in two patients with chronic lymphocytic leukemia. Br J Haematol. 2020 doi: 10.1111/bjh.16954. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favresse J., Eucher C., Elsen M., Marie T.H., Dogné J.M., Douxfils J. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies [published online ahead of print, 2020 Jun 2] Clin Chem. 2020 doi: 10.1093/clinchem/hvaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favresse J, Eucher C, Elsen M, Laffineur K, Dogné JM, Douxfils J. Response of anti–SARS-CoV-2 total antibodies to nucleocapsid antigen in COVID-19 patients: a longitudinal study. Clin Chem Lab Med. 2020c Jul 8:/j/cclm.ahead-of-print/cclm-2020-0962/cclm-2020-0962.xml. doi: 10.1515/cclm-2020-0962. [Epub ahead of print]. [DOI] [PubMed]
- Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. 2020. https://coronavirus.jhu.edu/map.html
- Serological population study of the coronavirus epidemic by Finnish Institute for Health and Welfare. 2020. https://thl.fi/fi/tutkimus-ja-kehittaminen/tutkimukset-ja-hankkeet/koronaepidemian-serologinen-vaestotutkimus
- Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick L, Rattigan SM, et al. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv 2020.04.14.20065771; doi: 10.1101/2020.04.14.20065771. [DOI] [PMC free article] [PubMed]
- Infantino M., Damiani A., Gobbi F.L., Grossi V., Lari B., Macchia D. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J. 2020;22(4):203–210. [PubMed] [Google Scholar]
- Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol. 2020;129:104480. doi: 10.1016/j.jcv.2020.104480. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki T., Metso S., Ylitalo R., Rontu R., Nikkilä M., Wuolijoki E. Microcrystalline chitosan is ineffective to decrease plasma lipids in both apolipoprotein E epsilon 4 carriers and non-carriers: a long-term placebo-controlled trial in hypercholesterolaemic volunteers. Basic Clin Pharmacol Toxicol. 2005;97(2):98–103. doi: 10.1111/j.1742-7843.2005.pto_111.x. [DOI] [PubMed] [Google Scholar]
- Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 2020;58(6):e00461–20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed]
- Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020 Jun;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [Epub 2020 Apr 29] [DOI] [PubMed] [Google Scholar]
- Merrill AE, Jackson JB, Ehlers A, Voss D, Krasowski MD. Head-to-head comparison of two SARS-CoV-2 serology assays. J Appl Lab Med. 2020 Jul 27:jfaa125. doi: 10.1093/jalm/jfaa125. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Montesinos I., Gruson D., Kabamba B., Dahma H., Van den Wijngaert S., Reza S. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti–SARS-CoV-2 antibodies. J Clin Virol. 2020;128:104413. doi: 10.1016/j.jcv.2020.104413. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plebani M, Padoan A, Negrini D, Carpinteri B, Sciacovelli L. Diagnostic performances and thresholds: the key to harmonization in serological SARS-CoV-2 assays? Clin Chim Acta. 2020a:S0009–8981(20)30263–1. doi: 10.1016/j.cca.2020.05.050. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Plebani M, Padoan A, Sciacovelli L, Basso D. Towards the rational utilization of SARS-CoV-2 serological tests in clinical practice. Clin Chem Lab Med. 2020b Jul 3:/j/cclm.ahead-of-print/cclm-2020-0880/cclm-2020-0880.xml. doi: 10.1515/cclm-2020-0880. [Epub ahead of print]. [DOI] [PubMed]
- Seow J, Graham C, Merrick B, Acors S, Steel KJA, Hemmings O, et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection; medRxiv 2020 2020.07.09.20148429; doi: 10.1101/2020.07.09.20148429. [DOI]
- Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa120. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W. Clinical performance of the Roche SARS-CoV-2 serologic assay. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa132. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tré-Hardy M, Wilmet A, Beukinga I, Dogné JM, Douxfils J, Blairon L. Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin Chem Lab Med. 2020:/j/cclm.ahead-of-print/cclm-2020-0594/cclm-2020-0594.xml. doi: 10.1515/cclm-2020-0594. [Epub ahead of print]. [DOI] [PubMed]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]