Abstract
We evaluated the effect of systemic antibiotics (azithromycin, amoxicillin, cotrimoxazole, or placebo) on the gut resistome in children aged 6 to 59 months. Azithromycin and cotrimoxazole led to an increase in macrolide and sulfonamide resistance determinants. Resistome expansion can be induced with a single course of antibiotics.
Keywords: resistome, antimicrobial resistance, randomized controlled trial
Antibiotic use selects for antibiotic resistance in individuals and populations [1, 2]. Pediatric infection is treated based on clinical symptoms in many rural sub-Saharan African settings, potentially resulting in inappropriate antibiotic use [3]. Lack of laboratory facilities and symptom-based treatment results in a high false-positive rate for use of antibiotics [4, 5]. Although the World Health Organization recommends tailoring empiric treatment to local resistance patterns, such a strategy is difficult with limited availability of resistance data [4]. In rural Burkina Faso, children receive nearly 2 antibiotic courses per year, most commonly including amoxicillin, cotrimoxazole, and erythromycin [6], which may have important implications for resistance selection. We used data from a randomized, controlled trial that evaluated the effect of 3 pediatric antibiotics compared to placebo on the gut microbiome [7] to assess the relationship between antibiotic use and the gut resistome, defined as the collection of resistance gene determinants in a given environment [8].
METHODS
Complete methods for the trial have been previously described (clinicaltrials.gov NCT03187834) [7]. Children were enrolled in 2 rural communities of the Health and Demographic Surveillance Site in Nouna District, Burkina Faso [9]. The study occurred in July 2017. The institutional review boards at the University of California, San Francisco, and the Centre de Recherche en Sante de Nouna provided ethical approval. Written informed consent was obtained from each child’s caregiver.
Households with at least 2 children aged 6 to 59 months were eligible for inclusion and randomized in a 1:1:1:1 fashion to amoxicillin (25 mg/kg/day in twice-daily doses), azithromycin (10 mg/kg on day 1 and then 5 mg/kg once daily for 4 days), cotrimoxazole (240 mg once daily), or placebo (powdered milk and sugar in bottled water). All study arms received 5 days of treatment. Within each household, children were randomly assigned to either the household’s treatment arm or placebo. In the placebo households, children assigned to “treatment” and “placebo” received the same drug (placebo). Sample processing for resistome analysis was restricted to children who were randomized to treatment to assess the direct effect of antibiotic use on selection for resistance determinants. Medications were prepared as pediatric oral suspension, prepared fresh each day and packaged in opaque syringes that were prelabeled with the child’s name and study identification number. Treatments were administered at a central point in the community and were directly observed.
Rectal samples were collected 5 days after the last antibiotic treatment. Swabs were immediately placed into Stool Nucleic Acid Collection and Transport Tubes with Norgen Stool Preservative (Norgen, Ontario, Canada). Samples were collected at ambient temperature in the field, then stored at –80°C in Nouna until they were shipped to San Francisco. Samples were deidentified in the field and placed in a random order for processing. All laboratory personnel were masked.
DNA was extracted from the fecal samples using the Norgen Stool DNA Isolation Kit (Norgen, Ontario, Canada) per manufacturer’s instructions. Double-stranded DNA was fragmented, size selected, and converted to Illumina libraries using the NEBNext Ultra II DNA Library Prep Kit (E7645) according to the manufacturer’s recommendation and then amplified with 11 polymerase chain reaction (PCR) cycles. Samples were sequenced on the Illumina Novaseq using 150-nucleotide paired-end sequencing. An initial human-sequence removal step was accomplished as previously described [10]. Another round of human reads removal was performed using the very-sensitive-local mode of Bowtie2 (v2.2.4) with the same hg38 and panTro4 reference genomes. The remaining nonhost read pairs were then aligned to the MEGARes reference antimicrobial database using Burrows-Wheeler alignment (BWA) with default settings [11]. In order to decrease false-positive antimicrobial resistant determinant (ARD) identification, only ARDs with a gene fraction of >80% were identified as present in the sample and included for further analyses. Each identified ARD was classified at the class and gene level. For each ARD in each sample, the total number of aligned reads was summed to create a count matrix with samples in rows and classes or gene in columns. A sample was determined to be resistant even if 1 ARD was detected.
We analyzed the effect of antibiotics on selection for genetic resistance determinants at the class and gene level. At the class level, we used modified Poisson models to estimate risk ratios for the presence of genetic resistance determinants by antibiotic arm. At the gene level, we calculated Chao1 total resistance gene determinant richness and compared posttreatment richness across arms using an analysis of variance (ANOVA) and pairwise comparisons for each antibiotic compared to placebo with a t test. We used permutational multivariate analysis of variance (PERMANOVA) to assess differences in genetic resistance determinant composition across study arms using Euclidean distance to avoid overweighting of rare determinants, with principal coordinates analysis used to graphically depict the centroids for each group. All analyses were conducted in R version 3.4.3 (R Foundation for Statistical Computing).
RESULTS
Of 124 children randomized, 120 had a rectal swab collected posttreatment (Supplementary Figure 1). Baseline characteristics have been previously reported [7] and were broadly similar across study arms (Supplementary Table 1). Antibiotic completion was high in all study arms [12]. Supplementary Table 1 displays the posttreatment prevalence of genetic resistance determinants to each antibiotic class by study arm. In the placebo arm, the most common genetic resistance determinants were to beta-lactams (73.3%), whereas sulfonamide resistance was the least common (3.3%).
Beta-lactam resistance was not significantly different in any antibiotic arm compared to placebo (Supplementary Tables 2 and 3). Azithromycin samples were more than twice as likely to have genetic resistance determinants to macrolides (risk ratio [RR], 2.61; 95% confidence interval [CI], 1.55 to 4.42; P = .0003) compared to placebo. Sulfonamide resistance was higher in samples from children in all antibiotic arms compared to placebo. Samples from children randomized to cotrimoxazole were more than 3 times as likely to have genetic resistance determinants to trimethoprim (RR, 3.29; 95% CI, 1.08 to 9.95; P = .04) compared to placebo. Trimethoprim resistance did not differ in samples from children randomized to azithromycin or amoxicillin.
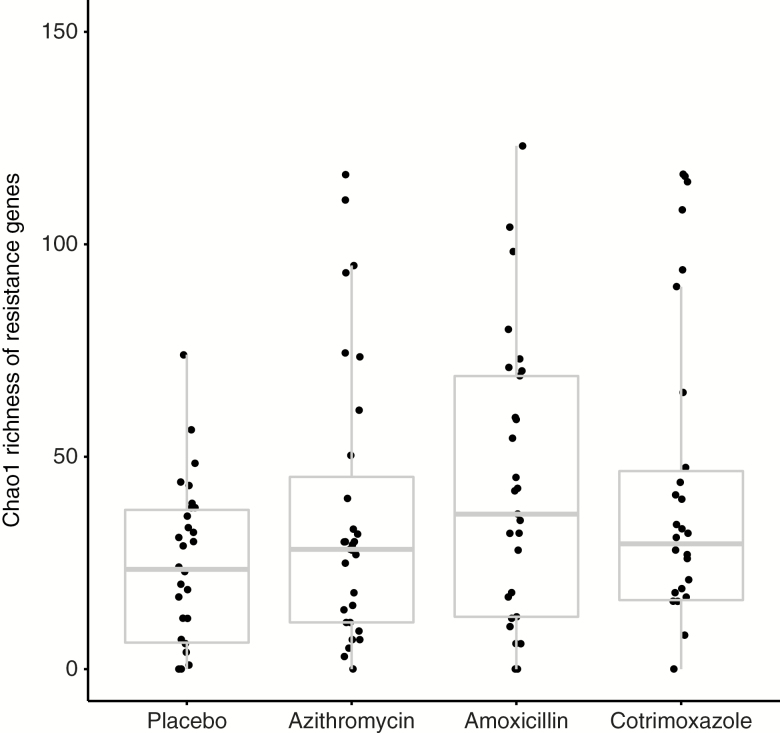
Chao1 richness at the gene level was not significantly different across study arms (P = .10). Mean richness was higher in amoxicillin-treated (42.6 vs 23.9, P = .02) and cotrimoxazole-treated (40.1 vs 23.9, P = .049) children compared to placebo-treated children (Figure 1). Richness was not significantly different between azithromycin-treated children and placebo-treated children (P = .15). The composition of genetic resistance determinants did not significantly differ by PERMANOVA in children randomized to amoxicillin (P = .26), azithromycin (P = .47), or cotrimoxazole (P = .053; Supplementary Figure 2).
Figure 1.
Box plot of Chao1 richness of resistance genes by antibiotic arm. Individual points represent individual samples.
DISCUSSION
We observed a rapid and dramatic increase in the prevalence of macrolide resistance determinants following a 5-day course of azithromycin. The preferential effects of antibiotics on susceptible strains compared to resistant strains leads to the enrichment of resistant organisms. In these same children treated with azithromycin, the composition of the gut microbiome was significantly altered with a reduction in bacterial diversity compared to children treated with placebo [7]. We were unable to detect a significant increase in resistance determinants of other classes of antibiotics with a single course of either amoxicillin or cotrimoxazole in the studied pediatric population. The prevalence of beta-lactam resistance was high, which could be explained by high levels of background amoxicillin use outside of the study [6]. Taken together, these results demonstrate that azithromycin may lead to more prominent microbiological changes in the gut than other antibiotic classes.
Selection pressure on antibiotic-sensitive organisms with antibiotic use predicts a reduction in the richness of antibiotic resistance gene determinants. However, none of the antibiotics evaluated in this study led to a decrease in resistome richness. Instead, the administration of both amoxicillin and cotrimoxazole led to an expansion of the number of antibiotic resistance gene determinants. Such observation can be explained if some or most of the bacteria being affected by these antibiotics harbor multiple resistance genes. This is a mechanism that can give rise to cross-resistance. Azithromycin administration did not lead to an expansion of the resistome. This finding is consistent with the results seen in prior antibiotic resistance studies where cross-resistance has not been observed despite multiple mass distributions of azithromycin to children for either trachoma or childhood mortality [1].
Several limitations of this study must be considered. The presence of genetic resistance determinants does not necessarily correlate with functional resistance. We could not assess if resistance genes occur in potentially pathogenic organisms and if they confer functional resistance to antibiotics. The sample size included in the study was relatively small, and CIs were wide. CIs for sulfonamides were particularly wide, due to the low prevalence of sulfonamide resistance genes in the placebo arm. The prevalence of beta-lactam resistance genes was high, limiting statistical power to evaluate differences across arms. The follow-up duration of this study was short and suggests that antibiotic use selects for resistance in the short term but does not provide data on longer-term effects on the resistome. The prevalence of genetic resistance determinants may return to normal over time.
Systemic antibiotic use may rapidly select for class-specific resistance genes after treatment. Resistome expansion can be induced with a single course of antibiotics. These results highlight the importance of regional antimicrobial resistance surveillance programs to inform antibiotic use policy.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by Research to Prevent Blindness Career Development awards to C. E. O. and T. D.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. O’Brien K, Emerson P, Hooper PJ, et al. . Antimicrobial resistance following mass azithromycin distribution for trachoma: a systematic review. Lancet Infect Dis 2019; 19:e14–25. [DOI] [PubMed] [Google Scholar]
- 2. Costelloe C, Metcalfe C, Lovering A, Mant D, Hay AD. Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ 2010; 340:c2096. [DOI] [PubMed] [Google Scholar]
- 3. Zetts RM, Stoesz A, Smith BA, Hyun DY. Outpatient antibiotic use and the need for increased antibiotic stewardship efforts. Pediatrics 2018; 141. [DOI] [PubMed] [Google Scholar]
- 4. Williams PCM, Isaacs D, Berkley JA. Antimicrobial resistance among children in sub-Saharan Africa. Lancet Infect Dis 2018; 18:e33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee ACC, Chandran A, Herbert HK, et al. . Treatment of infections in young infants in low- and middle-income countries: a systematic review and meta-analysis of frontline health worker diagnosis and antibiotic access. Byass P, ed. PLoS Med 2014; 11:e1001741–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sié A, Coulibaly B, Adama S, et al. . Antibiotic prescription patterns among children younger than 5 years in Nouna District, Burkina Faso. Am J Trop Med Hyg 2019; 100:1121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oldenburg CE, Sie A, Coulibaly B, et al. . Effect of commonly-used pediatric antibiotics on gut microbial diversity in preschool children in Burkina Faso: a randomized clinical trial. Open Forum Infect Dis 2018; In press. doi: 10.1093/ofid/ofy289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 2017; 15:422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sie A, Louis VR, Gbangou A, et al. . The Health and Demographic Surveillance System (HDSS) in Nouna, Burkina Faso, 1993–2007. Glob Health Action 2010; 3:5284 Available at: http://www.globalhealthaction.net/index.php/gha/article/view/5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doan T, Hinterwirth A, Arzika AM, et al. . Mass azithromycin distribution and community microbiome: a cluster-randomized trial. Open Forum Infect Dis 2018; 5:ofy182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lakin SM, Dean C, Noyes NR, et al. . MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 2017; 45:D574–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sié A, Dah C, Ouermi L, et al. . Effect of antibiotics on short-term growth among children in Burkina Faso: a randomized trial. Am J Trop Med Hyg 2018; 99:789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.