Abstract
Mitochondria play multiple important cellular functions. The purpose of this study was to evaluate whether leukocyte mitochondrial DNA copy number (mtDNAcn) is associated with aggressive prostate cancer (PCa) in African American (AA) men. We measured the mtDNAcn in peripheral blood leukocytes from 317 localized AA PCa patients and evaluated its associations with aggressive disease features at diagnosis and biochemical recurrence (BCR) after treatments. There was no significant difference in mtDNAcn among the clinical features at diagnosis, including age, prostate-specific antigen level, Gleason score and clinical stage under analysis of variance test. However, mtDNAcn was significantly associated with BCR in multivariate Cox analysis. Dichotomized into low and high mtDNAcn groups by the median value of mtDNAcn, patients with low mtDNAcn exhibited a significantly lower risk of BCR (hazard ratio = 0.32, 95% confidence interval: 0.13–0.79) compared to those with high mtDNAcn. There was a significant dose–response in tertile and quartile analyses (P for trend = 0.012 and 0.002, respectively). In Kaplan–Meier survival analyses, patients with higher mtDNAcn exhibited significantly shorter BCR-free survival time than those with lower mtDNAcn in dichotomous, tertile and quartile analyses, with long-rank P values of 0.017, 0.024 and 0.019, respectively. Our results showed for the first time that high leukocyte mtDNAcn was associated with worse prognosis in AA PCa patients.
Patients with higher leukocyte mtDNA copy number had significantly higher risks of biochemical recurrence and shorter disease-free survival time in African American prostate cancer patients. There was a significant dose–response for higher mtDNAcn and worse prognosis.
Introduction
Prostate cancer (PCa) is the most prevalent cancer and second most common cause of cancer death in US men (1). Due to the widespread use of prostate-specific antigen (PSA) screening, 90% of PCa patients are currently diagnosed at locoregional stages (78% confined to prostate and 12% spread to regional lymph nodes) (2). African American (AA) PCa patients have the highest incidence and mortality rates among all the racial/ethnic groups in the USA (2,3). The median age of diagnosis for PCa is 63 years for AA men compared to 66 years for white men (3). The incidence rate of PCa is 76% higher and mortality rate is 2.3 times higher in AA than in white men (3). The overall 5-year relative survival rate for PCa is 96% among AA and 98% among whites; 86% of AA PCa patients are diagnosed at local or regional stages with a 5-year survival rate of nearly 100%, which drops to 30% for patients diagnosed at distant metastatic stage (3). Socioeconomic, cultural and biological factors, as well as variations in care delivery and treatment selection, contribute to the higher incidence and mortality of AA PCa patients (4,5).
The majority of localized PCa are slow-growing and patients rarely die from these cancers. Despite the typically indolent nature of localized PCa and excellent prognosis, most such patients still undergo aggressive therapies, including radical prostatectomy and radiotherapy, which are often associated with significant side effects, causing clinical issues of over-diagnosis and over-treatment (6). Given the highly heterogeneous clinical course of PCa patients, prognostic factors are greatly needed to stratify patients and provide the best treatment options. Clinical factors, such as PSA and its derivatives, and pathological factors, such as Gleason score (GS) and tumor stage, are well-established prognostic factors, which, however, is not sufficient to accurately predict patients’ outcomes (7–12). Patients with similar clinical characteristics at diagnosis could have markedly different prognosis. Independent biomarkers that reflect the intrinsic biologic differences between indolent and aggressive PCa could improve risk stratification of localized PCa patients, leading to better-informed clinical decision making. A plethora of biomarkers are commercially available (10,13), for example, SelectMDX (using urinary HOXC6 and DLX1 mRNA levels as a predictor for the detection of clinically significant PCa); ExoDX (urinary exosome-gene expression of ERG and PCA3 for distinguishing insignificant from aggressive disease); Decipher (using expression of 22 genes in tumor tissues, plus clinical variables, for predicting metastasis after prostatectomy) and Prolaris (expression of 31 cell cycle progression genes and 15 housekeep genes in prostate biopsy for predicting PCa death, biochemical recurrence (BCR) and metastasis after prostatectomy and radiation therapy). Although all these biomarkers were initially developed in white patients, a few have been tested in AA PCa patients, for example, Prolaris has shown prognostic value in AA patients (14).
Mitochondria are the primary source of energy generation in eukaryotic cells via respiration and oxidative phosphorylation and play multiple diverse cellular functions such as energy metabolism, proliferation, apoptosis and autophagy (15). There are considerable variations of mitochondria number in different types of cells or tissues, ranging from several hundreds to more than 10 000 (16,17). Mitochondrion contains its own DNA, mtDNA, a 16.6-kb double-stranded circular DNA and each mitochondrion has 1–15 mtDNA molecules (18,19). Somatic mutations and deletions of mtDNA have been found in multiple human cancers and clearly play important roles in carcinogenesis (20–24). In addition, mtDNA copy number (mtDNAcn) in peripheral blood leukocytes (PBLs) has been extensively investigated as a risk factor for various cancers, including PCa (25–35). A classic twin study estimated a high genetic heritability (65%) for the leukocyte mtDNAcn (26), supporting the utilization of leukocyte mtDNAcn as a cancer risk assessment marker. Moreover, there were several reports showing leukocyte mtDNAcn might predict prognosis of cancer patients (36–40). A previous study suggested that leukocyte mtDNAcn was associated with disease progression in white PCa patients: patients with lower mtDNAcn exhibited a 56% increased risk of disease progression, although the association did not reach statistical significance (41). No study has evaluated whether leukocyte mtDNAcn is associated with aggressive disease phenotypes at diagnosis and prognosis of AA PCa patients.
In this study, we measured leukocyte mtDNAcn from 317 AA PCa patients and analyzed its association with aggressive phenotypes at diagnosis. In addition, we investigated whether leukocyte mtDNAcn could predict BCR independent of clinical variables.
Methods
Ethics statement
This study was approved by the MD Anderson Cancer Center Institutional Review Board, and written consent forms were obtained from each patient.
Study population and data collection
This study included a total of 317 AA men with histologically confirmed adenocarcinoma of prostate from the University of Texas MD Anderson Cancer Center. Patients’ blood specimens were collected at diagnosis before any treatments, followed by DNA and plasma isolation. Patients were followed up by PSA monitoring every 3–6 months, and imaging was performed if PSA arose. Clinical and follow-up data, including date of diagnosis, performance status, clinical stage, histological grade and pathological stage, treatment (active surveillance, prostatectomy, radiotherapy and hormone therapy) and progression (BCR and metastasis), were abstracted from patient medical records. MD Anderson Tumor Registry conducts annual vital status follow-ups for all cancer patients. BCR was defined as a serum PSA level of at least 0.2 ng/ml with a second confirmatory PSA level of at least 0.2 ng/ml for patients who undergo a radical prostatectomy or with a rise in PSA level by at least 2 ng/ml above the nadir PSA for patients receiving external-beam radiotherapy. If patients received radical prostatectomy and adjuvant radiotherapy, then the prostatectomy definition of BCR was used.
Measurements of mtDNA copy number
The relative mtDNAcn in PBLs was determined by a real-time quantitative PCR method as previously described (26). Briefly, two pairs of primers were used in two steps of relative quantification of mtDNAcn. One primer pair (ND1-F and ND1-R) was used to amplify the mitochondrial ND1 gene, and the other primer pair (HGB-1 and HGB-2) was used to amplify the single-copy nuclear HGB gene. In the first step, the ratio of the copy number of mitochondrial ND1 gene to the nuclear HGB gene was determined for each sample from standard curves. This ratio is proportional to the mtDNAcn in each cell. The ratio for each sample was then normalized to a calibrator DNA in order to standardize between different runs. The calibrator DNA is from a genomic DNA sample of a healthy control. The PCR mixture of 14 μl included 1 × SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 215 nM ND1-F and ND1-R (or HGB-1 and HGB-2) primers and 4 ng of genomic DNA. The thermal cycling conditions for the mitochondrial ND1 gene amplification were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min; for the nuclear HGB gene amplification, the cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 56°C for 1 min. All samples were plated in duplicates on 384-well plates and run in the Applied Biosystems ViiA 7 Real-Time PCR System. The PCR runs for ND1 and HGB genes were performed on exclusive 384-well plates with the same samples in the same well positions to avoid possible position effects.
A standard curve of a serially diluted reference DNA, one negative control and one calibrator DNA were included in each run. For each standard curve, one reference DNA sample was serially diluted 1:2 to produce a seven-point standard curve between 0.3125 and 20 ng of DNA. The R2 correlation for each standard curve was ≥0.99. Standard deviations for the cycle of threshold value were acceptable at 0.25. Otherwise, the test was repeated. The mean intra-assay coefficient of variation for all samples was 5%.
Statistical analysis
All statistical analyses were performed in the Stata 10.1 statistical software package (StataCorp, College Station, TX). We first used analysis of variance to compare the mean mtDNAcn among patients with different clinical characteristics at baseline. We then determined the association between mtDNAcn and BCR by calculating the hazard ratio (HR) and corresponding 95% confidence interval (95% CI) using multivariate Cox proportional hazards model, adjusting for age, body mass index (BMI), smoking status, pack-year, D’Amico risk groups and initial treatment. MtDNAcn was dichotomized at the median value of mtDNAcn or classified into three or four groups based on the tertile or quartile distribution of mtDNAcn. Finally, we used Kaplan–Meier survival function and log-rank test to evaluate BCR-free survival based on mtDNAcn level in patients. BCR-free survival was calculated from the date of the treatment to the date of the diagnosis of the BCR. All statistical tests were two sided, and P <0.05 was considered statistically significant.
Results
The distribution of selected characteristics of the 317 PCa of AA men and mtDNAcn copy number stratified by their characteristics are shown in Table 1. The median time between blood collection and BCR was 37.4 months. The follow-up rate was 98.1%. The median follow-up time was 45.1 months the total follow-up time was 1166.1 person-years. Eleven patients (3.47%) died during the follow-up. Almost 70% of patients were diagnosed at age 55 and older. There were 181 (57.1%) non-smokers, 95 (30.0%) former smokers and 41 (12.9%) current smokers. The majority of patients were either overweight (31.2%) or obese (40.4%). According to the biopsy GS before the treatment, 93 (29.3%) had GS of ≤6, 182 (57.4%) had GS of 7, and 42 (13.3%) had GS of ≥8. Among these patients, 179 (56.5%) were in T1 stage of tumor, 118 (37.2%) in T2 stage and only 20 (6.3%) in T3–T4 stage. Almost 77% of patients had PSA levels <10 ng/ml. The D’Amico risk stratification system classified 79 patients (24.9%) into the low-risk group, 164 patients (51.7%) into the intermediate-risk group and 74 patients (23.3%) in the high-risk group. As for their initial primary treatments, 53.6% received definitive radical prostatectomy and 20.5% received definitive radiotherapy. It appears that higher GS, higher tumor stage and higher D’Amico risk were associated with lower mean mtDNAcn, but these associations did not reach statistical significance.
Table 1.
mtDNA copy number in PBLs by selected characteristics
| Characteristics | N (%) | Mean (SD) | P value |
|---|---|---|---|
| Age at diagnosis, years | |||
| <55 | 100 (31.55) | 2.45 (1.37) | Ref. |
| 55–65 | 140 (44.16) | 2.37 (1.58) | 0.369 |
| >65 | 77 (24.29) | 2.16 (1.15) | 0.194 |
| Smoking status at diagnosis | |||
| Non-smoker | 181 (57.10) | 2.43 (1.56) | Ref. |
| Former smoker | 95 (29.97) | 2.22 (1.13) | 0.673 |
| Current smoker | 41 (12.93) | 2.26 (1.40) | 0.510 |
| BMI at diagnosis, kg/m2 | |||
| <25 | 37 (11.67) | 2.30 (1.26) | Ref. |
| 25–29.99 (overweight) | 99 (31.23) | 2.44 (1.72) | 0.901 |
| ≥30 (obese) | 128 (40.38) | 2.35 (1.38) | 0.932 |
| Total GS | |||
| ≤6 | 93 (29.34) | 2.43 (1.59) | Ref. |
| 7 | 182 (57.41) | 2.35 (1.40) | 0.464 |
| ≥8 | 42 (13.25) | 2.13 (1.09) | 0.274 |
| Clinical tumor stage | |||
| T1 | 179 (56.47) | 2.45 (1.60) | Ref. |
| T2 | 118 (37.22) | 2.22 (1.15) | 0.446 |
| T3–T4 | 20 (6.31) | 2.05 (1.11) | 0.285 |
| PSA at diagnosis | |||
| <10 ng/ml | 244 (76.97) | 2.29 (1.42) | Ref. |
| 10–20 ng/ml | 46 (14.51) | 2.43 (1.32) | 0.301 |
| >20 ng/ml | 27 (8.52) | 2.65 (1.60) | 0.170 |
| D’Amico risk group | |||
| Low | 79 (24.92) | 2.42 (1.54) | Ref. |
| Intermediate | 164 (51.74) | 2.40 (1.40) | 0.743 |
| High | 74 (23.34) | 2.14 (1.33) | 0.078 |
| Initial primary treatment | |||
| Radical prostatectomy | 170 (53.63) | 2.38 (1.62) | Ref. |
| Radiotherapy | 65 (20.50) | 2.28 (1.22) | 0.693 |
| Surveillance or unknown | 72 (22.71) | 2.27 (1.08) | 0.691 |
| Other treatment | 10 (3.15) | 2.60 (1.33) | 0.410 |
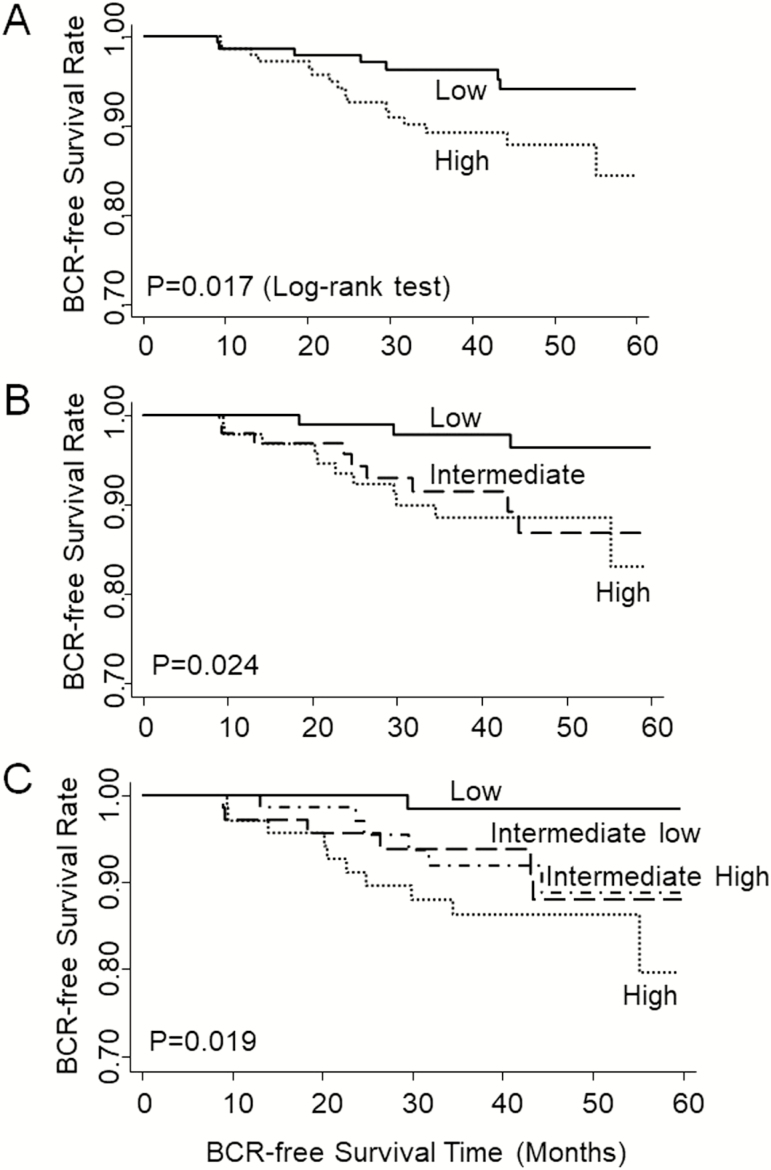
We then evaluated the association of mtDNAcn with the risk of BCR using multivariate Cox proportional hazards model adjusting for age only, or age, smoking status, pack-year, BMI, D’Amico risk groups and primary treatment (Table 2). Dichotomized into low and high mtDNAcn groups by the median (50th percentile) value of mtDNAcn, patients with low mtDNAcn exhibited a significantly lower risk of BCR (HR = 0.32, 95% CI: 0.13–0.79) compared to those with high mtDNAcn. There was a significant dose–response. When patients were classified into three groups based on the tertile values of mtDNAcn, those in the intermediate and low mtDNAcn groups exhibited reduced risks of BCR with HRs of 0.97 (95% CI: 0.40–2.36) and 0.19 (95% CI: 0.05–0.69), respectively (P for trend = 0.012) compared to patients with high mtDNAcn. In quartile analysis, the HRs for the third (intermediate high), second (intermediate low) and first quartile (lowest) of mtDNAcn were 0.67 (95% CI: 0.25–1.84), 0.58 (95% CI: 0.21–1.61) and 0.06 (95% CI: 0.01–0.47), respectively (P for trend = 0.002), compared to fourth quartile (highest) mtDNAcn (Table 2). In Kaplan–Meier survival analyses, patients with higher mtDNAcn exhibited significantly shorter BCR-free survival time than those with lower mtDNAcn in dichotomous, tertile and quartile analyses, with long-rank P values of 0.017, 0.024 and 0.019, respectively (Figure 1).
Table 2.
The association of mtDNA copy number with BCR
| No BCR | BCR | Adjusted HRa (95% CI) | P value | Adjusted HRb (95% CI) | P value | |
|---|---|---|---|---|---|---|
| mtDNAcn | N (%) | N (%) | ||||
| By median | ||||||
| High | 138 (86.79) | 21 (13.21) | Reference | Reference | ||
| Low | 150 (94.94) | 8 (5.06) | 0.35 (0.14–0.86) | 0.022 | 0.32 (0.13–0.79) | 0.014 |
| By tertile | ||||||
| High | 92 (86.79) | 14 (13.21) | Reference | Reference | ||
| Intermediate | 95 (89.62) | 11 (10.38) | 0.96 (0.41–2.28) | 0.932 | 0.97 (0.40–2.36) | 0.943 |
| Low | 101 (96.19) | 4 (3.81) | 0.21 (0.06–0.75) | 0.016 | 0.19 (0.05–0.69) | 0.012 |
| 0.017c | 0.012c | |||||
| By quartile | ||||||
| Highest | 68 (85.00) | 12 (15.00) | Reference | Reference | ||
| Intermediate high | 70 (88.61) | 9 (11.39) | 0.66 (0.25–1.76) | 0.41 | 0.67 (0.25–1.84) | 0.441 |
| Intermediate low | 73 (92.41) | 6 (7.59) | 0.60 (0.22–1.67) | 0.33 | 0.58 (0.21–1.61) | 0.294 |
| Lowest | 77 (97.47) | 2 (2.53) | 0.07 (0.01–0.54) | 0.011 | 0.06 (0.01–0.47) | 0.008 |
| 0.004c | 0.002c |
aAdjusted for age only.
bAdjusted for age, smoking status, pack-year, BMI, D’Amico risk groups and treatment.
c P for trend.
Figure 1.
Kaplan–Meier curve comparing the probability of the BCR-free survival in AA PCa patients based on mtDNAcn in PBLs. (A) Dichotomized at the median value of mtDNAcn. (B) Tertile analysis. (C) Quartile analysis.
We also performed exploratory stratified analyses by age and D’Amico risk groups (Table 3). The association between mtDNAcn and BCR was similar among young (HR = 0.32, 95% CI: 0.07–1.14, P = 0.08) and old-aged patients (HR = 0.35, 95% CI: 0.09–1.35, P = 0.102), although neither reached statistical significant due to small sample size. The association of mtDNAcn with BCR appeared to be stronger in high D’Amico risk group (HR =0.29, 95% CI: 0.07–1.22, P = 0.092) than in low- and intermediate-risk groups (HR = 0.52, 95% CI: 0.13–2.07, P = 0.362), but larger sample size study is needed for these stratified analyses.
Table 3.
Stratified analyses of mtDNA copy number with BCR
| No BCR | BCR | Adjusted HRa (95% CI) | ||
|---|---|---|---|---|
| mtDNAcn | N (%) | N (%) | P value | |
| Ageb | ||||
| Young | ||||
| High | 69 (87.34) | 10 (12.66) | Reference | |
| Low | 74 (93.67) | 5 (6.33) | 0.32 (0.07–1.14) | 0.080 |
| Old | ||||
| High | 69 (86.25) | 11 (13.75) | Reference | |
| Low | 76 (96.20) | 3 (3.80) | 0.35 (0.09–1.35) | 0.102 |
| D’Amico risk groups | ||||
| Low and intermediate | ||||
| High | 115 (95.04) | 6 (4.96) | Reference | |
| Low | 116 (95.08) | 6 (4.92) | 0.52 (0.13–2.07) | 0.362 |
| High | ||||
| High | 23 (62.16) | 14 (37.84) | Reference | |
| Low | 34 (91.89) | 3 (8.11) | 0.29 (0.07–1.22) | 0.092 |
aAdjusted for age, smoking status, pack-year, BMI, D’Amico risk groups and treatment.
bDichotomized using the median age as the cut-off age.
Discussion
In this study, we found that patients with low mtDNA copy number in PBLs exhibited a significant lower risk of BCR in localized AA PCa patients and there was a significant dose–response correlation between lower mtDNAcn and lower risk of BCR. To our knowledge, this is the first study to report a significant association between leukocyte mtDNAcn and BCR in AA PCa patients.
Given the essential roles of mitochondria playing in diverse cellular functions such as energy metabolism, proliferation, apoptosis and autophagy (15), mitochondrial dysfunction is an important cancer driving event. Earlier studies have focused on somatic mutations and deletions of mtDNA and demonstrated high prevalence of such events in many tumors including prostate tumors (20–24). In vitro and in vivo studies provided functional evidence that mtDNA mutations promoted PCa proliferation and tumorigenesis (23,24). MtDNAcn alterations have also been frequently observed in most cancer types compared to normal tissues (42,43). A recent study surveyed mtDNAcn variation in different tumor types using TCGA whole exome and whole genome sequencing data and found predominant reduction of mtDNAcn in tumor tissues compared to adjacent normal tissues (43), consistent with many previous studies based on quantitative real-time PCR measurement (42).
In contrast to the consistent observation of mtDNAcn reduction in most tumor tissues, the studies of leukocyte mtDNAcn of cancer patients and its association with cancer risk have been heterogeneous. Both high and low mtDNAcn in PBLs have been associated with increased cancer risks (44). The reasons for the heterogeneous results are multi-fold: different study design (prospective and retrospective), different cancer types and etiology, heterogeneous population, small sample size and different assays.
There have also been numerous studies evaluating the associations of mtDNAcn with cancer outcomes. A recent meta-analysis summarized the results of 18 studies comprising 3961 cancer patients and concluded that elevated mtDNAcns in PBLs and tumor tissues predict the opposite outcome of cancer (37). Specifically, high mtDNAcn in PBLs predicted a poor cancer prognosis (overall survival: HR = 1.624, 95% CI: 1.211–2.177, P = 0.001; disease-free survival: HR = 1.582, 95% CI: 1.026–2.439, P = 0.038), whereas high mtDNAcn in tumor tissue exhibited better outcomes (overall survival: HR = 0.604, 95% CI: 0.406–0.899, P = 0.013; disease-free survival: HR = 0.593, 95% CI: 0.411–0.857, P = 0.005). These findings were further substantiated in detailed analyses in blood or tissue subgroup (37). All the studies included in that meta-analysis were Caucasians and Asians. Our results were in line with that meta-analysis since we observed high leukocyte mtDNAcn was associated with a poor prognosis (worse BCR-free survival). Also consistent with the meta-analysis results, several previous reports showed that lower mtDNAcn in prostate tumors was associated with worse prognosis (45–47). Previously, there was only one study evaluating the prognostic role of mtDNAcn in PBLs in Caucasian PCa patients. Tu et al. (41) reported that mtDNAcn at diagnosis was significantly lower in patients with GS of ≥8 than in patients with GS of ≤7 and might predict disease progression, although the latter test did not reach statistical significance in multivariate Cox analysis. In an earlier Chinese study, Zhou et al. reported that high leukocyte mtDNAcn at diagnosis was significantly associated with high GS and advanced tumor stage (48). For the first time, we reported a significant association between higher level of leukocyte mtDNAcn and higher risk of BCR in localized PCa patients. Future studies are warranted to confirm our observations in AA and other races/ethnicities.
Biologically, mtDNAcn is specific to different tissue types, which reflects differing energy requirements (49,50). There is generally no correlation of mtDNAcn between different tissues. In tumor tissues, hypoxia inducible factor induced by aggravating hypoxia inhibits mitochondrial biogenesis and disrupts mitochondria by mitophagy, leading to lower mitochondrial activity as an advantage for cancer progression. Therefore, lower miDNAcn in prostate tumors favors cancer progression and is associated with worse prognosis. On the other hand, leukocyte mtDNAcn reflects the pathophysiological status of various immune cells. Previous studies have demonstrated that immune cell regulation may explain the mechanisms underlying the association of high leukocyte mtDNAcn with worse prognosis of cancer. In one study (39), glioma patients with high leukocyte mtDNAcn had significantly lower frequency of natural killer (NK) cells, which play important roles in the first-line defense against tumor (51), thus low frequency of NK cells may impair the immune defense against tumor cells and lead to poor prognosis. In another study (40), colorectal cancer patients with high leukocyte mtDNAcn exhibited enhanced immunosuppressive phenotypes, such as higher CD4+ T cells and higher percentage of CD4+CD25+FOXP3+ regulatory T (Treg) cells in CD4+ T cells, higher IL-2 and TGF-β1, and lower TNF-α concentration in the plasma. In a third study, liver cancer patients with high leukocyte mtDNAcn exhibited a higher frequency of CD4+CD25+FOXP3+ Treg cells and lower frequency of NK cells, higher TGF-β1 and lower TNF-α and IFN-γ plasma concentration, indicating an immunosuppressive status. Furthermore, high leukocyte mtDNAcn significantly enhanced the reactive oxygen species-mediated secretion of TGF-β1, which facilitates the differentiation and expansion of Treg (52). Collectively, these studies suggest that elevated leukocyte mtDNAcn promotes cancer progression at least partially through reactive oxygen species-induced immunosuppressive effects. Future studies are needed to determine whether leukocyte mtDNAcns are associated with the frequencies of Treg and NK cell.
There are several strengths of our study. This is the first report linking leukocyte mtDNAcn with worse prognosis in AA PCa patients. The observed association is biologically plausible. The blood samples were obtained before any treatments. All the patients were treated at MD Anderson Cancer Center with comprehensive clinical data and long-term follow-up. MtDNAcn was measured using the well-established, quantitative real-time PCR method. There are also a couple of limitations. First, the sample size was modest. Second, we only analyzed BCR as a prognosis endpoint, but not mortality, due to the small number of death events of localized PCa patients. Third, a variety of lifestyle factors including diet and physical activity, have been shown to influence mtDNAcn. We did not have data on these lifestyle factors and could not account for these variables in our analyses. There may be other residual confounding factors.
In summary, our study reported for the first time that high mtDNAcn in PBLs is associated with a significantly increased risk of BCR in localized AA PCa patient in a dose–response way. Future studies are warranted to validate our observations in independent patient cohorts and study the biological mechanism underlying the association between leukocyte mtDNAcn and worse prognosis in PCa patients.
Funding
This study was financially supported by an individual researcher award (RP140556) from the Cancer Prevention and Research Institute of Texas (CPRIT), a National Cancer Institute Specialized Program of Research Excellence (SPORE) grant (CA140388) and an MD Anderson Cancer Center start-up fund to J.G.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AA
African American
- BCR
biochemical recurrence
- BMI
body mass index
- CI
confidence interval
- GS
Gleason score
- HR
hazard ratio
- mtDNAcn
mitochondrial DNA copy number
- NK
natural killer
- PBL
peripheral blood leukocyte
- PCa
prostate cancer
- PSA
prostate-specific antigen
References
- 1. Siegel R.L., et al. (2019) Cancer statistics, 2019. CA. Cancer J. Clin., 69, 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Howlader N., et al. SEER Cancer Statistics Review, 1975–2016 National Cancer Institute, Bethesda, MD: https://seer.cancer.gov/csr/1975_2016/, based on November 2018 SEER data submission, posted to the SEER web site, April 2019 (16 July 2019, date last accessed). [Google Scholar]
- 3. DeSantis C.E., et al. (2019) Cancer statistics for African Americans, 2019. CA. Cancer J. Clin., 69, 211–233. [DOI] [PubMed] [Google Scholar]
- 4. McGinley K.F., et al. (2016) Prostate cancer in men of African origin. Nat. Rev. Urol., 13, 99–107. [DOI] [PubMed] [Google Scholar]
- 5. Karakas C., et al. (2017) Molecular mechanisms involving prostate cancer racial disparity. Am. J. Clin. Exp. Urol., 5, 34–48. [PMC free article] [PubMed] [Google Scholar]
- 6. Welch H.G., et al. (2009) Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986-2005. J. Natl Cancer Inst., 101, 1325–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodrigues G., et al. (2012) Pre-treatment risk stratification of prostate cancer patients: a critical review. Can. Urol. Assoc. J., 6, 121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mesko S., et al. (2018) Targeted prostate biopsy Gleason score heterogeneity and implications for risk stratification. Am. J. Clin. Oncol., 41, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanda M.G., et al. (2018) Clinically localized prostate cancer: AUA/ASTRO/SUO guideline. Part I: risk stratification, shared decision making, and care options. J. Urol., 199, 683–690. [DOI] [PubMed] [Google Scholar]
- 10. Chistiakov D.A., et al. (2018) New biomarkers for diagnosis and prognosis of localized prostate cancer. Semin. Cancer Biol., 52(Pt 1), 9–16. [DOI] [PubMed] [Google Scholar]
- 11. D’Amico A.V., et al. (1998) Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. J. Am. Med. Assoc., 280, 969–974. [DOI] [PubMed] [Google Scholar]
- 12. Pugliese D., et al. (2016) Clinical, pathological and molecular prognostic factors in prostate cancer decision-making process. Urologia, 83, 14–20. [DOI] [PubMed] [Google Scholar]
- 13. Cucchiara V., et al. (2018) Genomic markers in prostate cancer decision making. Eur. Urol., 73, 572–582. [DOI] [PubMed] [Google Scholar]
- 14. Rayford W., et al. (2018) Improving risk stratification in a community-based African American population using cell cycle progression score. Transl. Androl. Urol., 7(Suppl. 4), S384–S391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nunnari J., et al. (2012) Mitochondria: in sickness and in health. Cell, 148, 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Veltri K.L., et al. (1990) Distinct genomic copy number in mitochondria of different mammalian organs. J. Cell. Physiol., 143, 160–164. [DOI] [PubMed] [Google Scholar]
- 17. Clay Montier L.L., et al. (2009) Number matters: control of mammalian mitochondrial DNA copy number. J. Genet. Genomics, 36, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan D.C., et al. (2006) Mitochondria: dynamic organelles in disease, aging, and development. Cell, 125, 1241–1252. [DOI] [PubMed] [Google Scholar]
- 19. Satoh M., et al. (1991) Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res., 196, 137–140. [DOI] [PubMed] [Google Scholar]
- 20. Lightowlers R.N., et al. (1997) Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet., 13, 450–5. [DOI] [PubMed] [Google Scholar]
- 21. Schon E.A., et al. (2012) Human mitochondrial DNA: roles of inherited and somatic mutations. Nature reviews. Genetics, 13, 878–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wallace D.C., et al. (2012) Mitochondria and cancer. Nat. Rev. Cancer, 12, 685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arnold R.S., et al. (2009) Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment. Prostate, 69, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petros J.A., et al. (2005) mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl Acad. Sci. USA, 102, 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lan Q., et al. (2008) A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood, 112, 4247–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xing J. et al. (2008) Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J. Natl Cancer Inst., 100, 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thyagarajan B., et al. (2012) Mitochondrial copy number is associated with colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev., 21, 1574–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y., et al. (2014) Increased leukocyte mitochondrial DNA copy number is associated with oral premalignant lesions: an epidemiology study. Carcinogenesis, 35, 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofmann J.N., et al. (2014) A nested case-control study of leukocyte mitochondrial DNA copy number and renal cell carcinoma in the prostate, lung, colorectal and ovarian cancer screening trial. Carcinogenesis, 35, 1028–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Y., et al. (2014) Genetic and intermediate phenotypic susceptibility markers of gastric cancer in Hispanic Americans: a case-control study. Cancer, 120, 3040–3048. [DOI] [PubMed] [Google Scholar]
- 31. Lemnrau A., et al. (2015) Mitochondrial DNA copy number in peripheral blood cells and risk of developing breast cancer. Cancer Res., 75, 2844–2850. [DOI] [PubMed] [Google Scholar]
- 32. Maragh S., et al. (2015) Evaluation of two mitochondrial DNA biomarkers for prostate cancer detection. Cancer Biomark., 15, 763–773. [DOI] [PubMed] [Google Scholar]
- 33. Sun Y., et al. (2016) Lower mitochondrial DNA copy number in peripheral blood leukocytes increases the risk of endometrial cancer. Mol. Carcinog., 55, 1111–1117. [DOI] [PubMed] [Google Scholar]
- 34. Moore A., et al. (2017) A prospective study of mitochondrial DNA copy number and the risk of prostate cancer. Cancer Causes Control, 28, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campa D., et al. (2018) Mitochondrial DNA copy number variation, leukocyte telomere length, and breast cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Breast Cancer Res., 20, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., et al. (2018) Increased copy number of mitochondrial DNA predicts poor prognosis of esophageal squamous cell carcinoma. Oncol. Lett., 15, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen N., et al. (2016) Elevated mitochondrial DNA copy number in peripheral blood and tissue predict the opposite outcome of cancer: a meta-analysis. Sci. Rep., 6, 37404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao D., et al. (2016) Alterations of telomere length and mtDNA copy number are associated with overall survival in hepatocellular carcinoma patients treated with transarterial chemoembolization. Cancer Chemother. Pharmacol., 78, 791–799. [DOI] [PubMed] [Google Scholar]
- 39. Chen Y., et al. (2015) High leukocyte mitochondrial DNA content contributes to poor prognosis in glioma patients through its immunosuppressive effect. Br. J. Cancer, 113, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qu F., et al. (2015) Leukocyte mitochondrial DNA content: a novel biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Carcinogenesis, 36, 543–552. [DOI] [PubMed] [Google Scholar]
- 41. Tu H., et al. (2015) Mitochondrial DNA copy number in peripheral blood leukocytes and the aggressiveness of localized prostate cancer. Oncotarget, 6, 41988–41996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu M., et al. (2011) Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci., 89, 65–71. [DOI] [PubMed] [Google Scholar]
- 43. Reznik E., et al. (2016) Mitochondrial DNA copy number variation across human cancers. Elife, 5, e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Gisbergen M.W., et al. (2015) How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? challenges, opportunities and models. Mutat. Res. Rev. Mutat. Res., 764, 16–30. [DOI] [PubMed] [Google Scholar]
- 45. Kalsbeek A.M.F., et al. (2018) Altered mitochondrial genome content signals worse pathology and prognosis in prostate cancer. Prostate, 78, 25–31. [DOI] [PubMed] [Google Scholar]
- 46. Koochekpour S., et al. (2013) Reduced mitochondrial DNA content associates with poor prognosis of prostate cancer in African American men. PLoS One, 8, e74688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moro L., et al. (2008) Mitochondrial DNA depletion reduces PARP-1 levels and promotes progression of the neoplastic phenotype in prostate carcinoma. Cell. Oncol., 30, 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou W., et al. (2014) Peripheral blood mitochondrial DNA copy number is associated with prostate cancer risk and tumor burden. PLoS One, 9, e109470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. D’Erchia A.M., et al. (2015) Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion, 20, 13–21. [DOI] [PubMed] [Google Scholar]
- 50. Fernández-Vizarra E., et al. (2011) Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion, 11, 207–213. [DOI] [PubMed] [Google Scholar]
- 51. Vivier E., et al. (2008) Functions of natural killer cells. Nat. Immunol., 9, 503–510. [DOI] [PubMed] [Google Scholar]
- 52. He X., et al. (2016) High leukocyte mtDNA content contributes to poor prognosis through ROS-mediated immunosuppression in hepatocellular carcinoma patients. Oncotarget, 7, 22834–22845. [DOI] [PMC free article] [PubMed] [Google Scholar]