Abstract
Background
Vancomycin-resistant enterococci (VRE) are a major cause of hospital-acquired infections. The risk of infection from interventional radiology (IR) procedures is not well documented. Whole-genome sequencing (WGS) surveillance of clinical bacterial isolates among hospitalized patients can identify previously unrecognized outbreaks.
Methods
We analyzed WGS surveillance data from November 2016 to November 2017 for evidence of VRE transmission. A previously unrecognized cluster of 10 genetically related VRE (Enterococcus faecium) infections was discovered. Electronic health record review identified IR procedures as a potential source. An outbreak investigation was conducted.
Results
Of the 10 outbreak patients, 9 had undergone an IR procedure with intravenous (IV) contrast ≤22 days before infection. In a matched case-control study, preceding IR procedure and IR procedure with contrast were associated with VRE infection (matched odds ratio [MOR], 16.72; 95% confidence interval [CI], 2.01 to 138.73; P = .009 and MOR, 39.35; 95% CI, 7.85 to infinity; P < .001, respectively). Investigation of IR practices and review of the manufacturer’s training video revealed sterility breaches in contrast preparation. Our investigation also supported possible transmission from an IR technician. Infection prevention interventions were implemented, and no further IR-associated VRE transmissions have been observed.
Conclusions
A prolonged outbreak of VRE infections related to IR procedures with IV contrast resulted from nonsterile preparation of injectable contrast. The fact that our VRE outbreak was discovered through WGS surveillance and the manufacturer’s training video that demonstrated nonsterile technique raise the possibility that infections following invasive IR procedures may be more common than previously recognized.
Keywords: outbreak detection, vancomycin-resistant Enterococcus, healthcare-associated infections, interventional radiology, whole genome sequencing
An outbreak of vancomycin-resistant Enterococcus faecium infections in interventional radiology (IR) was detected through whole-genome sequencing surveillance. Epidemiologic investigation revealed nonsterile preparation of contrast medium as the transmission route. Infection after IR procedures is an unstudied area of infection prevention.
(See the Editorial Commentary by Hayden on pages 2344–6.)
Vancomycin-resistant enterococci (VRE) are enteric gram-positive organisms that are common causes of infections in hospitalized patients. In the United States in 2011–2014, 84% of Enterococcus faecium and 7% of Enterococcus faecalis infections were caused by vancomycin-resistant strains [1]. The predominant disease-causing VRE genetic lineage belongs to the globally distributed multidrug-resistant E. faecium clonal complex 17 (CC-17). Strains that belong to this clonal complex have adapted to spread within the hospital environment through acquisition of numerous drug-resistance genes, a pathogenicity island, and other mobile genetic elements [2].
Whole-genome sequencing (WGS) has become the gold standard for the investigation of suspected outbreaks in hospital settings [3–6]. The majority of studies have used reactive WGS to characterize bacterial genetic relatedness for investigation of outbreaks that were first detected using traditional epidemiologic methods [7]. In contrast, routine, prospective WGS surveillance could potentially lead to earlier detection of hospital outbreaks as well as outbreaks that might otherwise not be identified [8]. WGS surveillance is defined as prospective sequencing of all clinical isolates of selected bacterial species that are commonly transmitted in the hospital as a primary approach to outbreak detection. WGS surveillance, when coupled with epidemiologic assessment of patient exposures, is especially relevant for organisms such as VRE where high numbers of epidemiologically unrelated cases can make detection of smaller hospital outbreaks challenging.
Interventional radiology (IR) provides minimally invasive therapies and diagnostics while reducing hospital stays and healthcare costs [9]. Image-guided biopsy, abscess drainage, and transjugular intrahepatic portosystemic shunt placements are examples of common IR procedures [9]. A recent review of medical errors in IR suggest that adverse events in IR are comparable to adverse events in invasive surgery and that patient safety has lagged behind other invasive procedures [10]. To our knowledge, however, there are no published studies that measure the rate of infection after IR procedures.
Following the initiation of WGS surveillance in our hospital, we identified a cluster of infections caused by vancomycin-resistant E. faecium strains that were genetically highly related, suggesting a common route of transmission. We conducted an epidemiologic investigation to identify the route of transmission and implemented interventions to prevent further infections.
METHODS
Study Setting and Identification of an Outbreak
This study was conducted at the University of Pittsburgh Medical Center–Presbyterian Hospital (UPMC), an adult medical/surgical tertiary care hospital with 762 beds, 150 critical care unit beds, more than 32 000 yearly inpatient admissions, and more than 400 solid organ transplants per year. Ethics approval was obtained from the University of Pittsburgh Institutional Review Board.
In November 2016, we initiated a project called Enhanced Detection System for Hospital-Acquired Transmission (EDS-HAT). Our goal was to use a combination of WGS surveillance of hospital-associated, mostly multidrug-resistant bacterial pathogens (including VRE) and automated data mining of the electronic health record (EHR) to identify outbreaks and determine routes of transmission. During the developmental and validation phase of EDS-HAT, WGS was performed and analyzed with a multimonth lag period after the culture date. This analysis lag period allowed for the development of analytic algorithms and comparison of outbreaks detected by EDS-HAT with our standard infection prevention (IP) practice, which involves WGS of bacterial isolates suspected of belonging to outbreaks that were detected based on traditional hospital epidemiologic methods. VRE WGS data were initially analyzed for clinical culture dates between November 2016 and November 2017.
Eligibility of bacterial isolates for WGS under EDS-HAT required positive clinical culture for selected pathogens with either of the following criteria: >3 hospital days after admission and/or any procedure or prior inpatient stay in 30 days prior to isolate collection date. Eligible isolates were identified using TheraDoc software (version 4.6, Premier, Inc, Charlotte, NC).
Genomic Methods
Genomic DNA was extracted from pure overnight cultures of single bacterial colonies using a Qiagen DNAeasy Tissue Kit according to manufacturer’s instructions (Qiagen, Germantown, MD). Library construction and sequencing were conducted using the Illumina Nextera DNA Sample Prep Kit with 150 bp paired-end read length, and libraries were sequenced on the NextSeq WGS platform (Illumina, San Diego, CA). The average read coverage across the 10 VRE genomes was 100X. SPAdes v3.11 was used for de novo assembly from filtered short-read sequences [11]. Phylogenetic relationships between genomes were assessed by aligning reads to a VRE ST (sequence type)-1471 (which belongs to CC-17) reference genome using snippy [12]. Patients with isolates that differed by ≤15 single-nucleotide polymorphisms (SNPs) compared to any other case isolate were considered to be part of the outbreak. This SNP cutoff was chosen based on previous experience with outbreaks at our institution as well as other investigations that used WGS surveillance [13, 14]. The SNP cutoff was increased to 30 SNPs to evaluate potential detection of additional related isolates. A blastn analysis with a threshold of 80% sequence identity and query coverage was performed to determine whether there were gene differences among the 10 identified outbreak isolates.
Case-control Study
A matched case-control study was performed to identify exposures associated with the outbreak that might infer a putative pathway that was responsible for transmission. A case was defined as a patient with infection with an outbreak isolate. Four randomly selected control patients, defined as patients without infection with an outbreak isolate, with length of stay ≥3 days were matched by inpatient units from the positive VRE culture date or the prior discharging/transferring inpatient unit if the patient’s positive VRE culture was a day after transfer or present on admission to each of the 10 outbreak patients. Matching by inpatient unit was performed because the 10 patients were housed on 8 units. Review of the EHR, including information on room location, procedures, microbiology results, and clinical findings, was performed for cases and controls. Conditional logistic regression was performed using SAS (version 9.3, SAS Institute, Cary, NC) procedure LOGISTIC to calculate univariate matched odds ratios (MORs), 95% confidence intervals (CIs), and P values. Multivariable conditional logistic regression was not performed because of the limited number of case patients.
RESULTS
Description of the Outbreak
There were 439 clinical VRE isolates sequenced during the study period, of which 10 (2.3%) were genetically highly related ST-1471 strains by WGS and were therefore suspected to be part of an outbreak involving a common exposure. An initial EHR review of these patients revealed that 9 had undergone IR procedures within the past 22 days (Table 1). No other common exposures among the 10 patients were identified. Only 2/10 (20%) patients had a shared inpatient unit while the remaining 8 patients were housed on separate units with separate healthcare worker staff. There were no common procedures among the patients other than IR. The subsequent outbreak investigation therefore focused on determining whether IR procedures represented the most likely transmission route. Twenty-two days from IR procedure to infection was used as the maximum time from IR procedure to infection in the case-control study given the EHR review findings.
Table 1.
Clinical and Epidemiologic Characteristics of 10 Case Patients With Vancomycin-resistant Enterococci Infection
| Patient | Admit Daya | IR Procedure Daya | Culture Daya | Specimen Source | Days From IR to Culture | IR Procedure(s) | Contrast Used? | Staff Group | Charlson Comorbidity Index | Other Organisms Isolated From Same Specimen |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 0 | 22 | Jackson-Pratt drainage | 22 | Angiogram, chemoembolization of hepatocellular carcinoma | Yes | A | 8 | None |
| 2 | 36 | 15 | 36 | Pancreatic tissue | 21 | Venogram, angioplasty with central line placement | Yes | A | 1 | Candida dubliniensis |
| 3 | 68 | 48 | 68 | Blood | 20 | Cholangiogram, biliary catheter exchange | Yes | A | 4 | None |
| 4 | 170 | NA | 170 | Blood | NA | Noneb | NA | NA | 6 | None |
| 5 | 177 | 184 | 188 | Biliary pigtail | 4 | Cholangiogram, biliary catheter exchange | Yes | A | 10 | Klebsiella pneumoniae, Hafnia alvei, Candida tropicalis, Candidaglabrata |
| 6 | 187 | 187 | 199 | Blood | 12 | Cerebral arteriogram | Yes | B | 5 | Candida parapsilosis |
| 7 | 195 | 196 | 204 | Lower abdominal | 8 | Celiac and mesenteric arteriogram | Yes | A | 7 | Escherichia coli, Pseudomonas aeruginosa |
| 8 | 187 | 203 | 206 | Urine | 3 | GJ tube replacement | Yes | A | 5 | None |
| 9 | 243 | 249 | 251 | Hepatic fluid | 2 | Cholangiogram | Yes | A | 7 | Klebsiella variicola, Staphylococcus epidermidis |
| 10 | 260 | 241 | 260 | Blood | 19 | Lower extremity angiogram | Yes | A | 7 | None |
Abbreviations: GJ, gastrostomy-jejunostomy; IR, interventional radiology; NA, not applicable.
aDays are calculated as the number of days since the IR procedure day of patient 1.
bThis patient had no IR exposure and had a vancomycin-resistant enterococci (VRE) isolate with substantial genomic differences compared to the VRE genomes from the other 9 patients (see text).
Case-control Study
Age, gender, Charlson comorbidity index, use of immunosuppressive therapy, and rates of previous VRE colonization were not significantly different between case patients and their matched control patients (Table 2). Cases had a higher rate of prior solid organ malignancy (MOR, 4.45; 95% CI, 1.01 to 19.71; P = .05).
Table 2.
Results of the Matched Case-control Study
| Variable | Case Patients (N = 10) | Control Patients (N = 40) | Matched Odds Ratio | 95% Confidence Interval | P Value |
|---|---|---|---|---|---|
| Demographics | |||||
| Median age, y (range) | 64 (51 to 85) | 64 (24 to 87) | … | … | 1.00b |
| Non-white race | 0 (0.0%) | 7 (17.5%) | 0.36a | (0 to 2.08) | .37 |
| Male gender | 6 (60%) | 20 (50.0%) | 1.55 | (.35 to 6.82) | .57 |
| Comorbidities | |||||
| Median Charlson comorbidity index (range) | 6.5 (1 to 10) | 5 (0 to 15) | … | … | .22b |
| Solid organ transplant | 3 (30%) | 9 (23.1%) | 1.97 | (.25 to 15.73) | .52 |
| Immunosuppression | 3 (30%) | 10 (25.6%) | 1.46 | (.22 to 9.54) | .70 |
| Diabetes mellitus | 4 (40%) | 15 (37.5%) | 1.12 | (.26 to 4.84) | .88 |
| History solid organ malignancy | 5 (50%) | 7 (17.5%) | 4.45 | (1.01 to 19.71) | .05 |
| History hematologic malignancy | 0 (0%) | 2 (5.0%) | 1.66a | (0 to 13.89) | .99 |
| End-stage liver disease | 3 (30%) | 9 (22.5%) | 2.31 | (.25 to 21.11) | .46 |
| Chronic renal insufficiency | 2 (20%) | 10 (25.0%) | 0.73 | (.12 to 4.39) | .73 |
| VRE status | |||||
| VRE infection | 10 (100%) | 1 (2.5%) | 52.18a | (10.67 to infinity) | <.001 |
| Prior VRE colonization | 3 (30%) | 10 (25.0%) | 1.33 | (.26 to 6.80) | .73 |
| Prior clinical VRE infection (5 y) | 0 (0%) | 1 (2.5%) | 4 | (0 to 76.00) | >.99 |
| IR procedure | |||||
| IR | 9 (90%) | 12 (30.0%) | 16.72 | (2.01 to 138.73) | .009 |
| IR with contrast | 9 (90%) | 3 (7.5%) | 39.35a | (7.85 to infinity) | <.001 |
| Median days from IR to positive VRE culture (range) | 12 (2 to 22) | 8 (6 to 6) | … | … | .32b |
Abbreviations: IR, interventional radiology; VRE, vancomycin-resistant enterococci.
aMedian unbiased estimate calculated.
bWilcoxon test performed.
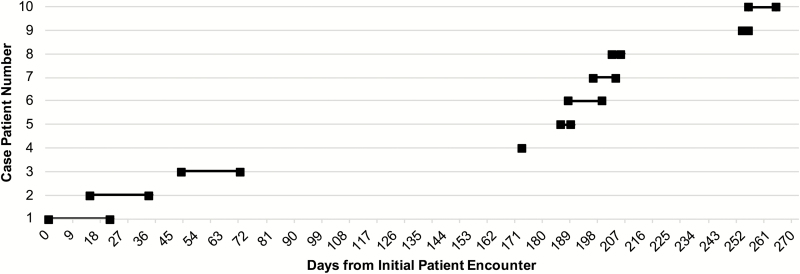
Nine (90%) of the 10 case patients had an IR procedure ≤22 days prior to their positive VRE culture compared to 12 (30%) of 40 control patients (MOR, 16.72; 95% CI 2.01 to 138.73; P = .009; Table 2). An IR procedure with contrast was performed in 9 of 10 case patients and 3 of 40 matched control patients (MOR, 39.35; 95% CI, 7.85 to infinity; P < .001). Median time from IR procedure to infection was 12 days (range, 2 to 22 days; Table 2 and Figure 1).
Figure 1.
Timeline of the outbreak displaying case patients’ interventional radiology procedure day to culture day.
Epidemiologic Investigation
IR procedures at UPMC are mainly performed in a suite of 4 rooms or, less commonly, in select operating rooms. Hepatobiliary procedures are performed by a group of dedicated physicians, nurses, and technicians (staff group A) in 2 adjacent procedure rooms; neurology-related IR procedures are performed by a second dedicated group of physicians, nurses, and technicians (staff group B) in the other 2 rooms located across a hall.
IR practices and staffing schedules were reviewed by the study team. An on-site audit of IR procedures (A. J. S.) was performed on 26 April 2018, including observation of aseptic technique throughout the procedure, intravenous contrast preparation, and use of a sterile syringe for contrast injection (Bayer-Medrad Mark 7 Arterion with quick-fill tube [QFT]) and automatic contrast injector (Bayer-Medrad Mark V ProVis).
Environmental sampling of the IR area was performed on 16 May 2018 using replicate organism detection and counting plates with Tween 80 medium or culture swabs with transport medium. The automatic contrast injector control panels and injector pressure jackets were cultured from 2 rooms used by different IR staff groups (A and B). In addition, keyboards, common rooms, and a toilet seat located in the staff A and B communal area were sampled. All environmental cultures in the IR area were also negative for VRE.
Review of IR Procedures and Practices
IR procedures on 8 of the 9 case patients were performed by group A staff in either 2 of the 4 IR suite rooms or an operating room. The remaining patient had an IR procedure performed by group B staff in 1 of the 2 remaining IR rooms located across the hall from the other IR suite rooms. Based on staff member interviews, a group A technician may have assisted with this patient’s procedure, although this could not be confirmed. Among the IR procedures for the 9 patients, there were no shared physicians or nurses scheduled on any of the patient procedure days. However, only 1 IR technician from group A (IR technician A) was assigned to work on all 9 patient procedure days. IR technician A was scheduled to work on procedure days of only 1 of the 3 control patients with IR contrast procedures. Eight of the 9 case patients had the procedure performed with 1 brand of contrast; for the remaining patient, another brand of contrast was used.
Two staff group A IR technicians demonstrated the use of the sterile disposable syringe (Bayer-Medrad Mark 7 Arterion with QFT) and automatic injector (Bayer-Medrad Mark V ProVis) for contrast injection to IP investigators for an upcoming patient procedure. Neither technician performed hand hygiene before preparation nor did they don gloves or other personal protective equipment (PPE) during preparation. Other observed breaches in hygiene included eating food during preparation, touching sterile portions of the contrast bottle, touching sterile portions of the syringe, inserting the nonsterile portions of the QFT into the contrast bottle, and nonsterile handling of the prepared contrast solution. Both technicians indicated that this preparation practice was standard procedure in the department. Because of immediate patient safety concerns, the loaded syringe was discarded, and the technicians were instructed to set up a new syringe using sterile gloves and sterile technique. The bottle of contrast and QFT for the prepared injection were cultured and negative for VRE.
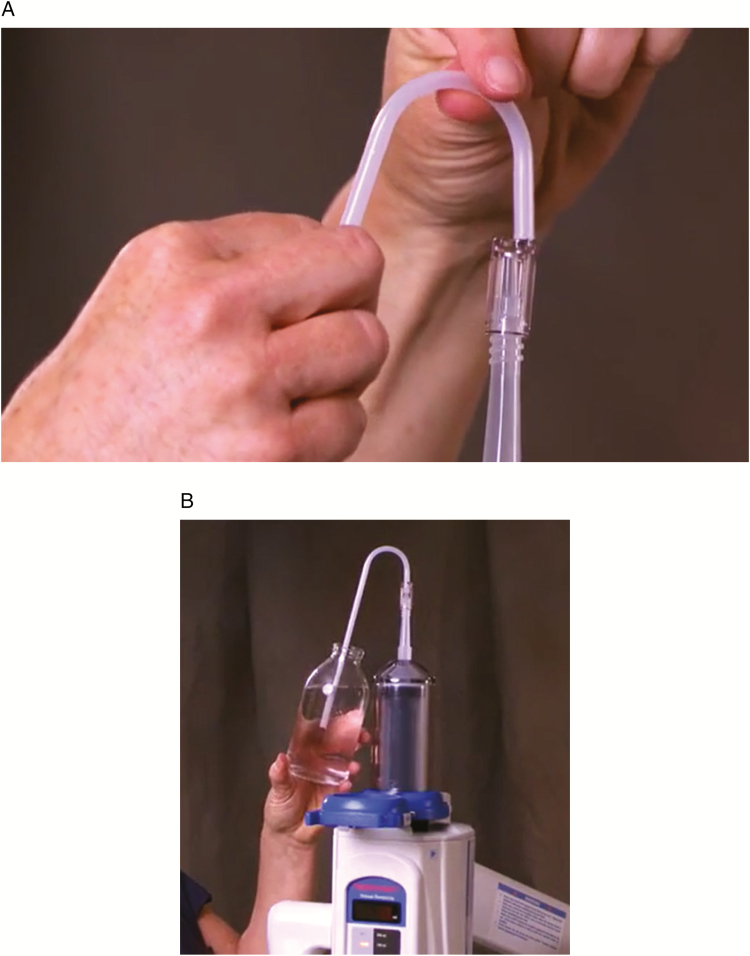
The manufacturer’s operations manual and training video were also investigated. While the manual of the ProVis Mark V notes, “The QFT is sterile, so do not touch either end. Hold the QFT at the curve to avoid touching the leg that will go into the bottle” [15], the manual does not explicitly direct the use of PPE during the handling of the QFT device. Moreover, the training video on the manufacturer’s website depicts a user preparing the injector without PPE and touching portions of the QFT that may enter the open bottle of contrast with ungloved hands (Figure 2) [16]. The standard procedure was changed to include use of sterile gloves and sterile technique during contrast preparation.
Figure 2.
Still shots of the manufacturer training video for the Medrad Mark V ProVis. A, Insertion of the sterile quick-fill tube (QFT) into the sterile contrast syringe (Bayer-Medrad Mark 7 Arterion). B, Filling of the sterile contrast syringe using the sterile QFT.
Genomic Relatedness of Outbreak Strains
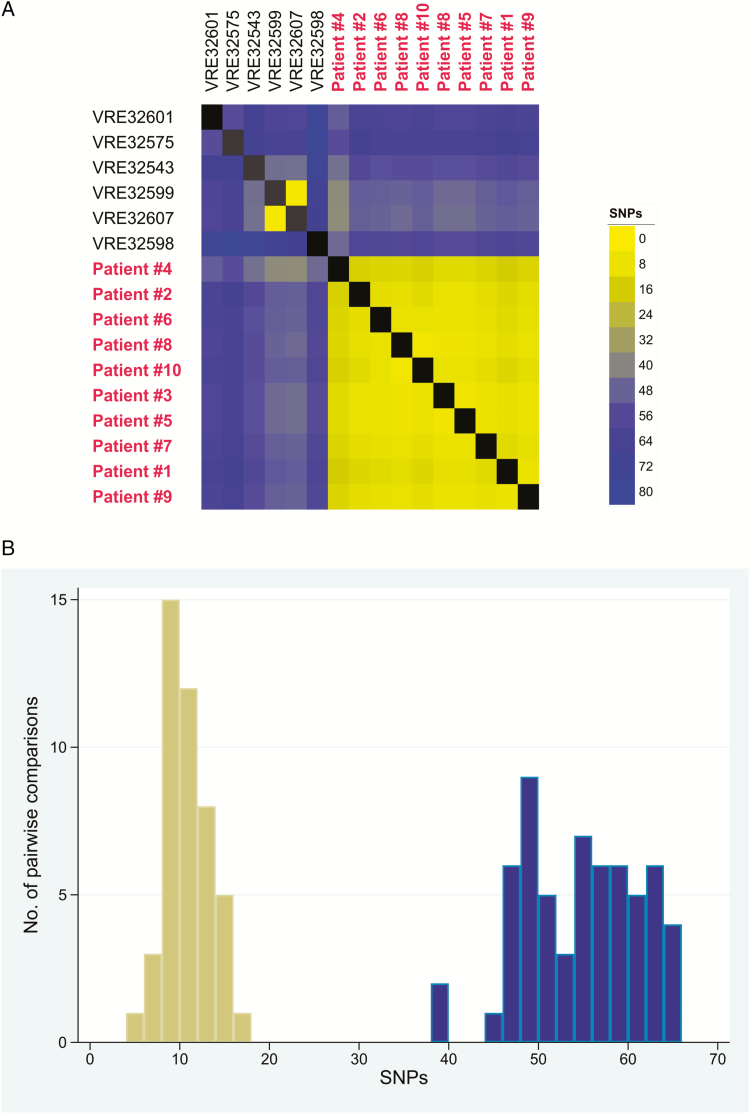
No additional IR procedure-related isolates were detected at a 30-SNP cutoff. The VRE isolate from the patient without IR exposure (patient 4) contained a 48-kb chromosomal sequence (genomic island) that was not found among other sequenced VRE isolates collected at our hospital (Figure 3). In addition, this patient’s VRE isolate had greater SNP differences than the isolates from patients with IR exposure (11 unique SNPs vs 2–7 unique SNPs, respectively). Together, these findings suggest that patient 4 may not have a direct transmission link with the IR outbreak. However, because this genomic relatedness analysis was performed post hoc, we included patient 4 in the case-control study.
Figure 3.
Genomic similarity of interventional radiology (IR)-related outbreak strains to one another and to contemporary VRE Enterococcus faecium ST1471 Enhanced Detection System for Hospital-Acquired Transmission (EDS-HAT) strains from our hospital not associated with IR procedures. A, Heat map of core genome single-nucleotide polymorphism (SNP) differences between all ST1471 EDS-HAT strains. Strains are ordered by their phylogenetic relatedness, with strains in red belonging to the outbreak cluster. Shading indicates genome similarity as measured by SNP distance, with yellow indicating higher similarity (ie, fewer SNPs) and blue indicating lower similarity. B, Histogram of pairwise comparisons of genome similarity among IR-related outbreak strains. Pairwise SNP differences were calculated for all outbreak strains compared to one another (yellow bars) and compared to all nonoutbreak ST1471 EDS-HAT strains (blue bars). Abbreviations: SNP, single-nucleotide polymorphism; VRE, vancomycin-resistant enterococci.
DISCUSSION
We describe a previously unrecognized VRE outbreak related to IR procedures with contrast. Our investigation strongly suggests that use of nonsterile technique during invasive IR procedures was responsible for the outbreak. This conclusion is based on WGS results that suggest the outbreak had a common transmission route; the results of the investigation that indicate IR procedures using contrast were the major risk factor, with a markedly high odds ratio; the evidence that the outbreak strain was transmitted over a period of at least 10 months and that 1 technician was associated with 9 of the 10 cases; and the observed breaches in sterile technique during IR procedures. The outbreak was not previously recognized because of the high background incidence of VRE infections at our institution and the fact that the case patients were not housed together geographically in the hospital.
Epidemiologic data support the possibility that an IR technician in staff group A may have been a VRE carrier because this technician was the only healthcare worker possibly present at all IR procedures. However, we did not culture any staff members to investigate this hypothesis. Further, we were unable to explain the 4-month gap between case 3 and case 5. Healthcare worker colonization and transmission to patients have been previously reported [17].
A number of IP interventions were instituted as a result of this investigation. First, IR staff members were reeducated on proper hand hygiene practices. Second, all IR technicians at our institution are required to use sterile gloves when preparing the sterile contrast and disposable syringe. Finally, daily ultraviolet light disinfection of the IR procedural suites was implemented. There have been no further VRE infections with the outbreak strain detected since implementation of these interventions.
The practice of loading disposable syringes for contrast injections and handling of sterile sections of the apparatus without gloves was considered to be standard practice among the IR technicians at our institution. Indeed, this practice was demonstrated in training materials provided by the manufacturer, which suggests that the practices that we observed could be standard practice at other healthcare institutions. Moreover, the QFT is open to the environment, which increases the likelihood of inadvertent contamination of contrast.
Our study has limitations. First, the polymicrobial nature of some of the infections suggests that other organisms could also have been transmitted, but we have no direct evidence of this. Second, the presence of other pathogens limited our ability to assess the clinical significance of the VRE infection for some of the patients. Third, we did not culture the IR staff members for VRE carriage. However, the observations of IR technician practice, breaches in sterility, prolonged transmission of the outbreak strain, and the epidemiologic data suggest a VRE carrier as a likely outbreak source. Alternatively, contamination of environmental surfaces could have contributed to transmission, especially because VRE can survive on surfaces for months [18]. Fourth, our investigation was limited to a single hospital. However, we believe that our findings have implications for other hospitals. Finally, because we initiated WGS surveillance in November 2016, we were unable to determine when the outbreak began.
Our study provides proof of concept that WGS surveillance can be used to detect otherwise undetected outbreaks and, in this case, to identify a transmission route (ie, IR procedures) that to our knowledge has not been definitively identified. We suspect that this outbreak would never have been identified in the absence of WGS surveillance. Although we performed manual EHR review for the present study, we have developed EHR data-mining tools for automated identification of routes of transmission that we have recently begun to use in conjunction with the WGS surveillance we described here [12, 19]. Our goal is for EDS-HAT to eventually run in real-time so that we may promptly detect, intervene, and stop outbreaks such as the one described in this study. EDS-HAT pathogens currently include major bacterial species that are commonly transmitted in the hospital, including Klebsiella pneumoniae, Clostridioides difficile, methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium, and many others.
We suspect that infections following invasive IR procedures may not be restricted to our institution because of the widespread use of contrast injectors for these procedures. Moreover, some clinical practice guidelines do not specifically address contrast preparation [20]. Based on the manufacturer’s training materials, it seems likely that the lapses in sterile technique that we observed may exist at other hospitals. Our experience suggests that other institutions should observe their current practices of contrast preparation for breaches of sterility. Interventions should be implemented to ensure patient safety and better outcomes as the standard for IR procedures. Remarkably, there are no published data on the risk of infection following invasive IR procedures, suggesting that this has been a neglected area of IP research.
In conclusion, we have described an outbreak of VRE infections related to invasive IR procedures that was detected using WGS surveillance. We believe that IP practices for invasive IR procedures need to be reexamined globally, including development of sound IP procedures, new educational materials, as well as new engineering controls.
Notes
Acknowledgments. The authors thank Maria M. Brooks for her thoughtful review of the manuscript, Sara Ohm for her contributions to the whole-genome sequencing, and Casey Lewis and Jill Bertoty for their assistance in the epidemiologic investigation.
Disclaimer. The National Institutes of Health (NIH) played no role in data collection, analysis, or interpretation; study design; writing of the manuscript; or decision to submit for publication.
Financial support. This study was funded in part by the National Institute of Allergy and Infectious Diseases, NIH (R21Al109459 and R01AI127472).
Potential conflicts of interest. All authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol 2016; 37:1288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willems RJ, Top J, van Santen M, et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 2005; 11:821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Köser CU, Holden MTG, Ellington MJ, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 2012; 366:2267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGann P, Bunin JL, Snesrud E, et al. Real time application of whole genome sequencing for outbreak investigation— what is an achievable turnaround time? Diagn Microbiol Infect Dis 2016; 85:277–82. [DOI] [PubMed] [Google Scholar]
- 5. Raven KE, Reuter S, Reynolds R, et al. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res 2016; 26:1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brodrick HJ, Raven KE, Harrison EM, et al. Whole-genome sequencing reveals transmission of vancomycin-resistant Enterococcus faecium in a healthcare network. Genome Med 2016; 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becker L, Fuchs S, Pfeifer Y, et al. Whole genome sequence analysis of CTX-M-15 producing Klebsiella isolates allowed dissecting a polyclonal outbreak scenario. Front Microbiol 2018; 9:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peacock SJ, Parkhill J, Brown NM. Changing the paradigm for hospital outbreak detection by leading with genomic surveillance of nosocomial pathogens. Microbiology 2018; 164:1213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charalel RA, McGinty G, Brant-Zawadzki M, et al. Interventional radiology delivers high-value health care and is an Imaging 3.0 vanguard. J Am Coll Radiol 2015; 12:501–6. [DOI] [PubMed] [Google Scholar]
- 10. Rawf F, Alsafi A, Zia A, Darzi A, Bicknell CD, Hamady MS. Medical errors in IR: where are we? A systematic review. J Vasc Interv Radiol 2015; 26:1741–3. [DOI] [PubMed] [Google Scholar]
- 11. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seemann T. Rapid haploid variant calling and core genome alignment. Snippy 2019. Available at: https://github.com/tseemann/snippy [Google Scholar]
- 13. Sundermann AJ, Miller JK, Marsh JW, et al. Automated data mining of the electronic health record for investigation of healthcare-associated outbreaks. Infect Control Hosp Epidemiol 2019; 40:314–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward DV, Hoss AG, Kolde R, et al. Integration of genomic and clinical data augments surveillance of healthcare-acquired infections. Infect Control Hosp Epidemiol 2019; 40:649–55. [DOI] [PubMed] [Google Scholar]
- 15. Medrad, Inc. Mark V ProVis operation manual KMP 910E. Operation manual 98411-T-167 Rev F. USA: Medrad; 1999, section 3–4. [Google Scholar]
- 16. Loading the Syringe. Medrad Mark V ProVis Angiographic Injection System. Available at: https://www.radiologysolutions.bayer.com/products/angiography/injection/provis/ Accessed 25 May 2018. [Google Scholar]
- 17. Berkelman RL, Martin D, Graham DR, et al. Streptococcal wound infections caused by a vaginal carrier. JAMA 1982; 247:2680–2. [PubMed] [Google Scholar]
- 18. Neely AN, Maley MP. Survival of enterococci and staphylococci on hospital fabrics and plastic. J Clin Microbiol 2000; 38:724–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller JK, Chen J, Sundermann A, et al. Statistical outbreak detection by joining medical records and pathogen similarity. J Biomed Inform 2019; 91:103126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan D, Downing D, Keough CE, et al. ; Society of Interventional Radiology; Association of periOperative Registered Nurses Joint practice guideline for sterile technique during vascular and interventional radiology procedures: from the Society of Interventional Radiology, Association of periOperative Registered Nurses, and Association for Radiologic and Imaging Nursing, for the Society of Interventional Radiology [corrected] Standards of Practice Committee, and Endorsed by the Cardiovascular Interventional Radiological Society of Europe and the Canadian Interventional Radiology Association. J Vasc Interv Radiol 2012; 23:1603–12. [DOI] [PubMed] [Google Scholar]