Abstract
Twenty-nine Lactobacillus plantarum strains isolated from different types of Polish regional cheeses (Oscypek and Korycinski) were assessed for selected probiotic properties and anti-staphylococcal activity. Most of the tested L. plantarum strains were considered safe. Whole bacterial cultures (WBC) and cell-free supernatants (CFSs) of L. plantarum strains inhibited growth of Staphylococcus aureus (average inhibition growth zones were 2.8 mm ± 1.2 and 2.8 mm ± 1.1 respectively). Moreover, almost all neutralized, catalase-treated cell-free supernatants (CFN) of L. plantarum cultures also exhibited slight anti-staphylococcal activity in vitro. The most promising strains Os4 and Kor14 were selected for further study. Both strains were able to survive during digestive gastro-intestinal passage model. Live cells of L. plantarum Os4 and Kor14 caused the strongest inhibition of S. aureus adhesion to Caco-2 cells comparing with CFN and heat-killed bacterial cells. S. aureus and L. plantarum (Os4 or Kor14) co-cultured in skim milk resulted in growth inhibition of S. aureus in both 8 °C and 37 °C incubation temperatures. Observed abilities, demonstrated for L. plantarum Os4 and Kor14, confirms that these strains could be used in the food industry as protective cultures.
Electronic supplementary material
The online version of this article (10.1007/s12602-019-09587-w) contains supplementary material, which is available to authorized users.
Keywords: Anti-staphylococcal activity, Lactobacillus plantarum, Protective cultures, Regional cheese
Introduction
Lactic acid bacteria (LAB) are ubiquitous microorganisms that give food-specific functional and sensory properties. Furthermore, numerous studies indicate that their presence positively affects the human health by colonizing the digestive tract, lowering of blood cholesterol, or competing with pathogens [1]. Traditional food in different regions of the world is a particularly rich source of LAB, which is connected with the natural process of production and often the spontaneous fermentation reaction. LAB in traditional foods are responsible for the flavor characteristics as well as prevent the development of undesirable microbes, which directly prolongs the sustainability and preservation of the microbiological safety of the product [2].
Staphylococcus aureus is a Gram-positive bacterium commonly found on the skin and also in the nasopharynx of humans and animals. In a food product, this bacterium produces heat-stable enterotoxin, in which consumption of this causes the symptoms of intoxication such as diarrhea, abdominal pain, nausea, and fever [3, 4].
Staphylococcal intoxication is often connected with the consumption of cheeses. Only in 2015 in the EU, it was observed that cheese was the vehicle of 33% of strong-evidence outbreaks [5]. Overall, in 2015, 39 strong-evidence outbreaks and 395 weak-evidence ones were reported. Moreover, a high rate of hospitalization was observed (113 patients hospitalized per 758 overall number of cases) in strong-evidence outbreaks. It shows that the special attention should be paid on presence of S. aureus in food products, mainly in raw milk cheeses, because of the high risk for food safety [5].
Numerous studies show that LAB isolated from different kinds of food can effectively inhibit the growth of S. aureus [2, 6–9]. Anti-bacterial activity of LAB is mainly connected with the pH lowering and organic acid production and also with the possibility of bacteriocin synthesis and other antimicrobial agents such as hydrogen peroxide, reuterin or reutericyclin, and peptidoglycan hydrolases [10]. These LAB cultures which show unique features could be possibly used in food technology in the future, especially when chemical preservation is undesirable. It has been already shown that LAB isolated from Polish regional cheeses exhibit strong anti-bacterial activity against foodborne pathogens such as Listeria monocytogenes, Salmonella enteritidis, and Escherichia coli [11, 12]. If they also inhibit the growth of S. aureus and survive in gastro-intestinal tract to trigger a positive impact for the consumer, it would be possible to use these strains as probiotic and protective cultures.
The aim of this study was the evaluation of anti-staphylococcal activity, as well as preliminary safety assessment of Lactobacillus plantarum strains isolated from Polish regional cheeses and recognition the possibilities of using these strains as a probiotic and protective cultures in food technology. The strains were screened in vitro for their anti-staphylococcal activity and the selection was made. The best strains were analyzed to check their ability to survive in human gastro-intestinal tract and compete for in vitro adhesion to human gut cells as well as anti-staphylococcal activity in food matrix.
Material and Methods
Strains and Culture Conditions
The 29 strains of L. plantarum were isolated and genetically identified in the previous study of Ołdak et al. [11]. Strain cultures were stored at – 80 °C with 20% glycerol (v/v) addition in MRS broth (LabM, UK) and the fresh cultures were made prior the assays. The probiotic strain L. plantarum 299v was used as a reference.
S. aureus ATCC 25923 and S. aureus 12.21 and 4.4—food origin pathogens—were used as the indicator strains. The strains S. aureus 12.21 and 4.4 were kindly provided by the collection of Chair of Industrial and Food Microbiology, University of Warmia and Mazury, Poland.
To determine the count of S. aureus, serial decimal dilutions were plated onto Baird-Parker Agar supplemented with the Egg Yolk Tellurite (Merck, Germany). Enumeration of L. plantarum was performed on MRS agar (LabM, UK). Chosen media were selective for mentioned microorganisms used in this study. The plates were incubated for 24–48 h at 37 °C. After incubation, colonies observed on each solid medium were counted and results were converted to logarithmic scale.
Antibiotic Resistance
The L. plantarum strains were analyzed to evaluate their resistance to the most common antibiotics using the E-test (HiMedia, India) in accordance with the manual provided by the producer. To determine the minimal concentration of antibiotic (MIC) inhibiting the growth of the microorganism, the inhibition zones were checked, and the antibiotic concentration were read from strips. Bacterial strains were categorized as susceptible or resistant to antimicrobials according to EFSA Scientific Opinion [14]:
Susceptible (S): a bacterial strain is defined as susceptible when it is inhibited at a concentration of a specific antimicrobial equal or lower than the established cut-off value (S ≤ x mg/l).
Resistant (R): a bacterial strain is defined as resistant when it is not inhibited at a concentration of a specific antimicrobial higher than the established cut-off value (R > x mg/l)
The susceptibility or resistance were evaluated to gentamycin (0.016–256 μg/ml), streptomycin (0.016–256 μg/ml), ampicillin (0.016–256 μg/ml), vancomycin (0.016–256 μg/ml), tetracycline (0.016–256 μg/ml), ciprofloxacin (0.016–256 μg/ml), chloramphenicol (0.016–256 μg/ml), kanamycin (0.016–256 μg/ml), penicillin (0.002–32 μg/ml), and erythromycin (0.016–256 μg/ml). Each time, 200 μl of culture in MRS broth medium (0.5 McFarland standard) was inoculated on Petri plates with Mueller-Hinton agar (LabM, UK). Then, the antibiotic strips were placed. Strains were incubated at 37 °C for 48 h under anaerobic conditions in the GENBox anaer (bioMérieux, France). The results were compared with MIC value suggestions of Danielsen and Wind [13, 14].
Enzymatic Profile
Enzymatic profiles of the L. plantarum strains were determined using API ZYM (bioMerieux, France) in accordance with the manufacturer’s protocols. Strains were incubated at 37 °C for 24 h. Five groups of enzyme activities were analyzed: lipolytic activity (including enzyme activity of esterase, esterase lipase, lipase), proteolytic activity (including enzyme activity of leucine arylamidase, valine arylamidase, cysteine arylamidase, trypsin, and α-chymotrypsin), saccharolytic activity (including enzyme activity of α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-mannosidase, and α-fucosidase), phosphatase activity (including enzyme activity of alkaline phosphatase, acid phosphatase, naphthol-AS-BI-phosphohydrolase), and harmful enzyme activity (production of α-chymotrypsin, β-glucuronidase and N-acetyl-β-glucosaminidase).
Screening for Anti-Staphylococcal Activity
Anti-staphylococcal effectiveness of whole bacterial cultures (WBC), cell-free supernatants (CFS), and neutralized, catalase-treated cell-free supernatants (CFN) of the 29 L. plantarum strains were screened using well diffusion method (each in triplicate). Briefly, the 24 h cultures (in stationary phase of growth) were centrifuged (6000×g, 10 min) and sterilized by filtration (0.2 μm) and the anti-staphylococcal activities of CFS were tested. Subsequently, the obtained supernatants were treated with catalase (300 IU/ml, Sigma Aldrich, Poland) and neutralized with 1 M NaOH for the exclusion of the influence of organic acids, lowered pH, or hydrogen peroxide synthesis on the antagonistic activity of tested L. plantarum strains.
Sterile Petri dishes were aseptically poured with Mueller-Hinton Agar (LabM, UK), dried, and inoculated with 200-μl culture of each S. aureus strain (concentration 6 log CFU/ml, measured by plate count method). After that, 5.5-mm-diameter wells were cut out and subsequently filled with 50 μl of WBC, CFS, and CFN. Afterwards, plates were incubated at 37 °C for 48 h and zones of growth inhibition were observed and measured. Anti-staphylococcal activity of WBC and CFS (x) was converted as follows: x = D − d, where D is the zone diameter and d is the cut well diameter. Antagonistic effect of CFN was, in turn, evaluated qualitatively as “+” which means slight (< 1 mm of diameter inhibition zone) and “−” which means none.
Two strains showing the highest overall anti-staphylococcal activity, L. plantarum Os4 isolated from Oscypek and L. plantarum Kor14 isolated from Korycinski cheese, were chosen to further studies.
Survival Ability in the Gastro-Intestinal Tract
Gastric and intestinal juices were prepared according to Rzepkowska et al. [15] and suggestions of Minekus et al. [16]. Gastric juice consisted of the following: distilled water, NaCl (5 g/l), and pepsin (3 g/l) adjusted to pH 2.0 with HCl, whereas intestinal juice consisted of the following: distilled water, NaHCO3 (6.4 g/l), KCl (0.239 g/l), NaCl (1.28 g/l), bile salts (3.0 g/l), and pancreatin (1.0 g/l), adjusted to pH 8.0 with NaOH.
Briefly, 1 ml overnight culture of L. plantarum (concentration approx. 109–1010 CFU/ml) was centrifuged 10,000×g for 5 min. Therefore, supernatant was removed and 5 ml of gastric juice was added. Samples were vortexed and incubated for 2 h at 37 °C. After incubation period, samples were centrifuged (10,000×g/5 min.), supernatant was removed, and 10 ml of intestinal juice was added. Samples were vortexed and additionally incubated for 3 h at 37 °C. L. plantarum count was assessed before (0 h) and after incubation in gastric juice (2 h) and after incubation in gastric and intestinal juice (5 h) by a plate count technique. All tests were done in triplicate.
Competition for Adhesion to Caco-2 Cells
Caco-2 epithelial cells, originating from human colorectal adenocarcinoma (ATCC HTB-37), were used in their terminally differentiated state to mimic small intestine mature enterocytes. The cells were cultured in Dulbecco’s modified Eagle’s minimal essential medium (DMEM, Biowest) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Biowest), 2 mM l-glutamine (Biowest), 100 U/ml penicillin, and 100 mg/ml streptomycin (Biowest) at 37 °C in an atmosphere containing 5% CO2. The culture medium was changed every other day and replaced by fresh DMEM. Cells were used between 26 and 40 cell passages.
The adherence of tested strains to Caco-2 cell line was tested according to Delgado et al. [17] with slight modifications. Caco-2 cells were seeded in 24-well tissue culture plates at 5 × 105 cells per well and grown during 14 days to obtain a monolayer of differentiated and polarized cells. The culture medium was changed every 2 days. Bacterial cultures (1 ml) were grown until they reached the late exponential phase and they were sedimented by centrifugation (10,000×g for 10 min). Then, they were washed twice and resuspended in fresh DMEM at a concentration of 108 CFU/ml and added to each well of the tissue culture plate containing a Caco-2 monolayer. In competition tests, S. aureus ATCC 25923 and L. plantarum Os4 or Kor 14 were co-cultured in 1:1 ratio. L. plantarum were used as WBC or as heat-inactivated (80 °C per 20 min) bacterial cultures (HK). Addition of L. plantarum CFNs (100 μl per well) was also tested as a competition factor. The plates were incubated for 2 h at 37 °C in a 5% CO2 atmosphere. After incubation, unattached bacteria were removed by washing the monolayers four times with sterile PBS. Additionally, 200 μl of trypsin (2.5%, w/v) was added to detach the monolayer. The cells were incubated for 15 min at room temperature and recovered by repeated pipetting with 800 μl of sterile ultra-pure water and bacteria were enumerated by plating serial dilutions on solid medium. Adhesion values (%) were calculated as follows:
where V0 is the initial viable count of bacteria tested, and V1 is the viable bacterial count obtained from Caco-2 cells, at the end of the experiment.
Each adhesion assay was conducted in triplicate.
Co-Culture in Skim Milk
Considering that L. plantarum strains were isolated from dairy products, the assessment of their anti-staphylococcal activity in milk matrix was conducted.
Ten milliliters of skim milk (Mlekovita, Poland) was inoculated with ~ 102 cells of indicator strain (S. aureus ATCC 25923). Afterwards, ~ 106 bacterial cells of L. plantarum (Os4 or Kor14) were added as WBC. Control samples were inoculated only with the indicator strain. Milk samples were incubated at 37 °C and 8 °C for 24 h, 72 h, and 120 h. The initial bacterial count and the enumeration in each time point were performed by a plate count technique. Each experiment was conducted in triplicate.
Statistical Analysis
Data are expressed as a mean ± standard error (SE) calculated over three independent experiments performed in triplicate. Analysis of statistical significance was performed by ANOVA with Student’s t test. Statistical analyses were performed using STATISTICA13PL (StatSoft Inc.).
Results
Antibiotic Resistance
Table 1 shows the number of L. plantarum strains isolated from regional cheeses that have been considered as resistant to antibiotics. Most of the tested strains were susceptible to gentamicin, ampicillin, tetracycline, chloramphenicol, and kanamycin. Total resistance of tested strains to streptomycin and vancomycin was demonstrated. Almost all bacterial strains isolated from the Oscypek were resistant to ciprofloxacin, which was less noticeable in the strains isolated from the Korycinski cheese. On the other hand, resistance to penicillin and erythromycin was found among the strains isolated from Korycinski cheese.
Table 1.
Antibiotic susceptibility or resistance of Lactobacillus strains
| Type of antibiotic | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain symbol | GN1 | ST1 | AM1 | VA1 | TR1 | CPH2 | CHL1 | KN1 | PN2 | ER1 |
| Os1 | S | R | R | R | S | S | S | S | R | S |
| Os2 | S | R | R | R | S | R | S | R | R | S |
| Os3 | S | R | S | R | S | R | S | S | R | S |
| Os4 | S | R | S | R | S | R | S | S | R | S |
| Os5 | S | R | S | R | S | R | R | S | R | R |
| Os6 | S | R | S | R | S | R | R | S | S | R |
| Os7 | S | R | S | R | S | R | R | S | S | S |
| Os8 | S | R | S | R | S | R | S | S | R | S |
| Os9 | S | R | S | R | S | R | R | S | S | S |
| Os10 | S | R | S | R | S | R | S | S | R | S |
| Os11 | S | R | S | R | S | R | S | S | S | R |
| Os12 | S | R | S | R | S | R | S | S | R | R |
| Os13 | S | R | S | R | S | R | R | S | S | S |
| Os14 | S | R | S | R | S | R | S | S | S | S |
| Os15 | S | R | S | R | S | R | S | R | R | S |
| Os16 | S | R | S | R | S | R | S | S | S | R |
| Os17 | S | R | S | R | S | R | R | S | S | R |
| Kor1 | S | R | S | R | R | S | S | S | R | R |
| Kor2 | S | R | S | R | S | R | R | S | R | R |
| Kor8 | S | R | R | R | R | S | S | S | R | R |
| Kor11 | S | R | S | R | S | S | R | S | R | R |
| Kor12 | S | R | R | R | S | S | S | S | R | R |
| Kor13 | S | R | S | R | S | S | S | S | R | R |
| Kor14 | S | R | S | R | R | S | S | S | R | R |
| Kor15 | S | R | S | R | R | R | S | S | R | R |
| Kor18 | S | R | S | R | S | R | S | S | R | R |
| Kor19 | S | R | S | R | R | R | S | S | R | S |
| Kor22 | S | R | R | R | R | R | S | S | R | R |
| Kor23 | S | R | S | R | S | R | S | S | R | S |
1MIC according to EFSA (2012)
2MIC according to Danielsen and Wind (2003)
R resistant, S susceptible, GN gentamicin, ST streptomycin, AM ampicillin, VA vancomycin, TR tetracycline, CPH ciprofloxacin, CHL chloramphenicol, KN kanamycin, PN penicillin, ER erythromycin
Enzymatic Profile
L. plantarum strains isolated from Oscypek and Korycinski cheeses showed weak lipolytic and proteolytic activity (excluding L. plantarum strains Os8, Os15, and Kor23 which showed high esterase and esterase lipase activity). Average saccharolytic activity was observed in the tested strains, especially in the case of α-galactosidase and β-galactosidase, which are technologically valuable enzymes (Table 2). High activity of β-galactosidase was observed in the case of L. plantarum strains Os6, Os8, Os11, Os14, Os15, Kor11, Kor12, Kor19, Kor22, and Kor23.
Table 2.
Enzymatic profiles of the Lactobacillus strains
| Enzyme | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin | Strain symbol | Control | Alkaline phosphatase | Esterase | Esterasel ipase | Lipase | Leucine arylamidase | Valine arylamidase | Cystine arylamidase | Trypsin | α-Chymotrypsin | Acid phosphatase | Naphthol-AS-BI-phosphohydrolase | α-Galactosidase | β-Galactosidase | β-Glucuronidase | α-Glucosidase | β-Glucosidase | N-acetyl-β-glucosaminidase | α-Mannosidase | α-Fucosidase |
| Oscypek | Os1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 0 | 0 | 5 |
| Os2 | 0 | 1 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 5 | 5 | 2 | 2 | 0 | 0 | 4 | 0 | 0 | 1 | |
| Os3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 5 | |
| Os4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Os5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Os6 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | |
| Os7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 2 | 3 | |
| Os8 | 0 | 5 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 2 | 4 | 0 | 5 | 5 | 0 | 4 | 5 | 0 | 5 | 5 | |
| Os9 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 2 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | |
| Os10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 5 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | |
| Os11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 5 | 0 | 5 | 4 | 0 | 0 | 0 | |
| Os12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | |
| Os13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Os14 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 5 | 5 | 0 | 5 | 5 | 0 | 5 | 0 | |
| Os15 | 0 | 5 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 5 | 0 | 5 | 5 | 0 | 5 | 4 | |
| Os16 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 5 | 0 | 5 | 0 | |
| Os17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 5 | 2 | 0 | 5 | 2 | 0 | 0 | 0 | |
| Korycinski cheese | Kor1 | 0 | 5 | 0 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| Kor2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kor8 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 4 | 5 | 0 | 0 | 0 | 5 | 0 | 5 | 0 | 0 | |
| Kor11 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Kor12 | 0 | 5 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | |
| Kor13 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Kor14 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 5 | 5 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | |
| Kor15 | 0 | 5 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 5 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | |
| Kor18 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Kor19 | 0 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 5 | 0 | 0 | 4 | 1 | 0 | 0 | |
| Kor22 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 | 5 | 0 | 5 | 3 | 1 | 5 | 5 | |
| Kor23 | 0 | 5 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 4 | 5 | 0 | 0 | 3 | 3 | 5 | 5 | |
Explanatory: 0–3 negative reaction; 4–5 positive reaction
Among the tested strains isolated from both sources, the average activity of following enzymes was found: alkaline phosphatase, naphthyl-AS-BI phospholipidase, and acid phosphatase (L. plantarum strains Os8, Os9, Os14, Os15, Kor1, Kor2, Kor8, and Kor15). Only two of all strains produce harmful enzymes: Os17 showed the ability of α-chymotrypsin production and Kor12, which produced β-glucuronidase.
Screening for Anti-Staphylococcal Activity
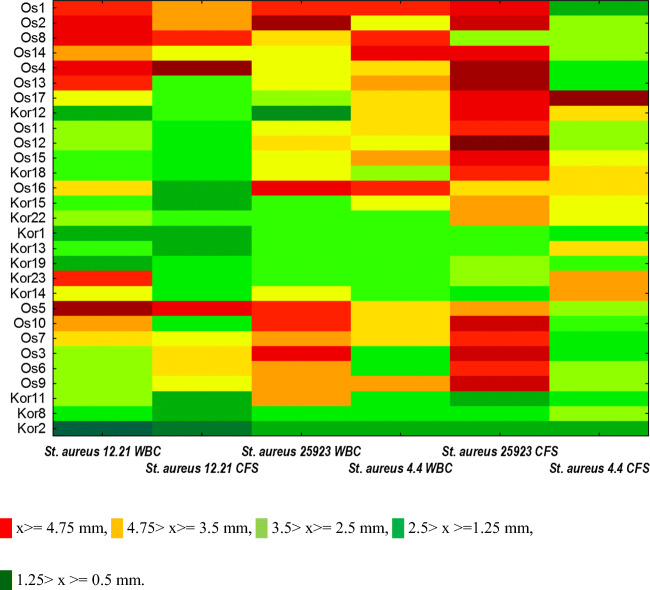
All of the tested, L. plantarum strains exhibited the antagonistic activity against S. aureus strains in the experiment (Fig. 1). The anti-staphylococcal WBC activity was assessed as slight with the average diameter of inhibition growth zone of 2.82 mm ± 1.23 (Supplementary material, Figs. S1–S3). The differences (P < 0.05) between the WBC (average) antagonism of examined L. plantarum strains were found, although the influence on three S. aureus strains was similar (P > 0.05). The greatest zones of inhibition were observed for L. plantarum Os5 and Os2 against S. aureus 12.21 (4.8 mm ± 1.2; 4.2 mm ± 0.2 respectively), for L. plantarum Os2 and Os4 against S. aureus ATCC 25923 (4.3 mm ± 1.5; 4.2 mm ± 0.7 respectively) and for L. plantarum Os14 against S. aureus 4.4 (4.2 mm ± 1.2). The lowest antagonistic activity was observed for L. plantarum Kor2 and Kor12 against all S. aureus strains.
Fig. 1.
Inhibition zones map including all tested L. plantarum whole bacterial cultures (WBC) and cell-free supernatant (CFS) in combination with three S. aureus indicators strains. x antimicrobial activity, inhibition zone (mm). x ≥ 4.75 mm,
x ≥ 4.75 mm,  4.75 > x ≥ 3.5 mm,
4.75 > x ≥ 3.5 mm,  3.5 > x ≥ 2.5 mm,
3.5 > x ≥ 2.5 mm,  2.5 > x ≥ 1.25 mm
2.5 > x ≥ 1.25 mm
 1.25 > x > = 0.5 mm.
1.25 > x > = 0.5 mm.
Differences between anti-staphylococcal effectiveness of CFS obtained from L. plantarum cultures against S. aureus strains were noted. S. aureus ATCC 25923 was more sensitive than S. aureus 12.21 and 4.4 and the observed average diameters of inhibition growth zones were 3.5 mm ± 1.3, 2.4 mm ± 1.3, and 2.6 mm ± 0.9 respectively. Significant differences (P < 0.05) between antagonistic activities of each L. plantarum strains were also observed (Fig. 1, Supplementary material, Figs. S4–S6). Low pH 4.27 ± 0.35 of tested CFSs was found.
Although, in the present study, the measured growth inhibition zones of S. aureus caused by WBC and CFS seem to be quite low, most of tested L. plantarum strains also exhibited anti-staphylococcal activities in neutralized and catalase-treated supernatant (CFN) (Supplementary material, Table S1). It was found that 11 out of 17 strains isolated from Oscypek cheese and 11 out of 12 strains isolated from Korycinski cheese slightly inhibited the growth of at least one S. aureus strain. The pH value of CFNs was approx. 6.5.
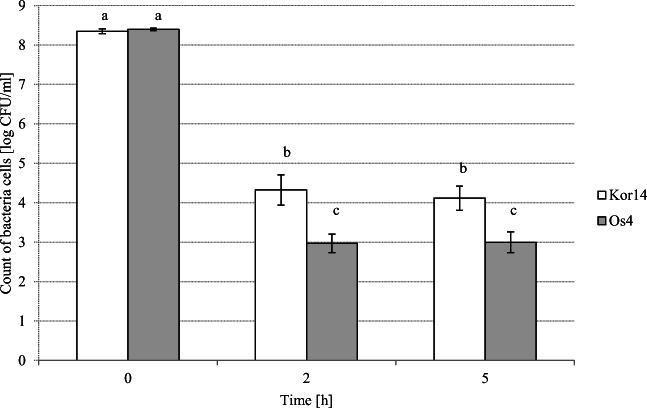
Survival Ability in the Gastro-Intestinal Tract
The survival of two selected L. plantarum strains, Os4 and Kor14, during 5 h of gastro-intestinal passage are presented on Fig. 2. It was found that after 2-h incubation at gastric conditions, the number of live bacterial cells was significantly decreased (P < 0.05) in both samples (4–5 log units), whereas after the next 3 h of intestinal passage, the count of bacteria were stable, and at the end of passage number of L. plantarum, Kor14 was 4.12 ± 0.25 log CFU/ml, and it was significantly higher (P < 0.05) in comparison with L. plantarum Os4 (3.0 ± 0.23 log CFU/ml).
Fig. 2.
Survival of the L. plantarum Kor14 and Os4 strains under simulated gastro-intestinal conditions. a, b, and c The same letters indicate no statistically significant differences (P < 0.05)
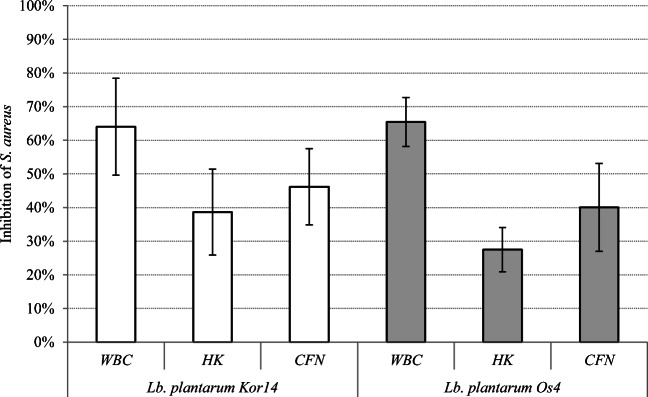
Competition for Adhesion to Caco-2 Cells
Presence of the live L. plantarum Os4 and Kor14 strains (WBC) caused the strongest inhibition of S. aureus adherence to Caco-2 cells (65.43 ± 7.3% and 64.03 ± 14.4% respectively). The results obtained in the experiment are shown on Fig. 3. Adhesion values of L. plantarum in co-culture with S. aureus cells were 99.78 ± 6.87% for Os4 and 91.76 ± 16.75% for Kor14, which confirms the high affinity of live lactic acid bacterial cells to human enterocytes. Our results show that the adherence inhibition caused by inactivated cells (HK) occurs, but the level of the phenomenon is lower (P < 0.05) than the inhibition caused by live cells (WBC) of Lactobacillus. Presence of heat-killed L. plantarum Os4 and Kor14 resulted in S. aureus adherence inhibition to Caco-2 layer (27.51 ± 6.5% and 38.66 ± 12.8% respectively). It was observed that the inhibition of S. aureus adhesion in the presence of L. plantarum CFN was stronger comparing with HK (P < 0.05) but weaker comparing with WBC (P < 0.05). The noted inhibition of 40.06 ± 13.1% and 46.19 ± 11.3% in the presence of CFNs of L. plantarum Os4 and Kor14 respectively was lower (P < 0.05) comparing with the control. Moreover, no adherence inhibition (P < 0.05) caused by sterile MRS broth was observed.
Fig. 3.
In vitro adhesion inhibition of S. aureus cells onto the enterocytes Caco-2 surface in tested combinations with L. plantarum whole bacterial cultures (WBC), L. plantarum heat-killed (HK) cells, and neutralized, catalase-treated cell-free supernatants (CFN) from L. plantarum cultures
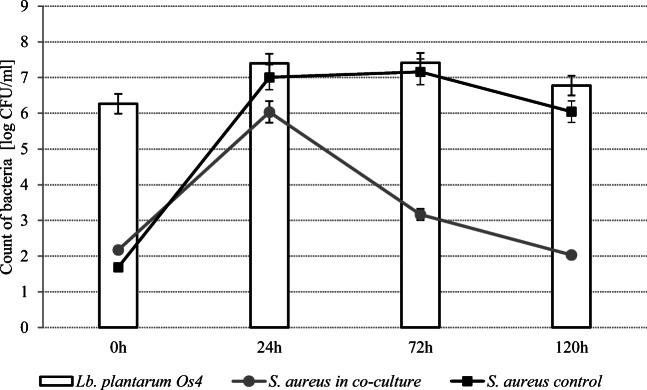
Co-Cultures in Skim Milk
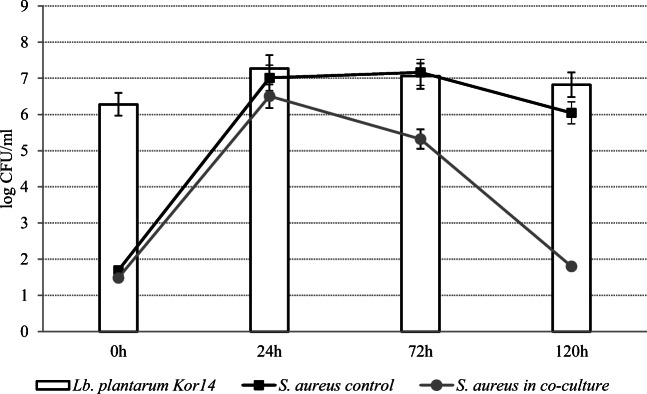
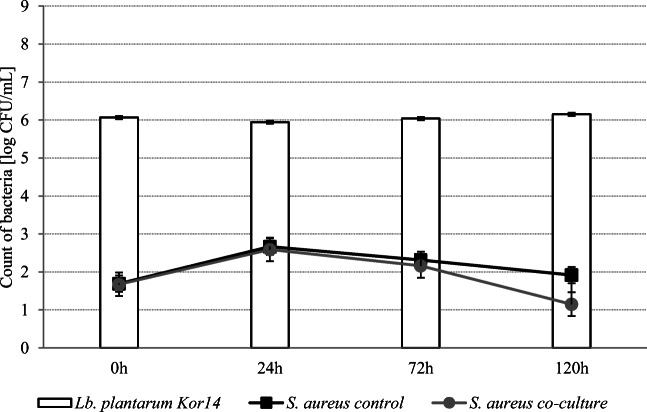
Figures 4 and 5 show the survival curves of L. plantarum strains in co-cultures with S. aureus ATCC 25923 at 37 °C. The reduction of S. aureus count in the samples inoculated with Os14 was greater after 72 h (P < 0.05) than in the case of L. plantarum Kor14. Generally, the number of S. aureus cells in skim milk samples decreased by more than 4 log CFU/ml after 120 h of incubation with L. plantarum Os4 and Kor14 addition. It was found that the number of L. plantarum Os4 was higher (P < 0.05) than L. plantarum Kor14 after 72 h of incubation.
Fig. 4.
Growth of L. plantarum Os4 and S. aureus ATCC 25923 at 37 in co-culture in skim milk
Fig. 5.
Growth of L. plantarum Kor14 and S. aureus ATCC 25923 at 37 in co-culture in skim milk
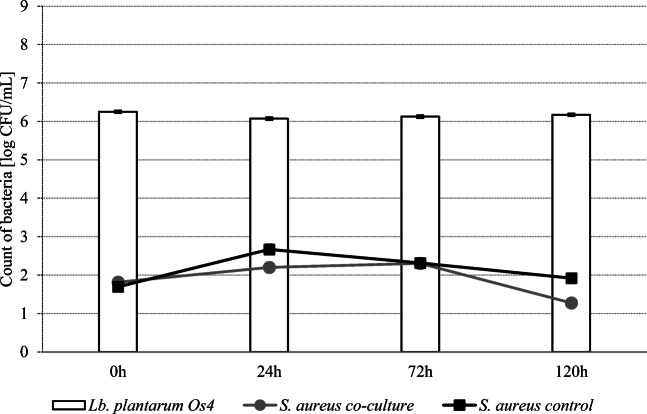
Figures 6 and 7 show the survival curves of L. plantarum strains in co-culture with S. aureus ATCC 25923 at 8 °C. A slight increase in S. aureus count was observed after 24 h of incubation (0.97 log CFU/ml for the control, 0.38 log CFU/ml in the presence of L. plantarum Os4, and 0.91 log CFU/ml in the presence of L. plantarum Kor14). There were no differences between the samples (P < 0.05) in S. aureus count after 72 h of incubation, but after 120 h of incubation at 8 °C was significant (P < 0.05), decrease of S. aureus count for both the control sample and the one inoculated with L. plantarum strains was observed. In samples inoculated with Os4 and Kor14, S. aureus count decreased by more than 1 log CFU/ml comparing with the control sample at 72-h incubation time. During the whole experiment, there were no differences (P < 0.05) between Lactobacillus counts, which suggest that the cells could multiply.
Fig. 6.
Growth of L. plantarum Os4 and S. aureus ATCC 25923 at 8 in co-culture in skim milk
Fig. 7.
Growth of L. plantarum Kor14 and S. aureus ATCC 25923 at 8°C in co-culture in skim milk
Discussion
One of the main concerns regarding technological, as well as probiotic usage of lactic acid bacteria and its safety, is resistance to antibiotics because of the possible transfer of antibiotic resistance to pathogenic bacteria. It was found that tested strains isolated from cheeses showed resistance or susceptibility to common antibiotics in regard to source of isolation. However, in some cases, the antibiotic resistance was similar. For example, resistance to vancomycin for the L. plantarum species is generally considered to be an intrinsic resistance that is associated with the construction of the bacterial cell wall. Resistance to streptomycin could be the reason of chromosomal mutations and therefore does not represent a potential risk of transferability [18]. According to EFSA Scientific Opinion (The FEEDAP Panel) [14] where all strains within a given taxonomic group show phenotypic resistance to an antimicrobial, such resistance can be considered intrinsic to the taxonomic group. Strains carrying intrinsic resistance present a minimal risk for horizontal spread and can be used as a feed additive [14]. On the other hand, susceptibility to gentamicin for L. plantarum strains has been observed previously in the study of Guo et al. [19]. Various studies have shown that the levels of susceptibility of Lactobacillus spp. to antibiotics were species-dependent or even strain-dependent. For example, in the Plessas et al. [20] study, strains isolated from Feta-type cheese have been demonstrated to be resistant to vancomycin, tetracycline, and erythromycin. Also, other authors confirmed Lactobacillus genus resistance to vancomycin and tetracycline [21]. The widespread incidence of antibiotic resistance in dairy food isolates is caused by the extensive use of antibiotics over the past few years in both animals and humans feeding [22].
The results obtained in the present study confirm the safety of tested L. plantarum strains in regard to antibiotic resistance; however, future study should be focused on identification and localization of antibiotic resistance genes. In the absence of information on the genetic nature of a demonstrated resistance, the strain should not be used as a feed additive [14].
The enzymatic profile of LAB used in cheese production is one of the factors which affect cheese flavor the most [21]. Lipolytic and proteolytic activity of Lactobacillus is species related, which was previously reported [23] and was observed in our study. Production of β-galactosidase is important for the production of dairy products and for consumers of dairy products who have lactose intolerance [24]. High β-galactosidase activity was observed in several of our strains (Os6, Os8, Os11, Os14, Os15, Kor11, Kor12, Kor19, Kor22, and Kor23). Among the L. plantarum strains, the average activities of phosphatase enzymes were found, which are the essential enzymes for the hydrolysis of phosphopeptides prevalent in cheese ripening [21]. From the safety point of view, it is important that bacterial strains do not produce harmful enzymes, i.e., α-chymotrypsin, β-glucuronidase, and N-acetyl-β-glucosaminidase, as they are known to be potential mediators of colon carcinogenesis [25]. Our results indicate that the enzymatic activity of tested strains should be considered as safe, except for Os17 and Kor12, which produced harmful enzymes.
An important element in the assessment of probiotic bacteria is their antimicrobial properties. All of strains tested in this study were previously assessed for their antagonistic activity against other pathogens such as L. monocytogenes, S. enteritidis, and E. coli as well as two food isolates Bacillus subtilis and Enterococcus faecium strains [11].
In the present study, anti-staphylococcal effectiveness of whole bacterial culture (WBC), cell-free supernatant (CFS), and neutralized, catalase-treated cell-free supernatant (CFN) was examined. The WBC contained live L. plantarum cells and all potential metabolites such as organic acids, hydrogen peroxide, and peptide substances. Anti-staphylococcal activity of CFSs was investigated in order to assess the influence of antimicrobial metabolites of L. plantarum cells, without the influence of live cells of bacteria. CFN used in the tests did not contain live or killed Lactobacillus cells and also organic acids and hydrogen peroxide activity were blocked by neutralization. The fact that neutralization of the supernatant caused the disappearance of antimicrobial activity for some of the strains, and its significant reduction for others, confirms that the low pH and organic acids present in CFS and WBC are the most important antimicrobial agents. This finding is in agreement with other studies [11, 15, 26].
Well diffusion method results of the present study showed generally much lower antagonistic activity of L. plantarum strains against S. aureus than for the other previously tested pathogens. In previous studies it was found that the L. plantarum Os1-Os17 strains exhibited strong anti-Listeria properties. In turn, L. plantarum strains isolated from Korycinski cheeses showed stronger antagonistic activity against E. coli in comparison with L. plantarum strains isolated from Oscypek [11]. Some other researchers did not observe the anti-staphylococcal activity of L. plantarum [27–29]. Arena [10] also assessed the antagonistic activity of L. plantarum strains as rather lower against S. aureus than L. monocytogenes, S. enteritidis, and E. coli. In turn, in other study, S. aureus growth inhibition zones were reported similar to the ones for other pathogens such as E. coli, Listeria innocua, and Salmonella typhimurium [7]. Karska-Wysocki et al. [30] reported similar to our study diameters of MRSA growth inhibition zones, which were enlarged when mixtures of few LAB strain cultures were tested. In turn, huge diversity of S. aureus growth inhibition zones ranging from 0.5 to 10 mm when plating with different strains of L. plantarum was observed by [10]. This confirms strain-dependent antimicrobial activity of lactic acid bacteria, demonstrated already by many studies [10–12, 29].
The tested L. plantarum strains have shown broad spectrum of antimicrobial activity, which in connection with present findings can be useful in food technology. Basing on our current results, we decided to select two most promising L. plantarum strains and test their possibility to compete for adherence to humans cells in vitro as well as in situ anti-staphylococcal activity in food matrix. L. plantarum Os4 and Kor14 strains have been chosen, based on average anti-staphylococcal activity for WBC and CFS, and according to their enzymatic activity profile as well as antibiotic resistance.
Although tested L. plantarum strains exhibited promising anti-bacterial properties, they should be at least resistant for gastro-intestinal conditions in the case of application as probiotic cultures. Generally, L. plantarum is known to be resistant to mucin, low gastric pH, and bile salts; however these features are in most cases strains related [31]. Similar results have been previously reported by others [32, 33], who highlighted the significant impact of low pH on the survival of lactobacilli. On the other hand, the stabilization of viability, observed under intestinal conditions, suggests that the tested L. plantarum strains could reach the intestine with significant concentrations of live cells, even after exposure to harmful acid challenge.
Many studies have shown that lactic acid bacteria compete with pathogens for adhesion to enterocytes which is an important feature of probiotic strains [1, 8, 9]. In the present study, it was decided the adhesion competition to Caco-2 cells as a good model for anti-staphylococcal activity test should be assessed. Strong adhesion competition of L. plantarum Os4 and Kor14 strains with S. aureus suggest that these strains are able to adhere, at least temporary, to the intestinal tract.
Many studies have shown that Lactobacillus adherence to Caco-2 varies among the genus [8, 34]. Some researchers have argued that even killed LAB cells can inhibit the adherence of pathogens to intestinal surfaces [35, 36]. In our study, also moderate adhesion inhibition caused by HK cells occurred. Inhibition of pathogen adhesion to Caco-2 cells may result from the biofilm formation by heat-killed Lactobacillus cells and the influence of bacterial metabolites on enterocyte layer [37]. Stronger adhesion inhibition of L. plantarum CFN was exhibited by Os4 and Kor14, which was probably caused by antimicrobial factors presented in supernatant. In other study, neutralized supernatant from Bifidobacterium cultures in MRS broth inhibits the adherence of Clostridium difficile in a similar scenario [38]. MRS did not cause adherence inhibition, which supports the thesis that tested CFNs containing other anti-adherent substances produced by examined L. plantarum strains.
Taking into account that the investigated LAB strains were isolated from dairy products, the skim milk matrix was chosen for the in situ studies. In the experiment, the inoculation with the S. aureus in approx. 102 CFU/ml and at the same time with L. plantarum cells (106 CFU/ml) reflected the situation of cross-contamination that may happen during the production of fermented milk products. The pathogenic effects of S. aureus are connected with enterotoxin production in food matrix. It has been proved that 5 × 105 cells of enterotoxin-producing S. aureus strains per ml is a threshold value resulting in the production of enterotoxin [39, 40]. Therefore, the key prevention should be to eliminate any factors and conditions that enhance the growth of the pathogen and the production of enterotoxin [41].
It has been found that L. plantarum strains Os4 and Kor14 successfully inhibited the growth of S. aureus at 37 °C in milk, but the dynamic of action was strain-dependent. Different growth kinetics of L. plantarum Os4 strain may be the reason for faster S. aureus count reduction in co-cultures in comparison with Kor14. In another study, S. aureus count decreased below the detection limit after 72 h of incubation when milk was inoculated with 3 log CFU/ml of both L. plantarum and S. aureus [42]. It was also shown that presence of S. aureus in co-culture with some L. plantarum strains slows down the pH lowering in the matrix, which could be the reason of overdue the effect of exhibited anti-staphylococcal efficiency [40].
The influence of the environmental conditions for the antimicrobial effectiveness of tested L. plantarum strains was also examined by the experiment conducted in a lower temperature. Slight inhibition of S. aureus growth was observed after 120 h of incubation. The temperature of 8°C was chosen to simulate the refrigeration conditions. Although there are many studies showing that the L. plantarum growth is inhibited in temperatures below 15 °C, the minimum temperature for S. aureus growth is 7 °C [43]. For example, the lag phase of Lactobacillus rhamnosus GG growth in 8 °C was long and lasted for 17.99 h. Moreover, this phenomenon results in a very slow lowering of the pH of the environment [44]. Many studies also show that in lower temperatures (e.g., 10–15 °C), synthesis of bacteriocins and other antimicrobial compounds with maximum yield was observed [45–47]. Lowering of S. aureus count in samples inoculated with L. plantarum Os4 and Kor14 strains can therefore be associated with other factors than organic acids, like bacteriocins. Moreover, used in this study, a high ratio of L. plantarum to S. aureus may facilitate Lactobacillus cells to the competition for nutrients in tested biological niche.
In conclusion, L. plantarum strains isolated from regional cheeses should generally be considered as safe; however, their antibiotic resistance was found to be source of isolation related. Most of L. plantarum strains isolated from the Oscypek were resistant to ciprofloxacin, whereas strains isolated from the Korycinski cheese were resistant to penicillin and erythromycin. According to the enzymatic profile study, the following strains should be excluded from further studies: Os17, which showed the production of α-chymotrypsin and Kor12, which produced β-glucuronidase.
All tested L. plantarum strains exhibited anti-staphylococcal activity, and almost all CFN of tested strains exhibit slight anti-staphylococcal activity. Chosen L. plantarum Os4 and Kor14 strains were able to survive in simulated human gastro-intestinal tract. S. aureus adhesion to Caco-2 cells was inhibited in the presence of L. plantarum live bacterial cells, as well as and dead bacterial cells and CFN. Therefore, tested L. plantarum strains of food origin could successfully compete with this pathogen in human gut. Moreover, the decrease in S. aureus counts in the milk matrix resulting from the presence of L. plantarum Os4 and Kor14 both at 37 and 8 °C confirms that these strains or their metabolites could be potentially used in the food industry as protective cultures to extend the shelf life of foodstuffs, in particularly fermented milk products.
Basing on our study, we recommend two strains of L. plantarum Os4 and Kor14 for use in food technology. Further studies are needed to evaluate the L. plantarum bacteriocin production and also to develop an efficient method for L. plantarum Os4 and Kor14 application as probiotic and protective cultures.
Electronic Supplementary Material
(DOCX 354 kb)
Funding Information
This research was financed by the Polish Ministry of Science and Higher Education within funds of the Faculty of Human Nutrition and Consumer Sciences, Warsaw University of Life Sciences (WULS), for scientific research.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Campana R, van Hemert S, Baffone W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017;9:12. doi: 10.1186/s13099-017-0162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pisano MB, Viale S, Conti S, Fadda ME, Deplano M, Melis MP, Deiana M, Cosentino S (2014) Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from sardinian dairy products. Biomed Res Int:286390–286399. 10.1155/2014/286390 [DOI] [PMC free article] [PubMed]
- 3.FDA . Bad bug book: foodborne pathogenic microorganisms and natural toxins handbook. 2. Silver Spring: US Food and Drug Administration; 2012. pp. 87–92. [Google Scholar]
- 4.Montville TJ, Matthews KR. Food microbiology: an introduction. 2. Washington: ASM Press; 2008. [Google Scholar]
- 5.EFSA, ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA Journal. 2016;14(12):4634–1–231. doi: 10.2903/j.efsa.2016.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Madboly LA, Abdullah AK. Potent antagonistic activity of Egyptian Lactobacillus plantarum against multiresistant and virulent food-associated pathogens. Front Microbiol. 2015;6:347. doi: 10.3389/fmicb.2015.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heredia-Castro PY, Méndez-Romero JI, Hernández-Mendoza A, Acedo-Félix E, González-Córdova AF, Vallejo-Cordoba B. Antimicrobial activity and partial characterization of bacteriocin-like inhibitory substances produced by Lactobacillus spp. isolated from artisanal Mexican cheese. J Dairy Sci. 2015;98:8285–8293. doi: 10.3168/jds.2015-10104. [DOI] [PubMed] [Google Scholar]
- 8.Ramos CL, Thorsen L, Schwan RF, Jespersen L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013;36:22–29. doi: 10.1016/j.fm.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Shokryazdan P, Sieo CC, Kalavathy R, Liang JB, Alitheen NB, Jahromi MF, Ahmed M. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int. 2014;2014(2):1–16. doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arena MP, Caggianiello G, Fiocco D, Russo P, Torelli M, Spano G, Capozzi V. Barley β-glucans-containing food enhances probiotic performances of beneficial bacteria. Int J Mol Sci. 2014;15:3026–3039. doi: 10.3390/ijms15023025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ołdak A, Zielińska D, Rzepkowska A, Kołożyn-Krajewska D (2017) Comparison of antibacterial activity of Lactobacillus plantarum strains isolated from two different kinds of regional cheeses from Poland: Oscypek and Korycinski cheese. Biomed Res Int (6820369):10. 10.1155/2017/6820369 [DOI] [PMC free article] [PubMed]
- 12.Sip A, Więckowicz M, Olejnik-Schmidt A, Grajek W. Anti-Listeria activity of lactic acid bacteria isolated from golka, a regional cheese produced in Poland. Food Control. 2012;26:117–124. doi: 10.1016/j.foodcont.2012.01.014. [DOI] [Google Scholar]
- 13.Danielsen M, Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int J Food Microbiol. 2003;82(1):1–11. doi: 10.1016/S0168-1605(02)00254-4. [DOI] [PubMed] [Google Scholar]
- 14.EFSA Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. The EFSA Journal. 2012;10:2740–2749. doi: 10.2903/j.efsa.2012.2740. [DOI] [Google Scholar]
- 15.Rzepkowska A, Zielińska D, Ołdak A, Kołożyn-Krajewska D. Organic whey as a source of Lactobacillus strains with selected technological and antimicrobial properties. Int J Food Sci Technol. 2017;52(9):1983–1994. doi: 10.1111/ijfs.13471. [DOI] [Google Scholar]
- 16.Minekus M, Alminger M, Alvito P, Balance S, Bohn T, Bourlieu C, et al. A standardised static in vitro digestion method suitable for food–an international consensus. Food &Function. 2014;5(6):1113–1124. doi: 10.1039/c3fo60702j. [DOI] [PubMed] [Google Scholar]
- 17.Delgado S, O’Sullivan E, Fitzgerald G, Mayo B. Subtractive screening for probiotic properties of Lactobacillus species from the human gastro-intestinal tract in the search for new probiotics. J Food Sci. 2007;72(8):310–315. doi: 10.1111/j.1750-3841.2007.00479.x. [DOI] [PubMed] [Google Scholar]
- 18.Gueimonde M, Sánchez B, de los Reyes-Gavilán CG, Margolles A. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:202. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Pan L, Li L, Lu J, Kwok L, Menghe B, Zhang H, Zhang W. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J Food Sci. 2017;82(3):724–730. doi: 10.1111/1750-3841.13645. [DOI] [PubMed] [Google Scholar]
- 20.Plessas S, Nouska C, Karapetsas A, Kazakos S, Alexopoulos A, Mantzourani I, Bezirtzoglou E. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. 2017;226:102–108. doi: 10.1016/j.foodchem.2017.01.052. [DOI] [PubMed] [Google Scholar]
- 21.Domingos-Lopes MFP, Stanton C, Ross PR, Dapkevicius MLE, Silva CCG. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017;63:178–190. doi: 10.1016/j.fm.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization (2014) Antimicrobial resistance global report on surveillance: 2014 summary. (No. WHO/HSE/PED/AIP/2014.2)
- 23.Ammor MS, Mayo B. Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: an update. Meat Sci. 2007;76(1):138–146. doi: 10.1016/j.meatsci.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Casarotti SN, Carneiro BM, Todorov SD, Nero LA, Rahal P, Penna ALB. In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water buffalo mozzarella cheese. Ann Microbiol. 2017;67(4):289–301. doi: 10.1007/s13213-017-1258-2. [DOI] [Google Scholar]
- 25.Wedajo B. Lactic acid bacteria: benefits, selection criteria and probiotic potential in fermented food. J Probiotics Health. 2015;3:129–138. doi: 10.4172/2329-8901.1000129. [DOI] [Google Scholar]
- 26.Aguilar-Toalá JE, Santiago-López L, Peres CM, Peres C, Garcia HS, Vallejo-Cordoba B, González-Córdova AF, Hernández-Mendoza A. Assessment of multifunctional activity of bioactive peptides derived from fermented milk by specific Lactobacillus plantarum strains. J Dairy Sci. 2017;100(1):65–75. doi: 10.3168/jds.2016-11846. [DOI] [PubMed] [Google Scholar]
- 27.Cizeikiene D, Juodeikiene G, Paskevicius A, Bartkiene E. Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control. 2013;31(2):539–545. doi: 10.1016/j.foodcont.2012.12.004. [DOI] [Google Scholar]
- 28.González L, Sandoval H, Sacristán N, Castro JM, Fresno JM, Tornadijo ME. Identification of lactic acid bacteria isolated from Genestoso cheese throughout ripening and study of their antimicrobial activity. Food Control. 2007;18(6):716–722. doi: 10.1016/j.foodcont.2006.03.008. [DOI] [Google Scholar]
- 29.Leite AMO, Miguel MAL, Peixoto RS, Ruas-Madiedo P, Paschoalin VMF, Mayo B, Delgado S. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J Dairy Sci. 2015;98:3622–3632. doi: 10.3168/jds.2014-9265. [DOI] [PubMed] [Google Scholar]
- 30.Karska-Wysocki B, Bazo M, Smoragiewicz W. Antibacterial activity of Lactobacillus acidophilus and Lactobacillus casei against methicillin-resistant Staphylococcus aureus (MRSA) Microbiol Res. 2010;20(165(8)):674–686. doi: 10.1016/j.micres.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 31.de Vries MC, Vaughan EE, Kleerebezem M, de Vos WM. Lactobacillus plantarum -survival, functional and potential probiotic properties in the human intestinal tract. Int Dairy J. 2006;16:1018–1028. doi: 10.1016/j.idairyj.2005.09.003. [DOI] [Google Scholar]
- 32.Arena MP, Silvain A, Normanno G, Grieco F, Drider D, Spano G, Fiocco D. Use of Lactobacillus plantarum strains as a bio - control strategy against food - borne pathogenic microorganisms. Front Microbiol. 2016;7:464. doi: 10.3389/fmicb.2016.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gheziel C, Russo P, Arena MP, Spano G, Ouzari HI, Kheroua O et al (2018) Evaluating the probiotic potential of Lactobacillus plantarum strains from Algerian infant feces: towards the design of probiotic starter cultures tailored for developing countries. Probiotics Antimicrob proteins:1–11. 10.1007/s12602-018-9396-9 [DOI] [PubMed]
- 34.Lavilla-Lerma L, Pérez-Pulido R, Martínez-Bueno M, Maqueda M, Valdivia E. Characterization of functional, safety, and gut survival related characteristics of Lactobacillus strains isolated from farmhouse goat’s milk cheeses. Int J Food Microbiol. 2013;163:136–145. doi: 10.1016/j.ijfoodmicro.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa H, Kutsukake E, Fukui T, Sato I, Shirai T, Kurihara T, Okada N, Danbara H, Toba M, Kohda N, Maeda Y, Matsumoto T. Oral administration of heat-killed Lactobacillus plantarum strain b240 protected mice against Salmonella enterica serovar typhimurium. Biosci Biotechnol Biochem. 2010;74:7,1338–7,1342. doi: 10.1271/bbb.90871. [DOI] [PubMed] [Google Scholar]
- 36.Lin W-H, Yu B, Lin C-K, Hwang W-Z, Tsen H-Y. Immune effect of heat-killed multistrain of Lactobacillus acidophilus against Salmonella typhimurium invasion to mice. J Appl Microbiol. 2007;102:22–31. doi: 10.1111/j.1365-2672.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura S, Kuda T, An C, Kanno T, Takahashi H, Kimura B. Inhibitory effects of Leuconostoc mesenteroides 1RM3 isolated from narezushi, a fermented fish with rice, on Listeria monocytogenes infection to Caco-2 cells and A/J mice. Anaerobe. 2012;18:19–24. doi: 10.1016/j.anaerobe.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Trejo FM, Minnaard J, Perez PF, De Antoni GL. Inhibition of Clostridium difficile growth and adhesion to enterocytes by Bifidobacterium supernatants. Anaerobe. 2006;12(4):186–193. doi: 10.1016/j.anaerobe.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Barker GC, Goméz-Tomé N. A risk assessment model for enterotoxigenic Staphylococcus aureus in pasteurized milk: a potential route to source-level inference. Risk Anal. 2013;33:249–269. doi: 10.1111/j.1539-6924.2011.01667.x. [DOI] [PubMed] [Google Scholar]
- 40.Zdenkova K, Babek A, Karamonova L, Purkrtova S, Karpiskova R, Demnerova K. Transcriptomic and metabolic responses of Staphylococcus aureus in mixed culture with Lactobacillus plantarum, Streptococcus thermophilus and Enterococcus durans in milk. J Ind Microbiol Biotechnol. 2016;43(9):1237–1247. doi: 10.1007/s10295-016-1794-y. [DOI] [PubMed] [Google Scholar]
- 41.Trząskowska M, Gasentzer P. Effects of probiotic Lactobacillus rhamnosus LOCK900 on the Staphylococcus aureus survival in carrot juice. J Food Saf. 2016;36(4):571–576. doi: 10.1111/jfs.12278. [DOI] [Google Scholar]
- 42.Anas M, Rizk HA, Eddine HJ, Ahmed K, Mebrouk K. Anti-bacterial activity of Lactobacillus plantarum isolated from Algerian raw goat’s milk against Staphylococcus aureus. Technologies Laboratoire. 2010;5:26–33. [Google Scholar]
- 43.Stewart C M (2003) Staphylococcus aureus and staphylococcal enterotoxins. Ch 12 In: Hocking A.D. (ed) Foodborne microorganisms of public health significance. 6th ed, Australian Institute of Food Science and Technology (NSW Branch), Sydney, p. 359–380
- 44.Valík L, Medveďová A, Liptáková D. Characterization of the growth of Lactobacillus rhamnosus GG in milk at suboptimal temperatures. J Food Nutr Res. 2008;47:60–67. [Google Scholar]
- 45.Abbasiliasi S, Tan JS, Ibrahim TAT, Bashokouh F, Ramakrishnan NR, Mustafaab S, Arif AB. Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: a review. RSC Adv. 2017;7:29395–29420. doi: 10.1039/c6ra24579j. [DOI] [Google Scholar]
- 46.Janes ME, Nannapaneni R, Johnson MG. Identification and characterization of two bacteriocin-producing bacteria isolated from garlic and gingerroot. J Food Prot. 1999;62(8):899–904. doi: 10.4315/0362-028X-62.8.899. [DOI] [PubMed] [Google Scholar]
- 47.Keren T, Yarmus M, Halevy G, Shapira R. Immunodetection of the bacteriocin lacticin RM: analysis of the influence of temperature and Tween 80 on its expression and activity. App Environ Microbiol. 2004;70(4):2098–2104. doi: 10.1128/AEM.70.4.2098-2104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 354 kb)