Abstract
Survivin is one of the rare proteins that is differentially expressed in normal and cancer cells and is directly or indirectly involved in numerous pathways required for tumor maintenance. It is expressed in almost all cancers and its expression has been detected at early stages of cancer. These traits make survivin an exceptionally attractive target for cancer therapeutics. Even with these promising features to be an oncotherapeutic target, there has been limited success in the clinical trials targeting survivin. Only recently it has emerged that survivin was not being specifically targeted which could have resulted in the negative clinical outcome. Also, focus of research has now shifted from survivin expression in the overall heterogeneous tumor cell populations to survivin expression in cancer stem cells as these cells have proved to be the major drivers of tumors. Therefore, in this review we have analyzed the expression of survivin in normal and cancer cells with a particular focus on its expression in cancer stem cell compartment. We have discussed the major signaling pathways involved in regulation of survivin. We have explored the current development status of various types of interventions for inhibition of survivin. Furthermore, we have discussed the challenges involving the development of potent and specific survivin inhibitors for cancer therapeutics. Finally we have given insights for some of the promising future anticancer treatments.
Keywords: Survivin, Cancer stem cells, Cancer signaling, Anticancer therapy, Inhibitors
Introduction
Survivin, an evolutionarily conserved inhibitor of apoptosis protein (IAP) is a multi-tasking extraordinaire, with differential expression patterns in normal and cancer cells. The copious information available on this protein, since its emergence has been vibrantly captivating the attention of both cell biologists and oncologists. This protein, central to various cellular signaling pathways and instrumental in cell proliferation and programmed cell death, has transpired as a cardinal onco-therapeutic target in the last two decades [1–3]. This protein performs important roles in cell cycle, apoptosis, angiogenesis and cancer formation and progression. It is found in different subcellular fractions and the major pools in cancer cells are in nucleus, cytoplasm, mitochondria and extracellular space [4]. Nuclear survivin regulates cell division while cytoplasmic expression is associated with cytoprotection for tumors. The balance between these two pools of the protein is indicative of ‘active survivin’ and the relative intracellular expression level quantification helps in analyzing the prognostic significance of the marker [5]. Mitochondrial survivin is associated with tumor growth and apoptotic resistance while extracellular survivin can re-enter tumor cells and increase proliferation, apoptotic resistance and invasion. The extracellular survivin is localized on exosomes secreted by cancer cells. This pool plays a role in cell-cell communication [2, 6].
Structure and Functions of Survivin
The 142 amino acid long survivin, encoded by BIRC5 (Baculoviral IAP repeat (BIR) - containing 5) gene is the smallest of the eight IAPs in human genome and is located at the telomeric end of chromosome 17 (17q25). It consists of a 70 amino acid long N-terminal BIR domain associated with apoptotic function, a carboxy terminal α-helix region that binds with BIR domain involved in mitotic function and a dimer binding domain [7, 8]. Studies have shown an increased expression of the protein in the G2/M phase of cell cycle, thus aiding in cell division and mitosis. This protein attaches to mitotic spindle through the α-helix, during the short gap between metaphase and anaphase, and in association with tubulin and constituent elements of the mitotic apparatus regulates microtubule dynamics and nucleation [9, 10]. Another pool of survivin localizes to the kinetochores and with the help of cytokinesis regulators, Aurora B kinase and inner centromere protein (INCEP), helps in chromosome segregation and cytokinesis [11, 12]. Thus the protein is a member of the chromosome passenger complex (CPC) which has significant role in regulation of chromosome-microtubule attachment, bipolar spindle formation, spindle assembly checkpoint and cytokinesis at cell division [13].
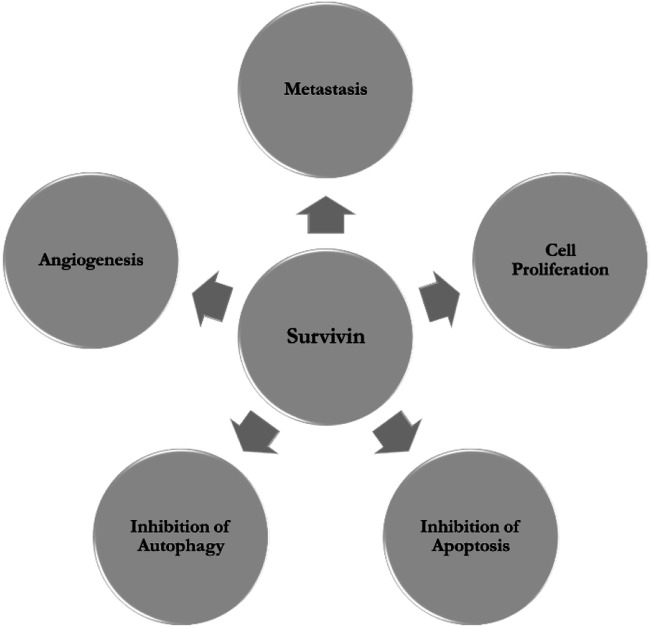
Survivin inhibits both apoptotic and autophagic cell death [14]. It inhibits extrinsic and intrinsic apoptotic pathways by both caspase independent and dependent mechanisms. While other IAPs bind to and directly degrade the active caspases 3, 7 and 9, survivin binds to and suppresses pro-caspase 9 with the help of hepatitis B X-interacting protein (HBXIP) cofactor, thus inhibiting death receptor signaling pathway [15, 16]. The interaction of mitochondrial survivin with X linked IAP (XIAP), resulting in the formation of an IAP-IAP complex can also suppress caspase 9 [17]. They also sequester and inactivate second mitochondria-derived activator of caspase/direct inhibitor of apoptosis-binding protein with low pI (Smac/DIABLO), that otherwise act as an IAP antagonist preventing the binding of XIAP to survivin thus releasing caspase 9 [8]. Even though increase in survivin expression has been reported in relation to inhibition of autophagy the exact mechanism and interacting proteins have not been determined yet [14]. The role of survivin in enhancing vascular endothelial growth factor (VEGF) transcription, synthesis and release and promoting vasculogenic mimicry in hypoxic conditions, substantiates its involvement in angiogenesis and subsequent upregulation in angiogenically stimulated cells [18]. Survivin also binds to c-Src (cellular sarcoma) establishing a bidirectional relationship that adversely affects focal adhesion dynamics and integrity of F-actin organization and disrupting cell-cell and cell-matrix interactions [19]. Moreover survivin is also involved in DNA damage repair, tissue response to injury and immune response [20]. Figure. 1 illustrates the major roles of survivin.
Fig. 1.
Survivin as a nodal protein
The protein has 3 introns and 4 exons and in addition to survivin wild type (WT) produces various isoforms, survivin 2b (S-2b), survivin 2a (S-2a), survivin 3b (S-3b), Survivin ΔEx3 (S-ΔEx3), survivin 2α (S-2α), survivin 3α (S-3α), survivin-3γ (S-3γ), survivin-3γV (S-3γV), survivin 2b + 32 (S-2b + 32) and survivin image (SI). Each of these variants has different intracellular localization, expression patterns and can form heterodimers with WT. Some splice variants with unknown biological functions, including survivin-ΔptEx1/2, survivin-ΔptEx1/2G/T, survivin-ΔptEx2/3 and survivin-ΔptEx2, have also been reported. Variants S-ΔEx3, S-3b, S-2α, S-3α, S-3γ and S-3γV are all antiapoptotic while S-2a and S-2b are pro-apoptotic due to the missing and truncated BIR domain. S-ΔEx3 is associated with cell mortality and often with angiogenesis [21–23]. S-3b is cytoprotective, associated with cell cycle regulation and helps in evading immune response[24].
Survivin in Non-malignant Non-Stem Cells
Even though a few human adult tissues like primitive hematopoietic cells, adult peripheral blood T cells, polymorphonuclear neutrophils, erythroid cells, megakaryocyte, vascular endothelial cells, cells of colonic and gastrointestinal mucosa, placenta, testes, ovary, neurons, melanocytes etc. express survivin, the presence is almost negligible in most terminally differentiated tissues [25–29]. The more mature the tissue the lesser the expression of the protein [20]. They have roles in vascular remodeling, angiogenic regulation, cell cycle regulation and proliferation, maintenance of normal adult hematopoiesis, erythropoiesis, megakaryocyte maturation, hepatocyte proliferation, apoptosis inhibition in neural development, self-renewal and differentiation of cryptic stem cells, spermatogenesis and oogenesis [27–29].
Even though most normal tissues do not express the splice variants, some do express minimal amounts of certain isoforms. S-ΔEx3 expression is observed in vascular endothelial cells. S-ΔEx3 is the only isoform expressed in adult brain cerebrum while fetal brain tissues showed the expression of WT and S-2b as well. Colon and soft sarcoma tissues did show trace amount of these variants in certain cases [23]. Minimal expression of S-2α has also been observed in normal brain cerebellum and breast tissues [30]. Expression of other splice variants are unknown till date.
Survivin in Stem Cells
Increased survivin expression has been reported in early embryonic stem cells (ES) wherein it reduces during fetal development and also some adult stem cells. Adida et al. reported survivin expression in apoptosis regulated human fetal tissues including thymic medulla, stem cell layer of stratified epithelia of skin, gastrointestinal tract and endocrine pancreas, preserving the viability of stem cells in a developmentally regulated fashion thus helping in tissue homeostasis and differentiation [31]. The analysis of the protein expression in umbilical cord blood cells and adult bone marrow CD34+ cells showed that the protein is hematopoietic cytokine regulated and hematopoietic growth factor dependent. The protein expression is observed in all stages of cell cycle and showed an inverse correlation with apoptosis in caspase dependent manner [32]. Survivin upregulation is a mandatory process for entry to cell cycle, self-renewal and maintenance in normal cord blood CD34+ cells and hematopoietic stem and progenitor cells (HSPCs) [33, 34]. A work by Filion et al. showed that in human embryonic stem cells (HES) subjected to ionizing radiation the expression of this IAP along with checkpoint kinase 2 (Chk2) helps to recuperate DNA damage without the dependency on p21 to support cell cycle arrest [35]. Higher survivin levels correlate to maintenance of the state of pluripotency in HES and induced pluripotent stem cells (iPSCs). The protein is involved in the regulation of cell proliferation and signaling pathways in stem cells [20]. A study conducted to understand the role of survivin in pluripotency showed a significantly higher expression of the protein in human embryonic stem cells (almost 9 fold) in comparison to that in differentiated cell types [36]. The IAP is involved in controlling basal and growth factor dependent survival, proliferation, differentiation and migration of mouse and human mesenchymal stem cells (MSCs). The expression is observed in all stages of interphase with exceptionally high expression in G2/M phase. Increased survivin expression protects MSCs that form HSCs supportive niche depleted post irradiation exposure [37]. The protein has been linked to survival in these cells subjected to stress induced by heat shock, UV or etoposide as well [38]. CD44+/CD105+ human amniotic fluid MSCs, that proliferate rapidly and are ideal for stem cell transplantation, also express high levels of survivin [39]. The involvement of hematopoietic adipocyte-derived stem cells (ASCs) in apoptotic resistance displayed by adipocyte tissue in obese subjects was correlated to upregulated survivin expression [40]. Keratinocyte Stem Cells (KSCs) show high expression of survivin, that plays a prominent role in preventing abnormal mitosis in these cells [41, 42]. The suitability of the IAP as a marker for KSCs, which helps in maintaining skin homeostasis has been explored [43]. The role of survivin in the regulation of cell physiology has also been demonstrated in neuronal stem cells and intestinal stem cells [44, 45].
HES expresses S-ΔEx3, S-2b, S-2α, S-3b along with WT in different subcellular locations. The expression is consistently high in HES in comparison to differentiated cell types like MSCs. S-ΔEx3 shows the highest expression levels followed by S-2b. The other two are expressed in minimal quantities [36]. S-3γ and S-3γV isoforms with expression higher than WT are responsible for imparting growth factor independent growth in HSPCs [46]. KSCs as well express S-ΔEx3 and S-2b at higher levels than S-2α, S-3b [42].
Survivin in Cancer
The overexpression of survivin in majority of carcinomas as well as in the early embryonic stages combined with its relative absence in most normal adult tissues, classifies it as a classic oncofetal gene [47]. A deregulation in the usual expression pattern, resulting in its overexpression throughout the cell cycle is detected in transformed cells and is found to be mediated by oncogenes or tumor suppressor genes, explaining the selective expression of survivin gene in cancers [48, 49]. The survivin pool in mitochondria was found to be involved in cancer [50]. Molecular profiling studies correlated increased survivin expression to aggressive and more invasive tumor behavior, reduced response to drugs, poor prognosis, abbreviated survival and enhanced recurrence [1, 13].
A clear majority of malignancies, including blood, breast, colon, ovarian, lung, liver, uterus, glioblastoma, astrocytoma, meningioma, bladder, prostate, gastrointestinal, non-melanoma skin cancer, melanoma, soft tissue sarcoma and with viral induced neoplasms show overexpression of survivin at different stages in tumor development [51–54]. Overexpression of the protein also acts as a key factor for radio and chemoresistance in various cancers.
Presence of survivin can be detected from body fluids of cancer patients or using survivin antibodies circulating in blood thus acting as an effective diagnostic marker [4]. Detection of survivin from urine of bladder cancer patients, proved to be a simple, yet sensitive diagnostic method for determining new or recurrent bladder cancer [55]. A study to identify survivin positive circulating breast cancer cells in peripheral blood, suggested its presence in more than half of breast cancer patients, while being absent in healthy controls. Similar studies involving survivin levels in serum also showed survivin to be a sensitive diagnostic marker for tumors [56]. Increased expression of the protein in hematological malignancies have suggested the possibility of using this as a common biomarker for detecting these types of diseases [57].
Isoform S-ΔEx3 has been expressed in a wide variety of cancers and is often found in association with malignant cancers and is often reported to be indicative of tumor grade and invasiveness. Studies conducted on S-2b report some contradictory results on the expression of the isoform. Some reports suggested the expression to be positively correlated with adverse clinical outcome while a few suggested an inverse correlation. In cancers the expression was reported to be more in benign or early stage tumors. Certain cases of breast cancer reported S-2α, S-3α and S-3b expression along with WT, S-ΔEx3 and S-2b. S-2α expression has also been observed in medulloblastoma. The expression of ΔptEx2/3 and ΔptEx1/2 are observed in acute myeloid leukemia (AML). S-3α and S-ΔEx3 have potentiality as diagnostic markers for breast and papillary thyroid cancers respectively [23, 30, 58].
Survivin in Cancer Stem Cells
Cancer stem cells (CSCs) within cancers self-renew, differentiate, accumulate mutations and extensively proliferate, regenerating tumors consisting of both tumorigenic and non-tumorigenic cells, thus sustaining cancer [59]. These cells disregard the normal rules of cell division and the pathways that regulate them, resist the intrinsic and extrinsic apoptotic pathways and improve metastasis. They are believed to be responsible for the relapse and recurrence of cancers and resistance to therapies [60].
To study the mechanisms underlying teratoma formation in HES Blum et al. analyzed the transcriptome of undifferentiated HES cells, teratoma and mature embryoid bodies that showed the upregulation of survivin in HES and teratoma. They reported it to be the strongest candidate gene and also suggested that the steady expression of survivin leads to teratoma formation by increasing apoptotic resistance [47].
Survivin upregulation has been reported to be responsible for the development of hematological malignancies and tumor formation. There is an increased risk of hematologic malignancies in transgenic mice overexpressing survivin due to increased protection from cell death [61]. Increased expression of the protein has been reported in CD34+38− AML stem/progenitor cells in comparison to bulk blasts and CD34− AML cells. The expression levels were higher in bone marrow samples in comparison to peripheral blood and the overexpression is often linked to expression of other proteins participating in proliferation and survival [62]. Fukuda et.al. reported the regulation of 137 gene by survivin in leukaemia stem cells (LSCs) of which 124 genes were exclusively expressed in LSCs and not in normal HSCs. Genes responsible for wide range of biological functions are regulated by the IAP and these are connected through epidermal growth factor receptor (EGFR) signalling network, suggesting the therapeutic potential of the IAP [63].
Upregulation has been observed in well characterized patient derived glioblastoma stem cell (GSC) lines on comparing the mRNA expression between GSC and non-GSC. The elevated expression of the IAP was observed in recurrent glioblastomas suggesting the possibility of the protein to contribute to therapy resistance in GSCs and its prognostic value in predicting postsurgical survival [64]. Similarly the analysis of gene mRNA expression of CD133+ GSCs, reported higher expression in comparison to CD133− cells. This indicates their role in modulation of cell cycle progression, cell division, signaling pathways and increased chemoresistance [65].
Flowcytometric analysis of colorectal CSCs (CR-CSC) revealed differential expression of survivin under different growth conditions. Sphere cells showed lower survivin levels than differentiated cells in adherence conditions, implying the slow growth rate of CSCs [66]. Li et al. observed elevated levels of survivin in CD133+ CR-CSCs than in small interfering ribonucleic acid (siRNA)-induced CD133− cells [67]. Similar elevations in levels have been reported in studies conducted in HCT-116 cell line, used as a model for colorectal cancer initiating cells with stem-like cells properties [68]. Transcriptome analysis of CD133+ CR-CSCs showed survivin regulated by Interleukin (IL)-4 to have prognostic value in tumor recurrence and patient survival [69]. Coexpression of Survivin and CD44+, in combination with epithelial mesenchymal transition (EMT) markers were indicative of recurrence and aggressive tumor behavior in rectal cancer patients subjected to preoperative radio and chemotherapy [70].
The significance of the expression of this protein in relation to EMT and metastasis has been studied using quiescent breast CSCs in vitro. Increased expression observed in the intermediate pre-metastatic phase helps in inhibiting apoptosis and surviving, under unfavorable and noxious conditions. After the cells attain mesenchymal phenotype and get adapted to the environment, the expression levels showed a significant decrease, thus giving sufficient evidence for the involvement of survivin in tumor development and progression and its potential as a good CSC marker [71].
Studies conducted by Yie S.M et al. reported the presence of survivin positive circulating cancer cells (CCCs) in peripheral blood samples of about 50% of patients with breast, gastric, lung, colorectal and esophageal squamous cell carcinomas, along with their association with various clinicopathological parameters like degree of tumor infiltration, nodal status and disease stages. These studies suggested the applicability of the protein in predicting metastasis and relapse, thus the possibility of using it as an ideal marker for migrating CSCs [56, 72, 73].
Increased survivin expression in association with increase in stemness has also been reported in prostate CSCs. Survivin was co-expressed with one of this transcriptional regulator Runt-related transcription factor 2 (Runx2) and both showed positive correlation with tumor growth in these CSCs [74]. The upregulated expression of stem cell marker, sex determining region Y box 2 (Sox 2) and survivin resulting in inhibited apoptotic pathway and drug resistance, was related to the presence of CSCs in salivary adenoid cystic carcinoma [75]. Radiation resistant CD44+/CD24+ cervical cancers also expressed survivin along with various other stem cell markers and drug resistant proteins [76].
Regulation of Survivin Expression in Cancers
Various signaling pathways and associated molecules play a key role in the positive and negative regulation of survivin. At transcriptional level the cell cycle-dependent element/ cell cycle genes homology region (CDE/CHE) elements in the promoter region control cell cycle dependent gene transcriptions. The survivin promoter has multiple sites for binding pro-oncogenic transcription factors including those that may be responsible for its differential expression in normal and cancer tissues, in combination with the multitude of tumorigenic pathways and signaling molecules [77].
Phosphoinositide-3 Kinase (PI3K)/Akt pathway is initiated on the activation of tyrosine kinase receptor that leads to activation of PI3K which then activates Akt molecule instigating cell proliferation and survival. Akt down regulates FOXO (Forkhead box O) subfamily FOXO1 and FOXO3a, that otherwise binds to and mediate acute silencing of survivin through the dual specificity phosphatase and tensin homolog (PTEN) [78]. VEGF, angiopoietin-1 (Ang-1), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-ĸB), cyclooxygenase-2 (COX-2), granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR), EGFR, human epidermal growth factor receptor 2 (HER 2) and 3 (HER 3) upregulate survivin through PI3K/Akt mediated pathway [79]. The stimulation of insulin like growth factor-1 (IGF-1) leads to elevated survivin expression with contributions from PI3K/Akt/mTOR (mammalian target of rapamycin) pathways or by inactivating transforming growth factor-β (TGF-β), a negative regulator of the protein [80, 81]. E2F family of transcription factors are involved with cyclin dependent kinases (CDKs) in cell cycle. E2F activators (E2F1, E2F2 and E2F3) bind to survivin promotor and cause transcriptional activation of the gene, mediated by CDE/CHR mechanisms, while E2F4 and E2F5 down regulate survivin [82, 83]. Downregulation of CDK4/Cyclin D complex by survivin allows hepatoma cells to evade apoptotic signaling due to the activation of CDK2/Cyclin E complex and release of p21 [84]. In HSPCs apoptosis inhibition by survivin is p21 dependent and cell cycle regulation is p21 independent [85].
Wild type-p53 tumor suppressor gene negatively regulates survivin at both mRNA and protein levels, thereby affecting p53 mediated apoptotic pathway [86]. p53 mediated survivin repression can be attributed to either modifications in the chromatin within survivin promoter or the elevation of p21 that results in the hypophosphorylation of retinoblastoma protein (pRB) family and transcriptional suppression by E2F family member [87]. It was found that p53 and RB, along with E2F2 that acts downstream of RB pathway, lead to negative regulation of survivin expression [82]. Mouse double minute 2 (MDM2), downstream of Akt, also downregulates p53, leading to survivin upregulation [88, 89]. Survivin along with p53, mitogen-activated protein kinase (MAPK) and c-Myc pathways play a role in the regulation of adult MSCs and thus help in maintaining the homeostasis of ASCs [90].
The binding of Wnt (Wingless/Integrated) to Frizzled receptor leads to adenomatous polyposis coli gene/ glycogen synthase kinase 3beta (GSK-3β) activation leading to build up of β-catenin that activates Wnt/β-catenin pathway. β-catenin/T cell factor transcriptional activator increases survivin levels aiding in inhibition of apoptosis and enhancing proliferation [91, 92]. The interaction between survivin and Wnt/β-catenin pathway aids in maintaining the pluripotent state of HES [20]. The improved efficiency of one factor (1F) octamer-binding transcription factor-4 (Oct-4) reprogramming of human neural progenitors to iPSCs can also be attributed to the Survivin/β-catenin interaction [93]. Hypoxia Inducible Factor (HIF-1α) directly binds to survivin promotor and shows a positive correlation specially under hypoxic conditions, while aiding in migration, survival and metastasis [94, 95].
Janus kinase (JAK)/ signal transducer and activator of transcription 3 (Stat3) pathway plays significant roles in cancer progression, migration, apoptosis and immunity. Stat3 oncogene downstream of JAK modulates transcriptional activation and repression of survivin in lymphoma, while the protein directly binds to survivin promotor and inhibit apoptosis in breast cancer cells [96]. The binding of IL-6 or IL-11 to the respective receptors or Bcr-abl activation initiates JAK2 in the cytosol that increases the DNA binding of Stat3 there by activating survivin[97]. Bcr-abl upregulates Survivin and the interaction has significant effect on telomerase activity in cells with high c-abl kinase activity [98, 99]. MAPK/extracellular-signal-regulated kinase (ERK) signaling cascade comprising of retrovirus-associated DNA sequences (RAS), rapidly accelerated fibrosarcoma (RAF), Mitogen-activated protein kinase (MEK) and ERK is another pathway leading to survivin gene regulation. Ras-signaling activates MAPK, mTOR and Akt pathways that increase the transcriptional activation of survivin [100]. Ras also plays significant role in the modulation of survivin expression in Ba/F3 hematopoietic cell line helping in their growth factor independent survival and proliferation [101].
Transforming growth factor β (TGF- β) transcriptionally downregulates survivin expression in association with SMADs 2 and 3 and CDE/CHR elements on survivin promoter or through inactivation of RB pocket. Notch signaling pathway induces cell differentiation or maintains cells in an undifferentiated state and aids in stemness in cells. It positively regulates survivin in association with HIF-α and Jagged-1 ligand (RPB-jĸ) in lung cancers [102]. Notch signaling with the help of c-Myc positively regulates survivin thus inducing T lymphocyte differentiation from HSCs [103].
There exists a functional link between survivin and Wee-1 kinase, that prevents the caspase 3 mediated degradation of Wee-1 kinase. This interaction enhances p34Cdc2 phosphorylation, leading to anti-apoptotic activity and improved cell survival in HSCs [104]. Prostaglandin E2 helps in survivin upregulation along with C-X-C motif chemokine receptor 4 (CXCR4) and stromal cell-derived factor (SDF 1), thus enhancing HSC survival and homing [105]. CXCR4/SDF-1 signaling in bone marrow stromal cell niche with crucial involvement of survivin, maintains cell function during hemostasis and promotes hematopoietic recovery [106]. Transcription factor ecotropic virus integration site 1 protein homolog (Evi-1) and its downstream targets Gata2, Pre-B cell leukemia transcription factor 1 (Pbx1) and Spalt Like Transcription Factor 2 (Sall2) are also correlated with survivin expression and associated functions in HSCs [107].
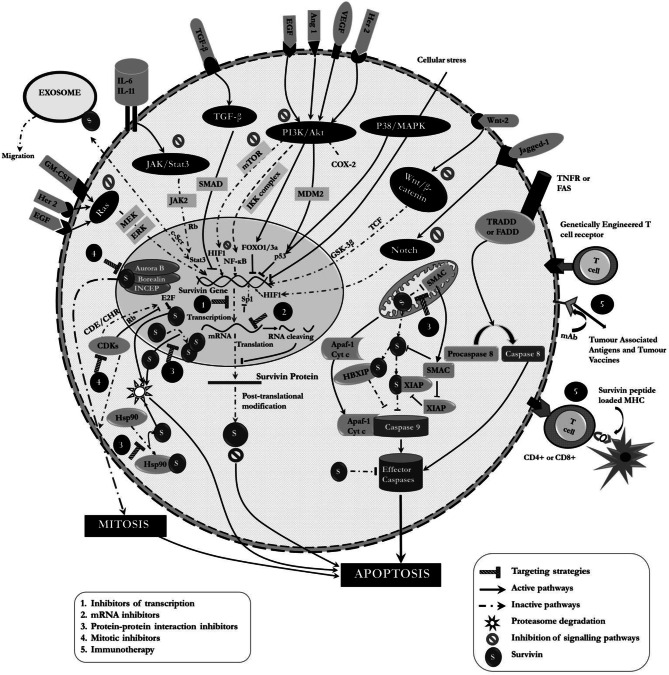
Survivin has also been implicated in CSC signaling paradigms. It is a major downstream target of the above mentioned pathways along with other genes like Sox-2, Oct-4, c-Myc, Nanog, cyclin D1 etc. [108]. Oct-4 with the help of Stat3 and survivin was significant in the survival of drug resistant CR-CSCs [109]. It is associated with unfavorable prognosis in leukemia stem/progenitor cells and modulation of sonic hedgehog (SHh) and TGF-β pathways, thereby promoting pancreatic and liver CSC growth respectively. It is involved in Stat 3 signaling favoring CSC progression in the niche [110]. Internal tandem duplication-Flt3 tyrosine kinase (ITD-Flt3) upstream of survivin helps in its regulation in a PI3K/Akt dependent manner in AML [111]. Survivin regulation via β-catenin was found to be responsible for radioresistance in PTEN(−) CSC-like cells of nasopharyngeal carcinoma [112]. Kruppel-like factor 5 (KLF5) is also capable of inducing survivin expression by binding to p53 and preventing p53 mediated down regulation of the protein acute lymphoblastic leukemia (ALL), colorectal and ovarian CSCs [77, 113]. Wnt/ β-catenin pathway regulates survivin with accompanied modulation of Cyclin-D1 and c-Myc in liver CSCs [114]. Transient receptor potential cation channel, subfamily M, member 7 (TRPM7) is indirectly involved in survivin regulation in CSCs through Notch and JAK2/Stat3 signaling pathways [115]. Figure 2 illustrates the various cellular networks that involve survivin.
Fig. 2.
Involvement of survivin in various cellular networks and associated current therapeutic interventions
Survivin Targeting Therapeutics
The inhibition of survivin expression can lead to sensitization of tumors to chemical and physical agents as this can inhibit tumor proliferation through a spontaneous increase in the rate of apoptosis. Various small molecule inhibitors, ribozymes, gene editing techniques, antisense oligonucleotides, cancer vaccines etc. have been developed with an aim to achieve the same (Fig. 2, Table 1).
Table 1.
Survivin therapeutics- Drug targets and mechanism of action in various cancers
| Drug | Target | Mechanism of action | Cancers |
|---|---|---|---|
| Inhibitors of Survivin Transcription | |||
| FL118 | Survivin Promotor | Multiple mechanisms leading to transcriptional inhibition | Head and Neck, Cervical, Ovarian, Pancreatic, Prostate, Leukaemia, Lung and CSCs [116, 117, 235, 236] |
| YM155 | SP1 binding site of Survivin Promotor | DNA damage and S phase arrest | Leukaemia, Neuroblastoma, Gastric, Pancreatic, Liver, Prostate, NSCLC [119] |
| Tolfenamic Acid | SP1 and SP3 | Loss of DNA binding specificity leading to transcriptional repression | Pancreatic, Ovarian and Ewing Sarcoma [122, 123] |
| Teremprocol | SP1 and cdc2 | Transcriptional Repression | Lung, Colorectal, Breast, Prostate, Liver [124, 125] |
| Mithramycin A | SP1 and SP3 | Loss of DNA binding specificity of SP1 | Ewing Sarcoma, LSCs [126, 127, 225] |
| Protein-Protein interaction inhibitors | |||
| Survivin-Survivin interaction inhibitors | |||
|
Abbot 8 LQZ7 LQZ7F |
Survivin | Binds to survivin dimer interface | Cervical, Pancreatic [128, 129] |
| Survivin-Smac interaction inhibitors | |||
|
Piperine Withanone UC112 4 g 10f |
Survivin | Bind to Survivin at Smac binding region, increases survivin expression | Melanoma, Pancreas, Breast and Colorectal [130–133] |
| Survivin-Hsp90 interaction inhibitors | |||
|
Shepherdin AICAR 17AAG NVP-922 |
HSP90/ Survivin complex | Disruption of survivin Hsp90 interaction and proteasomal degradation | Prostate, Cervical, Melanoma, Colorectal, Chronic myelogenous Leukaemia [133–140] |
| Mitotic Inhibitor | |||
| Indinavir | Survivin | Inhibit Aurora B phosphorylation and CPC complex formation | Breast [141] |
| S12 | Microtubule dynamics | Medulloblastoma, Cervical, Osteosarcoma, Breast [141] | |
| LLP3 | Affect survivin-Ran stability, and spindle formation | Colorectal, Cervical, GSC [64] | |
| Flavopiridol | CDKs | Hyperphosphorylation at 34th position, mitotic block | Ovarian, Breast, Cervical [142] |
| Purvalanol A | P34 cdc2 | Gastric, Ovarian [143] | |
| NU6140 | CDK2 | Transcriptional inhibition and caspase activation | Cervical, Ovarian [144] |
|
Cephalochro min |
CDK2/ Cyclin -E CDK4/ Cyclin-D1 |
Mitochondrial disruption and cell cycle arrest | Lung [145] |
| Inhibitors of Signalling molecules | |||
| Prodigiosin | P53 | Caspase activation | Breast, Liver, Colorectal, Leukaemia, CSCs [146] |
| Omega 3 fatty acids | Colon, Acute Myeloid Leukaemia, CSCs [147, 148, 232] | ||
| Nutlin 3a |
Integrin-α5β1 MDM2 |
P53 and mTOR dependent downregulation | Glioblastoma [149, 150] |
| RG7388 | MDM2 | Neuroblastoma [149] | |
| Rapamycin | mTOR | Binds to FKBP12 | Glioblastoma, Leukaemia, Lymphoma, Multiplemyeloma [151] |
| MK2206 | Akt | Inhibit Survivin-XIAP complex | Glioblastoma [152] |
| SH5 | Akt | Inhibit phosphatidyl inositol binding domain and Akti-X | Chronic myeloid leukaemia, lung and Prostate [153, 154] |
| TG101209 | JAK2 | – | CML, Lung [163, 164] |
| Gefitinib | EGFR | Dephosphorylation of EGFR | Lung, Breast [157] |
| Lapatinib | EGFR and HER-2 | Upregulation of BMI and attenuation of ErbB1 and ErbB2 phosphorylation | Pancreatic, Breast and Ovarian [155, 156] |
| Trastuzumab | ErbB2 | Transcriptional inactivation thorough β-Catenin/TCF | Breast [214] |
| Wnt-2Ab | Wnt | Lung [215] | |
| PD153035 | MAPK/ EGFR | H4 acetylation via TSA activation | Breast, Ovarian, Pancreatic, Lung, Colorectal and Blood [158–160] |
| AG1478 | |||
| PD98059 | |||
| CI1040 | MEK | Blocking phosphorylation of ERK | Leukaemia, Breast, Keratinocytes [161] |
| U0126 | |||
| Bay-11-7057 | IKK | Degradation of IĸB and inhibition of NF-ĸB | Pancreatic and Endothelial [182] |
| SD1029 | JAK2 | Combined inhibition of IAPs | Breast, Ovarian [168] |
|
Panepoxyd- one |
NF-ĸB | Regulation of IAPs | Breast [162] |
| Limonoids | Increase caspase 2 activity | Cervical [172] | |
| Arctigenin | Stat3 | Phosphorylation of Stat3, | NSCLC, Ovarian [165, 166] |
| Resveratrol | – | Cervical [169] | |
| Tambjamine | JAK2 and IL-6 | Stat3 phosphorylation | Lung [167] |
| GDP366 | Survivin | Chromosomal Instability, Senescence | Leukaemia, Colon, Cervical [183] |
| Curcumin |
NF-ĸB, Stat3, Akt |
Multiple mechanisms | Breast, Ovarian, Liver cholangiocarcinoma [170, 243] |
|
Etodolac Celecoxib |
Cox2 | PI3K/Akt suppression | Glioblastoma, Colon, Liver [173–175] |
| AKAP145 | TGF-β | Dissociates Survivin/XIAP complex | Colon [184] |
|
Metformin Phenformin |
Affects Stat3, TGF-β and SMAD | Rectal [185, 226] | |
| Salinomycin | Survivin downregulation | Renal cancer and CSCs [222] | |
| IGC-001 | β-catenin | Transcription inhibition and caspase activation | Colon [179], CSCs [237–239] |
| Emodin | Wnt /β-Catenin signalling regulation | Pancreatic [176, 177] | |
| WM-127 | Wnt |
Supress β-catenin /TCF signalling induces Bax expression |
Liver [178] |
|
Xanthohumol Bulbine Futescens |
Notch | Survivin suppression and affects stemness | Breast, Liver, Ovarian [180, 181] |
| SNS-032 | CDK7/9 | Downregulation of Survivin | Uveal melanoma [219] |
| Sabutoclax, | IL-6/Stat3 | Breast CSCs [229] | |
| CEP-1347 | P38/MAPK | Ovarian CSCs [234] | |
| Brexpiprazol | – | Reduction of Survivin | Pancreas, Lung, Brain CSCs [240] |
| AS602801 | – | Combined downregulation of survivin and MDR1 | Ovarian CSCs [248, 249] |
| Cardamonin | Stat 3 | Survivin downregulation | GSCs [244] |
| Apigenin | PI3K/Akt | p53 and Apaf1 upregulation | Pancreatic CSCs[242] |
| PD173074 | FGFR1 | FGFR1/Scr/NF-ĸB axis | Pancreatic ductal adenocarcinoma CSCs [228] |
| mRNA Inhibitors | |||
| LY2181301 (AO) | 3′ untranslated region | Target mRNA cleaving, caspase activation | Lymphoma, Leukaemia, Melanoma, Breast [187] |
| SPC3042 (AO) | Exon 4 | Target mRNA cleaving | Prostate, Lung [187, 189] |
| Oligo4003 (AO) | 232–251 nucleotides | Nuclear condensation, Apoptosome formation | Lung [190] |
| siRNA (RNAi) | Binding to RISC to Target RNA | Target RNA cleaving | Pancreatic, Breast, Colorectal, Glioblastomas [183, 233, 234] |
| miRNA (RNAi) | 3’-UTR | Target RNA cleaving | Prostate, Pancreatic, Lung, Gastric, Breast, Liver, Bladder, CSCs [194–198, 253–256] |
| Ribozymes | GUC(294) or CUA(110) of 3’ end | Hydrolysis of phosphodiester bond | Melanoma, Prostate [201, 202] |
Small Molecule Inhibitors
Inhibitors of Survivin Transcription
FL118, inhibits survivin and shows anti-tumor activity in p53 independent manner even at very low doses. It is known to possess excellent anti-tumor activity against colon, prostate, head and neck, ovarian and pancreatic cancers and in multiple myeloma in comparison to the already existing chemotherapeutics [116, 117]. It is also capable of selectively and independently inhibiting other anti-apoptotic genes including Mcl-1, XIAP and cIAP2 and has a favorable toxicity profile [118]. YM155 was identified as the first specific inhibitor of survivin that suppressed the activity of survivin promotor and showed promising results in human cancer cell lines and human hormone-refractory prostate tumor xenografts [119]. Even in phase I trials it was well tolerated and was effective against blood cancers to some extent. Surprisingly, in phase II trials on patients with refractory and advanced non-small-cell-lung-carcinoma (NSCLC) it was only as effective as other second-line agents [120]. It turned out that the suppression of survivin by YM155 was a secondary event and primarily it acts as a DNA damaging agent. Survivin repression was probably a result of reduced transcription [121].
Tolfenamic acid (TA) belonging to Nonsteroidal anti-inflammatory drugs (NSAIDs) causes post translational attenuation of SP1 and Sp3, which in turn results in the downregulation of survivin. TA in combination with Cisplatin has also been shown to reduce survivin levels in pancreatic and ovarian cancers [122, 123]. Copper II loaded complex of TA was found to be even more efficient in reducing survivin levels in pancreatic cancers[123]. Plant based Terameprocol which is in phase II studies, inhibits survivin expression in combination with cdc2 in cervical intraepithelial neoplasia. Their use has also shown positive effects on NSCLC, breast, colorectal, prostate, liver and erythroleukemia [124, 125]. Anti-neoplastic antibiotic Mithramycin A induces a similar effect and reduces cell viability in Ewing sarcoma and colon cancer [126, 127].
Inhibitors of Protein-Protein Interaction
Abbott-8, a set of small molecules developed computationally and identified using NMR, binds to survivin dimer interface inhibiting the symmetric interaction between two Survivin molecules. Inhibition of dimerization results in the exposure of hydrophobic interface of the protein resulting in conformational changes, causing selective degradation of survivin [79, 128]. In silco screening for compounds targeting the critical core residues for Survivin led to the identification of LQZ7 and its analogue LQZ7F, both of which could induce proteasome dependent degradation of Survivin. LQZ7F is less cytotoxic in comparison and has the potential to cause mitotic arrest, induce apoptosis and also inhibit tumor growth in mice [129].
Some compounds bind to survivin at Smac binding region, preventing the binding of Smac/DIABLO, making it available for inhibiting IAPs. Piperine, an alkaloid isolated from black pepper, works by this mechanism in colon and breast cancer and also in breast stem cells increasing bioavailability of drugs and decreasing drug metabolism [130, 131]. Leaf extract of Ashwagandha plant, Withanone, mimics Smac and inhibits survivin in cancers [132]. UC112 and its analogues 4 g and 10f, also compete with Smac to selectively downregulate Survivin through ubiquitin mediated pathway in melanoma, pancreatic and prostate cancers with low toxicity [133].
The disruption of the interaction between survivin and heat shock protein 90 (Hsp-90) destabilizes survivin, lowers antiapoptotic threshold and suppresses cell proliferation [134, 135]. Shepherdin, a peptidomimetic antagonist of Hsp90, leads to proteasomal degradation of survivin [136, 137]. AICAR destabilizes Survivin along with other HSP90 client proteins in tumors [133, 138]. A geldanamycin, 17AAG and a non-geldanamycin NYP-922 are two other Hsp90 inhibitors found to regulate survivin in myeloid leukemia, thyroid, lung, nasopharyngeal, cervical and colorectal cancers [139, 140].
Inhibitors of Mitosis
The in vitro studies on breast cancer cell lines showed the ability of an HIV treatment molecule Indinavir to cause mitotic arrest and apoptosis. They act by inhibiting chromosomal passenger complex (CPC) formation by phosphorylation of Aurora B kinase and by inhibiting survivin-XIAP binding. Another molecule S12 binds to survivin altering microtubule dynamics by specifically targeting mitotic check point and leads to cell death without being cytotoxic [141]. Computationally developed LLP3, disrupts the formation of Survivin-Ran complex, affecting microtubule stability and spindle formation in tumors [64].
Cyclin dependent kinase inhibitors like flavopiridol or purvalanol act by causing hypophosphorylation of survivin resulting in the loss of its function. This leads to inhibition of taxol-mediated mitotic block, activation of intrinsic and p53 independent apoptosis. Flavopiridol is in phase II clinical trial [142]. Sequence specific inhibition of CDK-1 with Taxol-Purvalanol A showed sequence and drug dependent pro-apoptotic efficacy that resulted from reduction in survivin expression [143]. A novel inhibitor Nu6140 suppresses survivin expression resulting in cell cycle arrest and increased apoptosis [144]. Cephalochromin, an isolate obtained from fermented fungi, has the potential to downregulate survivin, disrupt mitochondria and bring about cell cycle arrest in lung cancer cells [145].
Inhibitors Targeting Signaling Pathways
A secondary metabolite Prodigiosin and Omega-3 fatty acids (docosahexaenoic and eicosapentaenoic acid), downregulate survivin expression through p53 dependent pathways in ALL, AML, breast, liver and colorectal cancers [146–148]. MDM2 inhibitors, Nutlin 3a and RG7388, alone and in combination, suppress survivin at mRNA and protein levels, by activation of p53 and inhibition of mTOR pathways in neuroblastoma [149]. Inhibition of integrin-α5β1 by Nutlin 3a also results in induction of apoptosis in glioblastoma cell lines due to the proportional relation between survivin and integrin-α5 [150]. Rapamycin, a macrolide antibiotic, indirectly reduces survivin by inhibiting mTOR in glioblastoma, leukemia, lymphoma and multiple myeloma [151].
An allosteric kinase inhibitor of Akt, MK-2206 induces cell cycle arrest and apoptosis by preventing survivin/XIAP complex formation [152]. Similarly Akt specific SH5, also reduces survivin expression along with the significant reduction in other IAPs in chronic myeloid leukemia (CML), lung and prostate cancer [153, 154]. A dual kinase inhibitor, Lapatinib inhibits survivin in pancreatic, breast and ovarian cancers through the down regulation of ErbB1 and ErbB2 phosphorylation or upregulation in BMI1 expression [133, 155, 156]. EGFR inhibitors (Gefitinib, PD153035 and AG1478), MAPK antagonist (PD98059) and PI3K inhibitor (LY294002) have shown their potential to inhibit survivin in breast, lung, pancreatic, colon and ovarian cancer cell lines [157–160]. Specific MEK inhibitors CI1040 and U0126 were found to attenuate survivin levels in leukemia, with considerable reduction in cell viability through MAPK/ERK and PI3K/Akt pathways [161].
Panepoxydone isolated from edible mushroom causes combined downregulation of many IAPs in breast cancer cells without any cytotoxicity [162]. JAK2 inhibitor TG101209, suppresses survivin expression at nanomolar concentrations increasing susceptibility to chemotherapy in CML and radiotherapy in lung cancer [163, 164]. Arctigenin, isolated from the seeds of burdock and indole based Tambjamine, prevents Stat3 phosphorylation [165–167]. SD-1029 inhibits anti-apoptotic proteins downstream of JAK2 and increases sensitivity to paclitaxel in breast and ovarian cancer [168]. Resveratrol downregulates survivin and induces TRAIL based therapy in cervical cancers [169].
Curcumin, a broad spectrum anticancer drug was found to downregulate NF-ĸB, Akt and Stat3 by directly or indirectly reducing levels of survivin, thus inducing both intrinsic and extrinsic apoptosis [170, 171]. The neem limonoids, azadirachtin and nimbolide, reduce survivin levels through NF-ĸB in human cervical cancers [170, 172]. Etodolac and celecoxib, that selectively inhibit COX-2, are found to bring about significant survivin reduction in gliomas, colon and liver cancers thus improving efficacy of chemotherapy drugs [173–175]. Plant based compounds like WM-127, physodic acid, caperatic acid and Emodin induce Wnt mediated survivin reduction in cancers of liver colon and pancreas [176–178]. IGC-001, a β-Cantenin/ T Cell Factor mediated transcription antagonist, downregulates survivin expression leading to selective caspase activation in tumor cells [179]. Silencing Notch, using peptide antibiotic echinomycin, natural phytochemicals Xanthohumol and Bulbine frutescens, silences survivin in breast, ovary, liver and cholangiocarcinoma and has significant effects on stemness [180, 181]. Other small molecules including Bay-11-7057, AKAP145, Metformin, Phenformin, GDP366, etc. have also been found to be useful survivin inhibitors [182–186].
mRNA Inhibitors
Antisense Oligonucleotides (AO)
Ely Lilly & Co. produced the first AO, LY2181308, a second-generation oligonucleotide, that binds to 3’-UTR region of survivin transcript, leading to destruction of survivin RNA by RNAse H [187]. This AO has been discontinued after phase II trials due to disappointing results [188]. Another AO that has reached clinical trials is SPC3042 (EZN-3042), targeting exon 4 of the survivin transcript. They act by initiating cell cycle arrest at the short window between metaphase and anaphase and activating caspases 3 and 7 [187, 189]. Oligonucleotide 4003, targets the mRNA (232–251 nucleotides) of survivin. This results in nuclear condensation and DNA fragmentation along with caspase activation [190]. It was withdrawn after phase I trials due to intolerable dose-limiting toxicity levels when in combination with chemotherapeutic drugs [191].
RNA Interference (RNAi)
Silencing of survivin in human androgen independent prostate cancer using RNAi reduced the proliferative potential and elevated the rate of apoptosis [192]. siRNA directed against survivin in neuroblastoma cell lines completely suppressed the expression at both mRNA and protein levels [193]. Micro RNAs (miRNAs), including miR-203, miR-34a, miR-218, miR-138p, miR-485-p, miR-542-3p and miR-214-3p, delivered to malignant cells using viral or non-viral based delivery systems, bind to survivin and induce gene silencing. miR-203 is capable of direct targeting in prostate, laryngeal and pancreatic cancers and inhibiting proliferation [194, 195]. Overexpression of miR-34a in head and neck squamous cell carcinoma, NSCLC, laryngeal squamous cell carcinoma and gastric cancer caused survivin downregulation [196, 197]. miR-214-3p tetrahedral framework nucleic acids complex, reduces survivin expression in lung cancer inducing apoptosis [198]. Survivin repression by other miRNAs resulted in reduction in proliferation, invasion and reversal of chemosensitivity in cancers of bladder, liver and breast [199, 200].
Ribozymes
Small RNA molecules with endonucleolytic activity that cleaves target RNA sequence by catalyzing hydrolysis of phosphodiester bonds are regarded as ribozymes. They can act as transcriptional inhibitors of survivin activity [190]. Studies on human melanoma cell lines transfected with hammerhead ribozymes like CUA110 (RZ7) and GUC294 (RZ1), result in reduction of survivin expression that contributes to cell death and sensitivity to drugs and radiation. Attenuation of survivin expression with ribozymes improves susceptibility of melanoma cells to anti-tumor activity of cisplatin and topotecan triggering apoptosis [201, 202].
Immunotherapy
Survivin Derived Peptides
Survivin stimulates cytolytic T cell response, when corresponding peptides are loaded to Major Histocompatibility Complex (MHC). The MHC-peptide complex when presented to CD 4+ or CD8+ T cell through dendritic cells (DC) or other antigen presenting cells (APC), causes subsequent lysis of the human leukocyte class I or II antigen (HLA) matched target cancer cell [49, 203–205]. Autologous DC pulsed with survivin derived epitopes has been able to instigate CD4+ and CD8+ T-cell responses in patients with ALL reducing the chances of relapse [206]. Recombinant melanoma-associated antigen and recombinant survivin when used in combination was found to be effective for lung cancer [207]. Similarly survivin derived peptides were useful immunotherapeutic targets in prostate carcinoma, chronic lymphatic leukemia and malignant melanoma [208]. Adoptive T cell therapy that identifies survivin specific clone and discriminate cancerous and non-cancerous cell has been identified to be an effective therapeutic method [209]. Combination therapy using adoptive T cell transfer and tumor antigen vaccine, derived from peptides of human telomerase reverse transcriptase and survivin, after autologous stem cell transplantation, produced enhanced cellular and humoral host antitumor immunity [210].
Cancer Vaccines
The possibility of utilizing survivin based vaccines as a potential immunotherapeutic agent in colorectal, pancreatic and lung cancer, renal cell carcinoma and neuroblastoma has been reported [125, 211]. Survivin DPX-Survivac, a depot-based cancer vaccine is derived from survivin and contains multiple CD8 epitopes with significant HLA applicability. This vaccine in combination with metronomic cyclophosphamide in a low oral dose, was able to generate polyfunctional antigen specific immune response in ovarian cancer patients [212]. Subunit vaccine formulation containing survivin and costimulatory SA-4-1BBL has been found to eradicate lung carcinoma, with increased efficiency when given in a prime-boost dose. Combination of peptide vaccines of survivin and indoleamine 2,3-dioxygenase with temozolomide was able to initiate specific immunity in glioma patients [213]. Many of the survivin based vaccines have entered Phase I or II of clinical trials and are reported to be relatively safe, with improved specificity for tumor cells.
Monoclonal Antibodies
The potential of monoclonal antibodies to target survivin has also been reported in several cancers. They are HLA independent and have more immediate action in comparison to cancer vaccines [14]. Trastuzumab targets ErbB2 and prevents survivin transcription through the binding of β-Cantenin/T cell factor to survivin promotor in breast cancer [214]. You et.al. developed a monoclonal antibody Wnt-2 Ab against Wnt protein that attenuates survivin through the suppression of the same pathway in lung cancers [215].
Therapeutic Strategies Targeting Survivin in CSCs
A multitude of strategies have been developed and explored to target CSCs from bulk tumors. Targeting the factors helping in self-renewal, pathways promoting stemness in CSCs and pathways helping in CSC proliferation are approaches capable of reducing the frequency of CSCs considerably [216, 217]. A form of immunotherapy called oncolytic virotherapy, using competent replicating viruses, kills CSCs by hijacking the cell death machinery, inducing anti-tumor immunity, destroying tumor vasculature or using their toxic proteins to kill the cells directly [218]. Use of mRNA inhibitors and targeted drug delivery using nanoparticles are also extremely promising approaches. The most well established one is combination therapy utilizing the synergistic effect of survivin inhibitor and standard treatment modalities in eliminating these cells. Other modalities including the targeting of DNA repair pathways, CSC niche and differentiation therapy have also been explored [216].
Targeting Factors Promoting Stemness and Key Signaling Pathways
Treatment with SNS-032, a selective inhibitor of CDK7/9 showed dramatic decrease in the IAP levels in uveal melanoma and subsequent elimination of CSCs and reduction in liver metastasis [219]. TmSm (T34A), a recombinant protein targeting survivin had prominent effect reducing cell growth and proliferation and inducing apoptosis in breast CSCs. The protein downregulates Cyclin D1 and Rb, and interferes with survivin/β-catenin interaction [220]. Mitotic inhibitor LLP3 was found to be useful in targeting survivin in GSCs by decreasing their proliferation [64]. Aspirin was found to reduce proliferation and invasion in GSCs with downregulation of survivin along with Notch 1, Sox 2 and Stat 3 [221].
Salinomycin downregulates survivin through TGF-β signaling in renal cell carcinomas and at the same time was found to reduce stemness in the CSCs [222]. Aripiprazole, an antipsychotic with excellent safety profile, inhibits survivin probably through Wnt/β-Catenin mediated pathway in CSCs, sensitizing them to standard chemotherapeutic drugs. Similar effect is also produced by another antipsychotic drug, Olanzapine [223, 224]. Mithramycin A was found to be effective in reducing stemness of LSCs with the help of SP1 and c-Myc with significant downregulation of survivin expression as well [225]. Low concentration of Metformin inhibits EMT and chemosensitizes CSCs of breast, pancreas and ovary by downregulating the IAP [226]. Inhibitor of nuclear factor kappa-B kinase subunit beta (IKK β) inhibitor Cmpd4 inhibits IKK β/NF-ĸB signaling pathway with downregulation of survivin, along with stem cell and EMT markers in prostate CSCs [227]. Fibroblast Growth Factor Receptor (FGFR) 1 inhibitor PD173074 suppresses the FGFR1/Scr/NF-ĸB signaling in pancreatic ductal adenocarcinoma CSCs with significant reduction in expression of stem cell markers as well as IAPs [228].
An antagonist of Bcl-2 family proteins, Sabutoclax, downregulates survivin through IL-6/Stat3 signaling pathway, to improve chemosensitivity in breast cancer CSCs [229]. Disulfiram, a drug capable of de-regulating various factors associated with tumorigenesis, induced apoptosis via caspase 3 activation in presence of copper in triple negative breast CSCs. This drug downregulated Stat 3 signaling with subsequent effects of survivin and cyclin D1 leading to CSC elimination [230]. n-3 Polyunsaturated fatty acids induce growth inhibition in drug resistant CR-CSC through survivin downregulation and caspase-3 activation [231]. p53 activators, Prodigiosin and Omega-3 fatty acids were found to be useful survivin suppressors in breast and colon cancer CSCs [147, 232].
Synergistic effect of growth factor Midkine and natural flavonoid quercetin was able to bring about cell cycle arrest and apoptosis in prostate CSCs by downregulating survivin along with p38, ABCG2 and NF-ĸB via PI3K/Akt and MAPK/ERK pathways [233]. A small molecule kinase inhibitor CEP-1347, which has proven anti-tumor and more specifically anti-CSC activity, chemo-sensitizes ovarian CSCSs by downregulation of survivin through c-Jun N-terminal kinase (JNK) and p38/MAPK pathway [234]. FL118 has been used to reduce migration and invasion in NSCLC and breast cancer derived CSCs by downregulating survivin expression and drug resistance inducing proteins, thus increasing the sensitivity to chemotherapeutic treatments [235, 236].
Combination of Small Molecule Inhibitors with Standard Chemo or Radiotherapy
Combination of ICG-001 with standard chemotherapy eliminates CSCs in leukemia, salivary gland squamous cell carcinoma and pancreatic cancers. This Wnt/β-catenin modulator induces forced symmetric differentiation to safely eliminate drug resistant CSCs [237–239]. Synergistic effect of FH535 and sorafenib attenuated survivin levels that subsequently brought about inhibition of liver CSCs via Wnt/β-catenin pathway inhibition [114]. Brexpiprazol, a serotonin-dopamine activity modulator reduces survivin expression along with significant reduction in Sox-2 and sensitizes CSCs of lung, pancreas and brain to chemotherapy [240, 241].
A natural flavonoid apigenin increases sensitivity to cisplatin in prostate CSCs. Its combination with cisplatin decreases the migratory and proliferative potential via PI3K/Akt pathway and induces apoptosis by upregulating caspase-8, Apaf-1 and p53 [242]. Curcumin in low doses upregulates MAPK pathway, downregulate Stat3 signaling and inhibit self-renewal and survival in GSCs in reactive oxygen species dependent mechanism [243]. Cardamonin, a plant based dietary compound, has significant effect on downregulation of Stat3 signaling followed by subsequent downregulation of survivin in CD133+ GSCs, increasing their sensitivity to temozolomide [244]. Bortezomib also increases the sensitivity of GSC’s to temozolomide in a dose and time-dependent manner by downregulating the FOXM1/survivin signaling pathway [245]. Diuretic, spironolactone sensitizes CSCs to Gemcitabine and Osimertinib [246]. Rho-assisted protein kinase inhibitors sensitize pancreatic CSCs to Gemcitabine by downregulating survivin [247].
AS602801 sensitizes ovarian CSCs to carboplatin and paclitaxel through combined reduction in survivin and multi drug resistant protein 1 (MDR 1). This is a candidate drug for targeting tumor initiation and invasion in CSCs [248, 249]. Synergistic effect of suberoylanilide hydroxamic acid with Imatinib in LSC, downregulates co-expressing Bcr/Abl, Survivin, MDR-1 and histone deacetylases, reduces resistance to Imatinib and induces cell death signaling [250].
RNAi Based Treatment Modalities
Aptamer-siRNA chimera has been used to suppress survivin expression in doxorubicin resistant breast CSCs and this combination was capable of targeting both CSCs and the bulk cancer cell population [251]. Elimination of CSCs in CR-CSCs was achieved by aptamer mediated delivery of survivin siRNA in combination with chemotherapeutic agent 5-fluorouracil which otherwise is ineffective in targeting CSCs [252].
miR-136 was found to downregulate survivin in Notch dependent pathway inhibiting cell migration, angiogenesis and chemoresistance in ovarian CSCs and enhance the chemosensitivity to paclitaxel [253]. Survivin is a direct target of miR-203 and an indirect target of miR-29b, hence miR-203/survivin/Bmi1 and miR-29b/SP1/fucosyltransferase 4 axes are useful in targeting LSCs [254, 255]. Downregulation of c-Myc, Survivin and β-Catenin by miR-147 leads to inhibition of EMT and subsequent loss of stem like traits in colon CSCs [256].
Oncolytic Virotherapy
Oncolytic Virotherapy using survivin responsive conditionally replicating adenoviruses (CRAs) regulated with multiple tumor specific is another effective approach with proven ability to target chemoresistant CSCs and tumorigenic pluripotent stem cells. These viruses specifically replicate in and induce cell death in cancers and CSCs. The high survivin promoter activity, improved selectivity and specificity of survivin responsive CRAs, in comparison with other CRAs, make them more effective in treatment, alone or in synergy with radio or chemotherapy [257]. Survivin utilizing CRAs, that bind to heparan sulfate proteoglycans, improve sensitivity to radiotherapy and specifically target CD133+ GSCs [258]. They effectively target FGFR3 positive Rhabdomyosarcoma CSCs as well [259]. In an attempt to improve the efficiency of delivery of Survivin responsive CRAs, neuronal stem cells were used as carriers. This was effective in targeting ovarian CSCs in combination with cisplatin with minimal immunogenicity and clinical safety [260]. Many of these CRAs are under clinical trials.
Targeted Drug Delivery Using Nanocomplexes
Nanocomplexes with increased bioavailability, biocompatibility, sustained and controlled drug releasing potential and high efficacy are promising targeted drug delivery systems. Considerable downregulation in survivin expression was obtained using iron-saturated bovine lactoferrin nanocarriers in colorectal CSCs [261]. Specific drug delivery to target GSCs has been accomplished with a cationic liposome nanocomplex delivering a combination of survivin siRNA and Paclitaxel with relatively less toxicity [262]. Co-delivery NF-ĸB inhibitor, IMD-0354 and chemotherapeutic agent Doxorubicin, encapsulated in ligand targeted nanoparticle, effectively targeted tumor microenvironment by increasing sensitivity and reducing cytotoxicity of Doxorubicin [263]. Chitosan nanoparticle encapsulated dominant negative survivin, competitively inhibited the endogenous protein, specifically in survivin overexpressing CSCs [264]. They have been used in delivery of survivin inhibitors, alone or in combination with chemotherapeutic agents for synergistic downregulation of survivin. They have also been used in survivin based gene therapy. Drug delivery using these nanocomplexes have been reported in cancers of breast, brain, lung, ovary and prostate [265, 266].
Conclusions
Survivin will be an indispensable molecule in cancer therapeutics due to its differential expression in almost every cancer and its near absence from adult differentiated normal tissues. Despite survivin having this attractive distinction and strong preclinical data there has been only a partial success with anti-survivin treatments.
One of the possible reasons could be the lack of true survivin-specific inhibitors as observed in the case of small molecule inhibitor YM155 and AO LY2181308. Small molecules are required to disrupt the protein-protein interactions between survivin and other proteins. This entails significant challenges as most interfaces in such protein-protein interactions do not show effective small molecule binding [267]. This has resulted in the development of only a few survivin-specific inhibitors whereas majority of the inhibitors reduce survivin expression by indirectly acting on survivin. These indirect inhibitors show their effect by the combination of survivin inhibition along with other mechanisms of action [157, 268].
In case of AO therapy, an important factor impacting its efficacy is the target specificity which is difficult to attain. This is because RNA exists as a three-dimensional structure and the efficacy of an AO depends on the extent to which it is able to bind to the exposed regions of the RNA.
The limited success with these inhibitors may also be due to tumor heterogeneity. Lack of suitable assays for screening various drug candidates is also a hindrance to the development of clinically relevant small-molecule survivin inhibitors. Survivin being a nodal protein interferes with numerous processes like autophagy, mitosis and apoptosis and with a range of molecules like caspases, Hsp90, Smac/DIABLO etc. This makes it difficult to measure the target specificity of any drug [269]. Overcoming this would require optimized assay development.
Overall the current major challenge would be to design strategies that directly and precisely target survivin. This may be possible with homo-dimerization inhibitors or with immunotherapeutic methods. Nanocomplexes would be very promising for the delivery of such anti-survivin therapeutics. Another important avenue that needs to be explored is the possibility of combinatorial approach which would include survivin inhibitors along with current standard treatment modalities for synergistic effect. Molecules capable of targeting stemness promoting factors and the related signaling pathways are very promising. Combining RNAi or immunotherapeutic methods with conventional therapy also seems reassuring. Collectively, survivin inhibition in CSCs with the current standard treatment regimens hold the maximum possibility.
Acknowledgements
The authors would like to thank Dr. Vijendra Prabhu and Dr. Naresh Kumar Mani for their much appreciated technical and overall help in preparing this manuscript.
Abbreviations
- IAP
Inhibitor of Apoptosis Protein
- BIRC5
Baculoviral IAP Repeat (BIR) - Containing 5
- INCEP
Inner Centromere Protein
- HSCs
Hematopoietic Stem Cells
- HSPCs
Hematopoietic Stem and Progenitor Cells
- MSCs
Mesenchymal Stem Cells
- ASCs
Adipocyte-Derived Stem Cells
- ES
Embryonic Stem Cells
- HES
Human Embryonic Stem Cells
- CSCs
Cancer Stem Cells
- CCCs
Circulating Cancer Cells
- NSCLC
Non-Small Cell Lung Cancer
- GSC
Glioblastoma Stem Cells
- CR-CSCs
Colorectal CSCs
- ALL
Acute Lymphoblastic Leukemia
- AML
Acute Myeloid Leukemia
- CLL
Chronic Lymphoblastic Leukemia
- CML
Chronic Myeloid Leukemia
- LSCs
Leukemia Stem Cells
- SP1
Specificity Protein 1
- CPC
Chromosomal Passenger Complex
- XIAP
X Linked IAP
- Smac/DIABLO
-
Second Mitochondria-Derived Activator of Caspase/Direct Inhibitor of Apoptosis
Binding Protein With Low pI
- HSP
Heat Shock Protein
- VEGF
Vascular Endothelial Growth Factor
- CDE/CHE
Cell Cycle-Dependent Element/ Cell Cycle Genes Homology Region
- PI3K
Phosphoinositide-3 Kinase
- FOXO
Forkhead Box O
- PTEN
Phosphatase and Tensin Homolog
- Ang-1
Angiopoietin-1
- NF-ĸB
Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B Cells
- COX-2
Cyclooxygenase-2
- GM-CSFR
Granulocyte-Macrophage Colony-Stimulating Factor Receptor
- EGFR
Epidermal Growth Factor Receptor
- HER
Human Epidermal Growth Factor Receptor
- IGF
Insulin-like Growth Factor
- mTOR
Mammalian Target of Rapamycin
- CDK
Cyclin Dependent Kinases
- Runx2
Runt-Related Transcription Factor 2
- MDM2
Mouse Double Minute 2
- Rb
Retinoblastoma Gene
- GSK-3
Glycogen Synthase Kinase-3
- HIF
Hypoxia Inducible Factor
- JAK
Janus Kinase
- Stat
Signal Transducer and Activator of Transcription
- MAPK
Mitogen-Activated Protein Kinase
- ERK
Extracellular-Signal-Regulated Kinase.
- RAS
Retrovirus-Associated DNA Sequences.
- RAF
Rapidly Accelerated Fibrosarcoma.
- TGF-Β
Transforming Growth Factor Beta.
- CXCR4
C-X-C Motif Chemokine Receptor 4.
- SDF-1
Stromal Cell-Derived Factor.
- Sox-2
SRY (Sex Determining Region Y)-Box 2.
- Oct-4
Octamer-Binding Transcription Factor 4.
- SHh
Sonic Hedgehog.
- ITD-Flt3
Internal Tandem Duplication-Flt3 Tyrosine Kinase.
- TRPM7
Transient Receptor Potential Cation Channel, Subfamily M, Member 7.
- IKKβ
Inhibitor of Nuclear Factor Kappa-B Kinase Subunit Beta.
- IL
Interleukin.
- HIV
Human Immunodeficiency Viruses.
- NMR
Nuclear Magnetic Resonance.
- Nsaids
Nonsteroidal Anti-Inflammatory Drugs.
- FGFR
Fibroblast Growth Factor Receptor.
- MDR1
Multi Drug Resistant Protein 1.
- miRNA
Micro Ribonucleic Acid.
- siRNA
Small Interfering Ribonucleic Acid.
- MHC
Major Histocompatibility Complex.
- APC
Antigen-Presenting Cell.
- DC
Dendritic Cells.
- HLA
Human Leukocyte Antigen.
- TCF
T Cell Factor.
- TA
Tolfenamic Acid.
Funding Information
Open access funding provided by Manipal Academy of Higher Education, Manipal.
Compliance with Ethical Standards
Conflict of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Prasoon Agarwal, Email: prasoon@kth.se.
Praveen Kumar, Email: kumar.praveen@manipal.edu.
References
- 1.Altieri DC. Targeting survivin in cancer. Cancer Letters. 2013;332(2):225–228. doi: 10.1016/j.canlet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheatley SP, Altieri DC. Survivin at a glance. Journal of Cell Science. 2019;132(7):jcs223826. doi: 10.1242/jcs.223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature Medicine. 1997;3(8):917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 4.Duffy MJ, Donovan NO, Brennan DJ, Gallagher WM. Survivin : A promising tumor biomarker. Cancer Letters. 2007;249(1):49–60. doi: 10.1016/j.canlet.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Stauber RH, Mann W, Knauer SK. Nuclear and cytoplasmic survivin : molecular mechanism, prognostic, and therapeutic potential. Cancer Research. 2007;67(13):5999–6003. doi: 10.1158/0008-5472.CAN-07-0494. [DOI] [PubMed] [Google Scholar]
- 6.Khan S, Jutzy JMS, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16(1):1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapinas K, Kim H, Mandeville M, Martin-Buley LA, Croce CM, Lian JB, van Wijnen AJ, Stein JL, Altieri DC, Stein GS. microRNA-mediated survivin control of pluripotency. Journal of Cellular Physiology. 2015;230(1):63–70. doi: 10.1002/jcp.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal PK, Goel A, Mittal RD. Survivin : A molecular biomarker in cancer. Indian Journal of Medical Research. 2015;141(4):389–397. doi: 10.4103/0971-5916.159250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conte MS, Altieri DC. Survivin regulation of vascular injury. Trends in Cardiovascular Medicine. 2006;16(4):114–117. doi: 10.1016/j.tcm.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: Key regulator of mitosis and apoptosis and novel target for cancer therapeutics. Clinical Cancer Research. 2008;14(16):5000–5005. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 11.Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of survivin to the centromeres and the anaphase spindle during mitosis. Current Biology. 2001;11(11):886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- 12.Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by usrvivin binding and phosphorylation. Molecular Biology of the Cell. 2002;13(9):3064–3077. doi: 10.1091/mbc.e02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altieri DC. Survivin – The inconvenient IAP. Seminars in Cell & Developmental Biology. 2015;39:91–96. doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenstermaker RA, Figel SA, Qiu J, Barone TA, Dharma SS, Winograd EK, Galbo PM, Wiltsie LM, Ciesielski MJ. Survivin monoclonal antibodies detect survivin cell surface expression and inhibit tumor growth in-vivo. Clinical Cancer Research. 2018;24(11):2642–2652. doi: 10.1158/1078-0432.CCR-17-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marusawa H, Matsuzawa SI, Welsh K, Zou H, Armstrong R, Tamm I, Reed JC. HBXIP functions as a cofactor of survivin in apoptosis suppression. The EMBO Journal. 2003;22(11):2729–2740. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson ME, Howerth EW. Survivin: A bifunctional inhibitor of apoptosis protein. Veterinary Pathology. 2004;41(6):599–607. doi: 10.1354/vp.41-6-599. [DOI] [PubMed] [Google Scholar]
- 17.Peery RC, Liu JY, Zhang JT. Targeting survivin for therapeutic discovery: past, present, and future promises. Drug Discovery Today. 2017;22(10):1466–1477. doi: 10.1016/j.drudis.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Sanhueza C, Wehinger S, Bennett JC, Valenzuela M, Owen GI, Quest AFG. The twisted survivin connection to angiogenesis. Molecular Cancer. 2015;14:198. doi: 10.1186/s12943-015-0467-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunajova L, Cash E, Markus R, Rochette S, Townley AR, Wheatley SP. The N-terminus of survivin is a mitochondrial-targeting sequence and Src regulator. Journal of Cell Science. 2016;129(14):2707–2712. doi: 10.1242/jcs.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil-Kulik P, Krzyżanowski A, Dudzińska E, Karwat J, Chomik P, Świstowska M, Kondracka A, Kwaśniewska A, Cioch M, Jojczuk M, Nogalski A, Kocki J. Potential Involvement of BIRC5 in Maintaining Pluripotency and Cell Differentiation of Human Stem Cells. Oxidative medicine and cellular longevity. 2019;2019:8727925. doi: 10.1155/2019/8727925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sah NK, Seniya C. Survivin splice variants and their diagnostic significance. Tumor Biology. 2015;36(9):6623–6631. doi: 10.1007/s13277-015-3865-5. [DOI] [PubMed] [Google Scholar]
- 22.Pavlidou A, Kroupis C, Dimas K. Association of survivin splice variants with prognosis and treatment of breast cancer. World Journal of Clinical Oncology. 2014;5(5):883–894. doi: 10.5306/wjco.v5.i5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampath J, Pelus L. Alternative splice variants of survivin as potential targets in cancer. Current Drug Discovery Technologies. 2007;4(3):174–191. doi: 10.2174/157016307782109652. [DOI] [PubMed] [Google Scholar]
- 24.Knauer SK, Bier C, Schlag P, Fritzmann J, Dietmaier W, Rödel F, Klein-Hitpass L, Kovács AF, Döring C, Hansmann ML, Hofmann WK, Kunkel M, Brochhausen C, Engels K, Lippert BM, Mann W, Stauber RH. The survivin isoform survivin-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle. 2007;6(12):1502–1509. doi: 10.4161/cc.6.12.4305. [DOI] [PubMed] [Google Scholar]
- 25.Gianani R, Jarboe E, Orlicky D, Frost M, Bobak J, Lehner R, Shroyer KR. Expression of survivin in normal, hyperplastic, and neoplastic colonic mucosa. Human Pathology. 2001;32(1):119–125. doi: 10.1053/hupa.2001.21897. [DOI] [PubMed] [Google Scholar]
- 26.Li F, Brattain MG. Role of the Survivin gene in pathophysiology. The American Journal of Pathology. 2006;169(1):1–11. doi: 10.2353/ajpath.2006.060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dallaglio K, Petrachi T, Marconi A, Truzzi F, Lotti R, Saltari A, Morandi P, Puviani M, Maiorana A, Pincelli C. Expression of nuclear Survivin in normal skin and squamous cell carcinoma: A possible role in tumour invasion. British Journal of Cancer. 2014;110(1):199–207. doi: 10.1038/bjc.2013.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiou, S. K., Jones, M. K., & Tarnawski, A. S. (2003). Survivin - an anti-apoptosis protein: its biological roles and implications for cancer and beyond. Medical Science Monitor, 9(4), P125-P129. [PubMed]
- 29.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Molecular Cancer Therapeutics. 2006;5(5):1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 30.Caldas H, Honsey LE, Altura RA. Survivin 2alpha: a novel Survivin splice variant expressed in human malignancies. Molecular Cancer. 2005;4(1):11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adida C, Crotty PL, Mcgrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene Survivin in human and mouse differentiation. American Journal of Pathology. 1998;152(1):43–49. [PMC free article] [PubMed] [Google Scholar]
- 32.Fukuda S, Pelus LM. Regulation of the inhibitor-of-apoptosis family member survivin in normal cord blood and bone marrow CD34(+) cells by hematopoietic growth factors: Implication of survivin expression in normal hematopoiesis. Blood. 2001;98(7):2091–2100. doi: 10.1182/blood.v98.7.2091. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda S, Foster RG, Porter SB, Pelus LM. The antiapoptosis protein Survivin is associated with cell cycle entry of normal cord blood CD34(+) cells and modulates cell cycle and proliferation of mouse hematopoietic progenitor cells. Blood. 2002;100(7):2463–2471. doi: 10.1182/blood.V100.7.2463. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda S, Pelus LM. Elevation of survivin levels by hematopoietic growth factors occurs in quiescent CD34+ hematopoietic stem and progenitor cells before cell cycle entry. Cell Cycle. 2002;1(5):322–326. doi: 10.4161/cc.1.5.149. [DOI] [PubMed] [Google Scholar]
- 35.Filion TM, Qiao M, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, Lian JB, Altieri DC, Stein GS. Survival responses of human embryonic stem cells to DNA damage. Journal of Cellular Physiology. 2009;220(3):586–592. doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mull AN, Klar A, Navara CS. Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Research. 2014;12(2):539–549. doi: 10.1016/j.scr.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Singh P, Fukuda S, Liu L, Chittei BR, Pelus LM. Survivin is required for mouse and human bone marrorw mesenchymal stromal cell function. Stem Cells. 2018;36(1):123–129. doi: 10.1002/stem.2727. [DOI] [PubMed] [Google Scholar]
- 38.Blum B, Benvenisty N. The tumorigenicity of diploid and aneuploid human pluripotent stem cells. Cell Cycle. 2009;8(23):3822–3830. doi: 10.4161/cc.8.23.10067. [DOI] [PubMed] [Google Scholar]
- 39.Liu T, Huang Y, Guo L, Cheng W, Zou G. CD44+/CD105+ human amniotic fluid mesenchymal stem cells survive and proliferate in the ovary long-term in a mouse model of chemotherapy-induced premature ovarian failure. International Journal of Medical Sciences. 2012;9(7):592–602. doi: 10.7150/ijms.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ejarque M, Ceperuelo-Mallafré V, Serena C, Pachón G, Núñez-Álvarez Y, Terrón-Puig M, Calvo E, Núñez-Roa C, Oliva-Olivera W, Tinahones FJ, Peinado MA, Vendrell J, Fernández-Veledo S. Survivin, a key player in cancer progression, increases in obesity and protects adipose tissue stem cells from apoptosis. Cell Death & Disease. 2017;8(5):e2802. doi: 10.1038/cddis.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labarrade F, Botto JM, Domloge N. CRM1 and chromosomal passenger complex component survivin are essential to normal mitosis progress and to preserve keratinocytes from mitotic abnormalities. International Journal of Cosmetic Science. 2016;38(5):452–461. doi: 10.1111/ics.12311. [DOI] [PubMed] [Google Scholar]
- 42.Marconi A, Dallaglio K, Lotti R, Vaschieri C, Truzzi F, Fantini F, Pincelli C. Survivin Identifies Keratinocyte Stem Cells and Is Downregulated by Anti-β1 Integrin During Anoikis. Stem Cells. 2007;25(1):149–155. doi: 10.1634/stemcells.2006-0165. [DOI] [PubMed] [Google Scholar]
- 43.Dallaglio K, Marconi A, Pincelli C. Survivin: A dual player in healthy and diseased skin. Journal of Investigative Dermatology. 2011;132(1):18–27. doi: 10.1038/jid.2011.279. [DOI] [PubMed] [Google Scholar]
- 44.Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Research. 2014;74(10):2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 45.Feng R, Zhou S, Liu Y, Song D, Luan Z, Dai X, Li Y, Tang N, Wen J, Li L. Sox2 protects neural stem cells from apoptosis via up-regulating survivin. Biochemical Journal. 2013;450(3):459–468. doi: 10.1042/BJ20120924. [DOI] [PubMed] [Google Scholar]
- 46.Sampath J, Smith LM, Fukuda S, Pelus LM. Identification and characterization of novel survivin splice variants with anti-apoptotic activities. Journal of Immunology & Clinical Research. 2017;4(1):1039. [Google Scholar]
- 47.Blum B, Bar-nur O, Golan-lev T, Benvenisty N. The anti-apoptotic gene survivin contributes to teratoma formation by human embryonic stem cells. Nature Biotechnology. 2009;27(3):281–287. doi: 10.1038/nbt.1527. [DOI] [PubMed] [Google Scholar]
- 48.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22(53):8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 49.Altieri DC. Validating survivin as a cancer therapeutic target. Nature Reviews Cancer. 2003;3(1):46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 50.Owens, T. W., Gilmore, A. P., Streuli, C. H., & Foster, F. M. (2013). Inhibitor of apoptosis proteins: Promising targets for cancer therapy. Journal of Carcinogenesis & Mutagenesis, Suppl, 14, S14–S004. 10.4172/2157-2518.S14-004. [DOI] [PMC free article] [PubMed]
- 51.Ferrandina G, Legge F, Martinelli E, Ranelletti FO, Zannoni GF, Lauriola L, Gessi M, Gallotta V, Scambia G. Survivin expression in ovarian cancer and its correlation with clinico-pathological, surgical and apoptosis-related parameters. British Journal of Cancer. 2005;92(2):271–277. doi: 10.1038/sj.bjc.6602332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salz W, Eisenberg D, Plescia J, Garlick DS, Weiss RM, Wu XR, Sun TT, Altieri DC. A Survivin gene signature predicts aggressive tumor behavior. Cancer Research. 2005;65(9):3531–3534. doi: 10.1158/0008-5472.CAN-04-4284. [DOI] [PubMed] [Google Scholar]
- 53.Shariat SF, Lotan Y, Saboorian H, Khoddami SM, Roehrborn CG, Slawin KM, Ashfaq R. Survivin expression is associated with features of biologically aggressive prostate carcinoma. Cancer. 2004;100(4):751–757. doi: 10.1002/cncr.20039. [DOI] [PubMed] [Google Scholar]
- 54.Liu JL, Gao W, Kang QM, Zhang XJ, Yang SG. Prognostic value of survivin in patients with gastric cancer: A systematic review with meta-analysis. PloS One. 2013;8(8):e71930. doi: 10.1371/journal.pone.0071930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith SD, Wheeler MA, Plescia J, Colberg JW, Weiss RM, Altieri DC. Urine detection of survivin and diagnosis of bladder cancer. Journal of the American Medical Association. 2001;285(3):324–328. doi: 10.1001/jama.285.3.324. [DOI] [PubMed] [Google Scholar]
- 56.Yie SM, Luo B, Ye NY, Xie K, Ye SR. Detection of Survivin-expressing circulating cancer cells in the peripheral blood of breast cancer patients by a RT-PCR ELISA. Clinical & Experimental Metastasis. 2006;23(5):279–289. doi: 10.1007/s10585-006-9037-7. [DOI] [PubMed] [Google Scholar]
- 57.Sugahara K, Uemura A, Harasawa H, Nagai H, Hirakata Y, Tomonaga M, Murata K, Sohda H, Nakagoe T, Shibasaki S, Yamada Y, Kamihira S. Clinical relevance of survivin as a biomarker in neoplasms, especially in adult T-cell leukemias and acute leukemias. International Journal of Hematology. 2004;80(1):52–58. doi: 10.1532/ijh97.04031. [DOI] [PubMed] [Google Scholar]
- 58.Necochea-Campion R d, Chen CS, Mirshahidi S, Howard FD, Wall NR. Clinico-pathologic relevance of Survivin splice variant expression in cancer. Cancer letters. 2013;339(2):167–174. doi: 10.1016/j.canlet.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atena M, Reza AM, Mehran G. A review on the biology of cancer stem cells. Stem Cell Discovery. 2014;4(4):83–89. doi: 10.4236/scd.2014.44009. [DOI] [Google Scholar]
- 60.O’Brien CA, Kreso A, Jamieson CHM. Cancer stem cells and self-renewal. Clinical Cancer Research. 2010;16(12):3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 61.Small S, Keerthivasan G, Huang Z, Gurbuxani S, Crispino JD. Overexpression of survivin initiates hematologic malignancies in vivo. Leukemia. 2010;24(11):1920–1926. doi: 10.1038/leu.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carter BZ, Qiu Y, Huang X, Diao L, Zhang N, Coombes KR, Mak DH, Konopleva M, Cortes J, Kantarjian HM, Mills GB, Andreeff M, Kornblau SM. Survivin is highly expressed in CD34(+)38(−) leukemic stem/progenitor cells and predicts poor clinical outcomes in AML. Blood. 2012;120(1):173–180. doi: 10.1182/blood-2012-02-409888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukuda S, Abe M, Onishi C, Taketani T, Purevsuren J, Yamaguchi S, Conway EM, Pelus LM. Survivin selectively modulates genes deregulated in human leukemia stem cells. Journal of Oncology. 2011;2011:946936. doi: 10.1155/2011/946936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guvenc H, Pavlyukov MS, Joshi K, Kurt H, Banasavadi-Siddegowda YK, Mao P, Hong C, Yamada R, Kwon CH, Bhasin D, Chettiar S, Kitange G, Park IH, Sarkaria JN, Li C, Shakhparonov MI, Nakano I. Impairment of glioma stem cell survival and growth by a novel inhibitor for Survivin–Ran protein complex. Clinical Cancer Research. 2013;19(3):631–642. doi: 10.1158/1078-0432.CCR-12-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Molecular Cancer. 2006;5(1):67–79. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Stefano AB, Iovino F, Lombardo Y, Eterno V, Höger T, Dieli F, Stassi G, Todaro M. Survivin is regulated by interleukin-4 in colon cancer stem cells. Journal of Cellular Physiology. 2010;225(2):555–561. doi: 10.1002/jcp.22238. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Lee M, Choi E, Cho M. Clinicopathologic significance of survivin expression in relation to CD133 expression in surgically resected stage II or III colorectal cancer. Journal of Pathology and Translational Medicine. 2017;51(1):17–23. doi: 10.4132/jptm.2016.09.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sam S, Sam MR, Esmaeillou M, Safaralizadeh R. Effective targeting Survivin, Caspase-3 and microRNA-16-1 expression by Methyl-3-pentyl-6-methoxyprodigiosene triggers apoptosis in colorectal cancer stem-like cells. Pathology & Oncology Research. 2016;22(4):715–723. doi: 10.1007/s12253-016-0055-8. [DOI] [PubMed] [Google Scholar]
- 69.Kim ST, Sohn I, Do I-G, Jang J, Kim SH, Jung IH, Park JO, Park YS, Talasaz A, Lee J, Kim HC. Transcriptome analysis of CD133-positive stem cells and prognostic value of survivin in colorectal cancer. Cancer Genomics & Proteomics. 2014;11(5):259–266. [PubMed] [Google Scholar]
- 70.Siddharth S, Das S, Nayak A, Nath C. SURVIVIN as a marker for quiescent-breast cancer stem cells - An intermediate , adherent , pre-requisite phase of breast cancer metastasis. Clinical & Experimental Metastasis. 2016;33(7):661–675. doi: 10.1007/s10585-016-9809-7. [DOI] [PubMed] [Google Scholar]
- 71.Yie S, Lou B, Ye S, Cao M, He X, Li P, Hu K, Rao L, Wu S, Xiao H, Gao E. Detection of survivin-expressing circulating cancer cells (CCCs) in peripheral blood of patients with gastric and colorectal cancer reveals high risks of relapse. Annals of Surgical Oncology. 2008;15(11):3073–3082. doi: 10.1245/s10434-008-0069-x. [DOI] [PubMed] [Google Scholar]
- 72.Cao M, Yie SM, Wu SM, Chen S, Lou B, He X, Ye SR, Xie K, Rao L, Gao E, Ye NY. Detection of survivin-expressing circulating cancer cells in the peripheral blood of patients with esophageal squamous cell carcinoma and its clinical significance. Clinical & Experimental Metastasis. 2009;26(7):751–758. doi: 10.1007/s10585-009-9274-7. [DOI] [PubMed] [Google Scholar]
- 73.Liao CP, Adisetiyo H, Liang M, Roy-Burman P. Cancer stem cells and microenvironment in prostate cancer progression. Hormones and Cancer. 2010;1(6):297–305. doi: 10.1007/s12672-010-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo Y, Liu T, Fei W, Yue XG. Correlation between SOX2 and Survivin clinical features in patients with salivary adenoid cystic carcinoma. Journal of Infection and Public Health. 2019;12(6):847–853. doi: 10.1016/j.jiph.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J, Chen X, Bian L, Wang Y, Liu H. CD44 +/CD24 + −Expressing cervical cancer cells and radioresistant cervical cancer cells exhibit cancer stem cell characteristics. Gynecologic and Obstetric Investigation. 2019;84(2):174–182. doi: 10.1159/000493129. [DOI] [PubMed] [Google Scholar]
- 76.Kim J, Ahn S, Kim K, Cho MS, Kim KH, Lee RA, Nam EM. Prognostic significance of survivin expression and combined analysis with cancer stem cell and epithelial–mesenchymal transition-related markers in patients with rectal cancer undergoing preoperative chemoradiotherapy. Anticancer Research. 2018;38(12):6881–6889. doi: 10.21873/anticanres.13064. [DOI] [PubMed] [Google Scholar]
- 77.Vachtenheim J, Vlckova K. Insights into the regulation of Survivin expression in tumors. Single Cell Biology. 2016;5(2):139. doi: 10.4172/2168-9431.1000139. [DOI] [Google Scholar]
- 78.Guha M, Plescia J, Leav I, Li J, Languino LR, Altieri DC. Endogenous tumor suppression mediated by PTEN involves survivin gene silencing. Cancer Research. 2009;69(12):4954–4958. doi: 10.1158/0008-5472.CAN-09-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martínez-García D, Manero-Rupérez N, Quesada R, Korrodi-Gregório L, Soto-Cerrato V. Therapeutic strategies involving survivin inhibition in cancer. Medicinal Research Reviews. 2019;39(3):887–909. doi: 10.1002/med.21547. [DOI] [PubMed] [Google Scholar]
- 80.Vaira V, Lee CW, Goel HL, Bosari S, Languino LR, Altieri DC. Regulation of survivin expression by IGF-1/mTOR signaling. Oncogene. 2007;26(19):2678–2684. doi: 10.1038/sj.onc.1210094. [DOI] [PubMed] [Google Scholar]
- 81.Song K, Shankar E, Yang J, Bane KL, Wahdan-Alaswad R, Danielpour D. Critical role of a survivin/TGF-β/mTORC1 axis in IGF-I-mediated growth of prostate epithelial cells. PloS One. 2013;8(5):e61896. doi: 10.1371/journal.pone.0061896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raj D, Liu T, Samadashwily G, Grossman D. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis. 2008;29(1):194–201. doi: 10.1093/carcin/bgm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang Y, Saavedra HI, Holloway MP, Leone G, Altura RA. Aberrant regulation of Survivin by the RB/E2F family of proteins. The Journal of Biological Chemistry. 2004;279(39):40511–40520. doi: 10.1074/jbc.M404496200. [DOI] [PubMed] [Google Scholar]
- 84.Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19(10):1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- 85.Fukuda S, Mantel CR, Pelus LM. Survivin regulates hematopoietic progenitor cell proliferation through p21WAF1/Cip1-dependent and -independent pathways. Blood. 2004;103(1):120–127. doi: 10.1182/blood-2003-05-1756. [DOI] [PubMed] [Google Scholar]
- 86.Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Research. 2006;66(7):3392–3395. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- 87.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic Survivin gene by wild type p53. Journal of Biological Chemistry. 2002;277(5):3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 88.Mirza A, Mcguirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, Nielsen LL, Pickett CB, Liu S. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21(17):2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 89.Wang Z, Fukuda S, Pelus LM. Survivin regulates the p53 tumor suppressor gene family. Oncogene. 2004;23(49):8146–8153. doi: 10.1038/sj.onc.1207992. [DOI] [PubMed] [Google Scholar]
- 90.Yoon K, Lim YS, Yu SB, Kim DS, Ryu SJ, Kim KH, Jang TH, Kim SH. The expression of survivin and its related genes in adipocyte- derived stem cell by demethylation. Korean Journal of Anesthesiology. 2010;58(4):383–390. doi: 10.4097/kjae.2010.58.4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gedaly R, Galuppo R, Daily MF, Shah M, Maynard E, Chen C, Zhang X, Esser KA, Cohen DA, Evers BM, Jiang J, Spear BT. Targeting the Wnt/β-catenin signaling pathway in liver cancer stem cells and hepatocellular carcinoma cell lines with FH535. PloS One. 2014;9(6):e99272. doi: 10.1371/journal.pone.0099272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. Journal of the National Cancer Institute. 2014;106(1):djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 93.Zhou S, Liu Y, Feng R, Wang C, Jiang S, Zhang X, Lan F, Li Y. Survivin improves reprogramming efficiency of human neural progenitors by single molecule OCT4. Stem Cells International. 2016;2016:4729535. doi: 10.1155/2016/4729535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang YF, Ma SR, Wang WM, Huang CF, Zhao ZL, Liu B, Zhang WF, Zhao YF, Zhang L, Sun ZJ. Inhibition of Survivin reduces HIF-1α, TGF-β1 and TFE3 in salivary adenoid cystic carcinoma. PloS One. 2014;9(12):e114051. doi: 10.1371/journal.pone.0114051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen YQ, Zhao CL, Li W. Effect of hypoxia-inducible factor-1α on transcription of survivin in non-small cell lung cancer. Journal of Experimental & Clinical Cancer Research. 2009;28(1):29. doi: 10.1186/1756-9966-28-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carpenter RL, Lo HW. STAT3 target genes relevant to human cancers. Cancers. 2014;6(2):897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Enkemann S, Eschrich S, Lee J, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R. Persistent activation of Stat3 signaling induces Survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clinical Cancer Research. 2006;12(1):11–20. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z, Pelus LM. Disruption of Survivin in K562 cells elevates telomerase activity and protects cells against apoptosis induced by the Bcr-abl kinase inhibitor STI571. Cancer Therapy. 2008;6(B):603–610. [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Z, Sampath J, Fukuda S, Pelus LM. Disruption of the inhibitor of apoptosis protein survivin sensitizes Bcr-abl-positive cells to STI571-induced apoptosis. Cancer Research. 2005;65(18):8224–8232. doi: 10.1158/0008-5472.CAN-05-0303. [DOI] [PubMed] [Google Scholar]
- 100.Shinjyo T, Kurosawa H, Miyagi J-I, Ohama K, Masuda M, Nagasaki A, Matsui H, Inaba T, Furukawa Y, Takasu N. Ras-mediated up-regulation of survivin expression in cytokine-dependent murine pro-B lymphocytic Cells. Tohoku Journal of Experimental Medicine. 2008;216(1):25–34. doi: 10.1620/tjem.216.25. [DOI] [PubMed] [Google Scholar]
- 101.Fukuda S, Pelus LM. Activated H-Ras regulates hematopoietic cell survival by modulating Survivin. Biochemical and Biophysical Research Communications. 2004;323(2):636–644. doi: 10.1016/j.bbrc.2004.08.149. [DOI] [PubMed] [Google Scholar]
- 102.Palazzo E, Morandi P, Lotti R, Saltari A, Truzzi F, Schnebert S, Dumas M, Marconi A, Pincelli C. Notch cooperates with survivin to maintain stemness and to stimulate proliferation in human keratinocytes during ageing. International Journal of Molecular Sciences. 2015;16(11):26291–26302. doi: 10.3390/ijms161125948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haque R, Song J, Haque M, Lei F, Sandhu P, Ni B, Zheng S, Fang D, Yang JM, Song J. C-Myc-Induced survivin is essential for promoting the Notch dependent T cell differentiation from hematopoietic stem cells. Genes. 2017;8(3):97. doi: 10.3390/genes8030097. [DOI] [Google Scholar]
- 104.Guzman JR, Fukuda S, Pelus LM. Inhibition of caspase-3 by Survivin prevents Wee1 Kinase degradation and promotes cell survival by maintaining phosphorylation of p34Cdc2. Gene Therapy & Molecular Biology. 2009;13B:264–273. [PMC free article] [PubMed] [Google Scholar]
- 105.Hoggatt J, Singh P, Sampath J, Pelus LM. Prostaglandin E2 enhances hematopoietic stem cell homing, survival, and proliferation. Blood. 2009;113(22):5444–5455. doi: 10.1182/blood-2009-01-201335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Singh, P., Mohammad, K. S., & Pelus, L. M. (2020). CXCR4 expression in the bone marrow microenvironment is required for hematopoietic stem and progenitor cell maintenance and early hematopoietic regeneration after myeloablation. Stem Cells. 10.1002/stem.3174. [DOI] [PMC free article] [PubMed]
- 107.Fukuda S, Hoggatt J, Singh P, Abe M, Speth JM, Hu P, Conway EM, Nucifora G, Yamaguchi S, Pelus LM. Survivin modulates genes with divergent molecular functions and regulates proliferation of hematopoietic stem cells through Evi-1. Leukemia. 2015;29(2):433–440. doi: 10.1038/leu.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liskova A, Kubatka P, Samec M, Zubor P, Mlyncek M, Bielik T, Samuel SM, Zulli A, Kwon TK, Büsselberg D. Dietary phytochemicals targeting cancer stem cells. Molecules (Basel, Switzerland) 2019;24(5):889. doi: 10.3390/molecules24050899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wen K, Fu Z, Wu X, Feng J, Chen W, Qian J. Oct-4 is required for an antiapoptotic behavior of chemoresistant colorectal cancer cells enriched for cancer stem cells: Effects associated with STAT3/Survivin. Cancer Letters. 2013;333(1):56–65. doi: 10.1016/j.canlet.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 110.Kamalapuram SK, Kanwar RK, Kanwar JR. Survivin in cancer: A spider in the web. Journal of Nanomedicine & Nanotechnology. 2012;3(8):e118. doi: 10.4172/2157-7439.1000e118. [DOI] [Google Scholar]
- 111.Fukuda S, Singh P, Moh A, Abe M, Conway EM, Boswell HS, Yamaguchi S, Fu XY, Pelus LM. Survivin mediates aberrant hematopoietic progenitor cell proliferation and acute leukemia in mice induced by internal tandem duplication of Flt3. Blood. 2009;114(2):394–403. doi: 10.1182/blood-2008-11-188714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang G, Wang W, Yao C, Zhang S, Liang L, Han M, Ren J, Qi X, Zhang X, Wang S, Li L. Radiation-resistant cancer stem-like cell properties are regulated by PTEN through the activity of nuclear β-catenin in nasopharyngeal carcinoma. Oncotarget. 2017;8(43):74661–74672. doi: 10.18632/oncotarget.20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dong Z, Yang L, Lai D. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Proliferation. 2013;46(4):425–435. doi: 10.1111/cpr.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galuppo R, Maynard E, Shah M, Daily MF, Chen C, Spear BT, Gedaly R. Synergistic inhibition of HCC and liver cancer stem cell proliferation by targeting Ras/Raf/Mapk and Wnt/β-catenin pathways. Anticancer Research. 2014;34(4):1709–1713. [PMC free article] [PubMed] [Google Scholar]
- 115.Liu M, Inoue K, Leng T, Guo S, Xiong ZG. TRPM7 channels regulate glioma stem cell through STAT3 and Notch signaling pathways. Cellular Signalling. 2014;26(12):2773–2781. doi: 10.1016/j.cellsig.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ling X, Cao S, Cheng Q, Keefe JT, Rustum YM, Li F. A novel small molecule FL118 that selectively inhibits survivin, Mcl-1, XIAP and cIAP2 in a p53-independent manner, shows superior antitumor activity. PloS One. 2012;7(9):e45571. doi: 10.1371/journal.pone.0045571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ling X, Wu W, Fan C, Xu C, Liao J, Rich LJ, Huang RY, Repasky EA, Wang X, Li F. An ABCG2 non-substrate anticancer agent FL118 targets drug-resistant cancer stem-like cells and overcomes treatment resistance of human pancreatic cancer. Journal of Experimental & Clinical Cancer Research : CR. 2018;37(1):240. doi: 10.1186/s13046-018-0899-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Holthof LC, van der Horst HJ, van Hal-van Veen SE, Ruiter R, Li F, Buijze M, Andersen MN, Yuan H, de Bruijn J, van de Donk N, Lokhorst HM, Zweegman S, Groen R, Mutis T. Preclinical evidence for an effective therapeutic activity of FL118, a novel survivin inhibitor, in patients with relapsed/refractory multiple myeloma. Haematologica. 2020;105(2):e80–e83. doi: 10.3324/haematol.2018.213314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Kinoyama I, Matsuhisa A, Kudou M, Sasamata M. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Science. 2011;102(3):614–621. doi: 10.1111/j.1349-7006.2010.01834.x. [DOI] [PubMed] [Google Scholar]
- 120.Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, van Klaveren RJ, Chaudhary S, Gunther A, Shamsili S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. Journal of Clinical Oncology: Official Journal of the Americal Society of Clinical Oncology. 2009;27(27):4481–4486. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- 121.Glaros TG, Stockwin LH, Mullendore ME, Smith B, Morrison BL, Newton DL. The “survivin suppressants” NSC 80467 and YM155 induce a DNA damage response. Cancer Chemotherapy and Pharmacology. 2012;70(1):207–212. doi: 10.1007/s00280-012-1868-0. [DOI] [PubMed] [Google Scholar]
- 122.Sankpal UT, Ingersoll SB, Ahmad S, Holloway RW, Bhat VB, Simecka JW, Daniel L, Kariali E, Vishwanatha JK, Basha R. Association of Sp1 and survivin in epithelial ovarian cancer : Sp1 inhibitor and cisplatin, a novel combination for inhibiting epithelial ovarian cancer cell proliferation. Tumor Biology. 2016;37(10):14259–14269. doi: 10.1007/s13277-016-5290-9. [DOI] [PubMed] [Google Scholar]
- 123.Hurtado M, Sankpal UT, Kaba A, Mahammad S, Chhabra J, Brown DT, Gurung RK, Holder AA, Vishwanatha JK, Basha R. Novel Survivin Inhibitor for Suppressing Pancreatic Cancer Cells Growth via Downregulating Sp1 and Sp3 Transcription Factors. Cellular Physiology And Biochemistry : International Journal Of Experimental Cellular Physiology, Biochemistry, And Pharmacology. 2018;51(4):1894–1907. doi: 10.1159/000495715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kelly RJ, Lopez-Chavez A, Citrin D, Janik JE, Morris JC. Impacting tumor cell-fate by targeting the inhibitor of apoptosis protein survivin. Molecular Cancer. 2011;10(1):35. doi: 10.1186/1476-4598-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Athanasoula KC, Gogas H, Polonifi K, Vaiopoulos AG, Polyzos A, Mantzourani M. Survivin beyond physiology : Orchestration of multistep carcinogenesis and therapeutic potentials. Cancer Letters. 2014;347(2):175–182. doi: 10.1016/j.canlet.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 126.Shelake S, Sankpal UT, Paul Bowman W, Wise M, Ray A, Basha R. Targeting specificity protein 1 transcription factor and survivin using tolfenamic acid for inhibiting Ewing sarcoma cell growth. Investigational New Drugs. 2017;35(2):158–165. doi: 10.1007/s10637-016-0417-9. [DOI] [PubMed] [Google Scholar]
- 127.Estève PO, Hang GC, Pradhan S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. Journal of Biological Chemistry. 2007;282(4):2615–2625. doi: 10.1074/jbc.M606203200. [DOI] [PubMed] [Google Scholar]
- 128.Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target : what can we do after over two decades of study ? Journal of Experimental & Clinical Cancer Research. 2019;38(1):368. doi: 10.1186/s13046-019-1362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qi J, Dong Z, Liu J, Peery RC, Zhang S. Effective targeting of the Survivin dimerization interface with small-molecule inhibitors. Cancer Research. 2016;76(2):453–462. doi: 10.1158/0008-5472.CAN-15-1874. [DOI] [PubMed] [Google Scholar]
- 130.Yaffe PB, Power Coombs MR, Doucette CD, Walsh M, Hoskin DW. Piperine, an alkaloid from black pepper, inhibits growth of human colon cancer cells via G1 arrest and apoptosis triggered by endoplasmic reticulum stress. Molecular Carcinogenesis. 2015;54(10):1070–1085. doi: 10.1002/mc.22176. [DOI] [PubMed] [Google Scholar]
- 131.Abdelhamed S, Yokoyama S, Refaat A, Ogura K, Yagita H, Awale S, Saiki I. Piperine enhances the efficacy of TRAIL-based therapy for triple-negative breast cancer cells. Anticancer Research. 2014;34(4):1893–1900. [PubMed] [Google Scholar]
- 132.Wadegaonkar VP, Wadegaonkar PA. Withanone as an inhibitor of survivin : A potential drug candidate for cancer therapy. Journal of Biotechnology. 2013;168(2):229–233. doi: 10.1016/j.jbiotec.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 133.Xiao M, Li W. Recent advances on small-molecule survivin inhibitors. Current Medicinal Chemistry. 2015;22(9):1136–1146. doi: 10.2174/0929867322666150114102146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, Sessa WC, Altieri DC. Regulation of survivin function by Hsp90. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(24):13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Altieri DC. Coupling apoptosis resistance to the cellular stress response -The IAP-Hsp90 connection in cancer. Cell Cycle. 2004;3(3):255–256. doi: 10.4161/cc.3.3.711. [DOI] [PubMed] [Google Scholar]
- 136.Venkatesan N, Kanwar JR, Deepa PR, Navaneethakrishnan S, Joseph C, Krishnakumar S. Targeting HSP90/ Survivin using a cell permeable structure based peptido-mimetic shepherdin in retinoblastoma. Chemico-Biological Interactions. 2016;252(5):141–149. doi: 10.1016/j.cbi.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 137.Plescia J, Salz W, Xia F, Pennati M, Zaffaroni N, Daidone MG, Meli M, Dohi T, Fortugno P, Nefedova Y, Gabrilovich DI, Colombo G, Altieri DC. Rational design of shepherdin , a novel anticancer agent. Cancer Cell. 2005;7(5):457–468. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 138.Meli M, Pennati M, Curto M, Daidone MG, Plescia J, Toba S, Altieri DC, Zaffaroni N, Colombo G. Small-molecule targeting of heat shock protein 90 chaperone function: Rational identification of a new anticancer lead. Journal of Medicinal Chemistry. 2006;49(26):7721–7730. doi: 10.1021/jm060836y. [DOI] [PubMed] [Google Scholar]
- 139.Cheung CH, Chen HH, Cheng LT, Lyu KW, Kanwar JR, Chang JY. Targeting Hsp90 with small molecule inhibitors induces the over-expression of the anti-apoptotic molecule, survivin, in human A549, HONE-1 and HT-29 cancer cells. Molecular Cancer. 2010;9:77. doi: 10.1186/1476-4598-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J, Sun W, Dong W, Wang Z, Qin Y, Zhang T, Zhang H. HSP90 inhibitor NVP-AUY922 induces cell apoptosis by disruption of the survivin in papillary thyroid carcinoma cells. Biochemical and Biophysical Research Communications. 2017;487(2):313–319. doi: 10.1016/j.bbrc.2017.04.056. [DOI] [PubMed] [Google Scholar]
- 141.Sarvagalla S, Cheung HCA, Tsai J, Hsieh HP, Coumar MS. Disruption of protein–protein interactions: hot spot detection, structure-based virtual screening and in vitro testing for the anti-cancer drug target – survivin. RSC Advances. 2016;6:31947–31959. doi: 10.1039/C5RA22927H. [DOI] [Google Scholar]
- 142.Wall NR, Connor DSO, Plescia J, Pommier Y, Altieri DC. Suppression of survivin phosphorylation on Thr34 by flavopiridol enhances tumor cell apoptosis. Cancer Research. 2003;63(1):230–235. [PubMed] [Google Scholar]
- 143.O’Connor DS, Wall NR, Porter ACG, Altieri DC. A p34cdc2 survival checkpoint in cancer. Cancer Cell. 2002;2(1):43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 144.Pennati M, Campbell AJ, Curto M, Binda M, Cheng Y, Wang LZ, Curtin N, Golding BT, Griffin RJ, Hardcastle IR, Henderson A, Zaffaroni N, Newell DR. Potentiation of paclitaxel-induced apoptosis by the novel cyclin-dependent kinase inhibitor NU6140: a possible role for survivin down-regulation. Molecular Cancer Therapeutics. 2005;4(9):1328–1337. doi: 10.1158/1535-7163.MCT-05-0022. [DOI] [PubMed] [Google Scholar]
- 145.Hsiao CJ, Hsiao G, Chen WL, Wang SW, Chiang CP, Liu LY, Guh JH, Lee TH, Chung CL. Cephalochromin induces G0/G1 cell cycle arrest and apoptosis in A549 human non-small cell lung cancer cells by inflicting mitochondrial disruption. Journal of Natural Products. 2014;77(4):758–765. doi: 10.1021/np400517g. [DOI] [PubMed] [Google Scholar]
- 146.Sam MR, Pourpak RS. Regulation of p53 and survivin by prodigiosin compound derived from Serratia marcescens contribute to caspase-3-dependent apoptosis in acute lymphoblastic leukemia cells. Human and Experimental Toxicology. 2018;37(6):608–617. doi: 10.1177/0960327117718052. [DOI] [PubMed] [Google Scholar]
- 147.Sam MR, Tavakoli-mehr M, Safaralizadeh R. Omega-3 fatty acid DHA modulates p53 , survivin , and microRNA-16-1 expression in KRAS-mutant colorectal cancer stem-like cells. Genes and Nutrition. 2018;13:8. doi: 10.1186/s12263-018-0596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sam MR, Esmaeillou M, Sam S, Shokrgozar MA. Fish-oil-derived eicosapentaenoic acid decreases survivin expression and induces wt-p53 accumulation with caspase-3 activation in acute lymphoblastic leukemia cells. Human and Experimental Toxicology. 2017;37(7):714–724. doi: 10.1177/0960327117730879. [DOI] [PubMed] [Google Scholar]
- 149.Moreno-Smith M, Lakoma A, Chen Z, Tao L, Scorsone KA, Schild L, Aviles-Padilla K, Nikzad R, Zhang Y, Chakraborty R, Molenaar JJ, Vasudevan SA, Sheehan V, Kim ES, Paust S, Shohet JM, Barbieri E. p53 nongenotoxic activation and mTORC1 inhibition lead to effective combination for neuroblastoma therapy. Clinical Cancer Research : an official journal of the American Association for Cancer Research. 2017;23(21):6629–6639. doi: 10.1158/1078-0432.CCR-17-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Renner G, Janouskova H, Noulet F, Koenig V, Guerin E, Bär S, Nuesch J, Rechenmacher F, Neubauer S, Kessler H, Blandin AF, Choulier L, Etienne-Selloum N, Lehmann M, Lelong-Rebel I, Martin S, Dontenwill M. Integrin α5β1 and p53 convergent pathways in the control of anti-apoptotic proteins PEA-15 and survivin in high-grade glioma. Cell Death and Differentiation. 2016;23(4):640–653. doi: 10.1038/cdd.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Anandharaj A, Cinghu S, Park WY. Rapamycin-mediated mTOR inhibition attenuates survivin and sensitizes glioblastoma cells to radiation therapy. Acta Biochemica et Biophysica Sinica (Shanghai) 2011;43(4):292–300. doi: 10.1093/abbs/gmr012. [DOI] [PubMed] [Google Scholar]
- 152.Agarwal E, Chaudhuri A, Leiphrakpam PD, Haferbier KL, Brattain MG, Chowdhury S. Akt inhibitor MK-2206 promotes anti-tumor activity and cell death by modulation of AIF and Ezrin in colorectal cancer. BMC Cancer. 2014;14:145. doi: 10.1186/1471-2407-14-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Belyanskaya LL, Hopkins-Donaldson S, Kurtz S, Simoes-Wust AP, Yousefi S, Simon HU, Rolf S, Zangemeister-Wittke U. Cisplatin activates Akt in small cell lung cancer cells and attenuates apoptosis by survivin upregulation. International Journal of Cancer. 2005;117(5):755–763. doi: 10.1002/ijc.21242. [DOI] [PubMed] [Google Scholar]
- 154.Sethi G, Ahn KS, Sung B, Kunnumakkara AB, Chaturvedi MM, Aggarwal BB. SH-5 , an AKT inhibitor potentiates apoptosis and inhibits invasion through the suppression of anti-apoptotic , proliferative and metastatic gene products regulated by IkappaBalpha kinase activation. Biochemical Pharmacology. 2008;76(11):1404–1416. doi: 10.1016/j.bcp.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 155.Singla S, Pippin JA, Drebin JA. Dual ErbB1 and ErbB2 receptor tyrosine kinase inhibition exerts synergistic effect with conventional chemotherapy in pancreatic cancer. Oncology Reports. 2012;28(6):2211–2216. doi: 10.3892/or.2012.2053. [DOI] [PubMed] [Google Scholar]
- 156.Tanizaki J, Okamoto I, Fumita S, Okamoto W, Nishio K, Nakagawa K. Roles of BIM induction and survivin downregulation in lapatinib-induced apoptosis in breast cancer cells with HER2 amplification. Oncogene. 2011;30(39):4097–4106. doi: 10.1038/onc.2011.111. [DOI] [PubMed] [Google Scholar]
- 157.Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1α signal pathways increases resistance to apoptosis by up-regulating Survivin gene expression. Journal of Biological Chemistry. 2006;281(36):25903–25914. doi: 10.1074/jbc.M603414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, Kuramitsu Y, Baron B, Kitagawa T, Tokuda K, Akada J, Maehara S, Maehara Y, Nakamura K. PI3K inhibitor LY294002 , as opposed to wortmannin, enhances AKT phosphorylation in gemcitabine-resistant pancreatic cancer cells. International Journal of Oncology. 2017;50(2):606–612. doi: 10.3892/ijo.2016.3804. [DOI] [PubMed] [Google Scholar]
- 159.Siddiqa A, Long LM, Li L, Marciniak RA, Kazhdan I. Expression of HER-2 in MCF-7 breast cancer cells modulates antiapoptotic proteins Survivin and Bcl-2 via the extracellular signal-related kinase (ERK) and phosphoinositide-3 kinase (PI3K) signalling pathways. BMC Cancer. 2008;8:129. doi: 10.1186/1471-2407-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Okamoto K, Okamoto I, Okamoto W, Tanaka K, Takezawa K. Role of survivin in EGFR inhibitor-induced apoptosis in non-small cell lung cancers positive for EGFR mutations. Cancer Research. 2010;70(24):10402–10410. doi: 10.1158/0008-5472.CAN-10-2438. [DOI] [PubMed] [Google Scholar]
- 161.Carter BZ, Mak DH, Schober WD, Cabreira-hansen M, Beran M, Mcqueen T, Chen W, Andreeff M. Regulation of survivin expression through Bcr-Abl/ MAPK cascade : targeting survivin overcomes imatinib resistance and increases imatinib sensitivity in imatinib-responsive CML cells. Blood. 2006;107(4):1555–1564. doi: 10.1182/blood-2004-12-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Arora R, Yates C, Gary BD, Mcclellan S, Tan M, Xi Y, Reed E, Piazza GA, Owen LB, Dean-colomb W. Panepoxydone targets NF-kB and FOXM1 to inhibit proliferation, induce apoptosis and reverse epithelial to mesenchymal transition in breast cancer. PloS One. 2014;9(6):e98370. doi: 10.1371/journal.pone.0098370. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 163.Stella S, Tirro’ E, Conte E, Stagno F, Di Raimondo F, Manzella L, Vigneri P. Suppression of survivin induced by a BCR-ABL/JAK2/STAT3 pathway sensitizes imatinib-resistant CML cells to different cytotoxic drugs. Molecular Cancer Therapeutics. 2013;12(6):1085–1099. doi: 10.1158/1535-7163.MCT-12-0550. [DOI] [PubMed] [Google Scholar]
- 164.Sun Y, Moretti L, Giacalone NJ, Speirs CK, Carbone DP, Lu B. Inhibition of JAK2 signaling by TG101209 enhances radiotherapy in lung cancer models. Journal of Thoracic Oncology. 2011;6(4):699–706. doi: 10.1097/JTO.0b013e31820d9d11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang K, Li L, Meng Y, You Y, Fu X, Song L. Arctigenin promotes apoptosis in ovarian cancer cells via the iNOS/NO/STAT3/Survivin signalling. Basic & Clinical Parmacology & Toxicology. 2014;115(6):507–511. doi: 10.1111/bcpt.12270. [DOI] [PubMed] [Google Scholar]
- 166.Wang H, Jin J, Wang J. Arctigenin enhances chemosensitivity to cisplatin in human nonsmall lung cancer H460 cells through downregulation of survivin expression. Journal of Biochemical and Molecular Toxicology. 2014;28(1):39–45. doi: 10.1002/jbt.21533. [DOI] [PubMed] [Google Scholar]
- 167.Martínez-García D, Pérez-Hernández M, Korrodi-Gregório L, Quesada R, Ramos R, Baixeras N, Pérez-Tomás R, Soto-Cerrato V. The Natural-Based Antitumor Compound T21 Decreases Survivin Levels through Potent STAT3 Inhibition in Lung Cancer Models. Biomolecules. 2019;9(8):361. doi: 10.3390/biom9080361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duan Z, Bradner JE, Greenberg E, Levine R, Foster R, Mahoney J, Seiden MV. SD-1029 inhibits signal transducer and activator of transcription 3 nuclear translocation. Clinical Cancer Research. 2006;12(22):6844–6853. doi: 10.1158/1078-0432.CCR-06-1330. [DOI] [PubMed] [Google Scholar]
- 169.Nakamura H, Taguchi A, Kawana K, Baba S, Kawata A, Yoshida M, Fujimoto A, Ogishima J, Sato M, Inoue T, Nishida H, Furuya H, Yamashita A, Eguchi S, Tomio K, Mori-Uchino M, Adachi K, Arimoto T, Wada-Hiraike O, Oda K, Nagamatsu T, Osuga Y, Fuji T. Therapeutic significance of targeting survivin in cervical cancer and possibility of combination therapy with TRAIL. Oncotarget. 2018;9(17):13451–13461. doi: 10.18632/oncotarget.24413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jutooru I, Chadalapaka G, Lei P, Safe S. Inhibition of NFκB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein downregulation. The Journal of Biological Chemistry. 2010;285(33):25332–25344. doi: 10.1074/jbc.M109.095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kanwar JR, Kamalapuram SK, Kanwar RK. Targeting survivin in cancer: The cell-signalling perspective. Drug Discovery Today. 2011;16(11/12):485–494. doi: 10.1016/j.drudis.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 172.Priyadarsini RV, Murugan RS, Sripriya P, Karunagaran D, Nagini S. The neem limonoids azadirachtin and nimbolide induce cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells. Free Radical Research. 2010;44(6):624–634. doi: 10.3109/10715761003692503. [DOI] [PubMed] [Google Scholar]
- 173.Sakoguchi-okada N, Takahashi-yanaga F, Fukada K. Celecoxib inhibits the expression of survivin via the suppression of promoter activity in human colon cancer cells. Biochemical Pharmacology. 2007;73(9):1318–1329. doi: 10.1016/j.bcp.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 174.Seliger C, Meier CR, Becker C, Jick SS, Bogdahn U, Hau P, Leitzmann MF. Use of selective cyclooxygenase-2 inhibitors, other analgesics, and risk of glioma. PloS One. 2016;11(2):e0149293. doi: 10.1371/journal.pone.0149293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin J, Hsiao P, Chiu TH, Chao J. Combination of cyclooxygenase-2 inhibitors and oxaliplatin increases the growth inhibition and death in human colon cancer cells. Biochemical Pharmacology. 2005;70(5):658–667. doi: 10.1016/j.bcp.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 176.Yin H, Que R, Liu C, Ji W, Sun B, Lin X, Zhang Q, Zhao X, Peng Z, Zhang X, Qian H, Chen L, Yao Y, Su C. Survivin-targeted drug screening platform identifies a matrine derivative WM-127 as a potential therapeutics against hepatocellular carcinoma. Cancer Letters. 2018;425:54–64. doi: 10.1016/j.canlet.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 177.Hsiao P, Chang C, Liu H, Tsai C, Chiu TH, Chao J. Activation of p38 mitogen-activated protein kinase by celecoxib oppositely regulates survivin and gamma-H2AX in human colorectal cancer cells. Toxicology and Applied Pharmacology. 2007;222(1):97–104. doi: 10.1016/j.taap.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 178.Guo Q, Chen Y, Zhang B, Kang M, Xie Q, Wu Y. Potentiation of the effect of gemcitabine by emodin in pancreatic cancer is associated with survivin inhibition. Biochemical Pharmacology. 2009;77(11):1674–1683. doi: 10.1016/j.bcp.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 179.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo J, Wong S, Yeop H, Hwan S, Ryul J, Kahn M. A small molecule inhibitor of β-catenin/CREB-binding protein transcription. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kushwaha PP, Vardhan PS, Kapewangolo P, Shuaib M, Prajapati SK, Singh AK, Kumar S. Bulbine frutescens phytochemical inhibits notch signaling pathway and induces apoptosis in triple negative and luminal breast cancer cells. Life Sciences. 2019;234:116783. doi: 10.1016/j.lfs.2019.116783. [DOI] [PubMed] [Google Scholar]
- 181.Koprowski S, Sokolowski K, Kunnimalaiyaan S, Gamblin TC, Kunnimalaiyaan M. Curcumin-mediated regulation of Notch1/hairy and enhancer of split-1/survivin : molecular targeting in cholangiocarcinoma. Journal of Surgical Research. 2015;198(2):434–440. doi: 10.1016/j.jss.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 182.Modi S, Kir D, Banerjee S, Saluja A. Control of apoptosis in treatment and biology of pancreatic cancer. Journal of Cellular Biochemistry. 2016;117(2):279–288. doi: 10.1002/jcb.25284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chowdhury S, Howell GM, Rajput A, Teggart CA, Brattain LE, Weber HR, Chowdhury A, Brattain MG. Identification of a novel TGFβ/PKA signaling transduceome in mediating control of cell survival and metastasis in colon cancer. PloS One. 2011;6(5):e19335. doi: 10.1371/journal.pone.0019335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Leiphrakpam PD, Brattain MG, Black JD, Wang J. TGFβ and IGF1R signaling activates protein kinase A through differential regulation of ezrin phosphorylation in colon cancer cells. Journal of Biological Chemistry. 2018;293(21):8242–8254. doi: 10.1074/jbc.RA117.001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Park JH, Kim YH, Park EH, Lee SJ, Kim H, Kim A, Lee SB, Shim S, Jang H, Myung JK, Park S, Lee SJ, Kim MJ. Effects of metformin and phenformin on apoptosis and epithelial-mesenchymal transition in chemoresistant rectal cancer. Cancer science. 2019;110(9):2834–2845. doi: 10.1111/cas.14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shi X, Wang D, Ding K, Lu Z, Jin Y, Zhang J, Shi X, Wang D, Ding K, Lu Z, Jin Y, Zhang J, Pan J. GDP366 , a novel small molecule dual inhibitor of survivin and Op18, induces cell growth inhibition , cellular senescence and mitotic catastrophe in human cancer cells. Cancer Biology and Therapy. 2010;9(8):640–650. doi: 10.4161/cbt.9.8.11269. [DOI] [PubMed] [Google Scholar]
- 187.Coumar MS, Tsai FY, Kanwar JR, Sarvagalla S, Cheung CHA. Treat cancers by targeting survivin: Just a dream or future reality? Cancer Treatment Reviews. 2013;39(7):802–811. doi: 10.1016/j.ctrv.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 188.Wiechno P, Somer BG, Mellado B, Chłosta PL, Grau JMC, Castellano D, Reuterg C, Stöckle M, Kamradth J, Pikiel J, Durán I, Wedel S, Callies S, André V, Hurt K, Brown J, Lahn M, Heinrich B. A randomised phase 2 study combining LY2181308 sodium (survivin antisense oligonucleotide) with first-line docetaxel/prednisone in patients with castration-resistant prostate cancer. European Urology. 2014;65(3):516–520. doi: 10.1016/j.eururo.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 189.Jha K, Shukla M, Pandey M. Survivin expression and targeting in breast cancer. Surgical Oncology. 2012;21(2):125–131. doi: 10.1016/j.suronc.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 190.Zaffaroni N, Pennati M, Daidone MG. Survivin as a target for new anticancer interventions. Journal of Cellular and Molecular Medicine. 2005;9(2):360–372. doi: 10.1111/j.1582-4934.2005.tb00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huang J, Lyu H, Wang J, Liu B. microRNA regulation and therapeutic targeting of survivin in cancer. American Journal for Cancer Research. 2014;5(1):20–31. [PMC free article] [PubMed] [Google Scholar]
- 192.Paduano F, Villa R, Pennati M, Folini M, Binda M, Daidone MG, Zaffaroni N. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells. Molecular Cancer Therapeutics. 2006;5(1):179–186. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- 193.Zhang L, Liang H, Cao W, Xu R, Ju XL. Downregulation of survivin by siRNA inhibits invasion and promotes apoptosis in neuroblastoma SH-SY5Y cells. Brazilian Journal of Medical and Biological Research. 2014;47(7):548–553. doi: 10.1590/1414-431X20143459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Saini S, Majid S, Yamamura S, Tabatabai L, Suh SO, Shahryari V, Chen Y, Deng G, Tanaka Y, Dahiya R. Regulatory role of miR-203 in prostate cancer progression and metastasis. Clinical Cancer Research. 2011;17(16):5287–5298. doi: 10.1158/1078-0432.CCR-10-2619. [DOI] [PubMed] [Google Scholar]
- 195.Xu D, Wang Q, An Y, Xu L. MiR-203 regulates the proliferation, apoptosis and cell cycle progression of pancreatic cancer cells by targeting Survivin. Molecular Medicine Reports. 2013;8(2):379–384. doi: 10.3892/mmr.2013.1504. [DOI] [PubMed] [Google Scholar]
- 196.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PloS One. 2012;7(5):e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cao W, Fan R, Wang L, Cheng S, Li H, Jiang J, Geng M, Jin Y, Wu Y. Expression and regulatory function of miRNA-34a in targeting survivin in gastric cancer cells. Tumour Biology. 2013;34(2):963–971. doi: 10.1007/s13277-012-0632-8. [DOI] [PubMed] [Google Scholar]
- 198.Li S, Sun Y, Tian T, Qin X, Lin S, Zhang T, Zhang Q, Zhou M, Zhang X, Zhou Y, Zhao H, Zhu B, Cai X. MicroRNA-214-3p modified tetrahedral framework nucleic acids target survivin to induce tumour cell apoptosis. Cell Proliferation. 2019;53(1):e12708. doi: 10.1111/cpr.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Lian H, Yan X, Zhang S, Chen X, Fang F, Guo H, Zhang C. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Molecular Cancer. 2016;15(1):82. doi: 10.1186/s12943-016-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang M, Cai WR, Meng R, Chi JR, Li YR, Chen AX, Yu Y, Cao XC. miR-485-5p suppresses breast cancer progression and chemosensitivity by targeting survivin. Biochemical and Biophysical Research Communications. 2018;501(1):48–54. doi: 10.1016/j.bbrc.2018.04.129. [DOI] [PubMed] [Google Scholar]
- 201.Pennati M, Colella G, Folini M, Citti L, Daidone MG, Zaffaroni N. Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatin-induced apoptosis. The Journal of clinical investigation. 2002;109(2):285–286. doi: 10.1172/JCI14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Pennati M, Binda M, De Cesare M, Pratesi G, Folini M, Citti L, Daidone MG, Zunino F, Zaffaroni N. Ribozyme-mediated down-regulation of survivin expression sensitizes human melanoma cells to topotecan in vitro and in vivo. Carcinogenesis. 2004;25(7):1129–1136. doi: 10.1093/carcin/bgh107. [DOI] [PubMed] [Google Scholar]
- 203.Reed JC, Wilson DB. Cancer immunotherapy targeting survivin. Clinical Cancer Research. 2003;9(17):6310–6315. [PubMed] [Google Scholar]
- 204.Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin : a unique target for tumor therapy. Cancer Cell International. 2016;16:49. doi: 10.1186/s12935-016-0326-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Pisarev V, Yu B, Salup R, Sherman S, Altieri DC, Gabrilovich DI. Full-length dominant-negative Survivin for cancer immunotherapy. Clinical Cancer Research. 2003;9(17):6523–6533. [PubMed] [Google Scholar]
- 206.Weber G, Caruana I, Rouce RH, Barrett AJ, Gerdemann U, Leen AM, Rabin KR, Bollard CM. Generation of tumor antigen-specific T cell lines from pediatric patients with acute lymphoblastic leukemia--implications for immunotherapy. Clinical Cancer Research : An Official Journal Of The American Association For Cancer Research. 2013;19(18):5079–5091. doi: 10.1158/1078-0432.CCR-13-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li D, He S. MAGE3 and Survivin activated dendritic cell immunotherapy for the treatment of non-small cell lung cancer. Oncology Letters. 2018;15(6):8777–8783. doi: 10.3892/ol.2018.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Widenmeyer M, Griesemann H, Stevanovic S, Feyerabend S, Klein R, Attig S, Hennenlotter J, Wernet D, Kuprash DV, Sazykin AY, Pascolo S, Stenzl A, Ce’cile G, Rammensee HG. Promiscuous survivin peptide induces robust CD41 T-cell responses in the majority of vaccinated cancer patients. International Journal of Cancer. 2012;131(1):140–149. doi: 10.1002/ijc.26365. [DOI] [PubMed] [Google Scholar]
- 209.Arber C, Feng X, Abhyankar H, Romero E, Wu MF, Heslop HE, Barth P, Dotti G, Savoldo B. Survivin-specific T cell receptor targets tumor but not T cells. The Journal Of Clinical Investigation. 2015;125(1):157–168. doi: 10.1172/JCI75876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rapoport AP, Aqui NA, Stadtmauer EA, Vogl DT, Fang H, Cai L, Janofsky S, Chew A, Storek J, Akpek G, Badros A, Yanovich S, Tan MT, Veloso E, Pasetti MF, Cross A, Philip S, Murphy H, Bhagat R, Zheng Z, Milliron T, Cotte J, Cannon A, Levine BL, Vonderheide RH, June CH. Combination immunotherapy using adoptive T-cell transfer and tumor antigen vaccination on the basis of hTERT and survivin after ASCT for myeloma. Blood. 2011;117(3):788–798. doi: 10.1182/blood-2010-08-299396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ichiki Y, Hanagiri T, Takenoyama M, Baba T, Fukuyama T, Nagata Y, Mizukami M, So T, Sugaya M, Yasuda M, So T, Sugio K, Yasumoto K. Tumor specific expression of survivin-2B in lung cancer as a novel target of immunotherapy. Lung Cancer. 2005;48(2):281–289. doi: 10.1016/j.lungcan.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 212.Berinstein NL, Karkada M, Oza AM, Odunsi K, Villella JA, Nemunaitis JJ, Morse MA, Pejovic T, Bentley J, Buyse M, Nigam R, Weir GM, MacDonald LD, Quinton T, Rajagopalan R, Sharp K, Penwell A, Sammatur L, Burzykowski T, Stanford MM, Mansour M. Survivin-targeted immunotherapy drives robust polyfunctional T cell generation and differentiation in advanced ovarian cancer patients. OncoImmunology. 2015;4(8):e1026529. doi: 10.1080/2162402X.2015.1026529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nitschke, N. J., Bjoern, J., Iversen, T. Z., Andersen, M. H., & Svane, I. M. (2017). Indoleamine 2, 3-dioxygenase and survivin peptide vaccine combined with temozolomide in metastatic melanoma. Stem Cell Investigation, 4(77). 10.21037/sci.2017.08.06. [DOI] [PMC free article] [PubMed]
- 214.Zhu H, Zhang G, Wang Y, Xu N, He S, Zhang W, Chen M, Liu M, Quan L, Bai J, Xu N. Inhibition of ErbB2 by Herceptin reduces survivin expression via the ErbB2–β-cateninTCF4-survivin pathway in ErbB2-overexpressed breast cancer cells. Cancer Science. 2010;101(5):1156–1162. doi: 10.1111/j.1349-7006.2010.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, Reguart N, Moody TW, Kitajewski J, Mccormick F, Jablons DM. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23(36):6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 216.Desai A, Yan Y, Gerson SL. Concise reviews: Cancer stem cell targeted therapies: Toward clinical success. Stem Cells Translational Medicine. 2019;8(1):75–81. doi: 10.1002/sctm.18-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduction and Targeted Therapy. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chaurasiya S, Chen NG, Warner SG. Oncolytic virotherapy versus cancer stem cells: A review of approaches and mechanisms. Cancers. 2018;10(4):124. doi: 10.3390/cancers10040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhang J, Liu S, Ye Q, Pan J. Transcriptional inhibition by CDK7/9 inhibitor SNS-032 abrogates oncogene addiction and reduces liver metastasis in uveal melanoma. Molecular Cancer. 2019;18(1):140. doi: 10.1186/s12943-019-1070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ma X, Zhang Y, Kang Y, Li L, Zheng W. A recombinant protein TmSm(T34A) can inhibit proliferation and proapoptosis to breast cancer stem cells(BCSCs) by down-regulating the expression of Cyclin D1. Biomedicine and Pharmacotherapy. 2016;84:373–381. doi: 10.1016/j.biopha.2016.08.066. [DOI] [PubMed] [Google Scholar]
- 221.Pozzoli, G., Marei, H. E., Althani, A., Boninsegna, A., Casalbore, P., Marlier, L. N. J. L., Lanzilli, G., Zonfrillo, M., Petrucci, G., Rocca, B., Navarra, P., Sgambato, A., & Cenciarelli, C. (2019). Aspirin inhibits cancer stem cells properties and growth of glioblastoma multiforme through Rb1 pathway modulation. Journal of Cellular Physiology. 10.1002/jcp.28194, 10.1002/jcp.28194. [DOI] [PubMed]
- 222.Liu L, Wang Q, Mao J, Qin T, Sun Y, Yang J, Han Y, Li L, Li Q. Salinomycin suppresses cancer cell stemness and attenuates TGF-β-induced epithelial-mesenchymal transition of renal cell carcinoma cells. Chemico-Biological Interactions. 2018;296:145–153. doi: 10.1016/j.cbi.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 223.Sanomachi T, Suzuki S, Kuramoto K, Takeda H, Sakaki H, Togashi K, Seino S, Yoshioka T, Okada M, Kitanaka C. Olanzapine, an atypical antipsychotic, inhibits survivin expression and sensitizes cancer cells to chemotherapeutic agents. Anticancer Research. 2017;37(11):6177–6188. doi: 10.21873/anticanres.12067. [DOI] [PubMed] [Google Scholar]
- 224.Suzuki S, Okada M, Kuramoto K, Takeda H, Sakaki H, Watarai H, Sanomachi T, Seino S, Yoshioka T, Kitanaka C. Aripiprazole, an antipsychotic and partial dopamine agonist, inhibits cancer stem cells and reverses chemoresistance. Anticancer Research. 2016;36(10):5153–5161. doi: 10.21873/anticanres.11085. [DOI] [PubMed] [Google Scholar]
- 225.Zhang Y, Chen HX, Zhou SY, Wang SX, Zheng K, Xu DD, Liu YT, Wang XY, Wang X, Yan HZ, Zhang L, Liu QY, Chen WQ, Wang YF. Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Molecular Cancer. 2015;14:56. doi: 10.1186/s12943-015-0326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang R, Zhang P, Wang H, Hou D, Li W, Xiao G, Li C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44+CD117+ ovarian cancer stem cells. Stem Cell Research & Therapy. 2015;6:262. doi: 10.1186/s13287-015-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang Y, Lapidus RG, Liu P, Choi EY, Adediran S, Hussain A, Wang X, Liu X, Dan HC. Targeting IκB Kinase β/NF-κB Signaling in Human Prostate Cancer by a Novel IκB Kinase β Inhibitor CmpdA. Molecular Cancer Therapeutics. 2016;15(7):1504–1514. doi: 10.1158/1535-7163.MCT-15-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lai SW, Bamodu OA, Tsai WC, Chang YM, Lee WH, Yeh CT, Chao TY. The therapeutic targeting of the FGFR1/Src/NF-κB signaling axis inhibits pancreatic ductal adenocarcinoma stemness and oncogenicity. Clinical and Experimental Metastasis. 2018;35(7):663–677. doi: 10.1007/s10585-018-9919-5. [DOI] [PubMed] [Google Scholar]
- 229.Hu Y, Yagüe E, Zhao J, Wang L, Bai J, Yang Q, Pan T, Zhao H, Liu J, Zhang J. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Letters. 2018;423:47–59. doi: 10.1016/j.canlet.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 230.Kim YJ, Kim JY, Lee N, Oh E, Sung D, Cho TM, Seo JH. Disulfiram suppresses cancer stem-like properties and STAT3 signaling in triple-negative breast cancer cells. Biochemical and Biophysical Research Communications. 2017;486(4):1069–1076. doi: 10.1016/j.bbrc.2017.03.164. [DOI] [PubMed] [Google Scholar]
- 231.Sam MR, Ahangar P, Nejati V, Habibian R. Treatment of LS174T colorectal cancer stem-like cells with n-3 PUFAs induces growth suppression through inhibition of survivin expression and induction of caspase-3 activation. Cellular Oncology (Dordrecht) 2016;39(1):69–77. doi: 10.1007/s13402-015-0254-4. [DOI] [PubMed] [Google Scholar]
- 232.Ho TF, Peng YT, Chuang SM, Lin SC, Feng BL, Lu CH, Yu WJ, Chang JS, Chang CC. Prodigiosin down-regulates survivin to facilitate paclitaxel sensitization in human breast carcinoma cell lines. Toxicology and Applied Pharmacology. 2009;235(2):253–260. doi: 10.1016/j.taap.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 233.Erdogan S, Turkekul K, Dibirdik I, Doganlar O, Doganlar ZB, Bilir A, Oktem G. Midkine downregulation increases the efficacy of quercetin on prostate cancer stem cell survival and migration through PI3K/AKT and MAPK/ERK pathway. Biomedicine and Pharmacotherapy. 2018;107:793–805. doi: 10.1016/j.biopha.2018.08.061. [DOI] [PubMed] [Google Scholar]
- 234.Togashi K, Okada M, Yamamoto M, Suzuki S, Sanomachi T, Seino S, Yamashita H, Kitanaka C. A small-molecule kinase inhibitor, CEP-1347, inhibits survivin expression and sensitizes ovarian cancer stem cells to paclitaxel. Anticancer Research. 2018;38(8):4535–4542. doi: 10.21873/anticanres.12757. [DOI] [PubMed] [Google Scholar]
- 235.Wang J, Liu Z, Zhang D, Liu R, Lin Q, Liu J, Yang Z, Ma Q, Sun D, Zhou X, Jiang G. FL118, a novel survivin inhibitor, wins the battle against drug-resistant and metastatic lung cancers through inhibition of cancer stem cell-like properties. American Journal of Translational Research. 2017;9(8):3676–3686. [PMC free article] [PubMed] [Google Scholar]
- 236.Yang Z, Ji L, Jiang G, Liu R, Liu Z, Yang Y, Ma Q, Zhao H. FL118, a novel camptothecin analogue, suppressed migration and invasion of human breast cancer cells by inhibiting epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway. Bioscience Trends. 2018;12(1):40–46. doi: 10.5582/bst.2017.01288. [DOI] [PubMed] [Google Scholar]
- 237.Manegold P, Lai K, Wu Y, Teo JL, Lenz HJ, Genyk YS, Pandol SJ, Wu K, Lin DP, Chen Y, Nguyen C, Zhao Y, Kahn M. Differentiation therapy targeting the β-Catenin/CBP interaction in pancreatic cancer. Cancers. 2018;10(4):95. doi: 10.3390/cancers10040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhao Y, Masiello D, McMillian M, Nguyen C, Wu Y, Melendez E, Smbatyan G, Kida A, He Y, Teo JL, Kahn M. CBP/Catenin antagonist safely eliminates drug resistant leukemia initiating cells. Oncogene. 2016;35(28):3705–3717. doi: 10.1038/onc.2015.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Thomas PD, Kahn M. Kat3 coactivators in somatic stem cells and cancer stem cells: biological roles, evolution, and pharmacologic manipulation. Cell Biology and Toxicology. 2016;32(1):61–81. doi: 10.1007/s10565-016-9318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suzuki S, Yamamoto M, Sanomachi T, Togashi K, Sugai A, Seino S, Yoshioka T, Kitanaka C, Okada M. Brexpiprazole, a serotonin-dopamine activity modulator, can sensitize glioma stem cells to Osimertinib, a third-generation EGFR-TKI, via Survivin Reduction. Cancers. 2019;11(7):947. doi: 10.3390/cancers11070947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Suzuki S, Yamamoto M, Togashi K, Sanomachi T, Sugai A, Seino S, Yoshioka T, Kitanaka C, Okada M. In vitro and in vivo anti-tumor effects of brexpiprazole, a newly-developed serotonin-dopamine activity modulator with an improved safety profile. Oncotarget. 2019;10(37):3547–3558. doi: 10.18632/oncotarget.26949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Erdogan S, Turkekul K, Serttas R, Erdogan Z. The natural flavonoid apigenin sensitizes human CD44+ prostate cancer stem cells to cisplatin therapy. Biomedicine and Pharmacotherapy. 2017;88:210–217. doi: 10.1016/j.biopha.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 243.Gersey ZC, Rodriguez GA, Barbarite E, Sanchez A, Walters WM, Ohaeto KC, Komotar RJ, Graham RM. Curcumin decreases malignant characteristics of glioblastoma stem cells via induction of reactive oxygen species. BMC Cancer. 2017;17(1):99. doi: 10.1186/s12885-017-3058-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wu N, Liu J, Zhao X, Yan Z, Jiang B, Wang L, Cao S, Shi D, Lin X. Cardamonin induces apoptosis by suppressing STAT3 signaling pathway in glioblastoma stem cells. Tumor Biology. 2015;36(12):9667–9676. doi: 10.1007/s13277-015-3673-y. [DOI] [PubMed] [Google Scholar]
- 245.Tang JH, Yang L, Chen JX, Li QR, Zhu LR, Xu QF, Huang GH, Zhang ZX, Xiang Y, Du L, Zhou Z, Lv SQ. Bortezomib inhibits growth and sensitizes glioma to temozolomide (TMZ) via down-regulating the FOXM1-Survivin axis. Cancer Communications (London, England) 2019;39(1):81. doi: 10.1186/s40880-019-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sanomachi T, Suzuki S, Togashi K, Sugai A, Seino S, Okada M, Yoshioka T, Kitanaka C, Yamamoto M. Spironolactone, a classic potassium-sparing diuretic, reduces Survivin expression and chemosensitizes cancer cells to non-DNA-damaging anticancer drugs. Cancers. 2019;11(10):1550. doi: 10.3390/cancers11101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Takeda H, Okada M, Suzuki S, Kuramoto K, Sakaki H, Watarai H, Sanomachi T, Seino S, Yoshioka T, Kitanaka C. Rho-associated protein kinase (ROCK) inhibitors inhibit survivin expression and sensitize pancreatic cancer stem cells to gemcitabine. Anticancer Research. 2016;36(12):6311–6318. doi: 10.21873/anticanres.11227. [DOI] [PubMed] [Google Scholar]
- 248.Yamamoto M, Suzuki S, Keita T, Sanomachi T, Seino S, Kitanaka C, Okada M. AS602801 sensitizes ovarian cancer stem cells to paclitaxel by down-regulating MDR1. Anticancer Research. 2019;39(2):609–617. doi: 10.21873/anticanres.13154. [DOI] [PubMed] [Google Scholar]
- 249.Yamamoto M, Suzuki S, Togashi K, Sanomachi T, Seino S, Kitanaka C, Okada M. AS602801, an anticancer stem cell candidate drug, reduces survivin expression and sensitizes A2780 ovarian cancer stem cells to carboplatin and paclitaxel. Anticancer Research. 2018;38(12):6699–6706. doi: 10.21873/anticanres.13038. [DOI] [PubMed] [Google Scholar]
- 250.Bamodu OA, Kuo KT, Yuan LP, Cheng WH, Lee WH, Ho YS, Chao TY, Yeh CT. HDAC inhibitor suppresses proliferation and tumorigenicity of drug-resistant chronic myeloid leukemia stem cells through regulation of hsa-miR-196a targeting BCR/ABL1. Experimental Cell Research. 2018;370(2):519–530. doi: 10.1016/j.yexcr.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 251.Wang T, Gantier MP, Xiang D, Bean AG, Bruce M, Zhou SF, Khasraw M, Ward A, Wang L, Wei MQ, AlShamaileh H, Chen L, She X, Lin J, Kong L, Shigdar S, Duan W. EpCAM aptamer-mediated survivin silencing sensitized cancer stem cells to Doxorubicin in a breast cancer model. Theranostics. 2015;5(12):1456–1472. doi: 10.7150/thno.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Alshamaileh H, Wang T, Xiang D, Yin W, Tran PHL, Barrero RA, Zhang PZ, Li Y, Kong L, Liu K, Zhou SF, Hou Y, Shigdar S, Duan W. Aptamer-mediated survivin RNAi enables 5-fluorouracil to eliminate colorectal cancer stem cells. Scientific Reports. 2017;7(1):5898. doi: 10.1038/s41598-017-05859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Jeong J, Kang H, Kim T, Kim G. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Letters. 2016;386:168–178. doi: 10.1016/j.canlet.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 254.Zhang, Y., Zhou, S. Y., Yan, H. Z., Xu, D. D., Chen, H. X., Wang, X. Y., Wang, X., Liu, Y. T., Zhang, L., Wang, S., Zhou, P. J., Fu, W. Y., Ruan, B. B., Ma, D. L., Wang, Y., Liu, Q. Y., Ren, Z., Liu, Z., Zhang, R., & Wang, Y. F. (2016). miR-203 inhibits proliferation and self-renewal of leukemia stem cells by targeting survivin and Bmi-1. Scientific Reports, 6(19995). 10.1038/srep19995. [DOI] [PMC free article] [PubMed]
- 255.Liu B, Ma H, Liu Q, Xiao Y, Pan S, Zhou H, Jia L. MiR-29b/Sp1/FUT4 axis modulates the malignancy of leukemia stem cells by regulating fucosylation via Wnt/β-catenin pathway in acute myeloid leukemia. Journal Of Experimental & Clinical Cancer Research : CR. 2019;38(1):200. doi: 10.1186/s13046-019-1179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ning X, Wang C, Zhang M, Wang K. Ectopic expression of miR-147 inhibits stem cell marker and epithelial–mesenchymal transition (EMT)-related protein expression in colon cancer cells. Oncology Research. 2019;27(4):399–406. doi: 10.3727/096504018X15179675206495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Suzuki S, Kofune H, Uozumi K, Yoshimitsu M, Arima N, Ishitsuka K, Ueno SI, Kosai KI. A survivin-responsive, conditionally replicating adenovirus induces potent cytocidal effects in adult T-cell leukemia/lymphoma. BMC Cancer. 2019;19(1):516. doi: 10.1186/s12885-019-5730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nandi S, Ulasov IV, Tyler MA, Sugihara AQ, Molinero L, Han Y, Zhu ZB, Lesniak MS. Low-dose radiation enhances survivin-mediated virotherapy against malignant glioma stem cells. Cancer Research. 2008;68(14):5778–5784. doi: 10.1158/0008-5472.CAN-07-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Tanoue K, Wang Y, Ikeda M, Mitsui K, Irie R, Setoguchi T, Komiya S, Natsugoe S, Kosai KI. Survivin-responsive conditionally replicating adenovirus kills rhabdomyosarcoma stem cells more efficiently than their progeny. Journal of Translational Medicine. 2014;12:27. doi: 10.1186/1479-5876-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mooney R, Majid AA, Batalla-Covello J, Machado D, Liu X, Gonzaga J, Tirughana R, Hammad M, Lesniak MS, Curiel DT, Aboody KS. Enhanced delivery of oncolytic adenovirus by neural stem cells for treatment of metastatic ovarian cancer. Molecular Therapy Oncolytics. 2018;12:79–92. doi: 10.1016/j.omto.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kanwar JR, Mahidhara G, Roy K, Sasidharan S, Krishnakumar S, Prasad N, Sehgal R, Kanwar RK. Fe-bLf nanoformulation targets survivin to kill colon cancer stem cells and maintains absorption of iron, calcium and zinc. Nanomedicine. 2015;10(1):35–55. doi: 10.2217/nnm.14.132. [DOI] [PubMed] [Google Scholar]
- 262.Sun X, Chen Y, Zhao H, Qiao G, Liu M, Zhang C, Cui D, Ma L. Dual-modified cationic liposomes loaded with paclitaxel and survivin sirna for targeted imaging and therapy of cancer stem cells in brain glioma. Drug Delivery. 2018;25(1):1718–1727. doi: 10.1080/10717544.2018.1494225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Gomez-Cabrero A, Wrasidlo W, Reisfeld RA. IMD-0354 targets breast cancer stem cells: a novel approach for an adjuvant to chemotherapy to prevent multidrug resistance in a murine model. PloS One. 2013;8(8):e73607. doi: 10.1371/journal.pone.0073607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Roy K, Kanwar RK, Krishnakumar S, Cheung CHA, Kanwar JR. Competitive inhibition of survivin using a cell-permeable recombinant protein induces cancer-specific apoptosis in colon cancer model. International Journal of Nanomedicine. 2015;10:1019–1043. doi: 10.2147/IJN.S73916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mazur J, Roy K, Kanwar JR. Recent advances in nanomedicine and survivin targeting in brain cancers. Nanomedicine. 2017;13(1):105–137. doi: 10.2217/nnm-2017-0286. [DOI] [PubMed] [Google Scholar]
- 266.Xiao Y, Wang C, Xiong M, Yang Y, Zhang Q, Liu X, Zou B, Gou M. Self-assembled nanoparticle mediated Survivin-T34A for ovarian cancer therapy. Journal of Biomedical Nanotechnology. 2018;14(12):2092–2101. doi: 10.1166/jbn.2018.2641. [DOI] [PubMed] [Google Scholar]
- 267.Buchwald P. Small-molecule protein-protein interaction inhibitors: Therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life. 2010;62(10):724–731. doi: 10.1002/iub.383. [DOI] [PubMed] [Google Scholar]
- 268.Xia W, Bisi J, Strum J, Liu L, Carrick K, Graham KM, Treece AL, Hardwicke MA, Dush M, Liao Q, Westlund RE, Zhao S, Bacus S, Spector NL. Regulation of survivin by ErbB2 signaling: Therapeutic implications for ErbB2-overexpressing breast cancers. Cancer Research. 2006;66(3):1640–1647. doi: 10.1158/0008-5472.CAN-05-2000. [DOI] [PubMed] [Google Scholar]
- 269.Coumar MS, Tsai FY, Kanwar JR, Sarvagalla S, Cheung CHA. Treat cancers by targeting survivin: Just a dream or future reality? Cancer Treatment Reviews. 2013;39(7):802–811. doi: 10.1016/j.ctrv.2013.02.00. [DOI] [PubMed] [Google Scholar]