Abstract
Sorghum is a C4 cereal grain crop which is well adapted to harsh environment. It is a potential model for gaining better understanding of the molecular mechanism due to its wider adaptability to abiotic stresses. In this study, protein extraction was standardized using different methods to study the electrophoretic pattern of sorghum leaves under different salinity levels. The extraction of soluble protein with lysis buffer, followed by its clean-up was found to be the most effective method. The different profiles of salt-responsive proteins were analyzed in G-46 and CSV 44F sorghum genotypes based on their tolerance behavior towards salinity. The kafirin level also changed depending upon the concentration and exposure time to salts suggesting the stored proteins as energy source under stress conditions. The relative expression of salt-responsive genes was studied using Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) which might be used as a molecular screening tool for identification of salt-tolerant genotypes in affected areas. The validated responses were examined in terms of metabolic changes and the expression of stress-induced proteins—viz. heat shock proteins (hsp) via immunoblotting assay. The results showed that the two sorghum genotypes adopted distinct approaches in response to salinity, with G-46 performing better in terms of leaf function. Also, we have standardized different protein extraction methods followed by their clean-up for electrophoretic profiling.
Keywords: Electrophoretic profile, Heat shock protein, Polyacrylamide gel electrophoresis, Salinity, Sorghum
Introduction
Among abiotic stresses, salinity is the one of the major abiotic factors that limits the growth, development and productivity of plants (Ngara et al. 2012). The plants grown in saline regions have accumulated a diverse range of ionic compounds and their dissolved salts. Salt-affected soils are estimated to comprise 23% of the cultivated land, approximately 3.5108 ha, global extent of saline soils to be 412Mha, which closely agrees with the FAO. (Abbas et al. 2013; Corwin and Scudiero 2019). But there are no directly measured global inventories of soil salinity. For dry and semi-arid areas, the environmental stresses seem to be more prevalent. While irrigation is often used to offset low rainfall in arable land, over-irrigation can increase the salinity of the ground on a long-term basis and therefore exacerbate the situation (Flowers 2004). More than 50% of the arable land would be affected by rising soil salinization by 2050 (Wang et al. 2004). Salinity imposes oxidative and hyperosmotic stresses, nutrient deficiency, ion toxicity and loss of water retention capacity (Hurkman 1990; Zhu 2001). At cellular level (both cells and tissues), the physiological effect of salinity in plants is comparable to other hyperosmotic stresses such as cold, freezing, and heat (Kasuga et al. 1999; Xiong et al. 2002; Munns and Tester 2008; Sobhanian et al. 2011). Besides tissue and cell dehydration, salt stress also imposes ionic stress in plants. In gene expression trials, some biological and metabolic processes that include the dependence and tolerance on salt stress or those affected by salt stress are established. To keep the levels of active oxygen species under control, plants also have antioxidant defense systems (Gill and Tuteja 2010). It is consistent with the understanding that salt stress induces changes in gene expression like any abiotic stress, which eventually has an effect on the expression of gene products, the proteins (Hasegawa et al. 2000; Seki et al. 2003; Shinozaki et al. 2003; De Groot et al. 2003; Ngara et al. 2012).
Proteomics, the widespread analysis of proteins from a given organism, tissue or cell, is used for the study of the expression of salt stress-responsive proteins (Pandey et al. 2000; van Wijk 2001) in crops such as rice, potatoes and foxtail millet (Parker et al. 2006; Aghaei et al. 2008; Veeranagamallaiah et al. 2008). Several of these studies have been examined by Sobhanians et al. (2011) who identified the unusual proteome changes of many economically important food crops under salt stress. Despite these proteomics reports in food crops under salt-stress, comparative work is still very limited in sorghum; one of the most stress-tolerant commercial grain crop. Kumar Swami et al. (2011) documented changes in sorghum leaf tissues in response to salt stress in the initial cataloging of differential proteome expression.
Sorghum, a C4 plant of grass family, is well known for its adaptability in arid and semi-arid regimes, moderately drought tolerant and being highly biomass productive (Miemyk 1997; Mall et al. 2011). It possesses a diverse range of morphological, physiological, and anatomical characteristics that enable its tolerance to survive in stress conditions with modifications. The adaptability behavior of plant species towards tolerance may depend upon their genetic makeup depending upon the concentration of salts in the soil (Krishnamurthy et al. 2007). The survival of the salt-tolerant cultivars might be due to cellular adaptive mechanisms such as compartmentalization, synthesis and accumulation of organic solutes and osmolytes, ion transport and osmotic adjustments (Buchanan et al. 2005; Almodares et al. 2008; Paterson et al. 2009).
Many researchers have studied physiology and biochemistry specificities of the salt tolerance and also explored methods for controlling the plant's overall success in breeding programs (El Naim et al. 2012; Kausar et al. 2012). The cultivation of salt-tolerant crops in saline soils would allow the maximum use and restoration of soils damaged by salt, which would otherwise be non-productive. A few reports have been documented on the screening of sorghum varieties for salt tolerance, genomic changes in response to drought and high salinity, soluble carbohydrate changes and analysis of growth contributing traits and ion accumulation among various sorghum cultivars (Himani et al. 2019). Salinity induces oxidative stress by effecting plant’s ion homeostasis, thereby increase the formation of reactive oxygen species (ROS) due to imbalance of cellular homeostasis (Zhang et al. 2012). Development of salt-tolerant crops has been a major objective of plant breeding programs for decades to maintain crop productivity in semiarid and saline lands (Krishnamurthy et al. 2007). To cope with this problem, a prerequisite for the development of salt-tolerant crops is the identification of key genetic determinants and stress defense proteins/ salt-tolerant proteins (Ndimba et al. 2005; Sobhanian et al. 2011; Sekhwal et al. 2012). Under saline conditions, there is a change in the pattern of gene expression (qualitative and quantitative) as well as protein synthesis (Ngara et al. 2012; Jia et al. 2015). So, the proteomics study offers a new approach to discover proteins and pathways associated with crop physiological and stress responses and deals with determination, identification, expression profile and protein–protein interactions as well as complexity of biochemical processes under stress and non-stress conditions (Ghosh and Xu 2014; Ahmad et al. 2016; Van Emon 2016). Several studies have been reported in rice, Arabidopsis, and other plant species, but little information is available in sorghum till date (Sekhwal et al. 2012). Hence, it is highly desirable to standardize reliable methods for functional identification of proteins. The standardized methods in the paper potentially may have great application for further understanding of newly identified proteins that may help in plant development.
The proteomics studies provide valuable information concerning the impact of salt stress on gene expression, plant growth and the overproduction of soluble sugars which may work as osmoprotectants, but specific proteins that contribute to salt tolerance mechanisms in sorghum must be identified. Measuring the expression of proteins with proteomic tools would, therefore, provide a better indication of cellular activities under salt stress in sorghum. Thus, the present study aims to standardize and identify salt-stress responsive protein expression pattern using SDS-PAGE and RT-PCR in sorghum leaf extracts under salinity.
Materials and methods
Plant material and salt treatment
Seeds of two sorghum genotypes viz. G-46 and CSV 44F were procured from Forage Section, Department of Genetics and Plant Breeding, CCS Haryana Agricultural University Hisar, Haryana, India. The experiment was conducted at Animal Biotechnology Centre, National Dairy Research Institute, Karnal, Haryana, India. The characteristics of the sorghum genotypes used in the study are given in Table 1. The seeds were then first surface sterilized with 0.01% mercuric chloride (HgCl2) solution for 20 min followed by washing in distilled water. The seeds were planted in plastic trays containing cocopeat: vermiculite: perlite in 3:1:1 ratio. After 7 days of germination, geminated seedlings were exposed to 100 and 120 mM NaCl (treatment groups) using U.S saline laboratory staff solution (US 1954), and their leaves were harvested after time intervals of 24, 48, and 96 h (stressed leaves) and sorghum leaves without any salt treatment were defined as control. The experiment was performed in triplicates. Nutrient solution was given at different intervals (Hoagland and Arnon 1950). The trays were incubated at 25 °C under 16 h light/8 h dark regime for 14 days after which leaf was excised from the seedlings and immediately flash frozen in liquid nitrogen.
Table 1.
Characteristic and origin of sorghum genotypes
| Cultivars | Description | Source | Pedigree | Pericarp |
|---|---|---|---|---|
| G-46 | Germplasm 46 | CCS HAU Hisar, India | Selection from S 202 which is a selection from cross 10626B × 6090 M3-1–1 | Brown |
| S-713/CSV 44F | Haryana Jowar | CCS HAU, Hisar, India | Hybrid 308 × 437–1 | White |
Qualitative estimation by RT-PCR
Total RNA was extracted using SV Total RNA isolation system (Promega, USA). Total RNA was quantified and simultaneously quality was checked using Picodrop (Picodrop Ltd, Cambridge UK). Absorbance at 260 nm gave the concentration of total RNA with an average 500 ng/µl and the purity was evaluated by 260:280 ratio. The samples having a ratio of 1.9 and above had good RNA concentration which was used for further analysis. Single strand cDNA was synthesized from the DNase-treated RNA samples using iScript cDNA synthesis kit (BioRad, USA) with slight modifications. The reaction mixture for PCR was prepared using SybrGreen master mix (Biorad, USA). The reaction conditions were initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 45 s and extension at 72 °C for 60 s and the final extension at 72 °C for 10 min with hold at 4 °C. Twenty five microliter reaction mixture was set up in 0.2 ml PCR tubes for amplification in thermocycler (AB Master Cycler Gradient TM). The PCR products were then stored at − 20 °C for further analysis. The PCR products were analyzed on 1.5% agarose gel containing 0.5 µg/ml ethidium bromide. For estimation of amplicon size, 100 bp DNA ladder (MBI Fermentas) was run along with amplified products in agarose gel electrophoresis. The gels were analyzed using densitometric software Image J 1.51 K.
Primer designing
Primer-BLAST software from NCBI was used for designing the primers for P5CS1, BADH1, H+-PPase, Cu-ZnSOD, and Actin-1 (Table 2). The primers were then custom synthesized from Sigma.
Table 2.
Detail of primers
| Gene | Accession No. | Primers | Sequence (5′–3′) | Product size | Tm |
|---|---|---|---|---|---|
| P5CS1 | GQ377719.2 | Forward | CTGGTGGTATCTGCCATGTT | 496 bp | 62 °C |
| Reverse | TGTATGCGCCCTGTACTTATG | ||||
| BADH1 | U12195.1 | Forward | TCCTCTCCTGATGGCTACAT | 858 bp | 62 °C |
| Reverse | TGTGAGCAGTTTACCCAGATAC | ||||
| Actin-1 | XM_021463392.1 | Forward | CAACTGGGACGATATGGAGAAG | 571 bp | 62 °C |
| Reverse | AATGAAGGATGGCTGGAAGAG | ||||
| Cu–Zn SOD- | XM_002445626.2 | Forward | CCTCCACGAGTTTGGTGATAC | 296 bp | 62 °C |
| Reverse | CCAGTCTTCCACCAGCATTT | ||||
| H+-PPase | GQ469975.1 | Forward | CACCTCTCTGGTATCTGGTTTC | 629 bp | 62 °C |
| Reverse | GTGCGGGCTCAATTTCTTTC |
Sample preparation
Lysis buffer extraction
The protein was extracted from freshly collected sorghum leaves at different time intervals (24 h, 48 h, 96 h) of salt treatment. The leaf material was ground in liquid nitrogen (− 196 °C) using a pre-chilled pestle-mortar till fine powder and precipitated in lysis buffer (7 M urea, 2 M thiourea, 2% CHAPS and 30 mM Tris). The homogenate was kept on shaker for 10–15 min. and then sonicated in sonicator (UP200S, Hielscher) for 10 s. This was repeated four times. After sonication, the suspension was kept on rotator for 2.5 h for intermittent mixing. It was than centrifuged at 14,000 × g for 15 min. The supernatant was collected and stored at − 80 °C for further use.
Phenol–sulfuric extraction
Fresh leaves were crushed in liquid nitrogen, using a pre-chilled pestle- mortar. The crushed samples (1 gm) were then suspended in extraction buffer, vortexed, and incubated with shaking for 10 min on ice. After that, an equal volume of Tris-buffered phenol was added and the solution was incubated on a shaker for 10 min at room temperature and centrifuged for at 5500 g for 10 min, 4 °C for phase separation. The upper phenolic phase was collected carefully to avoid contact with the interphase and decanted into a new tube and back-extracted with extraction buffer. Thereafter, sample was shaken for 5 min, vortexed and again centrifuged for 10 min at 5500 g and 4 °C and for phase separation. The sample was mixed by inverting the tube and incubated overnight at − 20 °C. Incubated sample was centrifuged (10 min, 5500 g at 4 °C) and proteins were pelleted out.
TCA extraction methods
10% TCA containing 0.07% β-ME and 1 mM PMSF
Fresh leaves were crushed in liquid nitrogen using pre-chilled pestle- mortar and suspended in 10% TCA with 0.07% β-mercaptoethanol (β-ME) and 1 mM phenylmethylsulfonyl fluoride (PMSF) dissolved in acetone. After homogenization, the suspension was centrifuged at 15,000 g for 25 min at 4 °C and the pellet obtained was washed.
10% TCA containing 0.07% β-ME
In this method, leaves were crushed in liquid nitrogen and suspended in 10% TCA containing 0.07% β-ME in acetone. The homogenate was sonicated for 1 min and kept at − 20 °C for 1 h with intermittent mixing and vortexing and then centrifuged at 25,000 g for 15 min at 4 °C and the pellet obtained was washed.
10% TCA with 0.07% DTT
In this method, fresh leaves were ground in liquid nitrogen in pre-chilled pestle- mortar and suspended in 10% TCA with 0.07% dithiothreitol (DTT) in acetone. After suspending in the solubilizing buffers, the suspension was subjected to sonication followed by incubation of 1 h at − 20 °C. The suspension was centrifuged at 20,000 g for 20 min at 4 °C and the pellet was air dried.
Protein precipitation methods
The supernatant containing soluble protein samples as described above were precipitated with five different protein precipitation methods.
Acetone precipitation
One milliliter of supernatant and chilled acetone (− 20 °C) was mixed in a ratio of (1:1 v/v) and centrifuged at 5500 g for 15 min at 4 °C. The supernatant (from lysis buffer and phenol–sulfuric method extraction) was discarded and the pellet was air dried and re-dissolved in acetone. The process was repeated thrice. Finally, the pellet was dissolved in solubilizing buffer (7 M Urea, 2 M Thiourea, 4% CHAPS, 30 mM Tris) and 100 mM TEAB and stored at − 80 °C until further analysis.
Acetone precipitation with 0.07% β-ME and 1 mM PMSF
The pellet obtained was washed twice with pre-chilled acetone containing 0.07% β-mercaptoethanol and 1 mM PMSF and centrifuged at 15,000 g for 25 min at 4 °C and incubation of 1 h at − 20 °C. The pellet obtained was by centrifuged for 30 min at 25000 g. The pellet was then dissolved in lysis buffer and stored at 4 °C.
Acetone precipitation with 0.07% β-ME
The pellet was washed with acetone containing 0.07% (w/v) DTT at 15,000 g for 25 min at 4 °C and stored at − 20 °C for 1 h and again centrifuged at 25,000 g for 15 min. Finally, the pellet was air dried and re-suspended in lysis buffer by repeat pipetting and stored at 4 °C.
Acetone precipitation with 0.07% DTT
The pellet was washed thrice with acetone containing 0.07% (w/v) DTT at 20,000 g for 20 min. Finally, the pellet was air dried and re-suspended in lysis buffer by repeat pipetting and stored at 4 °C.
Clean up
The interfering substances such as salts, detergents, nucleic acids, and lipids were removed from the supernatant using 2D-Clean Up kit (Biorad, USA). 100 µl of supernatant (stored at − 80 °C) was precipitated using precipitation agents followed by centrifugation at 12,000 × g for 5 min. The pellet obtained was again washed with wash reagent and centrifuged at 12,000 × g for 5 min. The supernatant was discarded and pellet was incubated at − 20 °C for 1 h. After incubation, it was centrifuged at 12,000 × g for 5 min. The pellet was air dried for 5 min at room temperature and re-suspended in lysis buffer for at least 1 h with gentle shaking (avoid bubbling) at room temperature. The proteins were dissolved from the dried precipitate into lysis buffer (7 M Urea, 2 M Thiourea, 4% CHAPS, 30 mM Tris) by repeated pipetting.
Protein quantification
The total protein concentration present in the dissolved pellet was estimated using 2D Quant kit (GE Healthcare, USA) as per manufacturer’s instructions in Elisa Reader (M200 pro NanoQuant, TECAN). The protein concentration was calculated using bovine serum albumin (BSA) as standard.
SDS-PAGE
The individual proteins in leaf extract were analyzed by (10 × 10.5 cm) SDS-PAGE with 4% stacking and 12% resolving gel using MiniVE gel electrophoresis apparatus (GE healthcare, USA). Thirty microgram protein was loaded in each well as quantified by BSA standard. The gels were stained with Coomassie Brilliant Blue G 250 (BioRad Laboratories, USA) for 1 h and destained. Gel slab was scanned using gel proanalyzer ver. 3.3.
Immunoblotting assay for Heat shock protein 70 (Hsp70)
Immunoblotting analysis for Hsp70 in sorghum leaf protein extracts was performed as previously described by (Ndimba et al. 2010). The protein extracts from stressed and control leaves separated on SDS-PAGE gel were transferred onto PVDF transfer membrane (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden) using a Mini Trans-Blot Electrophoretic Transfer Cell (BioRad). The transfer was performed at 36 V, overnight at 4 °C with constant stirring. After transfer, the membrane was washed and incubated in blocking solution. The membrane was then incubated with primary antibody (human HeLa cells anti-Hsp70/Hsc70 monoclonal antibody) for 1 h. Hsp proteins were detected using a chemiluminescent substrate according to the manufacturer’s instructions.
Data analysis
The gel profiles were visually scored by assigning a number to each distinctive band. The presence or absence of bands were scored as 1 or 0, respectively. The data were statistically analyzed using Gel Doc 2000 BioRad system.
Results and discussion
Protein extraction
A preliminary experiment was conducted to explore the most effective protein extraction method in sorghum. The proteins extracted through different methods are shown in Fig. 1. The protein extracted from phenol–sulfuric method did not show desirable results (Fig. 1 lane 3 and 4) while the proteins extracted from lysis buffer showed significant results with clear and band high intensity (Fig. 1 lane 1 and 2). The protein extracted with 10% TCA containing 0.07% β-ME and 1 mM PMSF showed clear band when precipitated with clean-up kit (Fig. 1 lane 5 and 6) while washing with acetone did not showed clear band. TCA with 0.07% β-mercaptoethanol (Fig. 1 lane 7) and 0.07% DTT (Fig. 1 lane 8) also did not effectively extracted the proteins as no clear bands were observed. After extraction, the suspension was subjected to different precipitation methods in which acetone washing proteins exhibited smeared bands as being hydrophobic in nature, it decreased the solubility of protein and increased its non-polar nature. Protein precipitation with 2D-Clean up kit dissolved the protein pellets uniformly and the protein concentration also increased as reported when quantified. Thus, this study indicated that extraction of proteins with lysis buffer followed by its clean-up is the most effective method for protein extraction in sorghum. Similar results have also been reported by several researchers (Kumar Swami et al. 2011; Ngara et al. 2012; Khalil 2013). The standardization of protein extraction methods in sorghum under salinity may potentially have importance and applications in further understanding of newly identified proteins that may help in plant development.
Fig. 1.
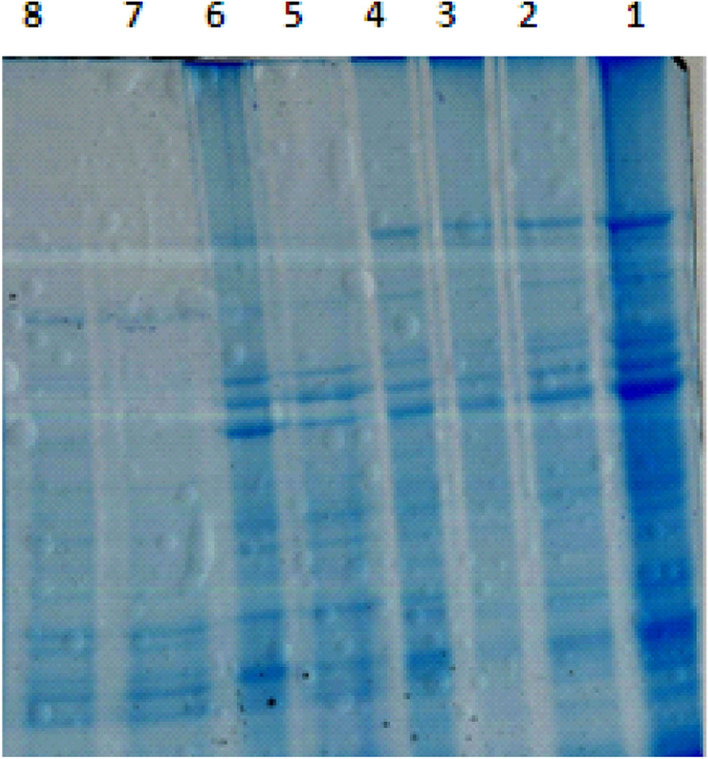
Gel showing proteins extracted through different methods. Lane 1: Lysis buffer with 2D-clean-up; Lane 2: lysis buffer with acetone precipitation; Lane 3: phenol–sulfuric extraction with 2D-clean-up; Lane 4: Phenol–sulfuric extraction with acetone precipitation; Lane 5: 10% TCA containing 0.07% β-ME and 1 mM PMSF with acetone washing (0.07% β-ME + 1 mM PMSF); Lane 6: 10% TCA containing 0.07% β-ME and 1 mM PMSF with 2D-clean-up; lane 7: 10% TCA containing 0.07% β-ME with acetone washing (0.07% β-ME); Lane 8: 10% TCA containing 0.07% DTT with acetone washing (0.07% DTT)
Gel showing proteins extracted through different methods. Lane 1: Lysis buffer with 2D-clean-up; Lane 2: lysis buffer with acetone precipitation; Lane 3: phenol–sulfuric extraction with 2D-clean-up; Lane 4: Phenol–sulfuric extraction with acetone precipitation; Lane 5: 10% TCA containing 0.07% β-ME and 1 mM PMSF with acetone washing (0.07% β-ME + 1 mM PMSF); Lane 6: 10% TCA containing 0.07% β-ME and 1 mM PMSF with 2D-clean-up; lane 7: 10% TCA containing 0.07% β-ME with acetone washing (0.07% β-ME); Lane 8: 10% TCA containing 0.07% DTT with acetone washing (0.07% DTT).
Differential expression of salt-responsive proteins using SDS-PAGE
In this study, two sorghum genotypes (G-46 and CSV 44F) which differ in their response to salinity based on the physiological differences of their photosynthetic apparatus were evaluated (Figs. 2 and 3). Salt-tolerant sorghum genotype (G-46) was germinated under saline (100 and 120 mM NaCl) and normal conditions. The electrophoretic analysis of proteins revealed a total of 90 protein bands in sorghum genotypes under investigation. Among these, some bands were characteristic and constant markers for each genotype which allowed the identification of their electrophoregrams. SDS-PAGE of the sorghum protein extracts showed that protein abundance, expression and loading across the biological replicates was relatively uniform in both the control and salt stressed leaves. Thirty micrograms of leaf protein extracts from three independent biological replicates of stressed and control sorghum genotypes were added in each well with 4% stacking and 12% resolving SDS-PAGE gels. Gel electrophoresed proteins were visualized after Coomassie Brilliant Blue (CBB) staining and imaged. The inclusion of biological replicates in proteomic comparison studies was important both to account for normal biologic variability and treatment groups and to minimize the chances of detecting variations between experiments with non-reproducible protein expression.
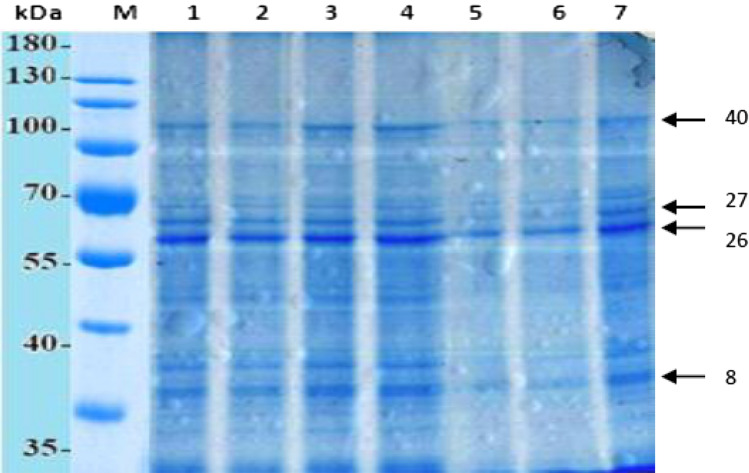
Fig. 2.
SDS-PAGE protein electrophoretic pattern of salt-responsive genes in G-46 genotype under salt stress. Lane 1: protein marker; Lane 2: 120 mM at 96 h*; Lane 3: 100 mM at 96 h; Lane 4: 120 mM at 48 h; Lane 5: 100 mM at 48 h; Lane 6: 120 mM at 24 h; Lane 7: 100 mM at 24 h; lane 8: control (without salt). *Leaves were harvested after time intervals
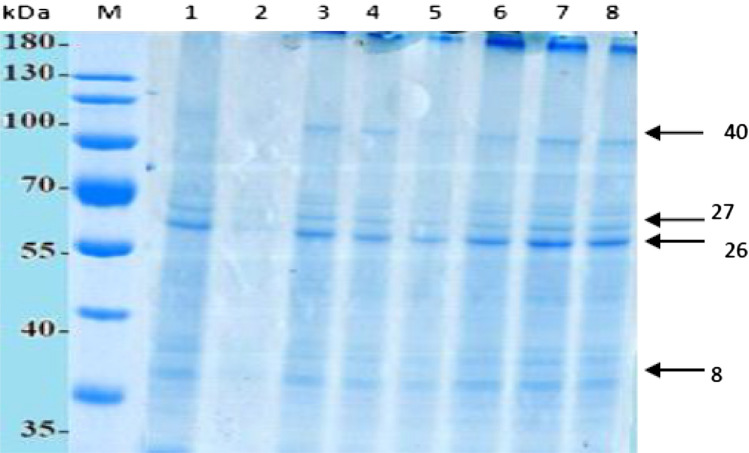
Fig. 3.
SDS-PAGE protein electrophoretic pattern of salt-responsive genes in CSV 44F genotype under salt stress. Lane 1 and 2: control (without salt); Lane 3: 100 mM at 96 h*; Lane 4: 100 mM at 48 h; Lane 5: 100 mM at 24 h; Lane 6: 120 mM at 24 h; Lane 7: 120 mM at 48 h; 120 mM at 96 h. *Leaves were harvested after time intervals
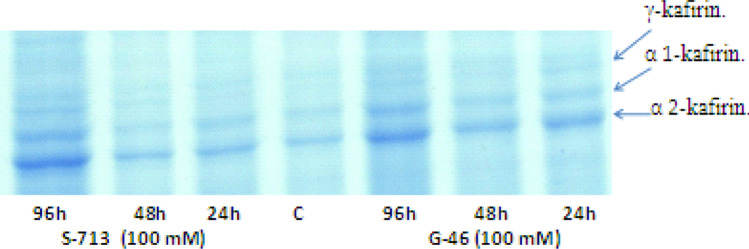
Figures 2 and 3 show the electrophoretic protein pattern in G-46 and CS 44F sorghum genotypes, respectively. The main variations were observed in band number 8, 26, 27 and 40 which differed in band intensity and appearance depending upon the exposure to salt treatments. The analysis of gel revealed that the molecular weight of protein subunits ranged between 30.84 and 130.05 kDa. The number of bands varied under different salt treatments and time intervals with the largest number (95) in G-46 at 96 h (120 mM) 4 °C (Fig. 2), and the lowest number (78) in CSV 44F at 24 h (Fig. 3). Data showed that G-46 was resolved into 19 bands, while CSV 44F was resolved into 20 bands, respectively under saline conditions. Also, the protein band having molecular weight 77.22, 99.05 and 130.56 kDa for G-46 revealed the same trend. Regarding polymorphism, there were 18 monomorphic bands (common bands), 6 polymorphic bands and 4 unique bands. A measurable band intensity shift for all sorghum genotypes grown under salt conditions was detected. G-46 recorded higher band intensity values as compared to S-713. For similar bands, the kafirin level also changed under salinity in both genotypes (Fig. 4). The maximum level was observed in G-46 at 100 mM (96 h) and minimum at 24 h. These results are also supported by Thongngam 2007 who examined the sorghum protein expression using SDS-PAGE. The proteins expressed consisted of α, β and γ forms of kafirin. α-kafirin represented the highest proportion among all the groups. In addition, Capouchová et al. 2006 evaluated 6 sorghum and 12 oats genotypes using SDS-PAGE and showed that the highest percentage of proteins was represented by glutelins and prolamins, between 74.96 and 53.43% of the total storage proteins (Sekhwal et al. 2012; Khalil 2013; Osman et al. 2013). The functional identification of many salt-tolerant proteins has been reported by molecular and genomic analyses in Arabidopsis, rice and other plants (Dai Yin et al. 2005). In this study, proteins analysis through SDS-PAGE elucidated strong correlation between accumulation of seed storage kafirin proteins and differential expression of salt-responsive proteins towards salinity tolerance in G-46 (salt-tolerant).
Fig. 4.
Kafirin spectra of sorghum genotypes at 100 mM salt concentration
Since the sorghum genome sequencing project has been completed, the most awaited sorghum gene products tend to be experimentally unidentified as hypothetical proteins (Paterson et al. 2009). Through nature, hypothetical proteins are a protein of genome sequences predicted, but their presence has not been proved at protein level experimentally (Lubec et al. 2005). As such, the present and other sorghum-proteomic studies have faced inadequate sorghum decoding data that need full characterization (Kumar Swami et al. 2011). This study would act as a baseline for detection and identification of salt-responsive proteins and genes which governs the salinity tolerance in sorghum genotypes and might be utilized in future breeding programs for developing high yielding, and tolerant sorghum genotypes. The detailed functional analysis of these proteins would provide further information regarding direct regulatory networks in sorghum.
Gene expression in response to salinity. In higher plants, glycine betain and proline acts as osmoprotectants and its expression is up regulated during stress conditions (Rhodes and Hanson 1993; Khalil 2013), while SOD acts as antioxidative enzyme to scavenge the toxic effects of reactive oxygen species under stress environment. The PCR product using specific primer of salt-tolerant genes viz. BADH1, P5CS1, H+-PPase, Cu-ZnSOD and Actin reported the appearance of single band (Fig. 5) and their up regulation during salinity which indicated that the antioxidative cellular defense mechanism combats the toxic effects of salt accumulation inside the cytoplasm. The expression was higher at 120 mM (96 h) after salt stress. The detection of salt-tolerant genes could be helpful towards the identification of salt-tolerant genotypes in sorghum.
Fig. 5.
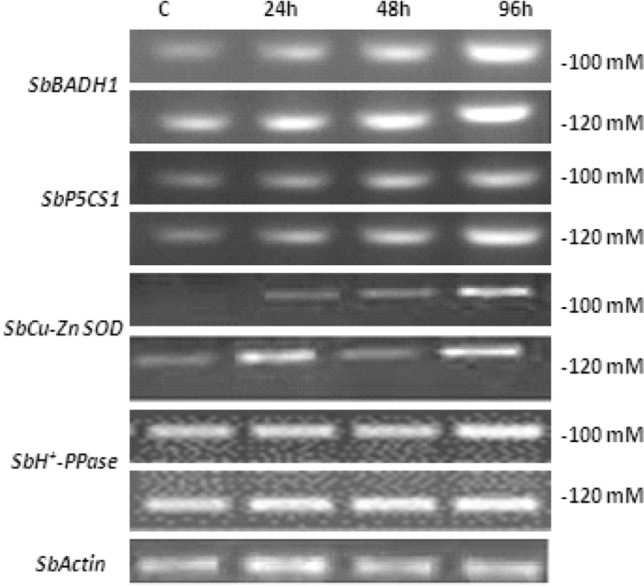
Expression of salt-responsive genes in sorghum under salinity. Act Actin, BADH1 betaine aldehyde dehydrogenase, H+-PPase Vacuolar hydrogen pyrophosphatase, P5CS1 Pyrroline-5 carboxylate synthetase 1, SOD Superoxide dismutase, Sb Sorghum bicolor
Validation of salt concentration and its effect on protein expression
In this experiment, surface sterilized seeds were plated and grown on plastic rays enriched with nutrient medium. After germination, the seedlings were supplemented with different salt concentrations (60, 80, 100, 120 and 140 mM NaCl) along with control at 14th day of germination for different time intervals. As illustrated in Fig. 6, the seeds were able to germinate and grow at different degrees across the treatment regime. At 120 mM NaCl, the efficiency of seed germination and seedling growth were greatly reduced. Therefore, on the basis of this preliminary study, 100 mM NaCl was selected as the concentration for use in subsequent salt treatment for different time intervals (24 h, 48 h, 96 h). Plant cells may alter their gene expression, leading to an increase, decrease, induction or overall suppression of some stress-responding proteins to adapt and/or withstand salt stress (Ho and Sachs 1989; Seki et al. 2003; Shinozaki et al. 2003). To assess whether or not 100 mM NaCl salt concentration was within the physiological range of the experimental method for 14 days, the expression of Heat shock protein (Hsp70) (stress-responsive protein/molecular chaperones) was examined using immunoblotting assay (Miemyk 1997). Hsp70 is a recognized stress-reactant protein and responds to abiotic stress including cold, drought, salinity and other oxidative stresses (Wang et al. 2004; Ngara et al. 2012). Immunoblotting was conducted on protein extracts from both salt stressed (100 mM NaCl) and control leaves using a human HeLa cells anti-Hsp70/Hsc70 monoclonal antibody. The levels of Hsp70 protein was enhanced in stressed plants as compared to control. The biological function of Hsp70 is to prevent the aggregation of denatured proteins as a result of oxidative stress and mediates the refolding of proteins to restore their native biological functions (Sung et al. 2001; Wang et al. 2004; Kumar Swami et al. 2011). The up regulated level of Hsp70 confirmed that growth of sorghum seedlings at 100 mM NaCl was within the physiological range, inducing known stress responses and indicated that further experiments may be performed to confirm other salt-stress responsive proteins using high-throughput techniques. To use the proteomic technologies for the expression of proteins, involved in various biological processes in sorghum are yet to be yet explored completely under different abiotic stresses. This will encourage scientific community worldwide to view sorghum as a possible model cereal plant and thus invest resources in further sorghum protein characterization.
Fig. 6.
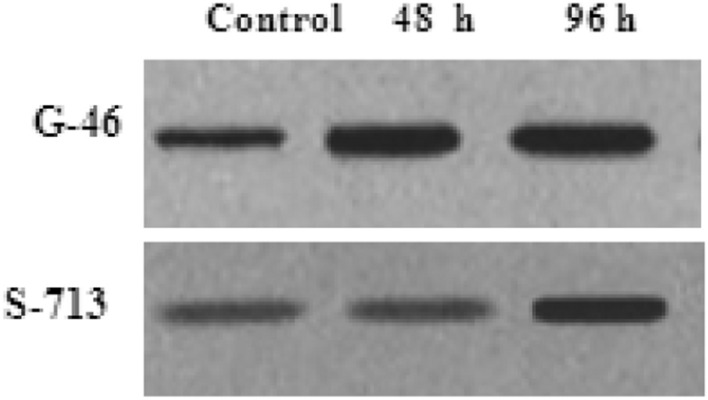
Immunoblotting assay of heat shock protein (Hsp70) to validate the effect of salt concentration and its effect on protein expression
Conclusion
The adverse effects of environmental stresses, such as salinity and drought are well-known threats to agricultural societies, particularly for the farmers in the developing world. The seedling growth, stress concentration and different protein extraction methods gave desirable and reproducible SDS-PAGE results. The expression of kafirin (seed storage protein) varied under different salinity levels. The expression of Hsp 70 at 100 mM was within the physiological range reflecting that the G-46 was behaving as tolerant genotype under this salinity level. Sorghum is still understudied at the molecular level in spite of its well-known natural stress-resistant characteristics. The current and future studies may aim to better understand the functional molecular interactions of the protein candidates, particularly those with specific sorghum characteristics, and the mechanisms that make this crop stress-tolerant in relation to other grains.
Acknowledgements
First Author Ms. Himani is thankful to Department of Science and Technology, SERB and CII under the Prime Minister’s Fellowship Scheme for Doctoral Research for providing financial assistance to carry out this research work. Authors are also thankful to Animal Biotechnology Centre, National Dairy Research Institute, Karnal Haryana, for providing for providing the research facilities to carry out the research work, and Forage Section, Department of Genetics and Plant Breeding, CCS HAU, Hisar for providing the seeds of sorghum genotypes and CCS HAU, Hisar for providing the necessary facilities and support to carry out the research.
Abbreviations
- Act
Actin
- β-ME
β-Mercaptoethanol
- BADH1
Betaine aldehyde dehydrogenase
- CBB
Coomassie Brilliant Blue
- DTT
Dithiothreitol
- H+-PPase
Vacuolar hydrogen pyrophosphatase
- HSP
Heat shock protein
- PAGE
Polyacrylamide gel electrophoresis
- P5CS1
Pyrroline-5 carboxylate synthetase 1
- PMSF
Phenylmethylsulfonylfluoride
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- SDS-PAGE
Sodium dodecyl sulfate-polyacrlyamide gel electrophoresis
- SOD
Superoxide dismutase
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by HP and JT. The first draft of the manuscript was written by HP under the guidance of Dr. JT (Nutritional Biochemistry and Molecular Biology). The research work was carried out under the supervision of Dr. AKM and Dr. PR. Dr. AKM (Proteomics and Structural Biology of Proteins) check the final draft and suggested corrections. Dr. S (Agronomist, Forage Section) provided the seeds. Mr. AM and Ms. SB helped in manuscript writing. All the authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Contributor Information
Himani Punia, Email: himanipunia91@gmail.com.
Jayanti Tokas, Email: jiyaccshau@gmail.com.
Surina Bhadu, Email: surinabhadu@gmail.com.
Ashok K. Mohanty, Email: ashokmohanty1@gmail.com
Preeti Rawat, Email: rawatpreeti1975@gmail.com.
Anurag Malik, Email: anuragmalikseed@hau.ac.in.
Satpal, Email: satpal.fpj@gmail.com.
References
- Abbas A, Khan S, Hussain N, et al. Characterizing soil salinity in irrigated agriculture using a remote sensing approach. Phys Chem Earth Parts A/B/C. 2013;55:43–52. [Google Scholar]
- Aghaei K, Ehsanpour AA, Komatsu S. Proteome analysis of potato under salt stress. J Proteome Res. 2008;7:4858–4868. doi: 10.1021/pr800460y. [DOI] [PubMed] [Google Scholar]
- Ahmad P, Abdel Latef AAH, Rasool S, et al. Role of proteomics in crop stress tolerance. Front Plant Sci. 2016;7:1336. doi: 10.3389/fpls.2016.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almodares A, Hadi MR, Ahmadpour H. Sorghum stem yield and soluble carbohydrates under different salinity levels. Afr J Biotechnol. 2008;7(22):4051–4055. [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, et al. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol. 2005;58:699–720. doi: 10.1007/s11103-005-7876-2. [DOI] [PubMed] [Google Scholar]
- Capouchová I, Petr J, Krejčířová L. Protein composition of sorghum and oat grain and their suitability for gluten-free diet. Žemdirbystė. 2006;93:271–284. [Google Scholar]
- Corwin DL, Scudiero E (2019) Review of soil salinity assessment for agriculture across multiple scales using proximal and/or remote sensors. In: Sparks DL (ed) Advances in agronomy, vol 158. ZoeKruzeAcquisition, pp 2–326
- Dai Yin C, Luo YH, Min SHI, et al. Salt-responsive genes in rice revealed by cDNA microarray analysis. Cell Res. 2005;15:796–810. doi: 10.1038/sj.cr.7290349. [DOI] [PubMed] [Google Scholar]
- De Groot CC, Marcelis LFM, Van Den Boogaard R, et al. Interaction of nitrogen and phosphorus nutrition in determining growth. Plant Soil. 2003;248:257–268. doi: 10.1023/A:1022323215010. [DOI] [Google Scholar]
- El Naim AM, Mohammed KE, Ibrahim EA, Suleiman NN. Impact of salinity on seed germination and early seedling growth of three sorghum (Sorghum biolor L. Moench) cultivars. Sci Technol. 2012;2:16–20. [Google Scholar]
- Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004;55:307–319. doi: 10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Xu J. Abiotic stress responses in plant roots: a proteomics perspective. Front Plant Sci. 2014;5:6. doi: 10.3389/fpls.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Himani BS, Tokas J, Satpal Variation in structural carbohydrates of forage sorghum [Sorghum bicolor (L.) Moench] under saline conditions. Forage Res. 2019;45(2):123–127. [Google Scholar]
- Ho THD, Sachs MM. Environmental control of gene expression and stress proteins in plants. In: Jones HG, Flowers TJ, Jones MB, editors. Plants under Stress. Biochemistry, Physiology and Ecology and Their Application to Plant Improvement. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ. 1950;347:1–32. [Google Scholar]
- Hurkman WJ. Use of two-dimensional gel electrophoresis to characterize changes in gene expression associated with salt stress of barley. In: Katterman F, editor. Environmental injury to plants. San Diego, CA: Academic Press; 1990. pp. 205–229. [Google Scholar]
- Jia H, Shao M, He Y, et al. Proteome dynamics and physiological responses to short-term salt stress in Brassica napus leaves. PLoS One. 2015;10:e0144808. doi: 10.1371/journal.pone.0144808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, et al. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kausar A, Ashraf MY, Ali I, et al. Evaluation of sorghum varieties/lines for salt tolerance using physiological indices as screening tool. Pak J Bot. 2012;44:47–52. [Google Scholar]
- Khalil RMA. Molecular and biochemical markers associated with salt tolerance in some sorghum genotypes. World Appl Sci J. 2013;22:459–469. doi: 10.5829/idosi.wasj.2013.22.04.5413. [DOI] [Google Scholar]
- Krishnamurthy L, Serraj R, Hash CT, et al. Screening sorghum genotypes for salinity tolerant biomass production. Euphytica. 2007;156:15–24. [Google Scholar]
- Kumar Swami A, Alam SI, Sengupta N, Sarin R. Differential proteomic analysis of salt stress response in Sorghum bicolor leaves. Environ Exp Bot. 2011;71:321–328. doi: 10.1016/j.envexpbot.2010.12.017. [DOI] [Google Scholar]
- Lubec G, Afjehi-Sadat L, Yang J-W, John JPP. Searching for hypothetical proteins: theory and practice based upon original data and literature. Prog Neurobiol. 2005;77:90–127. doi: 10.1016/j.pneurobio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Mall TK, Dweikat I, Sato SJ, et al. Expression of the rice CDPK-7 in sorghum: molecular and phenotypic analyses. Plant Mol Biol. 2011;75:467–479. doi: 10.1007/s11103-011-9741-9. [DOI] [PubMed] [Google Scholar]
- Miemyk J. The 70 kDa stress-related proteins as molecular chaperones. Trends Plant Sci. 1997;2:180–187. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Ndimba BK, Chivasa S, Simon WJ, Slabas AR. Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2005;5:4185–4196. doi: 10.1002/pmic.200401282. [DOI] [PubMed] [Google Scholar]
- Ndimba BK, Thomas LA, Ngara R. Sorghum 2-dimensional proteome profiles and analysis of Hsp70 expression under salinity stress. Kasetsart J (Nat Sci) 2010;44:768–775. [Google Scholar]
- Ngara R, Ndimba R, Borch-Jensen J, et al. Identification and profiling of salinity stress-responsive proteins in Sorghum bicolor seedlings. J Proteomics. 2012;75:4139–4150. doi: 10.1016/j.jprot.2012.05.038. [DOI] [PubMed] [Google Scholar]
- Osman G, Munshi, et al. Genetic variation and relationships of Zea mays and Sorghum species using RAPD-PCR and SDS-PAGE of seed proteins. Afr J Biotechnol. 2013;12:4269–4276. doi: 10.5897/ajb12.2644. [DOI] [Google Scholar]
- Pandey A, Soccol CR, Nigam P, Soccol VT. Biotechnological potential of agro-industrial residues. I: sugarcane bagasse. Bioresour Technol. 2000;74:69–80. [Google Scholar]
- Parker R, Flowers TJ, Moore AL, Harpham NVJ. An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J Exp Bot. 2006;57:1109–1118. doi: 10.1093/jxb/erj134. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Hanson AD. Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Biol. 1993;44:357–384. [Google Scholar]
- Sekhwal MK, Swami AK, Sarin R, Sharma V. Identification of salt treated proteins in sorghum using gene ontology linkage. Physiol Mol Biol Plants. 2012;18:209–216. doi: 10.1007/s12298-012-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K. Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol. 2003;14:194–199. doi: 10.1016/s0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Sobhanian H, Aghaei K, Komatsu S. Changes in the plant proteome resulting from salt stress: toward the creation of salt-tolerant crops? J Proteomics. 2011;74:1323–1337. doi: 10.1016/j.jprot.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Sung D, Kaplan F, Guy CL. Plant Hsp70 molecular chaperones: protein structure, gene family, expression and function. Physiol Plant. 2001;113:443–451. [Google Scholar]
- Thongngam M (2007) Characteristics and functional properties of sorghum protein (kafirin). In: 45. Kasetsart University Annual Conference, Bangkok (Thailand), 30 Jan-2 Feb 2007
- (US) RSL (1954) Diagnosis and improvement of saline and alkali soils. US Department of Agriculture. US Government Printing Office
- Van Emon JM. The omics revolution in agricultural research. J Agric Food Chem. 2016;64:36–44. doi: 10.1021/acs.jafc.5b04515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ. Challenges and prospects of plant proteomics. Plant Physiol. 2001;126:501–508. doi: 10.1104/pp.126.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranagamallaiah G, Jyothsnakumari G, Thippeswamy M, et al. Proteomic analysis of salt stress responses in foxtail millet (Setaria italica L. cv. Prasad) seedlings. Plant Sci. 2008;175:631–641. [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han B, Wang T, et al. Mechanisms of plant salt response: insights from proteomics. J Proteome Res. 2012;11:49–67. doi: 10.1021/pr200861w. [DOI] [PubMed] [Google Scholar]
- Zhu J-K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/s1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]