Abstract
The precise localization of CaV2 voltage-gated calcium channels at the synapse active zone requires various interacting proteins, of which, Rab3-interacting molecule or RIM is considered particularly important. In vertebrates, RIM interacts with CaV2 channels in vitro via a PDZ domain that binds to the extreme C-termini of the channels at acidic ligand motifs of D/E-D/E/H-WC-COOH, and knockout of RIM in vertebrates and invertebrates disrupts CaV2 channel synaptic localization and synapse function. Here, we describe a previously uncharacterized clade of RIM proteins bearing domain architectures homologous to those of known RIM homologs, but with some notable differences including key amino acids associated with PDZ domain ligand specificity. This novel RIM emerged near the stem lineage of metazoans and underwent extensive losses, but is retained in select animals including the early-diverging placozoan Trichoplax adhaerens, and molluscs. RNA expression and localization studies in Trichoplax and the mollusc snail Lymnaea stagnalis indicate differential regional/tissue type expression, but overlapping expression in single isolated neurons from Lymnaea. Ctenophores, the most early-diverging animals with synapses, are unique among animals with nervous systems in that they lack the canonical RIM, bearing only the newly identified homolog. Through phylogenetic analysis, we find that CaV2 channel D/E-D/E/H-WC-COOH like PDZ ligand motifs were present in the common ancestor of cnidarians and bilaterians, and delineate some deeply conserved C-terminal structures that distinguish CaV1 from CaV2 channels, and CaV1/CaV2 from CaV3 channels.
Keywords: Rab3-interacting molecule, RIM, synapse, evolution of the nervous system, voltage-gated calcium channels, CaV2
Introduction
The tight spatiotemporal regulation of cytoplasmic Ca2+ fluxes is integral to ensuring that Ca2+-dependent biological processes are effected with fidelity, and preventing the toxicity that arises with prolonged elevated levels of intracellular Ca2+ (Clapham 2007). A variety of differentially gated ion channels are the route for Ca2+ entry into the cytoplasm. Of these, voltage-gated calcium (CaV) channels mediate Ca2+ influx that underlies such fundamental processes as neurotransmitter release (Katz and Miledi 1965) and excitation–contraction coupling (Catterall 2011), and whose dysfunction is causal to variegated pathologies (Adams and Snutch 2007; Simms and Zamponi 2014). All CaVs are defined by a current-conducting α subunit comprised of four homologous domains each containing six transmembrane segments (S1–S6) connected by cytoplasmic linkers. These linkers, along with their cytoplasmic N- and C-termini, are largely disordered in structure (Catterall 2011). The high-voltage activated (HVA) L-type (CaV1.1–1.4), P/Q-type (CaV2.1), N-type (CaV2.2), and R-type (CaV2.3) channels associate with CaVα2δ and CaVβ ancillary subunits, the latter via the alpha interaction domain (AID) located in the domain I-II linker of the channel α subunit. Calmodulin, a Ca2+ sensor important for modulating CaV channel function, interacts with HVA α subunits at C-terminal IQ motifs (Catterall 2011; Ben-Johny and Yue 2014). Recently, low-voltage activated (LVA) T-type (CaV3) channels have also been shown to interact with the CaVβ subunit (Bae et al. 2010), as well as calmodulin (Chemin et al. 2017), though they lack AID and IQ motifs. Concomitant to phylogenetic, biophysical, and pharmacological distinctions between CaV1, CaV2 and CaV3 channels, it is apparent that distinct sets of interacting proteins are integral to their unique functions in different cell types that appear broadly conserved in the Metazoa (Senatore et al. 2016).
The presynaptic active zone is the locus for synaptic vesicle exocytosis mediated largely by CaV2 type calcium channels in vertebrate and invertebrate synapses (fig. 1A) (Spafford and Zamponi 2003; Südhof 2012). A leading functional model for CaV2 channel tethering at the active zone involves the tripartite interaction between Rab3-interacting molecule (RIM), RIM-binding protein (RIM-BP), and CaV2 channels, thought to ensure that depolarization-induced cytoplasmic Ca2+ plumes are close to Ca2+ sensors of the exocytotic machinery (Südhof 2012). Specifically, RIM is thought to selectively recruit N- and P/Q-type CaV2 channels in vertebrates, or the single CaV2 channel in invertebrates, via a PDZ (postsynaptic density 95 protein, Drosophila disk large tumor suppressor, and zonula occludens-1 protein) domain that interacts with amino acid motifs of D/E-D/E/H-WC-COOH located on the extreme C-termini of the calcium channels (Kaeser et al. 2011; Graf et al. 2012). In addition, RIM is involved in priming and docking of synaptic vesicles, binding to the vesicular protein Rab3 with an N-terminal alpha helical structure, and the SNARE-associated protein Munc-13 with an adjacent Zn2+-finger domain (fig. 1A) (Wang et al. 1997, 2001; Betz et al. 2001; Fukuda 2003; Dulubova et al. 2005; Lu et al. 2006; Quade et al. 2019). RIM-BP, which bears three Src homology 3 (SH3) protein interaction domains, binds proline-rich motifs in RIMs and the distal CaV2 C-terminus to strengthen the RIM–CaV2 interaction (Wang et al. 2000; Hibino et al. 2002; Kaeser et al. 2011) (fig. 1A). In vertebrates, RIMs have also been shown to suppress CaV2 channel voltage-dependent inactivation to potentiate neurotransmitter release, mediated by an interaction between the RIM C2B domain and the CaVβ subunit (Kiyonaka et al. 2007; Uriu et al. 2010). Vertebrate RIM1 and RIM2 also interact with two separate regions in the N- and P/Q-channel C-terminus (encoded by exons 44 and 47), which for RIM2, further suppresses CaV2 channel voltage-dependent inactivation (Hirano et al. 2017).
Fig. 1.
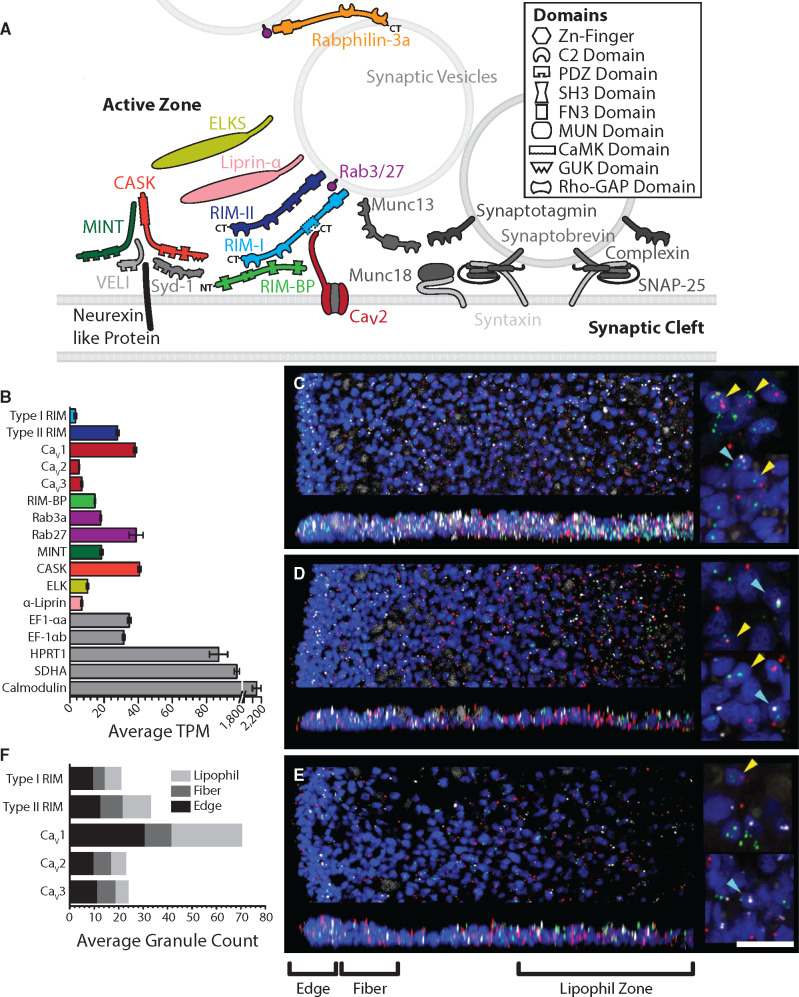
(A) Schematic of presynaptic active zone proteins identified in the transcriptome of T. adhaerens (Wong et al. 2019). Colored proteins represent key presynaptic scaffolding proteins that interact with N-, P/Q-, and R-type CaV2 channels. Presence of InterPro-predicted canonical domain structures for each protein is illustrated. The dashed line denotes the absence of a predicted PDZ domain for the Trichoplax RIM-I homolog. (B) Average TPM expression level analysis of the Trichoplax whole animal transcriptome (Wong et al. 2019) reveals expression of a rich set of active zone proteins, plus all three CaV channel paralogs; the color scheme follows that of the CaV-interactome depicted in panel A. The housekeeping genes eukaryotic translation elongation factor EF-1αa and EF-1αb, hypoxanthine phosphoribosyltransferase 1, and succinate dehydrogenase were used as standards for expression level. (C–E) FISH with RNAScope probes for RIM and CaV channel (CaV1, CaV2, or CaV3) genes in wholemounts of Trichoplax. (C) CaV1 (green), RIM-II (red), and RIM-I (white). (D) CaV2 (green), RIM-II (red), and RIM-I (white). (E) CaV3 (green), RIM-II (red), and RIM-I (white). Nuclei are blue. The top image in each part shows a horizontal (x, y) projection of a series of optical sections through a region beginning at one edge and extending halfway across the animal, and the lower image shows a vertical (x, z) projection of a 10 µm (C) or 15 µm (D, E) wide strip through the same region. Color-separated images of the same samples are shown in supplementary figure S1, Supplementary Material online. Insets (right) show enlarged views captured with an enhanced resolution detector. In each part, the top inset is a projection of five optical sections (0.185-µm interval) through the most ventral nuclei in a region within the lipophil zone and the lower inset is projection of five optical sections in a region at the edge. Nuclei that are in close apposition to labels for both a CaV channel and RIM-II or a CaV channel and RIM-I are indicated (yellow and cyan arrowheads, respectively). Scale, 20 µm (for left panels); 5 µm (for right panels). (F) Average fluorescent granules for RIM-I, RIM-II, and the CaV1–CaV3 channels counted within the edge, fiber zone and lipophil zone regions of fluorescently labeled Trichoplax from five (CaV1–CaV3) and seven (RIM-I and RIM-II) separate experiments (i.e. average grains per 10 µm2).
Ubiquitously conserved and unique to animals (Paps and Holland 2018), the presence of RIM genes has been tied to the origin of animal multicellularity and accords with the underrepresentation of active zone proteins in choanoflagellates, a sister group to the Metazoa otherwise harboring a rich set of synaptic proteins (Burkhardt 2015). RIMs likely also serve non-neuronal functions, given that homologs are found in animals lacking synapses (Paps and Holland 2018), and although predominantly expressed in the nervous systems of vertebrates, RIM expression has been reported in non-nervous tissues (Iezzi et al. 2000). Unfortunately, a paucity of data exists regarding the conservation of the RIM, RIM-BP, and CaV2 interaction among early-diverging metazoans, and more generally, invertebrate phyla, hampering progress in understanding the molecular evolution of the presynaptic active zone. Here, via maximum likelihood and Bayesian phylogenetic analyses, we show for the first time that various invertebrates possess a novel RIM homolog that emerged near the stem metazoan lineage, and has undergone multiple losses in cnidarians and bilaterians. We demonstrate that this homolog contains, with a single known phyletic exception, a protein domain architecture akin to that of vertebrate α-RIMs but is significantly shorter and possesses a PDZ domain that differs at amino acid positions associated with ligand specificity compared with previously characterized RIMs. Furthermore, we provide a systematic phylogeny of metazoan CaV channels, complete with annotations of predicted C-terminal PDZ and SH3 domain-binding motifs, to evaluate the interactomic potential of distinct CaV channel clades. On a more granular level, we examine the conservation of short linear motifs (SLiMs) in metazoan CaV channel C-termini, and identify putative structural distinctions between the ancestral CaV1 and CaV2 channels, and between CaV1/CaV2 channels and CaV3 channels.
Results
Characterization of Two RIM Homologs Identified in the Transcriptome of Trichoplax adhaerens
Previous research has reported that invertebrates possess a single gene encoding the active zone protein RIM (Wang and Südhof 2003; Südhof 2012). Work in our laboratory identified two RIM homologs in the transcriptome of Trichoplax adhaerens, a small sea water invertebrate that diverged from other animals roughly 600 Ma (Dos Reis et al. 2015), and that lacks a nervous system and synapses (Smith et al. 2014). One paralog was found to be considerably longer in protein sequence (2,487 residues) and to lack a predicted PDZ domain for interactions with the C-termini of CaV2 channels (fig. 1A) (Wong et al. 2019). We decided to name this particular homolog type I RIM (RIM-I), based on similarities with canonical RIM genes described in other animals. The other we named type II RIM (RIM-II), as it is considerably shorter in length (1,098 residues) yet bears the expected domain architecture of an N-terminal Zn2+-finger domain flanked by alpha helices, followed by a PDZ domain and two C2 domains (fig. 1A and supplementary file S1, Supplementary Material online). The differential lengths of the RIM-I and RIM-I proteins were found consistent with the lengths of corresponding sequences identified in the Trichoplax genome (Kamm et al. 2018) (i.e. 2,455 and 1,098 residues for NCBI accession numbers RDD47777.1 and RDD47753.1, respectively), and sequences predicted from the transcriptome of the placozoan Hoilungia hongkongensis (Eitel et al. 2018) (2,253 and 1,096 residues with respective protein sequence identifiers of m.47227 and m.66299). The apparent absence of synapses in Trichoplax is not reflected by its expressed gene set, which in addition to RIM includes homologs for key active zone proteins such as SNARE and associated proteins, and scaffolding proteins that interact with CaV2 calcium channels at nerve terminals such as Mint, CASK, Liprin-α, ELKS, and RIM-BP (fig. 1A and B). Trichoplax is also the most early-diverging animal with gene homologs for all three metazoan CaV channel types: CaV1, CaV2, and CaV3 (Senatore et al. 2012; Moran and Zakon 2014; Senatore et al. 2016; Smith et al. 2017).
Fluorescence in situ hybridization (FISH) with probes for RIM-I, RIM-II, and the CaV1–CaV3 channel mRNAs in wholemounts of Trichoplax (fig. 1C–E) confirmed that each gene is expressed. CaV1 expression was more abundant at the edge of the animal and in the central region starting ∼80 µm from the edge compared with the intervening region. CaV2, CaV3, RIM-I, and RIM-II had more uniform radial expression patterns (fig. 1C–E, supplementary fig. S1A, Supplementary Material online). Label for both RIMs and all three calcium channels was evident near the dorsal and ventral surfaces (fig. 1, vertical projections), suggesting expression in dorsal and ventral epithelial cells. Nuclei that were in close apposition to probes for RIM-I or RIM-II and one of the calcium channels (CaV1, CaV2, or CaV3) were present (fig. 1C–E, right insets), suggesting that RIM proteins are coexpressed with calcium channels. However, only a small number (generally <4) of probe labels were associated with individual nuclei. The small number of probe labels likely indicates that mRNA for these proteins is in low abundance, because much higher probe label densities have been observed by the same in situ hybridization technique with probes for highly expressed proteins, such as digestive enzymes (Mayorova et al. 2019). The higher abundance of CaV1 expressing cells near the edge and within the lipophil zone than in the intervening region is interesting because secretory cells are prevalent near the edge and in the lipophil zone but are rare in the intervening region. Mucocytes are the most prevalent secretory cell type near the edge and can be recognized by staining with a fluorescence conjugated lectin, wheatgerm agglutinin (WGA) (Mayorova et al. 2019). Combining FISH for calcium channels with WGA staining (supplementary fig. S1B, Supplementary Material online) showed that probe labels for CaV1 and CaV2 were often present inside WGA stained mucocytes, whereas only a few mucocytes had CaV3 probe labels in their interiors.
Average counts of fluorescent granules within regions of the animal characterized by distinct cell-type content (i.e. the edge, fiber cell zone, and lipophil cell zone [Mayorova et al. 2019]), revealed that RIM-II is more abundantly expressed than RIM-I overall and in each region (fig. 1F;P values for Tukey’s tests after one-way ANOVAs: edge <0.05; fiber zone <0.0005; lipophil zone <0.00005; sum of regions <0.00005; ANOVA for separate regions: df = 5, F = 17.1, P = 2.1E−15; ANOVA for regions combined: df = 1, F = 24.5, P = 2.1E−6). Notably, these patterns are consistent with mRNA expression levels measured as average transcripts per million (TPM) in the transcriptome data, where RIM-II is more abundantly expressed than RIM-I at the whole animal level (fig. 1B). Also consistent were the average TPM values and counted granules for in situ hybridization of the three Trichoplax CaV channels (CaV1–CaV3), where TPM and granule counts for CaV1 were significantly higher compared with CaV2 and CaV3 within edge and lipophil zones and all three regions combined (i.e. compare fig. 1B and fig. 1F) (P values for Tukey’s tests after one-way ANOVAs of granule counts: CaV1 vs. CaV2 edge <0.00005; CaV1 vs. CaV2 lipophil zone <0.00005; CaV1 vs. CaV2 total <0.00005; CaV1 vs. CaV3 edge <0.00005; CaV1 vs. CaV3 lipophil zone <0.00005; CaV1 vs. CaV3 total <0.00005; ANOVA for separate regions: df = 8, F = 86.8, P = 0; ANOVA for regions combined: df = 2, F = 159.0, P = 0). Indeed, in spite of low-level expression, the consistency between the transcriptome TPM and fluorescent granule count data indicates that fluorescent signals observed in the hybridization experiments reflect true mRNAs expression. Lastly, RIM-I, CaV2, and CaV3 each appear to be enriched in the edge compared with the lipophil zone, with average respective granule count ratios of 1.5, 1.7, and 2.1 (edge/lipophil zone), compared with RIM-II and CaV1 (ratios of 1.0 and 1.1, respectively).
Identification of a Novel Clade of Invertebrate RIM Homologs
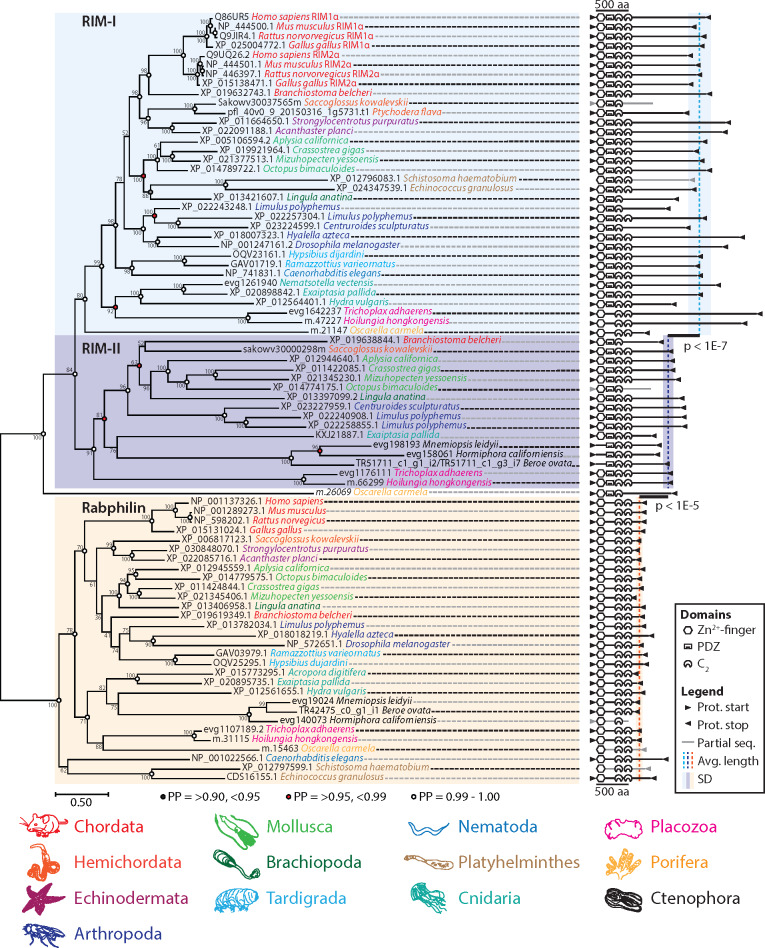
The existence of two RIM homologs in placozoans prompted us to determine whether the RIM-II gene is also found in other animals, and whether it is phylogenetically distinct from RIM-I. Both maximum likelihood and Bayesian protein phylogenetic inference demonstrated that various invertebrates possess RIM-IIs, which form a sister clade with canonical vertebrate RIM1/RIM2 and invertebrate RIM genes (i.e. RIM-Is) (fig. 2). Vertebrate RIM3 and RIM4 proteins were not included in the phylogeny due to their truncated nature, although they clustered with vertebrate RIM1 and RIM2 sequences in preliminary analyses (not shown), consistent with the notion that these evolved via gene duplication along the vertebrate stem lineage (Wang and Südhof 2003). The related protein, rabphilin 3A, a regulator of synaptic vesicle recruitment that like RIM-Is binds Rab3 in a GTP-dependent manner (Shirataki et al. 1993; Stahl et al. 1996; Burns et al. 1998; Wang et al. 2001), was used as an outgroup (fig. 2). Excluding sponge homologs, both type I and II RIMs cluster together 99% and 93% of the time, respectively, with the rabphilins as a sister clade. With a few notable exceptions, both I and II RIMs feature conserved domain architectures comprised of an N-terminal Zn2+-finger domain, a PDZ domain, and two C2 domains, including the vertebrate homologs RIM1αβ and RIM2α. That RIM-I, RIM-II, and rabphilin form distinct clades was also indirectly supported by analysis of average protein length. RIM-II (n = 15, 1,113 ± 156 aa) was found to be significantly shorter than RIM-I (n = 32, 1,604 ± 347 aa) but significantly longer than rabphilin (n = 27, 666 ± 100 aa) (fig. 2; P values for Tukey’s tests after one-way ANOVA of sequence lengths: RIM-I vs. RIM-II P = 2.45E−8; RIM-II vs. rabphilin P = 1.03E−6; ANOVA df = 2, F = 106.02, P = 0).
Fig. 2.
Maximum likelihood phylogenetic analysis reveals a novel RIM homolog (RIM-II), present in numerous invertebrate phyla, that forms a separate clade from previously identified RIM proteins (RIM-Is) and rabphilins. Bayesian posterior probabilities indicated on select nodes corroborate maximum likelihood estimation. RIM-I and RIM-II, while significantly different in length (one-way ANOVA of sequence lengths: df = 2, F = 106.02, P = 0; P values for Tukey’s post hoc tests are shown for pairwise comparisons), possess similar InterProScan-predicted domain architectures, comprised of an N-terminal Zn2+-finger domain, a PDZ domain, and two C2 domains. Although domain synteny is depicted faithfully, interdomain distances do not accord with the provided scale.
The role of RIM-I in both capacitating Ca2+ entry at the presynapse through selective recruitment of CaV2s and modulating synaptic vesicle fusion has been documented in rodents (Kaeser et al. 2011), fruit flies (Graf et al. 2012; Müller et al. 2012), and worms (Koushika et al. 2001; Kushibiki et al. 2019). The apparent ubiquity of this gene can be contrasted with what appear to be several independent losses of RIM-II in the Bilateria and Cnidaria. We could not identify RIM-II in the fruit fly Drosophila melanogaster or the nematode worm Caenorhabditis elegans, both of whose genomes and transcriptomes have been subject to considerable annotation. We also failed to identify RIM-II in the tardigrades Ramazzottius varieornatus and Hypsibius dujardini, which form a sister clade to arthropods. Instead, arthropod chelicerates Centroides sculpturatus and Limulus polyphemus were found to possess both I and II RIM genes, but RIM-II was absent in the gene data for the crustacean Hyalella azteca. Thus, it might be that the ancestral ecdysozoan possessed both RIM genes, and that type II was independently lost in select arthropods (e.g. Mandibulata), nematodes and tardigrades. Furthermore, in deuterostomes the RIM-II gene was not identified in the echinoderms Strongylocentrotus purpuratus and Acanthaster planci, but present in gene data for the cephalochordate Branchiostoma belcheri and the hemichordate Saccoglossus kowalevskii, despite being largely absent in gene data for chordates, suggesting independent losses in Echinodermata and clades within Chordata. We identified type I and II RIM genes in the cnidarian Exaiptasia pallida, but only type I in other cnidarians (i.e. Hydra vulgaris and Nematostella vectensis), suggesting that losses of RIM-II also occurred within Cnidaria, a sister taxon to the bilateria. In contrast to other metazoan phyla, both RIM homologs are highly conserved among molluscs, as identified in the gastropods Aplysia californica and Lymnaea stagnalis (the latter was not included in the tree due to fragmentation of assembled mRNA transcripts [Sadamoto et al. 2012]), the bivalves Crassostrea gigas and Mizuhopecten yessoensis, and the cephalopod Octopus bimaculoides. RIM-II was also identified in the brachiopod Lingula anatina, suggesting broad conservation among the Lophotrochozoa (fig. 2). Interestingly ctenophores, the most early-diverging animals that bear nervous systems and synapses, are unique in that they lack RIM-I and possess only RIM-II. In addition, their RIM-II lacks a predicted Zn2+-finger domain broadly conserved in other RIM homologs (fig. 2). This is significant because this particular domain is required for a direct interaction with the protein Munc-13, and hence for RIM to play a role in synaptic vesicle docking and priming (Betz et al. 2001; Dulubova et al. 2005; Lu et al. 2006; Quade et al. 2019). Nevertheless, ctenophores possess Munc-13 in their genomes (Ryan et al. 2013; Moroz et al. 2014). We note that although the absence of identifiable RIM-II genes for various metazoan species could be accounted for by incomplete genome/transcriptome sequencing data, we had little difficulty identifying RIM-I sequences for most of the species included in our analysis.
RIM-I and RIM-II Are Differentially Expressed in the Mollusc Snail L. stagnalis
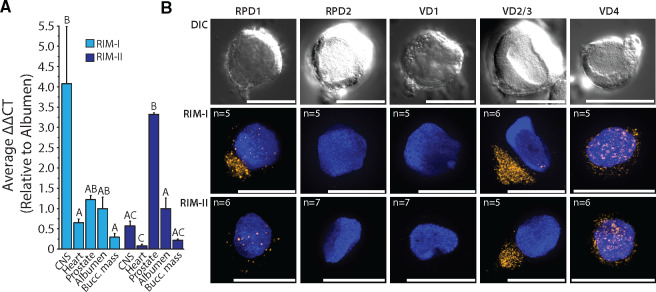
The obligate retention of RIMs across metazoan genomes (Paps and Holland 2018), coupled with the apparent frequent loss of RIM-II in various clades, suggests that this newly identified gene plays a secondary, redundant role to RIM-Is when both are present in the genome. Nevertheless, our ability to detect RIM-II broadly within Mollusca suggests that they utilize this gene nonredundantly to RIM-I. This compelled us to determine the expression of both genes in different tissues of the pulmonate gastropod L. stagnalis (pond snail). The known anatomy and large neurons of the snail have made it a key model organism for studying the neural correlates of behavior, and much is known about the properties of individual neurons and neural circuits in Lymnaea (Syed et al. 1990; Kemenes and Benjamin 2009). Although quantitative polymerase chain reaction (qPCR) experiments of RIM-I revealed relatively high expression in the central nervous system (CNS) of the snail, RIM-II had its highest expression in the prostate, an organ involved in peptidergic secretion and signaling for sexual reproduction (Koene et al. 2010) (fig. 3A). At lower levels, RIM-I was also detected in the heart, prostate, albumen gland, and buccal mass (used for feeding), whereas RIM-II was detected in the CNS and albumen gland and minimally in the buccal mass and heart.
Fig. 3.
(A) qPCR transcript quantification across various tissues of the pond snail L. stagnalis reveals RIM-I expression is most prominent in the CNS, whereas RIM-II is most abundantly expressed in the prostate gland. (B) Fluorescent in situ hybridization on cultured Lymnaea giant neurons from the visceral and right parietal ganglia using gene-specific LNA probes indicates coexpression of RIM-I and RIM-II mRNAs in select identified neurons. Differential interference contrast and fluorescence channels are shown for RPD1, RPD2, VD1, VD2/3 (indistinguishable), and VD4 neurons. Nuclei, labeled with DAPI stain, appear blue. Scale bars denote 50 μm.
Despite the dichotomy seen in Lymnaea CNS expression of the two RIMs, the genes exhibited overlapping cellular expression as evidenced by mRNA in situ hybridization on isolated and cultured neurons (fig. 3B). Various identifiable neurons chosen to study included: the giant right parietal dorsal 1 (RPD1), RPD2, visceral dorsal 1 (VD1), 2/3 (VD2/3), and 4 (VD4) neurons. Each of these exists as a single cell in the CNS (except for VD2/3, which are analogous neurons and hard to separately identify) and each has been previously subjected to detailed characterization of phenotype, function, and morphology (Benjamin and Winlow 1981; Syed et al. 1990; Beekharry et al. 2015). Although the right parietal ganglion (e.g. A group neurons) is immunoreactive to serotonin (5-HT) (Elekes et al. 1989), this has not been reported for RPD1, which is thought to express the neuropeptide FMRFamide (Bright et al. 1993), or RPD2, which harbors a rich peptidome (Jiménez et al. 2006). Neuropeptide expression has also been reported in the visceral ganglion neurons VD1 (Jiménez et al. 2006) and VD4 (Nesic et al. 1996). VD4 forms reciprocal inhibitory synapses with postsynaptic neuron right pedal dorsal 1 (RPeD1), where the VD4 neuron is reported to switch transmitters from a FMRFamide-like peptide to acetylcholine, the latter switching the postsynaptic response of RPeD1 to excitatory (Woodin et al. 2002). RPD1, VD2/3, and VD4 neurons all expressed both RIM homologs, however, RPD1 showed stronger expression of RIM-I than RIM-II, and whereas both RIM mRNAs clustered in discrete cytoplasmic foci in VD2/3, the expression in VD4 was considerably more diffuse (fig. 3B). Instead, neither RIM homolog was expressed in the electrically coupled RPD2 and VD1 neurons that innervate organs responsible for cardio-respiratory functions (Bogerd et al. 1991; Kerkhoven et al. 1993; Jiménez et al. 2006; Beekharry et al. 2015), indicating that these genes are not necessarily expressed in all neurons.
RIM-I and RIM-II PDZ Domains Diverged at Key Loci Associated with Ligand Specificity
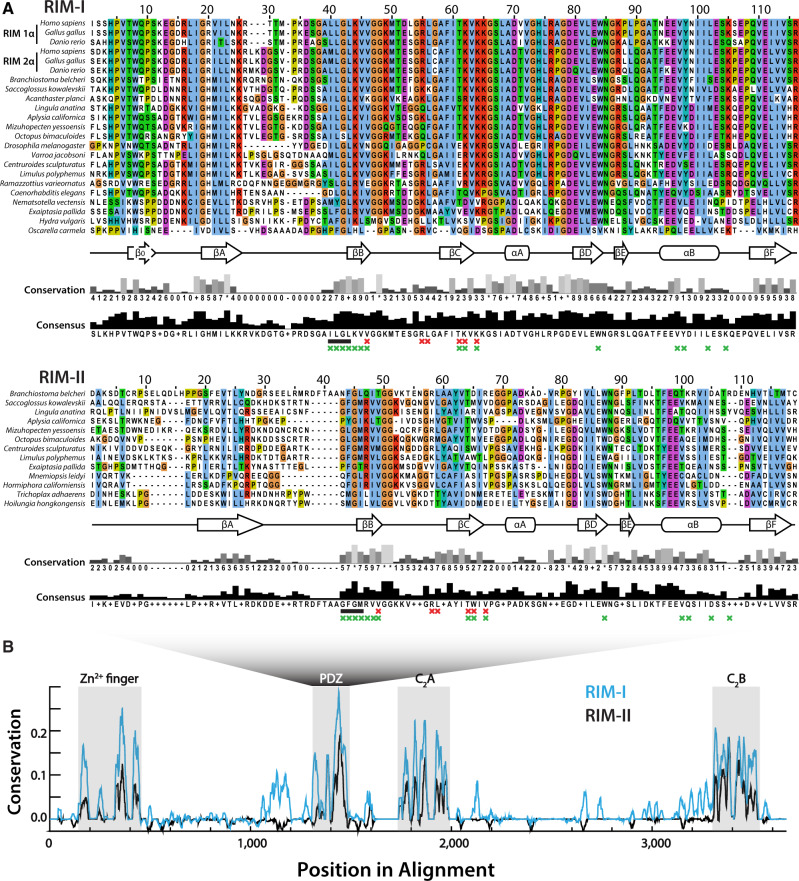
As noted, the synaptic interaction between RIM-Is and CaV2 channels is proposed to largely depend on the PDZ domain of RIM binding to D/E-D/E/H-WC-COOH motifs on the extreme C-termini of CaV2 channels (Kaeser et al. 2011; Graf et al. 2012; Hirano et al. 2017). Aiming to infer how RIM-I and RIM-II might compare in mediating PDZ-dependent protein interactions, we aligned representative PDZ domain sequences of both homologs (fig. 4A). The PDZ domains of both RIM types were found to be approximately 100 amino acids long and characterized by predicted stereotypical secondary structures of six beta strands (βA–F), a short alpha helix (αA), and a long alpha helix (αB). Predicted secondary structure was not conserved for the PDZ domain of the sponge Oscarella carmela, whose sequence failed to align with other metazoan PDZ domains due to divergence. Globally, the RIM-I PDZ domain shared higher sequence identity (52.6% ±17.7% average paired identity score ±SD), compared with PDZ domains of RIM-II (30.1 ± 9.7%), particularly toward the N-terminal side upstream of βB. In this region, RIM-Is bear an additional predicted beta strand (β0), consistent with NMR structures (Lu et al. 2005). Instead, the β0 strand is absent in most other PDZ domains (Lee and Zheng 2010) including RIM-II (fig. 4A). Both RIM-I and RIM-II PDZ domains showed high conservation of the ligand carboxylate-accommodating loop of consensus sequence X-φ-G-φ (where φ denotes a hydrophobic amino acid and X any amino acid) (Lee and Zheng 2010), located on the N-terminal side of βB (fig. 4A, black bars). An exception is the O. bimaculoides RIM-I ortholog, which contains an insert at this key ligand-binding locus that was removed from the alignment.
Fig. 4.
(A) Multiple sequence alignment of various RIM-I and RIM-II PDZ domains. The secondary structures are defined by six beta strands (βA–F), a short alpha helix (αA), and a long alpha helix (αB). The carboxylate accommodating loop with consensus sequence X-φ-G-φ is indicated by a black bar. Red Xs denote the six residues identified as unequivocal classifiers of distinct PDZ clades (Sakarya et al. 2010), and green Xs positions of the RIM-I PDZ domain that interact with bound ligands in NMR structures (Lu et al. 2005). (B) Graph depicting sequence conservation/entropy of aligned RIM proteins delineates conserved Zn2+-finger, PDZ, C2A, and C2B domains, and low homology within interdomain protein regions.
Early studies on the ligand specificities of different PDZ domains delineated three classes based on C-terminal ligand recognition sequences, the most common being class I PDZ domains (ligand motifs of X-S/T-X-φ-COOH, where X denotes any amino acid and φ a hydrophobe), as well as class II and class III domains (respective ligand motifs of X-φ-X-φ-COOH and X-E/D-X-φCOOH) (Songyang et al. 1997; Stricker et al. 1997; Nourry et al. 2003). More recently, a comprehensive analysis of PDZ domain ligand specificities through peptide-phage display of over 330 PDZ domains in human and nematode worm expanded the specificity classes of known PDZ domains to 16 distinct ligand classes (Tonikian et al. 2008). Apparent is that the ligand specificity of type I RIM does not neatly fit within the 3 or 16 type classification systems, indicative of a unique specialized selectivity for the CaV2 channel ligand of D/E-D/E/H-WC-COOH. By leveraging evolutionary divergence of distinct PDZ domains within the proteomes of numerous animals and closely related eukaryotes, a study identified six aligned amino acid positions that share high general entropy but low within-clade entropy, representing unequivocal classifiers of the clade to which a given PDZ domain belongs (Sakarya et al. 2010). Based on NMR structures of RIM-I PDZ domains interacting with C-terminal ligands of ELKS1b and CaV2.1, four of these six amino acids (i.e. βB4, βC4, βC5, and βC–αA-1, where βB4 denotes the fourth residue of the second beta strand) contact the entropic p-1 and p-3 residues of ligands (where p0 denotes the distal-most C-terminal residue), and are involved in ligand selectivity (Lu et al. 2005; Sakarya et al. 2010; Kaeser et al. 2011). In an effort to parse out potential differences in the protein-binding capabilities of type I and II RIMs, we labeled these key residues in our alignment (fig. 4A). Interestingly, βC5, and βC–αA-1 differ between the two RIM homologs: whereas RIM-I has a highly conserved basic region defined by K46 and K48 (i.e. TKVK motif), RIM-II features hydrophobic W46 and V48 (TWIV). These particular amino acids have been shown to contribute to the binding pocket of type I RIM1, which exhibit shifts in heteronuclear single quantum coherence spectra upon binding to CaV2.1 C-terminal peptides (Kaeser et al. 2011). Another key site associated with ligand specificity is the consensus amino acid in position αB1 of the PDZ domain, which interacts with p-2 ligand residues (Hung and Sheng 2002). For most RIM PDZ domains examined in this study, this position was occupied by a phenylalanine (F), albeit with considerable variation (fig. 4A). αB1 amino acids that form hydrogen bonds (i.e. Y, N, Q, K, R) preferentially bind hydroxy group-containing serine or threonine p-2 residues of class I ligands (X-S/T-X-φ-COOH), whereas hydrophobic amino acids select for hydrophobic p-2 residues of class II ligands (X-φ-X-φ-COOH), and tyrosines interact with acidic p-2 residues of class III ligands (X-[D/E]-X-φ-COOH). However, this model is inconsistent with the ligand-binding properties of RIM-I PDZ domains, because both coimmunoprecipitation and NMR experiments have demonstrated that type I RIM1 and RIM2 PDZ domains interact with the DDWC-COOH ligand on the CaV2.1 channel C-terminus (Kaeser et al. 2011; Hirano et al. 2017), despite RIM1 bearing an asparagine in position αB1 consistent with class I ligands, and RIM2 bearing a phenylalanine consistent with class II ligands. Thus, this locus might play a minimal role in RIM ligand specificity. In summary, although both type I and II RIM proteins bear canonical PDZ secondary structures, the two homologs have differences at key loci suggesting differences in ligand specificity.
We also compared the Zn2+-finger and C2 domain sequences conserved between RIM-I and RIM-II (fig. 4B), plus rabphilin (supplementary fig. S2A, Supplementary Material online). With few exceptions, the N-termini contained a predicted Zn2+-finger domain and α helical structures (αA) involved in Munc-13 and Rab3 binding, respectively (Wang et al. 1997, 2001; Betz et al. 2001; Fukuda 2003; Dulubova et al. 2005; Lu et al. 2006; Quade et al. 2019), two short β strands (βA and βB), and a second α helix (αB) (supplementary fig. S2A and file S1, Supplementary Material online). Notably, although the molecular determinants for RIM/rabphilin interactions with Munc-13 and Rab3 are considered separate (Ostermeier and Brunger 1999), mutations in the Zn2+-finger domain nevertheless disrupt interactions with Rab3 (McKiernan et al. 1996; Stahl et al. 1996), indicative of structural interdependence between these two regions. Of the eight Zn2+-finger cysteine (C) residues required for Zn2+ and Rab3 binding (Stahl et al. 1996), seven were very highly conserved across most orthologs. Furthermore, the αB helix SGAWFF motif, identified as a Rab complementarity-determining region that confers specificity to select Rab proteins (Ostermeier and Brunger 1999), had deep conservation across rabphilin sequences but was less conserved in RIM-I and RIM-II. The C2A and C2B domains of all three proteins were characterized by 8 predicted β strands (βA to βH), common to type I C2 domains (i.e. synaptotagmin family C2 domains), which form an eight-stranded antiparallel beta sandwich secondary structure (Biadene et al. 2006). As noted, the RIM C2B domain, present in all four vertebrate RIM-I paralogs (RIM1–RIM4), potentiates Ca2+ influx through CaV2 channels via an interaction with the CaVβ subunit, which attenuates voltage-dependent inactivation to prolong presynaptic Ca2+ influx (Kiyonaka et al. 2007; Uriu et al. 2010; Kaeser et al. 2012). Although the mechanisms for this protein–protein interaction have not yet been elucidated, it is likely not dependent on Ca2+, given that RIM-I C2 domains are degenerate in their Ca2+-binding capacity, lacking key residues including five aspartates that comprise Ca2+-binding sites in related proteins rabphilin and synaptotagmin (Wang et al. 1997; Ubach et al. 1998; Coudevylle et al. 2008). To assess whether the RIM-II C2 domains might bind Ca2+, we aligned RIM-I, RIM-II, and rabphilin C2A and C2B domains, identifying the five aspartate (D) residues that mediate Ca2+ binding (supplementary fig. S2B, Supplementary Material online). A high conservation of aspartate in all three proteins was seen exclusively at p110, suggesting that RIM-IIs, like RIM-Is, have degenerate C2 domains.
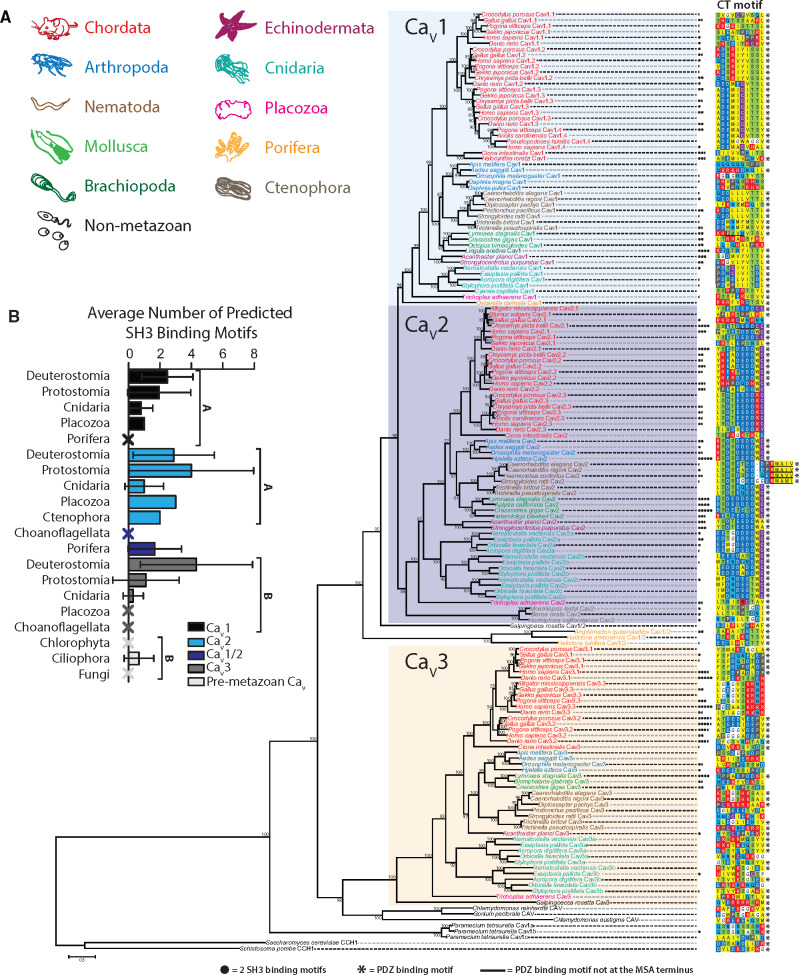
The Phylogeny of CaV Channels Informs on Conserved PDZ and SH3 Domain Ligand Motifs
The reported conservation of D/E-D/E/H-WC-COOH PDZ ligand motifs on the distal C-termini of CaV2 channels from vertebrates (Kaeser et al. 2011; Gardezi et al. 2013), fruit flies (Graf et al. 2012), and molluscs (Spafford et al. 2003) is notable given that at the phylum-level protein alignments of orthologous CaV channel intracellular linkers and N- and C-termini tend to show poor sequence homology (Spafford et al. 2003; Senatore and Spafford 2010; Tyson and Snutch 2013). In addition to binding RIM-Is, the CaV2 PDZ ligand motif also mediates interactions with a PDZ domain of the presynaptic scaffolding protein Mint, documented in rodents (Maximov et al. 1999), chick (Gardezi et al. 2013), and the gastropod mollusc L. stagnalis (Spafford et al. 2003). Thus, it seems likely that interactions between CaV2 channels and RIM-I/Mint-1 were present in the last common ancestor of the bilaterians. Nevertheless, a comprehensive analysis of CaV channel C-terminal sequences within the Metazoa, to explore conservation of C-terminal PDZ ligand motifs, has not been reported. Furthermore, recently sequenced genomes and transcriptomes permit re-exploration of the CaV channel phylogeny (Moran and Zakon 2014; Senatore et al. 2016). Hence, using sequences compiled from genomic and transcriptomic databases, we constructed a comprehensive maximum likelihood protein phylogeny of various CaV α subunits, and aligned their 10 distal-most C-terminal amino acid sequences which would bear putative PDZ ligand motifs (fig. 5A). Rooting the tree with fungal CCH1 CaV channel homologs (Saccharomyces cerevisiae and Schistosoma pombe) revealed three distinct clades of metazoan CaV channels, with LVA CaV3 channels forming a sister clade with the HVA CaV1 and CaV2 channels. Similar to CCH1, CaV channel homologs from the ciliate Paramecium tetraurelia, involved in regulating ciliary beating (Lodh et al. 2016), form a sister clade with metazoan CaV channels, whereas those from the green algae Chlamydomonas sp. and Gonium pectoral (also involved in regulating ciliary beating [Fujiu et al. 2009]) form a sister clade with CaV3 type channels. Our phylogenetic tree is consistent with previous reports that HVA and LVA channels existed in the last common ancestor of animals and choanoflagellates, where the choanoflagellate species Salpingoeca rosetta possesses a bona fide CaV3 channel homolog, as well a CaV1/CaV2 channel posited to be ancestral to CaV1 and CaV2 channels (Moran and Zakon 2014). Also consistent with previous reports, sponges Amphimedon queenslandica, Haliclona amboinensis, and Haliclona tubifera possess single CaV1/CaV2 channel homologs, and lack CaV3 channels, attributed to gene loss. It has been proposed that CaV1 and CaV2 channels emerged via gene duplication from a CaV1/CaV2-like channel, perhaps after sponges diverged from other animals (Moran et al. 2015). However, here we identify a CaV1 channel homolog in the gene sequences of the sponge O. carmela, suggesting instead that this event occurred prior to the divergence of sponges, and in turn, that most lineages of sponges lost CaV1 and CaV2 channels (fig. 5). Indeed, such a scenario would explain the presence of CaV2 channels in the gene sequences of ctenophore species Mnemiopsis leidyi, Beroe ovata, and Hormiphora californiensis, which based on the leading species phylogeny, are the most early-diverging group of animals (Ryan et al. 2013; Moroz et al. 2014; Whelan et al. 2017). As such, CaV1/CaV2, CaV1, and CaV2 channels, plus CaV3 channels, might have existed in the common ancestor of all animals, and these were differentially lost such that ctenophores retained only CaV2 channels, and sponges CaV1 or CaV1/CaV2 channels. Instead, the placozoan T. adhaerens, which forms a sister clade with cnidarians and bilaterians, is the most early-diverging animal to possess all three types of metazoan CaV channels (i.e. CaV1–CaV3), and along with cnidarians and bilaterians, they lack CaV1/CaV2 channels (fig. 5). Also evident are the two rounds of CaV gene duplication in the stem lineage of vertebrates, resulting in ten vertebrate CaV channels (i.e. CaV1.1–CaV1.4, CaV2.1–CaV2.3, and CaV3.1–CaV3.3), and independent duplications in Cnidaria resulting in six CaV channel homologs (CaV1, CaV2a–CaV2c, and CaV3a–CaV3b) (fig. 5A) (Jegla et al. 2009; Moran and Zakon 2014; Moran et al. 2015).
Fig. 5.
(A) Maximum likelihood phylogenetic tree of the alpha subunit of metazoan and premetazoan CaV channel homologs. Bootstrap values for 1,000 ultrafast replicates are indicated on corresponding nodes. The distal ten amino acids that contain putative PDZ domain ligand motifs are aligned for all sequences. PDZPepInt predictions of PDZ domain binding are annotated by black asterisks. SH3PepInt predictions of C-terminal SH3-binding motifs are denoted with filled black circles. (B) Quantitation of average number of predicted SH3-binding motifs per CaV paralog and clade. CaV1 and CaV2 have on average a significantly higher number of SH3 motifs compared to CaV3 channels (one-way ANOVA and Dun’s post hoc test with the Benjamini–Hochberg adjustment). Error bars denote standard deviation, and Xs denote zero predictions.
Pursuant to our characterization of the PDZ domains of RIM-I and RIM-II, we examined the sequences of putative CaV C-terminal PDZ ligands across all homologs (fig. 5). Four amino acids at the extreme C-termini typically participate in the β-strand complementation that mediates PDZ ligand-domain protein interactions (Hung and Sheng 2002), however, at least seven residues upstream of the carboxylate group are known to strengthen this interaction via intermolecular bonds and attractions (Tonikian et al. 2008; Ernst et al. 2014). In contrast to the hypervariable sequence that typifies the medial and distal thirds of CaV1 and CaV2 C-termini (fig. 6), our alignment evidences high conservation of the ten most distal amino acids within respective paralogs (fig. 5A). In large part, CaV1 orthologs possess class I PDZ ligands, and CaV2 orthologs possess noncanonical ligands. The CaV1–CaV3 channels from Trichoplax, and CaVs from the sponges, all possess class I PDZ ligands, the latter bearing ligand motifs of E-T-S/T-V-COOH identified as a consensus PDZ ligand sequence for Disks large homolog (DLG) scaffolding proteins of both humans and nematode worms (Tonikian et al. 2008). Importantly, we found motifs similar to D/E-D/E/H-WC-COOH to be conserved in CaV2 channels throughout Bilateria and Cnidaria (fig. 5), but absent in orthologs from Trichoplax and ctenophores. CaV3 channels largely lack distal C-terminal conservation across phyla (figs. 5A and 6A). On a more granular level, we observed apparent sequence divergence from a class I PDZ ligand among CaV1 channels in arthropods and CaV1.1 channels in vertebrates, and a conserved hydrophobic insert disrupting the D/E-D/E/H-WC-COOH like motif in nematodes of the clade Rhabditomorpha (Caenorhabditis, Haemonchus, and Strongyloides), but not Trichinellida. Furthermore, we note the apparent absence of D/E-D/E/H-WC-COOH like motifs in the CaV2 channel of early-diverging chordate Ciona intestinalis and avian P/Q-type CaV2.1 channels, the latter concomitant with the reported loss of C-terminal exon 47 (human equivalent) in the gene from Gallus gallus (Snidal et al. 2018). To corroborate our sequence data, we also predicted C-terminal PDZ ligands with the cluster-based prediction tool PDZPepInt (Kundu et al. 2014), which references a set of 226 PDZ domains from humans, mouse, fly, and nematode worm, finding that most metazoan and all premetazoan CaVs bear predicted PDZ ligands (fig. 5A).
Fig. 6.
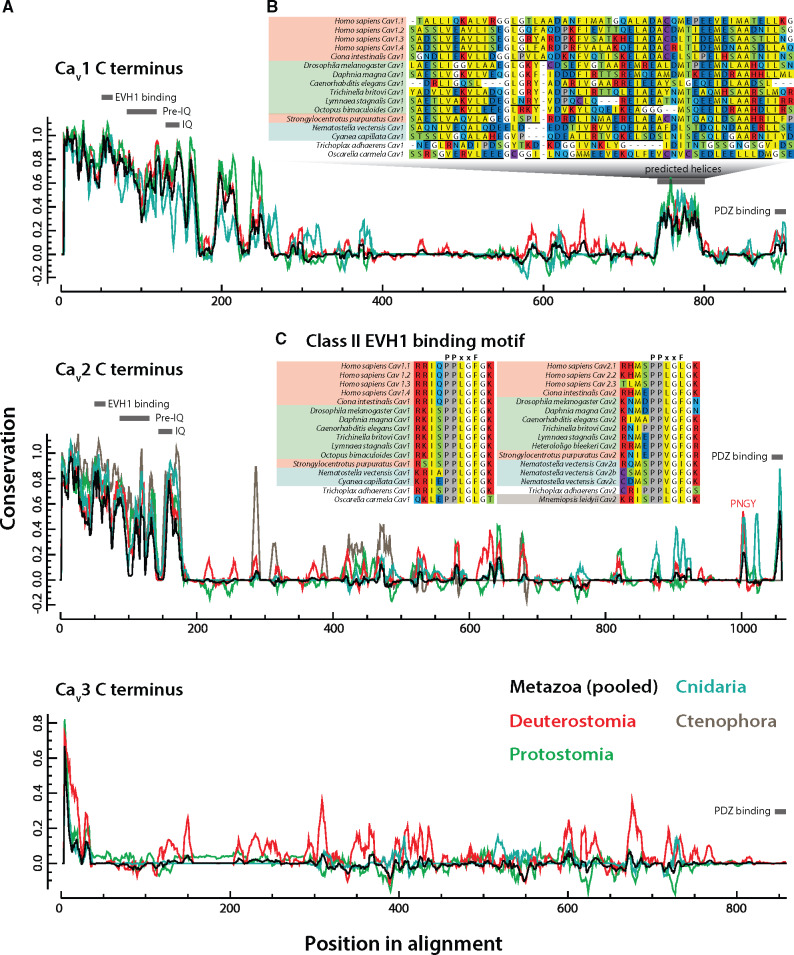
(A) Graph depicting sequence conservation/entropy of aligned CaV channel C-termini reveals distinct pockets of sequence conservation within the largely disordered protein sequences. Separate traces showing conservation among individual clades were made by pooling sequences constituting a given clade from the original multiple sequence alignment and normalizing to one scale. Locations of conserved SLiMs are indicated with grey bars. (B) A highly conserved region in the CaV1 distal C-terminus corresponds to a predicted helical structure conserved even in the identified homolog from the sponge O. carmela, absent in CaV2, CaV3, and CaV1/CaV2 channels (PSIPRED-predicted α helical secondary structures are denoted with black underlines, and β strands with orange underlines). The CaV1 channel from T. adhaerens uniquely lacked a predicted helix in this region. (C) Alignment of the identified class II EVH1 domain-binding motif of consensus sequence PPXXF reveals deep conservation of this motif in both CaV1 and CaV2 channels.
To expand our analysis of putative protein-interaction motifs on CaV channel C-termini, we also predicted SH3 domain ligands, present in CaV2 and RIM-I homologs, where the scaffolding protein RIM-BP binds to form a tripartite CaV2/RIM/RIM-BP complex at the synapse active zone (Südhof 2012) (fig. 1A). RIM-BP SH3 domains have also been shown to interact with proline-rich regions on the C-termini of CaV1 channels (Hibino et al. 2002), however such interactions have not been reported for CaV3 channels, nor has the presence of putative SH3 ligand motifs been studied systematically in CaV channels from divergent animal phyla. This paucity of data likely stems both from the sequence hypervariability that characterizes the C-termini of CaVs (fig. 6), and the fact that SH3 domains recognize numerous noncanonical binding motifs (Teyra et al. 2017) making predictions difficult. Nonetheless, we used three independent methods to predict the number of SH3-binding motifs within the C-termini of all examined CaV homologs (fig. 5 and supplementary S3, Supplementary Material online), finding that CaV1 and CaV2 channels contain on average significantly more SH3 motifs than either CaV3 or premetazoan CaV channels (fig. 5B). Intrachannel (within paralog) interclade comparisons revealed a significantly greater number of predicted SH3 motifs among CaV3 channels from deuterostomes compared with protostomes (P = 0.0010) or cnidarians (P = 0.0013) (Kruskal–Wallis test and Dunn’s post hoc test with Benjamini–Hochberg adjustment), whereas interclade differences were nonsignificant for CaV1 and CaV2 paralogs. Interestingly however, we note an enrichment in SH3 motifs among CaV2 channels from molluscs (fig. 5A). Lastly, intraclade interchannel comparisons revealed a significantly greater number of SH3 motifs among CaV2 versus CaV3 channels from protostomes (P = 0.00713), but no significant differences between CaV channel paralogs from deuterostomes or cnidarians.
Intrinsically Disordered CaV Channel C-Termini and Linker Regions Are Hubs for SLiMs
The noted sequence entropy within CaV channel cytoplasmic regions reflects lower evolutionary constraints, perhaps facilitating the emergence of novel motifs with novel interactomic functions in distinct channel clades. Accordingly, channel termini and linkers are important regions for differential CaV channel modulation by regulatory proteins (Tyson and Snutch 2013). To systematically characterize the cytoplasmic regions of the CaV channels included in our phylogenetic tree (fig. 5), we first performed a quantitation of the protein sequence length of the N- and C-termini, plus the I-II, II-III, and III-IV cytoplasmic linkers (supplementary fig. S4, Supplementary Material online). Although no significant differences were noted among N-termini lengths between calcium channel paralogs when all clades were pooled (Kruskal–Wallis χ2 = 5.9176, df = 3, P value = 0.1157), both CaV1 and CaV2 had significantly longer C-termini as compared with CaV3 and premetazoan channels (Kruskal–Wallis χ2 = 45.272, df = 3, P value = 8.1E−10) (supplementary fig. S4, Supplementary Material online). Notably, linkers among CaV3 channels were both significantly longer and more variable than those of CaV1 and CaV2 channels, and particularly so for the I-II and II-III linkers (I-II: Kruskal–Wallis χ2 = 113.82, df = 3, P value < 2.2E−16; II-III: Kruskal–Wallis χ2 = 60.884, df = 3, P value = 3.806E−13; III-IV: Kruskal–Wallis χ2 = 140.14, df = 3, P value < 2.2E−16). Intrachannel (within paralog)-interclade comparisons of termini and linker lengths were also performed. Despite the generally shorter II-III linker in CaV2 channels as compared with CaV3, the deuterostome CaV2 channels have significantly longer linkers relative to those found in protostomes and cnidarians (Kruskal–Wallis χ2 = 40.2965, df = 2, P value = 0). This is consistent with the observation that the II-III linker SYNPRINT motif, involved in interactions between CaV2 channels and exocytotic SNARE proteins, is a feature unique to vertebrate, and perhaps all deuterostome, channels (Spafford et al. 2003). Lastly, whereas a significant expansion in length was noted for bilaterian CaV2 C-termini as compared with cnidarian orthologs (Kruskal–Wallis χ2 = 22.822, df = 2, P value = 1.107E−05), deuterostome C-termini were significantly longer than those of protostomes for CaV1 and CaV3 (intra CaV1: Wilcoxon statistic W = 367.5, P value = 0.03867; intra CaV3 Kruskal–Wallis χ2 = 14.265, df = 2, P value = 0.0007986). Altogether the variability observed in these disordered structures likely reflects differential protein interactions and modulatory capacities for the different CaV channel types within and across different clades (Tyson and Snutch 2013).
Next, we sought to determine whether we could identify novel SLiMs, or concomitantly, evidence the lack of known motifs in these disordered protein regions by leveraging sequence conservation analysis (Spafford et al. 2003). A running window of the sequence conservation of representative bilaterian and cnidarian sequences was generated for all CaV paralogs, then visualized by pooling respective sequences from the original multiple sequence alignment to allow for identification of clade-specific SLiMs (fig. 6A). Generally, CaV3 channels were found to be more variable than either CaV1 or CaV2 channels, particularly in the proximal third of the C-terminus. Furthermore, we identified an island of conservation amid highly entropic sequences in the distal third of CaV1, found to possess helical character upon PSIPRED secondary structure prediction (fig. 6A and B). This locus has been characterized as a cAMP-dependent protein kinase-anchoring protein 15 (AKAP15) binding domain in CaV1.2 channels, required to effect β-adrenergic receptor mediated increase in calcium current (Hulme et al. 2003). In addition, proteolytic cleavage of the distal C-terminus bearing this motif produces an autoinhibitory peptide that binds a proximal region of the CaV channel C-terminus to inhibit its activation (Hulme et al. 2006), and can translocate to the nucleus to act as a transcription factor (Gomez-Ospina et al. 2006). Here, we show that this helical AKAP15-binding element is conserved across Bilateria and Cnidaria, and exists even in the identified CaV1 channel homolog from the sponge O. carmela (fig. 6B), structurally distinguishing it from CaV1/CaV2 channels from other sponge species (figs. 5A and 6A).
Next, we used the motif elicitation tools SIB MyHits (exhaustive database search) (Pagni et al. 2007) and Multiple Em for Motif Elicitation (MEME) (Bailey et al. 2009) to identify SLiMs hidden within poorly conserved regions of the CaV C-termini. Although MyHits returned questionable or weak matches, MEME, combined with manual analysis of proline-rich motifs in our CaV multiple sequence alignment, identified a highly conserved type II Drosophila enabled/vasodilator-stimulated phosphoprotein homology 1 (EVH1) domain-binding motif within the proximal C-termini of CaV1 channels (consensus of P-P-X-X-F), and CaV2 channels (P-P-X-X-φ; fig. 6C). Like the SH3 domain, EVH1 domains bind proline-rich regions on target proteins with low affinity, and feature prominently in signaling networks and at synapses (Ball et al. 2002). Notable among EVH1 domain-containing proteins is the postsynaptic scaffolding protein Homer (and related proteins). Homers have been reported to regulate excitation–contraction coupling through a physical interaction with CaV1.2 channels and the ryanodine receptor RyR2 (Huang et al. 2007), and to mediate the flux of extracellular Ca2+ between the plasma membrane and the endoplasmic reticulum though interactions with CaV1.2 and STIM1 (Dionisio et al. 2015). Notably, it has not yet been determined whether these specific interactions involve the conserved EVH1-binding site identified here for CaV1 channels. Nevertheless, the interaction between Homer and CaV1.2 requires a functional Homer EVH1 domain, because point mutations that disrupt its binding capacity disrupt binding with CaV1.2 channels (Huang et al. 2007). Much less is known about the potential interaction between Homer and CaV2 channels. In one study, G-protein inhibition of CaV2.2 channels by metabotropic glutamate receptors (mGluRs) was found to be disrupted by select Homer variants (Kammermeier et al. 2000), suggesting that Homer is either directly modulating mGluR function (Brakeman et al. 1997), or alternatively, affecting G-protein binding to the CaV channel. Indeed, here we show that although all human CaV2 subtypes possess a type II EVH1-like motif of P-P-X-X-L, the canonical P-P-X-X-F motif is present in CaV2 channels as early-diverging as Trichoplax (fig. 6C), which suggests that EVH1 domain-bearing proteins like Homer may regulate CaV2 channels broadly in the Metazoa.
Discussion
On the Phylogeny of RIM-I
The multifaceted nature of RIM is still being unraveled more than 20 years after its initial characterization as an effector of Rab3, a neuronal GTP-binding protein that regulates synaptic vesicle fusion (Wang et al. 1997). RIM protein isoforms that bear N-terminal Zn2+-finger domains and flanking alpha helical structures, which bind Munc-13 and Rab3, play important roles in regulating synaptic vesicle priming and docking (Gracheva et al. 2008; Deng et al. 2011). Separately, C-terminal regions of RIM interact with CaV2 channels (via the PDZ domain), the CaVβ subunit (via the C2B domain), and RIM-BP (via a proline-rich motif between C2A and C2B) (Kiyonaka et al. 2007; Uriu et al. 2010; Kaeser et al. 2012; Südhof 2012), allowing the protein to functionally link exocytosis-ready vesicles with the excitation-dependent Ca2+ signaling. Indeed, the broad conservation of this functionality across bilateria (Kaeser et al. 2011; Graf et al. 2012; Kushibiki et al. 2019), points to an early evolutionary adaptation of RIM-I for regulating fast, synchronous synaptic exocytosis that requires nanometer proximity between CaV2 channels and exocytotic vesicles (Eggermann et al. 2012; Wang and Augustine 2015; Stanley 2016). That the N- and C-terminal interactions/functions of RIM-I might be considered separate is suggested by a recent study on the mechanisms for exocytosis of large dense-core vesicles in isolated neurons from conditional RIM1/RIM2 knockout mice. Here, genetic reintroduction of RIM1 variants bearing disrupted N-terminal sequences failed to rescue dense-core vesicle exocytosis, whereas variants lacking the PDZ domain were successful at rescuing exocytosis (Persoon et al. 2019). Hence, in these neurons, the N-terminus-associated functions of RIM (priming and docking of vesicles) appear essential, whereas its role in CaV channel localization does not.
The recent characterization of RIM as one of only 25 genes that are unique to animals and that have broadly resisted genetic loss (Paps and Holland 2018), including in animals that lack nervous systems and synapses, points to a general functionality for this gene that is perhaps distinct from its role in synchronous neuronal Ca2+/excitation-dependent exocytosis. For example, RIM-I is present in the gene data for both placozoans (T. adhaerens, H. hongkongensis) and poriferans (O. carmela) (fig. 2), all of which lack synapses. Homologs from these early-diverging animals lack PDZ domains (fig. 2), likely lost from a common ancestor, and as a result, their putative capacity to interact with CaV2 channels (although Oscarella has a second RIM homolog that formed a sister clade with RIM-I and RIM-II and bears a PDZ domain). Poriferans also lost the majority of genes required for fast electrical neural signaling, including voltage-gated sodium and potassium channels (Moran et al. 2015), and thus lack the capacity for canonical electrical signaling in the form of action potentials, and by extension, fast excitation–secretion coupling. Perhaps, the bimodal functionality of RIM-I is phylogenetically conserved, where its regulation of vesicle-cell membrane interactions is widely conserved, whereas its roles in nanodomain coupling of CaV2 channels is restricted to select neurons in animals that utilize fast, synchronous synaptic transmission. Interestingly, we show here through in situ hybridization that only a subset of cultured neurons from the CNS of the mollusc snail L. stagnalis express RIM-I (fig. 4B), implying that this gene is not essential for synaptic exocytosis in all neuron types, or instead, that phylogenetically distinct proteins can carry out redundant functions in neurons that do not express RIM-I. Nonetheless, the importance of RIM-I is underscored by genetic disruption studies in vertebrates and invertebrates, where for example, double knockout of RIM1 and RIM2 in mouse is postembryonic lethal, attributed to disrupted neurotransmitter release (Schoch et al. 2006).
Notable is that Trichoplax is the most early-diverging animal to possess genes for RIM-I plus all three types of CaV channels found in cnidarians and bilaterians (CaV1–CaV3 channels). However, the Trichoplax CaV2 protein lacks a C-terminal D/E-D/E/H-WC-COOH like ligand motif, and as noted above, its RIM-I lacks a PDZ domain. All three cnidarian CaV2 channel subtypes possess D/E-D/E-WC-like motifs (CaV2c bears an atypical ETWC motif), and RIM-I is broadly conserved in these animals (fig. 2). Thus, based on current models, cnidarians might have the capacity for a RIM-I/CaV2 presynaptic interaction, akin to bilaterians. Indeed, CaV channels are known to drive synaptic transmission in cnidarians (Bullock 1943; Kerfoot et al. 1985). However, whether they similarly exhibit nanodomain and microdomain synapses, distinguished by the proximity between CaV channels and synaptic vesicle Ca2+ sensors, is not known (Senatore et al. 2016).
Identification of a Novel Clade of RIM Homologs
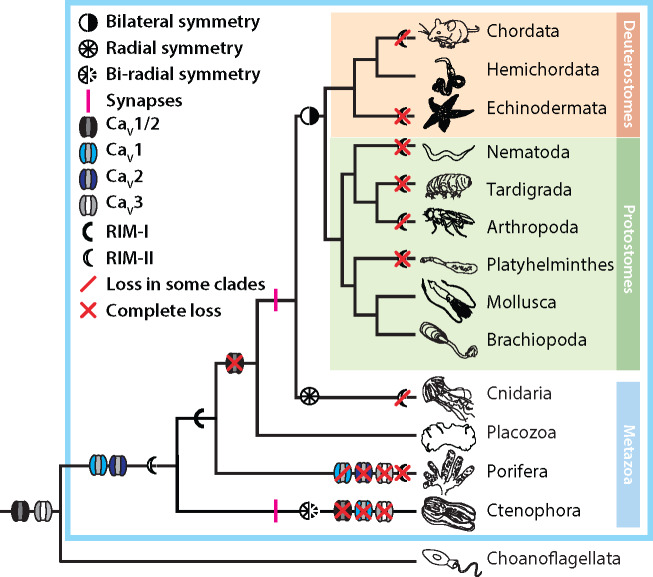
Here, we report the identification of a previously unknown clade of metazoan RIM genes (RIM-IIs), with similar protein domain architectures as RIM-Is, but generally shorter in length (fig. 2), and bearing sequence differences at key loci including the PDZ domain ligand interface (fig. 4). We acknowledge that our analysis is only as good as the available sequence data and, as we obtained sequences from both genomic and transcriptomic databases, we cannot say whether RIM-II is expressed in all of the organisms that harbor it within their genomes. Although we demonstrate that RIM-I is ubiquitously present in animals with the exception of ctenophores, RIM-II appears to have undergone independent losses in multiple lineages, including at the subphylum level (i.e. Chordata, Arthropoda, and Cnidaria; figs. 2 and 7). In the context of the hypothesis that ctenophores, and not sponges, are the most early-diverging group of animals (Ryan et al. 2013; Moroz et al. 2014; Whelan et al. 2017), the most parsimonious explanation of RIM evolution (based strictly on our sequence data) is that RIM-II emerged at the stem lineage of Metazoa, whereas RIM-I emerged in the common ancestor of poriferans, placozoans, cnidarians, and bilaterians (fig. 7). That RIM-II has been repeatedly lost, but no animal lineage has lost both RIM-I and RIM-II (Paps and Holland 2018), alludes to the importance of RIM genes in animals. This also suggests that these two genes exhibit some functional redundancy, where RIM-I might be more essential given its ubiquity in cnidarians and bilaterians (fig. 2). Molluscs, and perhaps other Lophotrochozoans, represent an interesting case in that they have broadly retained both genes (fig. 2). Based on our qPCR expression analysis of RIM-I and RIM-II in various tissues of the freshwater snail Lymnaea, it is evident that the two genes differ in their tissue expression levels (fig. 3A). Nevertheless, despite the observation that RIM-I is enriched in the CNS compared to RIM-II, the two RIM genes overlapped in their neuronal expression (fig. 3). Hence, in the CNS, it is possible that the two genes are functionally complementary. Instead, in different tissues, there might be differential requirements in terms of abundance for one gene over the other. Trichoplax also retains both homologs, but our mRNA expression and localization studies of RIMs and CaVs point to low-level expression (fig. 1 and supplementary S1, Supplementary Material online), making it difficult to interpret their cell-type expression profiles and possible roles. We note that in ongoing studies being carried out in our lab, all three Trichoplax CaV channels express in vitro to conduct voltage-sensitive Ca2+ that resemble those of CaV1–CaV3 homologs from other animals (Smith et al. 2017; Julia Gauberg, Salsabil Abdallah and Adriano Senatore, unpublished data). However, the role of CaV channels and transient membrane Ca2+ signaling in Trichoplax biology is unknown. Speculation of the roles RIM proteins might play in ctenophores is equally, if not more, intriguing. Ctenophores exclusively possess RIM-II, making them the only animals with synapses that lack RIM-I. The ctenophore RIM-II protein is also atypical in that it is predicted to lack an N-terminal Zn2+-finger domain, conserved in RIM-Is, RIM-IIs, and rabphilins (fig. 2). As noted previously, this particular domain is crucial for direct interaction with Munc-13 (Betz et al. 2001; Dulubova et al. 2005; Lu et al. 2006; Quade et al. 2019), and thus, RIM-II might not play a role in synaptic vesicle priming in ctenophore synapses. Nevertheless, we note broad conservation of predicted N-terminal alpha-helical structures associated with Rab3 binding (supplementary fig. S2 and file S1, Supplementary Material online), suggesting that RIM-IIs can interact with vesicles. To our knowledge, ctenophores are the only animals in which the requirement for presynaptic Ca2+ influx for exocytosis and synaptic transmission has not yet been confirmed, and little is known about the mechanisms for synaptic transmission in these animals (Senatore et al. 2016). However, microscopy studies have revealed structures with hallmark features of synapse active zones (Hernandez-Nicaise 1973), and ctenophores possess CaV2 channels. Given the proposal that ctenophores independently evolved the nervous system and synapses (Moroz et al. 2014; Moroz and Kohn 2016), it will be particularly interesting to decipher the roles that CaV2 and RIM-II play in this group of animals.
Fig. 7.
Parsimonious depiction of CaV channel and RIM-I/RIM-II evolution, based on our available sequence data, highlighting the putative emergence of CaV1 and CaV2 channel genes at the stem of Metazoa, and the frequent independent complete or partial loss of RIM-II in various animal phyla. Evident is that Ctenophora is the only animal phylum with synapses but lacking RIM-I, possessing only RIM-II without N-terminal Zn2+-finger motifs. Of note, the phylogenetic relationships depicted in the tree correspond to the hypothesis that ctenophores, and not sponges, are the most early-diverging animals. Furthermore, alternate scenarios are possible, such as gene duplication and differential loss along select nodes of the metazoan phylogeny.
The PDZ domain of RIM-I is the physical link for selectively recruiting presynaptic CaV2 channels to the active zone (Kaeser et al. 2011). We detailed the primary and secondary structures of RIM-I and RIM-II PDZ domains to gain insight into whether they might differ in their ligand binding. Generally, the PDZ domains of both RIM homologs shared a common secondary structure but RIM-II featured lower sequence conservation (fig. 4A). However, the two PDZ domains differed in residue positions involved in ligand selectivity. Specifically, of the three amino acids in the RIM PDZ domain that both interface with bound ligands (Lu et al. 2005), and are sites for evolutionary divergence in PDZ domain ligand specificity (Sakarya et al. 2010), two are different between RIM-I and RIM-II, with respective consensus sequences of TKVK and TWIV (fig. 4A). That they differ in this key locus, with RIM-I having positively charged lysines and RIM-II neutral tryptophan and valine, provides additional support for RIM-II being an independent clade. Given that these motifs interface with bound ligands, it is also tempting to speculate that the positively charged lysines in the RIM-I PDZ domain provide charge attraction for the conserved negatively charged glutamate and aspartate residues of CaV2 channel D/E-D/E-WC-COOH like motifs (fig. 5A), and hence by extension, that RIM-IIs select ligands with different chemical profiles.
One note that is important to mention with respect to the RIM-I/CaV2 interaction, is that although the ligand specificity of the RIM-I PDZ domains appears conserved between rodent and fruit fly (Kaeser et al. 2011; Graf et al. 2012), CaV2 channels from nematodes of the rhabditomorpha, including C. elegans, lost the D/E-D/E-WC-COOH like motif (fig. 5A). Furthermore, phage display screening of the C. elegans RIM-I PDZ domain identified a consensus ligand sequence of F-S/C/I/D-F-W-L/I-COOH (Tonikian et al. 2008), which is quite different from the C-terminal sequence of its corresponding CaV2 channel, and the acidic motif of other CaV2 channels. Despite these differences, genetic experiments have established that RIM and RIM-BP are redundantly required for active zone localization of CaV2 channels in C. elegans (Kushibiki et al. 2019), suggesting they both directly interact with the channel. A possible explanation for this inconsistency might be that, as has been shown for the human CaV2.1–RIM interaction, RIM can bind at secondary internal sites independently of the distal PDZ ligand motif (Hirano et al. 2017). Alternatively, the interaction between CaV2 and RIM might be indirect, mediated by shared interactions with RIM-BP (fig. 1A). Interestingly like C. elegans, cnidarian RIM-Is exhibit sequence divergence in the PDZ domain TKVK motif, but nevertheless conserved negatively charged D/E-D/E-WC-COOH like motifs in their CaV2 channel C-termini (fig. 5A). Clearly, future wet lab experiments aimed at characterizing these interactions in cnidarians, ctenophores, and other animal lineages will be essential toward our understanding of RIM evolution and function in animals.
Insights into CaV Channel Evolution
Our phylogenetic analysis of metazoan CaV channels revealed deep conservation of C-terminal PDZ ligand motifs for both CaV1 and CaV2 (fig. 5A). CaV1 channels, including the homolog from Trichoplax, bear hydrophobic C-termini that fall within the class I PDZ ligands with motifs of X-S/T-X-φ-COOH. Little is known about the conservation of PDZ-mediated interactions for CaV1 type channels, which in contrast to CaV2 tend to localize to postsynaptic sites in neurons and muscle. In vertebrates, the scaffolding protein Shank interacts with the CaV1.3 channel C-terminus via PDZ and SH3 domains, localizing the channels at appropriate postsynaptic locations (Zhang et al. 2005). Shank is also known to be important in invertebrate postsynaptic functions (Harris et al. 2016), and a direct interaction has been reported between the CaV1 C-terminus and Shank in C.elegans (Pym et al. 2017). Hence, CaV1 channels, like CaV2, might share deep conservation in C-terminus-dependent protein interactions.. Interestingly, the identified CaV1 homolog from Oscarella, and the CaV1/CaV2 homologs from fellow sponges Amphimedon and Haliclona sp., bear E-T-S/T-V-COOH motifs, corresponding to the consensus sequence for PDZ domains of DLG synaptic scaffolding proteins from human and nematode worm (Tonikian et al. 2008). This is in contrast to CaV homologs from premetazoan organisms, which have more variable (and positively charged) residues in their extreme C-termini. Based on these observations, it may be that animal-specific adaptations in CaV channel function occurred early and involved incorporation into specific PDZ domain-mediated interaction networks, a process that is proposed to have given rise to expansion and complexification of PDZ interactions networks in metazoan proteomes (Kim et al. 2012). The significance of the presence of these motifs in channel homologs from early-diverging animals is unclear, especially given how little is known about CaV channel function in these animals (Senatore et al. 2016). Nevertheless, CaV channel signaling functions are highly dependent on cellular localization and proximity to Ca2+-sensitive cytoplasmic proteins. This is because Ca2+ can be cytotoxic and tends to be quickly extruded and chelated once inside the cytoplasm (Clapham 2007), restricting high concentration zones to just micrometers from the channel pore (Rizzuto and Pozzan 2006).
Also interesting is that we identified a CaV1 channel in the gene data for the sponge O. carmela, significant because CaV1 channels were thought to be absent in sponges (Moran and Zakon 2014; Moran et al. 2015). Furthermore, we identified a structural feature that distinguishes CaV1 and CaV2 channels in the alpha helical structure predicted in C-termini of CaV1 channels including the Oscarella homolog (fig. 6A and B). We also identified an additional structural feature that phylogenetically distinguishes CaV1/CaV2 channels and CaV3 channels, the EVH1-binding motifs in the C-termini of CaV1/CaV2 channels upstream of the IQ motif (fig. 6C). Here, the potential for interactions with Homer and other EVH1 domain-bearing proteins further alludes to differential integration into membrane-localizing protein complexes as a mechanism for CaV channel adaptation for distinct cellular functions.
Our analysis of proline-rich SH3 ligands in CaV channel C-termini was less clear than for PDZ ligands. Our impetus for performing this analysis was the consideration of the tripartite interaction between CaV2, RIM-I, and RIM-BP conserved between protostome and deuterostome bilaterians, though we were aware that SH3 domain ligands exhibit a considerable degree of sequence entropy and are difficult to predict with confidence (Teyra et al. 2017). Indeed, although it is likely that the various linkers and N-/C-termini of CaV channels bear conserved binding sites for RIM-BP SH3 domains, we point out a flaw in this analysis where SH3 ligand motifs were not predicted for the C. elegans CaV2, despite its expected interactions with RIM-BP in vivo (Gracheva et al. 2008). Nevertheless, it is notable that SH3 ligands appear enriched in CaV1 and CaV2 channels relative to CaV3 and premetazoan CaVs, outside of CaV3.2 and Cav3.3 in chordates (fig. 5A and B).
Based on our presented analyses, CaV1/CaV2 channels appear to have emerged just prior to the divergence of animals from closely related eukaryotes (fig. 7), upon which they took on the capacity to interact with PDZ domain-bearing proteins, constraining their Ca2+ signaling functions to discrete subcellular locations. The identification of a CaV1 channel gene in sponges, CaV2 in ctenophores, and CaV3 channel in choanoflagellates, suggests that the last common ancestor to all animals possessed a full complement of CaV channels: CaV1–CaV3 plus CaV1/CaV2 channels. Under this model, and consistent with reports of substantial loss of ion channel genes in early-diverging groups (Liebeskind et al. 2015), ctenophores lost CaV1, CaV3, and CaV1/CaV2 channel genes, sponges lost CaV3, CaV2, and either CaV1 or CaV1/CaV2, and placozoans and the remaining animal groups lost CaV1/CaV2 channels but retained CaV1–CaV3 (fig. 7). This model supports the notion that CaV1 and CaV2 channels evolved from an ancestral CaV1/CaV2-like channel (Moran and Zakon 2014; Moran et al. 2015), but suggests that all three channel types coexisted in an ancestral species. If so, early in the divergence between CaV1 and CaV2 channels, they took on differential functional attributes, such as the pronounced Ca2+-dependent inactivation of CaV1 compared with CaV2 channels mediated by interactions with calmodulin at the C-terminal IQ motif (Catterall 2011; Taiakina et al. 2013). Included in the divergence between CaV1 and CaV2 channels, which are respectively specialized for post- and presynaptic functions (Senatore et al. 2016), might have been differential incorporation into distinct membrane complexes including those mediated by scaffolding proteins bearing PDZ domains.
Materials and Methods
mRNA Quantification and Localization
Trichoplax adhaerens animals were prepared for fluorescence in situ hybridization (FISH) by freezing in tetrahydrofuran (THF) overnight on dry ice followed by fixation in 3% acetic acid in methanol (MeOH) for 30 min at −20°C and then 4% paraformaldehyde in methanol for 30 min at room temperature, as described (Mayorova et al. 2019). In situ hybridization was performed with RNAscope probes for RIM-I (no. 72781-C3), RIM-II (no. 572791-C2), CaV1 (no. 442461), CaV2 (no. 442471), and CaV3 (no. 488711) and Multiplex Fluorescent Assay reagents (no. 320850) from Advanced Cell Diagnostics (Hayward, CA). For dual labeling with probes for CaV1, CaV2, or CaV3 and CF-405-conjugated WGA (no. 29027-1, Biotium, Freemont, CA), animals were frozen in THF as described above and then fixed in 4% formaldehyde in MEOH for 30 min at −20°C and 30 min at room temperature. Following in situ hybridization, the samples were incubated in CF-405 WGA diluted 1:200 in PBS for 1 h at room temperature. Fluorescence images were collected with a 63× NA 1.4 objective on a LSM880 laser scanning confocal microscope (Carl Zeiss Microscopy LLC, Thornwood, NY). Images in figure 1C–E, left panels, were collected with a 32-channel spectral detector in the lambda mode and processed by linear unmixing. Enlarged views in figure 1C–E insets were collected with an AiryScan detector. Projected images were generated with Zen software (Carl Zeiss Microscopy LLC).
For the qPCR experiments, young adult Lymnaea stagnalis albumen gland, buccal mass, brain, heart, and prostate gland were microdissected from anesthetized animals and pooled into triplicate tubes (5–10 individual tissues per tube), and total RNA extracted as previously reported (Senatore et al. 2014). Complimentary DNA (cDNA) libraries were prepared from each RNA isolate with SuperScript III reverse transcriptase (ThermoFisher Scientific, Canada) and an anchored oligo-dT18 primer (table 1). Gene-specific primers for Lymnaea RIM-I (NCBI accession FX186940.1), RIM-II (NCBI accession FX181400.1), and elongation factor-1α (EF-1α; NCBI accession DQ278441.1) (table 1) were used for quantitative PCR using the iQ SYBR Green Supermix (BioRad, Canada) and the following cycling conditions: denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s (denaturation) and 57–60°C for 30 s (extension). To ensure that single amplicons were produced in each PCR reaction, PCR products were electrophoresed on 1.5% agarose gels, and melting curve protocols were performed after each run. We also verified that the designed primers showed no cross-complementarity between the RIM-I and RIM-II cDNA sequences. Real-time PCR fluorescence measurement and melt curve analyses were done using a Bio-Rad C1000TM Thermal Cycler equipped with a CFX96TM System (Bio-Rad). Transcript expression levels were quantified and normalized relative to EF-1α using the ΔΔ cycle threshold (ΔΔCT) method (Andersen et al. 2004): ratio = (Etarget gene) ΔCTTarget gene/(EEF-1α) ΔCTEF-1α, where E denotes PCR efficiency for respective PCR primer pairs. Normalized transcript abundance of all tissues was standardized to the transcript abundance of the RIM-I and RIM-II in the albumen, which was set to 100%. One-way ANOVA was also performed to confirm that transcript abundance of EF-1α did not significantly differ between tissues (P = 0.217).
Table 1.
Sequence of Oligonucleotides Used for Lymnaea qPCR and In Situ Hybridization
| Primer Name | Sequence (5′–3′) |
|---|---|
| RIM-I LNA probe | ATGCAAGAGATTACGGATTGAA |
| RIM-II LNA probe | TGGCTGATCTTCTTGATAGCA |
| Control LNA probe | GTGTAACACGTCTATACGCCCA |
| Anchored oligo-dT18 | TTTTTTTTTTTTTTTTTTVN |
| Lymnaea RIM-I forward | GTGAGGAAGCAGGAAGTGGA |
| Lymnaea RIM-I reverse | CCAGCACAATAGACCCAACC |
| Lymnaea RIM-II forward | CACTACCAGCCACACAAAGC |
| Lymnaea RIM-II reverse | TGTTCCCACTCAGGATGACA |
| Lymnaea EF-1α forward | TGGCAAGTCAACCACAACTG |
| Lymnaea EF-1α reverse | TAATACCACGCTCACGCTCA |
For the FISH experiments on isolated neurons from the CNS of L. stagnalis, central ring ganglia (CNS) from young adult L. stagnalis (16–18 mm in length) were isolated and exposed to antibiotic washes prior to cell culture. Individual ganglia were then desheathed, enabling removal of specific, identified neurons using suction applied via a fire polished pipette. Individual cells were plated on poly-l-lysine (Millipore Sigma) coated Falcon dishes (VWR) following isolation. Cells were given 10–30 min to attach to the cell culture plates and were then fixed using 4% paraformaldehyde. Prior to staining, cells were treated with 3% H2O2 (v/v in 1 × PBS; Millipore Sigma, Canada) to eliminate endogenous peroxidase activity and dehydrated overnight in 70% ethanol. The next day, cells were incubated in hybridization buffer (25% formamide, 0.05 M EDTA, 4× saline-sodium citrate buffer [SSC], 10% dextran sulfate, 1× Denhardts solution, 0.5 mg/ml Escherichia coli tRNA, 20 mM ribonucleoside vanadyl complexes and 9.2 mM citric acid; Millipore Sigma) at 55°C for 2 h. Following prehybridization, cells were incubated in either 10 nM of LNA enhanced detection probes (Qiagen) targeting mRNAs for RIM-I, RIM-II, or a negative control probe (table 1) at 55°C for 1 h. Next, cells were washed in a series of stringency washes, including 4× SSC, 2× SSC, 1× SSC, and 0.1× SSC at 37°C for 30 min, incubated in blocking buffer (3% bovine serum, 4× SSC, 0.1× Tween-20) for 30 min, then horseradish peroxidase-conjugated streptavidin (Thermo-Fisher) for 30 min. Cells were then washed in TNT buffer (0.1 M Tris HCl, 0.15 M NaCl, 0.05% Tween-20), then incubated in tyramide (Perkin-Elmer) according to the manufacturer’s instructions for 1 h at room temperature. Following additional washes in TNT buffer, cells were mounted in FluoroshieldTM containing DAPI (Millipore Sigma) as a counterstain to label nuclei. Cells were imaged using a Carl Zeiss Axio Observer.Z1 inverted light/epifluorescence microscope, with Apotome.2 optical sectioning (Zeiss). For all images, Z-stack slices were taken at 0.29-µm intervals and were rendered into 2D maximum intensity projections using the Zeiss Zen 2 microscopy software.
Phylogenetic Inference
Both maximum likelihood and Bayesian inference strategies were used to infer the phylogenetic relationships of RIM-Is, RIM-IIs, and rabphilins. Briefly, candidate protein sequences were identified in select genomic/transcriptomic databases (supplementary table S1, Supplementary Material online) using protein BLAST (Altschul et al. 1990), with various vertebrate and invertebrate RIM-I, RIM-II, and rabphilin sequences as queries. This produced a list of candidate sequences with alignment score E values below 1E−6, which were subsequently analyzed via SmartBLAST (Coordinators 2016) to ensure homology with corresponding proteins, and InterPro (Jones et al. 2014) to determine the presence and organization of hallmark domains (i.e. Zn2+-finger, PDZ, C2A, and C2B). Sequences that did not match expected homologs with SmartBLAST, grossly lacked expected domain architectures, and/or were highly fragmented, were not used in subsequent analyses. Identified protein sequences (supplementary table S1, Supplementary Material online) were then aligned with MUSCLE (Edgar 2004) and trimmed with the trimAl (Capella-Gutiérrez et al. 2009) using a gap threshold of 0.6 (accession numbers listed below; raw sequences and trimmed alignment are provided in supplementary file S1, Supplementary Material online). The trimmed alignment was then used to infer a maximum likelihood phylogenetic tree using IQ-TREE (Nguyen et al. 2015) with default parameters and 1,000 ultrafast bootstrap replicates for estimating node support, and the best-fit substitution model of VT+F+G4 identified though the −m TEST parameter of IQ-TREE. Using the same alignment and model, Bayesian inference was done using MrBayes version 3.2.6 (Ronquist et al. 2012), with two runs, four Markov chains, 10,000,000 generations, a tree sampling frequency of 100, and a burn-in fraction 0.25. We estimated that the inference reached convergence after potential scale reduction factor (PSRF) statistics approached values of 1.0 (i.e. tree lengths: 1.000–1.085; alpha: 1.034). Phylogenetic trees were visualized with MEGA X (Kumar et al. 2018) and FigTree 1.4.3 (Rambaut 2007), and shared nodes with respectively strong bootstrap support and high posterior probability values were annotated on the maximum likelihood tree shown in figure 2.
To infer the phylogenetic relationships of CaV channels, a similar approach was used as described for the RIM/rabphillin maximum likelihood tree, with manual trimming of the protein alignment, and a best-fit substitution model of LG+I+G4 identified by IQ-TREE (sequences and trimmed alignment provided in respective supplementary files S2 and S3, Supplementary Material online). NCBI protein sequence accession numbers (unless otherwise indicated) are provided in supplementary table S2, Supplementary Material online.
Protein Alignments and Structural Predictions
Protein alignments were generated using MUSCLE (Edgar 2004) within the MEGA X software (Kumar et al. 2018), and visualized with JalView (Waterhouse et al. 2009). Jalview was also used to generate consensus and conservation plots (fig. 4A and supplementary fig. S2, Supplementary Material online). PROMALS3D was used to predict all secondary structures (Pei and Grishin 2014) (fig. 4A, supplementary figs. S2 and S6B), with the exception of alpha helices predicted at the N-termini of RIM and rabphilin homologs (supplementary file S1, Supplementary Material online). EMBOSS plotcon (Rice et al. 2000) was used to generate conservation versus position in alignment plots with a running amino acid alignment window of 15 (fig. 4B) or 6 (fig. 6). Protein domains, including PDZ, SH3, Zn2+, and C2A/C2B (figs. 1A and 2), were predicted with InterProScan (Jones et al. 2014), and secondarily with hmmscan (Finn et al. 2011). PDZ ligand motifs (fig. 5A) were predicted using PDZPepInt (Kundu et al. 2014), and SH3 ligand domains (fig. 5A and supplementary fig. S3, Supplementary Material online) were predicted using three separate algorithms: 1) Find Individual Occurrences (FIMO) (Grant et al. 2011); 2) LMDIPred (Sarkar et al. 2018), and 3) SH3PepInt (Kundu et al. 2014). FIMO, part of the MEME Suite of sequence analysis tools, identified all cases of the consensus SH3-binding motif PXXP in CaV C-terminal sequences, providing a liberal estimate of the actual number of motifs. SH3PepInt used a graph-kernel algorithm to predict SH3 motifs based on peptide-array data for 69 human SH3 domains and 31 regular expressions for canonical SH3 motifs (run using the default 15mer peptide window and a step size of 5 amino acids). Linear Motif Domain Interaction Prediction (LMDIPred) used four independent methods (support vector machine [SVM] prediction, position-specific scoring matrix, motif instance matching, and regular expression scanning) to predict 6-mer SH3-binding motifs. We counted a SH3 motif only if it was identified using three or more of the independent LMDIPred algorithms. Statistical analysis of SH3PepInt-predicted SH3 ligand motifs was performed first conducting normality assessment with Shapiro–Wilk tests, and homogeneity of variance with Levene’s test (ANOVA on residuals). Post hoc analysis was done using Kruskal–Wallis and Dunn’s tests with Benjamini–Hochberg P value adjustment. This same approach was used for comparing the three different algorithms used to predict SH3 ligand domains (supplementary fig. S3, Supplementary Material online). For comparing the lengths of intracellular regions of various CaV channels (supplementary fig. S4, Supplementary Material online), PSIPRED (McGuffin et al. 2000), TMHMM (Krogh et al. 2001), and ExPASy ProtScale Kyte-Doolittle plots (Gasteiger et al. 2005) were used to identify interfaces between transmembrane and cytoplasmic/extracellular regions. De novo motif identification (particularly SLiMs; fig. 6) was performed using Swiss Institute of Bioinformatics (SIB) MyHits Motif Scan (Pagni et al. 2007), using HAMAP, PROSITE, and Pfam HMM databases; and hits were cross-referenced with existing entries in the eukaryotic linear motifs (ELM) database (Dinkel et al. 2012); and manually inspected to identify tandem amino acid repeats.
Supplementary Material
Acknowledgments
This project was funded by an NSERC Discovery Grant (RGPIN-2016-06023), a Canadian Foundation for Innovation Grant (35297), an Ontario Early Researcher Award (ER17-13-247), and University of Toronto start-up funds to A.S., NSERC Canadian Graduate Scholarships to J.G., C.L. and S.E.W, an NSERC Discovery Grant (RGPIN-2015-03780) to G.E.S., and the Intramural Research Program of the NIH, NINDS to C.L.S. We would like to thank Dr Mark Martindale and Dr Joseph Ryan (Whitney Lab, Florida) for providing us with access to gene sequences from unpublished genome and transcriptome data of the ctenophore Beroe ovata.
Literature Cited
- Adams P, Snutch T.. 2007. Calcium channelopathies: voltage-gated calcium channels. In: Calcium signalling and disease. Dordrecht: Springer. p. 215–251. [DOI] [PubMed]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF.. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64(15):5245–5250. [DOI] [PubMed] [Google Scholar]
- Bae J, Suh EJ, Lee C.. 2010. Interaction of T-type calcium channel CaV3.3 with the β-subunit. Mol Cells 30(3):185–191. [DOI] [PubMed] [Google Scholar]
- Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server):W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LJ, Jarchau T, Oschkinat H, Walter U.. 2002. EVH1 domains: structure, function and interactions. FEBS Lett. 513(1):45–52. [DOI] [PubMed] [Google Scholar]
- Beekharry CC, Zhu GZ, Magoski NS.. 2015. Role for electrical synapses in shaping the output of coupled peptidergic neurons from Lymnaea. Brain Res. 1603:8–21. [DOI] [PubMed] [Google Scholar]
- Benjamin P, Winlow W.. 1981. The distribution of three wide-acting synaptic inputs to identified neurons in the isolated brain of Lymnaea stagnalis (L.). Comp Biochem Physiol A Physiol. 70(3):293–307. [Google Scholar]
- Ben-Johny M, Yue DT.. 2014. Calmodulin regulation (calmodulation) of voltage-gated calcium channels. J Gen Physiol. 143(6):679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A, et al. 2001. Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron 30(1):183–196. [DOI] [PubMed] [Google Scholar]
- Biadene M, Montaville P, Sheldrick GM, Becker S.. 2006. Structure of the C2A domain of rabphilin-3A. Acta Crystallogr Sect D Biol Crystallogr. 62(7):793–799. [DOI] [PubMed] [Google Scholar]
- Bogerd J, Geraerts W, Van Heerikhuizen H, Kerkhoven R, Joosse J.. 1991. Characterization and evolutionary aspects of a transcript encoding a neuropeptide precursor of Lymnaea neurons, VD1 and RPD2. Mol Brain Res. 11(1):47–54. [DOI] [PubMed] [Google Scholar]
- Brakeman PR, et al. 1997. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature 386(6622):284–288. [DOI] [PubMed] [Google Scholar]
- Bright K, et al. 1993. Mutually exclusive expression of alternatively spliced FMRFamide transcripts in identified neuronal systems of the snail Lymnaea. J Neurosci. 13(6):2719–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock TH. 1943. Neuromuscular facilitation in Scyphomedusae. J Cell Comp Physiol. 22(3):251–272. [Google Scholar]
- Burkhardt P. 2015. The origin and evolution of synaptic proteins—choanoflagellates lead the way. J Exp Biol. 218(4):506–514. [DOI] [PubMed] [Google Scholar]
- Burns ME, Sasaki T, Takai Y, Augustine G.. 1998. Rabphilin-3A: a multifunctional regulator of synaptic vesicle traffic. J Gen Physiol. 111(2):243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. 2011. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol. 3(8):a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin J, et al. 2017. Calmodulin regulates CaV3 T-type channels at their gating brake. J Biol Chem. 292(49):20010–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. 2007. Calcium signaling. Cell 131(6):1047–1058. [DOI] [PubMed] [Google Scholar]
- Coordinators NR. 2016. Database resources of the national center for biotechnology information. Nucleic Acids Res. 44:D7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudevylle N, Montaville P, Leonov A, Zweckstetter M, Becker S.. 2008. Structural determinants for Ca2+ and phosphatidylinositol 4,5-bisphosphate binding by the C2A domain of rabphilin-3A. J Biol Chem. 283(51):35918–35928. [DOI] [PubMed] [Google Scholar]
- Deng L, Kaeser PS, Xu W, Südhof TC.. 2011. RIM proteins activate vesicle priming by reversing autoinhibitory homodimerization of Munc13. Neuron 69(2):317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, et al. 2012. ELM—the database of eukaryotic linear motifs. Nucleic Acids Res. 40(D1):D242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio N, et al. 2015. Homer proteins mediate the interaction between STIM1 and CaV1.2 channels. Biochim Biophys Acta 1853(5):1145–1153. [DOI] [PubMed] [Google Scholar]
- Dos Reis M, et al. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 25(22):2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, et al. 2005. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 24(16):2839–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermann E, Bucurenciu I, Goswami SP, Jonas P.. 2012. Nanodomain coupling between Ca2+ channels and sensors of exocytosis at fast mammalian synapses. Nat Rev Neurosci. 13(1):7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitel M, et al. 2018. Comparative genomics and the nature of placozoan species. PLoS Biol. 16(9):e3000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elekes K, Hiripi L, Benjamin P.. 1989. A comparison of four techniques for mapping the distribution of serotonin and serotonin-containing neurons in fixed and living ganglia of the snail, Lymnaea. J Neurocytol. 18:193–208. [DOI] [PubMed] [Google Scholar]
- Ernst A, et al. 2014. A structural portrait of the PDZ domain family. J Mol Biol. 426(21):3509–3519. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(suppl):W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Yanagisawa A, Sokabe M, Yoshimura K.. 2009. Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr Biol. 19(2):133–139. [DOI] [PubMed] [Google Scholar]
- Fukuda M. 2003. Distinct Rab binding specificity of RIM1, RIM2, rabphilin, and Noc2. J Biol Chem. 278(17):15373–15380. [DOI] [PubMed] [Google Scholar]
- Gardezi SR, Li Q, Stanley EF.. 2013. Inter-channel scaffolding of presynaptic CaV2.2 via the C terminal PDZ ligand domain. Biol Open 2(5):492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, et al. 2005. Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Totowa (NJ): Humana press. p. 571–607.
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R.. 2006. The C terminus of the L-type voltage-gated calcium channel CaV1.2 encodes a transcription factor. Cell 127(3):591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Hadwiger G, Nonet ML, Richmond JE.. 2008. Direct interactions between C. elegans RAB-3 and RIM provide a mechanism to target vesicles to the presynaptic density. Neurosci Lett. 444(2):137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, et al. 2012. RIM promotes calcium channel accumulation at active zones of the Drosophila neuromuscular junction. J Neurosci. 32(47):16586–16596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS.. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7):1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KP, Akbergenova Y, Cho RW, Baas-Thomas MS, Littleton JT.. 2016. Shank modulates postsynaptic Wnt signaling to regulate synaptic development. J Neurosci. 36(21):5820–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Nicaise M-L. 1973. The nervous system of ctenophores III. Ultrastructure of synapses. J Neurocytol. 2(3):249–263. [DOI] [PubMed] [Google Scholar]
- Hibino H, et al. 2002. RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca2+ channels. Neuron 34(3):411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, et al. 2017. C-terminal splice variants of P/Q-type Ca2+ channel CaV2.1 α1 subunits are differentially regulated by Rab3-interacting molecule proteins. J Biol Chem. 292(22):9365–9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, et al. 2007. Ca2+ signaling in microdomains Homer1 mediates the interaction between RyR2 and CaV1.2 to regulate excitation–contraction coupling. J Biol Chem. 282(19):14283–14290. [DOI] [PubMed] [Google Scholar]
- Hulme JT, Lin T-C, Westenbroek RE, Scheuer T, Catterall WA.. 2003. β-adrenergic regulation requires direct anchoring of PKA to cardiac CaV1.2 channels via a leucine zipper interaction with A kinase-anchoring protein 15. Proc Natl Acad Sci. 100(22):13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme JT, Yarov‐Yarovoy V, Lin TWC, Scheuer T, Catterall WA.. 2006. Autoinhibitory control of the CaV1.2 channel by its proteolytically processed distal C‐terminal domain. J Physiol. 576(1):87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sheng M.. 2002. PDZ domains: structural modules for protein complex assembly. J Biol Chem. 277(8):5699–5702. [DOI] [PubMed] [Google Scholar]
- Iezzi M, Regazzi R, Wollheim CB.. 2000. The Rab3‐interacting molecule RIM is expressed in pancreatic β‐cells and is implicated in insulin exocytosis. FEBS Lett. 474(1):66–70. [DOI] [PubMed] [Google Scholar]
- Jegla TJ, Zmasek CM, Batalov S, Nayak SK.. 2009. Evolution of the human ion channel set. Comb Chem High Throughput Screen. 12(1):2–23. [DOI] [PubMed] [Google Scholar]
- Jiménez CR, et al. 2006. Peptidomics of a single identified neuron reveals diversity of multiple neuropeptides with convergent actions on cellular excitability. J Neurosci. 26(2):518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, Deng L, Fan M, Südhof TC.. 2012. RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci. 109(29):11830–11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, et al. 2011. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell 144(2):282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm K, Osigus H-J, Stadler PF, DeSalle R, Schierwater B.. 2018. Trichoplax genomes reveal profound admixture and suggest stable wild populations without bisexual reproduction. Sci Rep. 8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR.. 2000. Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 20(19):7238–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B, Miledi R.. 1965. The effect of calcium on acetylcholine release from motor nerve terminals. Proc R Soc Lond Ser B Biol Sci. 161:496–503. [DOI] [PubMed] [Google Scholar]
- Kemenes G, Benjamin PR.. 2009. Lymnaea. Curr Biol. 19(1):R9–11. [DOI] [PubMed] [Google Scholar]
- Kerfoot P, Mackie G, Meech R, Roberts A, Singla C.. 1985. Neuromuscular transmission in the jellyfish Aglantha digitale. J Exp Biol. 116:1–25. [DOI] [PubMed] [Google Scholar]
- Kerkhoven R, et al. 1993. Neurons in a variety of molluscs react to antibodies raised against the VD1/RPD2 α-neuropeptide of the pond snail Lymnaea stagnalis. Cell Tissue Res. 273(2):371–379. [DOI] [PubMed] [Google Scholar]
- Kim J, et al. 2012. Rewiring of PDZ domain–ligand interaction network contributed to eukaryotic evolution. PLoS Genet. 8(2):e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyonaka S, et al. 2007. RIM1 confers sustained activity and neurotransmitter vesicle anchoring to presynaptic Ca2+ channels. Nat Neurosci. 10(6):691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koene JM, et al. 2010. Male accessory gland protein reduces egg laying in a simultaneous hermaphrodite. PLoS One 5(4):e10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, et al. 2001. A post-docking role for active zone protein RIM. Nat Neurosci. 4(10):997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305(3):567–580. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu K, Mann M, Costa F, Backofen R.. 2014. MoDPepInt: an interactive web server for prediction of modular domain–peptide interactions. Bioinformatics 30(18):2668–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushibiki Y, Suzuki T, Jin Y, Taru H.. 2019. RIMB-1/RIM-binding protein and UNC-10/RIM redundantly regulate presynaptic localization of the voltage-gated calcium channel in C. elegans. J Neurosci. 0506–0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Zheng JJ.. 2010. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH.. 2015. Convergence of ion channel genome content in early animal evolution. Proc Natl Acad Sci U S A. 112(8):E846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodh S, Yano J, Valentine MS, Van Houten JL.. 2016. Voltage-gated calcium channels of Paramecium cilia. J Exp Biol. 219(19):3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li H, Wang Y, Südhof TC, Rizo J.. 2005. Solution structure of the RIM1α PDZ domain in complex with an ELKS1b C-terminal peptide. J Mol Biol. 352(2):455–466. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. 2006. Structural basis for a Munc13-1 homodimer to Munc13-1/RIM heterodimer switch. PLoS Biol. 4(7):e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A, Südhof TC, Bezprozvanny I.. 1999. Association of neuronal calcium channels with modular adaptor proteins. J Biol Chem. 274(35):24453–24456. [DOI] [PubMed] [Google Scholar]
- Mayorova TD, Hammar K, Winters CA, Reese TS, Smith CL.. 2019. The ventral epithelium of Trichoplax adhaerens deploys in distinct patterns cells that secrete digestive enzymes, mucus or diverse neuropeptides. Biol Open 8(8):bio045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT.. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405. [DOI] [PubMed] [Google Scholar]
- McKiernan CJ, Stabila PF, Macara IG.. 1996. Role of the Rab3A-binding domain in targeting of rabphilin-3A to vesicle membranes of PC12 cells. Mol Cell Biol. 16(9):4985–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Barzilai MG, Liebeskind BJ, Zakon HH.. 2015. Evolution of voltage-gated ion channels at the emergence of Metazoa. J Exp Biol. 218(4):515–525. [DOI] [PubMed] [Google Scholar]
- Moran Y, Zakon HH.. 2014. The evolution of the four subunits of voltage-gated calcium channels: ancient roots, increasing complexity, and multiple losses. Genome Biol Evol. 6(9):2210–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510(7503):109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kohn AB.. 2016. Independent origins of neurons and synapses: insights from ctenophores. Philos Trans R Soc Lond B Biol Sci. 371(1685):20150041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Liu KSY, Sigrist SJ, Davis GW.. 2012. RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J Neurosci. 32(47):16574–16585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic OB, et al. . 1996. Glutamate as a putative neurotransmitter in the mollusc, Lymnaea stagnalis Neuroscience 75(4):1255–1269. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourry C, Grant SG, Borg J-P.. 2003. PDZ domain proteins: plug and play. Sci Signal. 2003(179):re7. [DOI] [PubMed] [Google Scholar]
- Ostermeier C, Brunger AT.. 1999. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 96(3):363–374. [DOI] [PubMed] [Google Scholar]
- Pagni M, et al. 2007. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35(Web Server):W433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paps J, Holland PW.. 2018. Reconstruction of the ancestral metazoan genome reveals an increase in genomic novelty. Nat Commun. 9(1):1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV.. 2014. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. In: Multiple sequence alignment methods. Totowa (NJ): Humana press. p. 263–271. [DOI] [PMC free article] [PubMed]
- Persoon CM, et al. 2019. The Rab3-RIM pathway is essential for the release of neuromodulators. Neuron 104(6):1065–1080.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym E, et al. 2017. Shank is a dose-dependent regulator of Cav1 calcium current and CREB target expression. Elife 6:e18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade B, et al. 2019. Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8:e42806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2007. FigTree, a graphical viewer of phylogenetic trees. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16(6):276–277. [DOI] [PubMed]
- Rizzuto R, Pozzan T.. 2006. Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol Rev. 86(1):369–408. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342(6164):1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadamoto H, et al. 2012. De novo sequencing and transcriptome analysis of the central nervous system of mollusc Lymnaea stagnalis by deep RNA sequencing. PLoS One 7(8):e42546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarya O, et al. 2010. Evolutionary expansion and specialization of the PDZ domains. Mol Biol Evol. 27(5):1058–1069. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Jana T, Saha S.. 2018. LMDIPred: a web-server for prediction of linear peptide sequences binding to SH3, WW and PDZ domains. PLoS One 13(7):e0200430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S, et al. 2006. Redundant functions of RIM1α and RIM2α in Ca2+‐triggered neurotransmitter release. EMBO J. 25(24):5852–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Guan W, Boone AN, Spafford JD.. 2014. T-type channels become highly permeable to sodium ions using an alternative extracellular turret region (S5-P) outside the selectivity filter. J Biol Chem. 289(17):11952–11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Raiss H, Le P.. 2016. Physiology and evolution of voltage-gated calcium channels in early diverging animal phyla: Cnidaria, Placozoa, Porifera and Ctenophora. Front Physiol. 7:481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Spafford JD.. 2010. Transient and big are key features of an invertebrate T-type channel (LCaV3) from the central nervous system of Lymnaea stagnalis. J Biol Chem. 285(10):7447–7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Zhorov BS, Spafford JD.. 2012. CaV3 T‐type calcium channels. Wiley Interdisciplinary Reviews: Membrane Transport and Signaling 1(4):467–491. [Google Scholar]
- Shirataki H, et al. 1993. Rabphilin-3A, a putative target protein for smg p25A/rab3A p25 small GTP-binding protein related to synaptotagmin. Mol. Cell Biol. 13(4):2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms BA, Zamponi GW.. 2014. Neuronal voltage-gated calcium channels: structure, function, and dysfunction. Neuron 82(1):24–45. [DOI] [PubMed] [Google Scholar]
- Smith CL, et al. 2014. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr Biol. 24(14):1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, et al. 2017. Evolutionary insights into T-type Ca2+ channel structure, function, and ion selectivity from the Trichoplax adhaerens homologue. J Gen Physiol. 149(4):483–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snidal CA, et al. 2018. Molecular characterization of an SV capture site in the mid-region of the presynaptic CaV2.1 calcium channel C-terminal. Front Cell Neurosci. 12:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, et al. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275(5296):73–77. [DOI] [PubMed] [Google Scholar]
- Spafford JD, et al. 2003. Calcium channel structural determinants of synaptic transmission between identified invertebrate neurons. J Biol Chem. 278(6):4258–4267. [DOI] [PubMed] [Google Scholar]
- Spafford JD, Zamponi GW.. 2003. Functional interactions between presynaptic calcium channels and the neurotransmitter release machinery. Curr Opin Neurobiol. 13(3):308–314. [DOI] [PubMed] [Google Scholar]
- Stahl B, Chou J, Li C, Südhof T, Jahn R.. 1996. Rab3 reversibly recruits rabphilin to synaptic vesicles by a mechanism analogous to raf recruitment by ras. EMBO J. 15(8):1799–1809. [PMC free article] [PubMed] [Google Scholar]
- Stanley EF. 2016. The nanophysiology of fast transmitter release. Trends Neurosci. 39(3):183–197. [DOI] [PubMed] [Google Scholar]
- Stricker NL, et al. 1997. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat Biotechnol. 15(4):336–342. [DOI] [PubMed] [Google Scholar]
- Südhof TC. 2012. The presynaptic active zone. Neuron 75(1):11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed N, Bulloch A, Lukowiak K.. 1990. In vitro reconstruction of the respiratory central pattern generator of the mollusk Lymnaea. Science 250(4978):282–285. [DOI] [PubMed] [Google Scholar]
- Taiakina V, et al. 2013. The calmodulin-binding, short linear motif, NSCaTE is conserved in L-type channel ancestors of vertebrate CaV1.2 and CaV1.3 channels. PLoS One 8(4):e61765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyra J, et al. 2017. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure 25(10):1598–1610.e1593. [DOI] [PubMed] [Google Scholar]
- Tonikian R, et al. 2008. A specificity map for the PDZ domain family. PLoS Biol. 6(9):e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JR, Snutch TP.. 2013. Molecular nature of voltage‐gated calcium channels: structure and species comparison. Wiley Interdisciplinary Reviews: Membrane Transport and Signaling 2(5):181–206. [Google Scholar]
- Ubach J, Zhang X, Shao X, Südhof TC, Rizo J.. 1998. Ca2+ binding to synaptotagmin: how many Ca2+ ions bind to the tip of a C2‐domain. EMBO J. 17(14):3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriu Y, et al. 2010. Rab3-interacting molecule γ isoforms lacking the Rab3-binding domain induce long lasting currents but block neurotransmitter vesicle anchoring in voltage-dependent P/Q-type Ca2+ channels. J Biol Chem. 285(28):21750–21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L-Y, Augustine GJ.. 2015. Presynaptic nanodomains: a tale of two synapses. Front Cell Neurosci. 8:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu B, Zimmermann B, Kilimann MW.. 2001. RIM1 and rabphilin-3 bind Rab3-GTP by composite determinants partially related through N-terminal α-helix motifs. J Biol Chem. 276(35):32480–32488. [DOI] [PubMed] [Google Scholar]
- Wang Y, Okamoto M, Schmitz F, Hofmann K, Südhof TC.. 1997. RIM is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388(6642):593–598. [DOI] [PubMed] [Google Scholar]
- Wang Y, Südhof TC.. 2003. Genomic definition of RIM proteins: evolutionary amplification of a family of synaptic regulatory proteins. Genomics 81(2):126–137. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Südhof TC.. 2000. The RIM/NIM family of neuronal C2 domain proteins interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem. 275(26):20033–20044. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan NV, et al. 2017. Ctenophore relationships and their placement as the sister group to all other animals. Nat Ecol Evol. 1(11):1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y-Y, Le P, Elkhatib W, Piekut T, Senatore A.. 2019. Transcriptome profiling of Trichoplax adhaerens highlights its digestive epithelium and a rich set of genes for fast electrogenic and slow neuromodulatory cellular signaling. Res Square, Preprint. Available from: 10.21203/rs.2.14504/v1+. [DOI]
- Woodin MA, Munno DW, Syed NI.. 2002. Trophic factor-induced excitatory synaptogenesis involves postsynaptic modulation of nicotinic acetylcholine receptors. J Neurosci. 22(2):505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. 2005. Association of CaV1.3 L-type calcium channels with Shank. J Neurosci. 25(5):1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.