Abstract
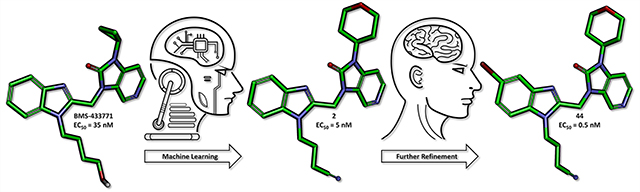
A series of five benzimidazole-based compounds were identified using a machine learning algorithm as potential inhibitors of the respiratory syncytial virus (RSV) fusion protein. These compounds were synthesized, and compound 2 in particular exhibited excellent in vitro potency with an EC50 value of 5 nM. This new scaffold was then further refined leading to the identification of compound 44, which exhibited a 10-fold improvement in activity with an EC50 value of 0.5 nM.
Keywords: RSV, fusion inhibitor, benzimidazole, machine learning
Graphical Abstract
Respiratory syncytial virus (RSV), an orthopneumovirus, has remained the global leading cause of lower respiratory tract infections in vulnerable patient populations since its discovery in 1956.1,2 It accounts for 68% of acute respiratory infections in infants during their first viral season and infects nearly all children by 2–3 years of age,3 40% of whom will develop a secondary respiratory infection.4 In 2015 alone, RSV-related acute lower respiratory infections resulted in the hospitalization of ~3.2 million children worldwide and 66 000 deaths.5,6 RSV infection of the small airways of the lungs (bronchiolitis) in children has been associated with a higher risk of developing asthma, but a direct causal link has yet to be established.7 RSV infection is also prevalent among the elderly population, where it is often misdiagnosed as influenza, and among immunocompromised patients8 as well those with cardiopulmonary diseases.9,10 Emerging epidemiological evidence has suggested that RSV has a morbidity burden in older adults similar to that of nonpandemic influenza.11,12 Unfortunately, despite its widespread occurrence, there are currently no direct-acting therapeutics for RSV and treatment is often limited to palliative care.13 A prophylactic agent, palivizumab (Synagis), is available for high-risk infants, but its use is typically limited to the developed world due to its cost.14–16 There is therefore a major unmet medical need for safe and effective therapeutics, both for treating active RSV infections and for prophylaxis.
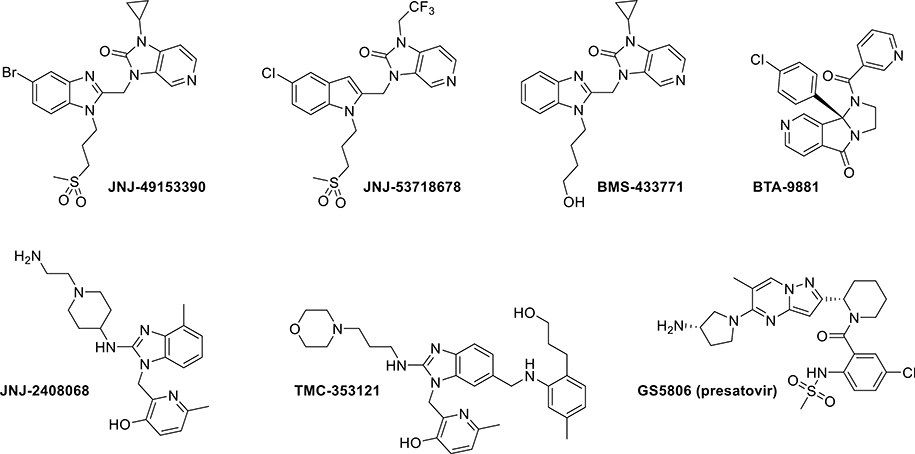
In recent years, attempts to combat RSV infection have mostly focused on the inhibition of viral polymerase activity or membrane fusion. A number of anti-RSV agents targeting the F protein in particular, the protein responsible for mediating membrane fusion,17 have been reported in the literature, some of which have advanced into clinical trials (Figure 1).3,16,18–22 This provided us with a promising opportunity to employ our machine learning algorithm in the form of Naive Bayes Networks (NBNs) to scan the known structure activity relationship (SAR) information and design new, potent RSV fusion protein inhibitors, an approach that has served us well in the past.23–25
Figure 1.
Examples of known RSV fusion inhibitors.
METHODS
Our machine learning approach focused on a series of RSV fusion inhibitors developed by Bristol-Myers Squibb (BMS) that resulted in the orally bioavailable clinical candidate BMS-433771 with nanomolar potency (Figure 1).18,26 The development of BMS-433771 was halted due to a change in the strategic focus of the company, which left this series available for further exploration.18
With the chemical space and biological target selected, we created a machine learning algorithm designed to predict highly active compounds. We employed the ChEMBL database as our source of information27 as well as the patent literature.28 We selected an NBN approach, as this algorithm is highly tolerant of any incidental noise in the data, is extremely fast to generate, and in our experience is good at predicting potencies.29 Additionally, we used PipelinePilot as our platform to perform both our informatics processing and machine learning. Having obtained a set of known RSV fusion inhibitors with associated EC50 values, we split the data into an inactive class (EC50 > 30 nM) and an active class (EC50 ≤ 30 nM). For our independent variable, we employed extended connectivity fingerprints, which are quick to compute and are often utilized for machine learning.23,24 We used a 70–30 split to perform training and testing and used the leave-one-out cross validation method as a training control. Using the 30% hidden data as a test of our classification algorithm, we found that a test set receiver operating characteristics area under the curve (ROC AUC) of 0.929 and the algorithm had a perfect enriching capability at the top 5% of predicted values (all of the top 5% structures were active).
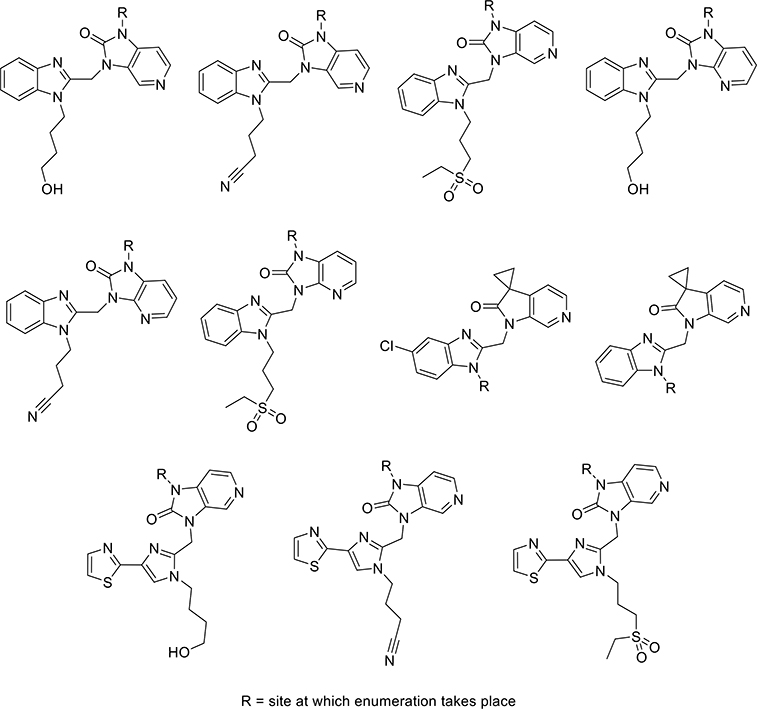
With a highly predictive algorithm in hand, we set out to explore the chemical space of several families of fusion inhibitors known in the literature. These structurally diverse core scaffolds (Figure 2), which had all been advanced to late stage development, were enumerated in PipelinePilot using the ZINC15 database of commercially available building blocks.30 This data set was then prioritized by our NBN and subsequently screened using several cheminformatics filters to generate a prioritized virtual library (>115 000 analogs). These filters removed all non-novel structures and structures containing reactive functional groups. It also included a Lipinski Rule of 5 filter and a cLogD < 1.8 filter (the evaluation of the BMS series showed a decrease in metabolic stability in human liver microsomes for compounds with cLogD > 1.8).23,24,26,31,32 The resulting top 50 list of compounds was further refined on the basis of their synthetic tractability.
Figure 2.
Examples of core scaffolds identified and sites of enumeration.
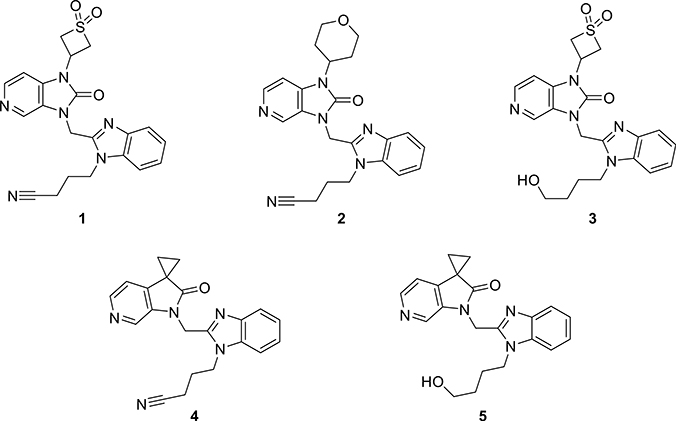
Using this workflow, we identified molecules 1–3 (Figure 3), which met all the criteria as candidates for synthesis. To validate the utility of the cLogD <1.8 filter, we also identified compounds 4 and 5, which were predicted to be potent but were removed by the cLogD filter.
Figure 3.
Candidates for synthesis identified by machine learning.
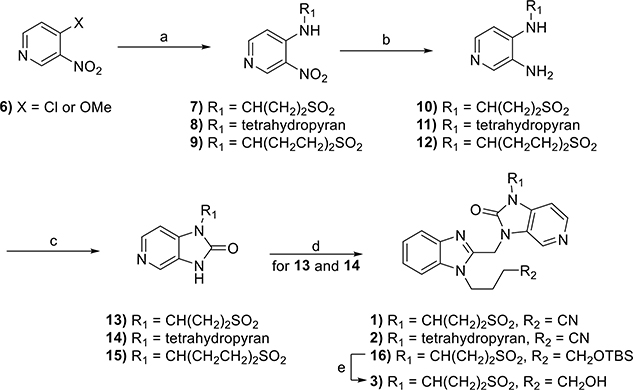
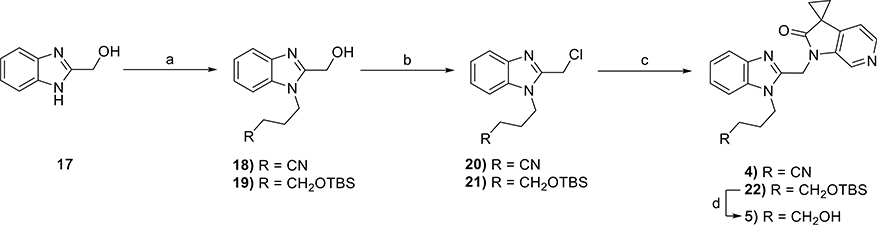
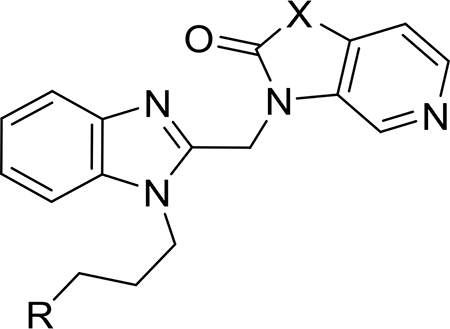
The synthesis of compounds 1–3 is shown in Scheme 1. Th nucleophilic aromatic substitution (SNAr) reaction of commercially available 4-chloro-3-nitropyridine or 4-methoxy-3-nitropyridine (6) with the requisite amines produced substituted pyridines 7–9. The reduction of nitro groups of each using classical hydrogenation conditions led to diaminopyridines 10–12, which were subsequently cyclized with carbonyl diimidazole (CDI)to yield the desired substituted imidazopyridine-2-ones 13–15. Each substituted imidazopyridin-2-one was then alkylated with the requisite chloromethyl benzimidazole 20 and 21 (vide infra) to afford the desired targets, 1 and 2, as well as precursor 16. Subsequent removal of the tert-butyldimethylsilyl (TBS) protecting group of 16 with tetrabutylammonium flouride (TBAF) afforded target compound 3.
Scheme 1.
Reagents and conditions: (a) tetrahydro-2H-pyran-4-amine, 3-aminothietane 1,1-dioxide hydrochloride, or 1,1-dioxothian-4-amine, Hünig’s base, EtOH, 90 °C, 18 h, 30–61%; (b) Pd/C, H2, MeOH, rt, 40 min to 3 h, 100%; (c) CDI, MeCN, 3–12 h, 21–77%; (d) Cs2CO3, MeCN, 70 °C, 18 h, 33–43% or Cs2CO3, KI, DMF, 80 °C, 8 h, 31%; (e) TBAF, THF, rt, 32 h, 65%.
The synthesis of 4 and 5 commenced with the generation of the benzimidazole-2-methanol intermediates 18 and 19 (Scheme 2) from commercially available 1H-benzimidazole-2- methanol 17. Subsequent chlorination resulted in the production of multigram quantities of 20 and 21, which were then individually coupled with commercially available spirocyclopropyl pyrrolopyridinone 11 to produce 4 and 22, respectively. The latter was subsequently deprotected to yield 5.
Scheme 2.
Reagents and conditions: (a) alkyl halide, NaH or K2CO3, DMF, 0 °C to rt, 18 h, 76–94%; (b) SOCl2, Et3N, DCM, rt, 3 h, 80–93%; (c) Cs2CO3, MeCN, rt, 4–18 h, 39–75%; (d) TBAF, THF, rt, 16 h, 84%.
Once in hand, target compounds 1–5 were evaluated in a whole-cell (Hep-2 or BEAS-2B) RSV replication assay using recombinant RSV strain A2-L19F expressing renilla luciferase or nano luciferase as additional transcription units, respectively.33 In parallel, all candidates were tested against recombinant RSV-A2-L19FD489E, carrying a previously described point mutation in the F protein that provides pan-resistance to all current RSV entry inhibitor classes.33 The resulting antiviral activities are listed in Tables 1 and S1. Most candidates identified by our machine learning algorithm were found to exhibit antiviral activities that were comparable to BMS-433771. Our best compound 2 exhibited an EC50 value of 5 nM, an approximately 7-fold improvement in potency compared to BMS-433771. Moreover, all our compounds possessed CC50 values greater than 300 μM, indicating good to excellent therapeutic indices for all. When the compounds were incubated with human liver microsomes (HLMs), they all exhibited t1/2 > 30 min (see Table 1). Compounds 1 and 3 exhibited the greatest stability with t1/2 values of >2 h, while BMS-433771 had a t1/2 of 64 min. Our most potent compound 2 exhibited a t1/2 of 84 min. We note that compounds 4 and 5 had poorer metabolic stability than compounds 1–3, consistent with our hypothesis that compounds with cLogD > 1.8 produce unfavorable metabolic stability.
Table 1.
Antiviral Activities and Stability Data of Compounds Identified by Our Machine Learning Algorithm
 | |||||||
|---|---|---|---|---|---|---|---|
| compd | R | X | EC50 (nM) | EC90 (nM) | CC50 (nM) | therapeutic index | HLM (t1/2) (min) |
| 1 | CN | NCH(CH2)2SO2 | 11 | 1180 | >300 000 | >27 272 | 1380 |
| 2 | CN | N-THP | 5 | 470 | >300 000 | >60 000 | 84.4 |
| 3 | CH2OH | NCH(CH2)2SO2 | 13 | 1010 | >300 000 | >23 076 | 433 |
| 4 | CH2OH | C(CH2)2 | 1940 | >2000 | >300 000 | >154 | 27.8 |
| 5 | CN | C(CH2)2 | 22 | 1530 | >300 000 | >13 636 | 30.1 |
| BMS-433771 | CH2OH | N-cPr | 34.5 | >2000 | >300 000 | >8695 | 64.2 |
These results successfully demonstrated the utility of employing machine learning to efficiently identify a series of novel and potent inhibitors of RSV fusion.
While we were performing our studies, Roymans et al. disclosed the structures and binding modes of a series of RSV fusion inhibitors with structural similarity to BMS-433771 (JNJ-53718678 and JNJ-49153390, shown in Figure 1) but with significantly improved antiviral activities.34 In this work, strategic incorporation of a chlorine or bromine atom facilitated a halogen bonding interaction with the backbone of Thr397 and is the likely reason for the improved potency.34 We envisaged that the binding affinity of our compounds for the F protein could similarly be improved with the introduction of a halogen at the 5-position on the benzimidazole portion of our compounds. To validate this, we generated another small library of analogues based on compound 2, now featuring a halogen at the 5-position on the benzimidazole core.
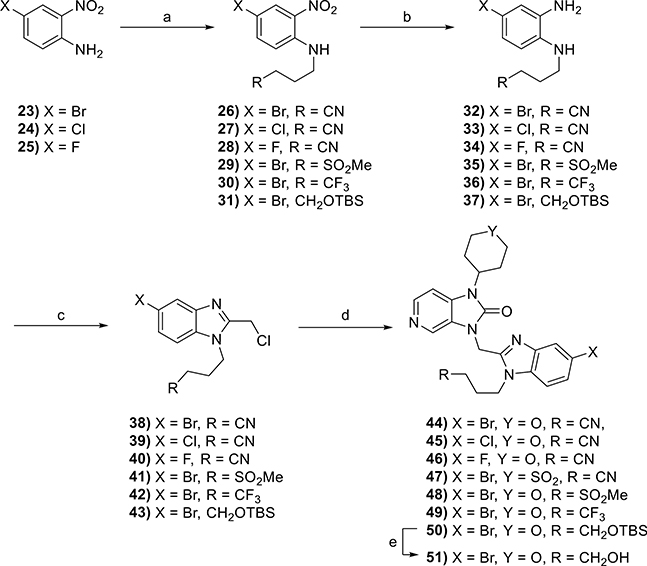
The synthesis of these compounds started with the alkylation of commercially available 4-bromo-, 4-chloro-, or 4-fluoro-2-nitroanilines 23–25 to give intermediates 26–31 (Scheme 3). This was followed by the reduction of the nitro group in the presence of iron with ultrasonic irradiation or hydrogenation using Pt/C to give 32–37 and a ring-closing reaction with 2-chloro-1,1,1-trimethoxy-ethane to yield the desired benzimidazole precursors 38–43. These were subsequently coupled with substituted imidazopyridine-2-ones 14 or 15 (the synthesis of which is described in Scheme 1) to give the desired products 44–49 and precursor 50. A final TBS deprotection reaction of 50 using HF-pyridine produced compound 51.
Scheme 3.
Reagents and conditions: (a) alkyl halide, NaH, DMF, 0 °C to rt, 15–51%; (b) Fe(s), EtOH, AcOH, H2O, rt, 2 h, 48–67% or Pt/C, H2, EtOAc, rt, 18 h, 70%; (c) 2-chloro-1,1,1-trimethoxy-ethane, p-TsOH·H2O, DCM, rt, 4 h, 26–87%; (d) 14 or 15, K2CO3, DMF, 50 °C, 18 h, 16–86%; (e) HF-pyridine, THF, rt, 3 h, 58%.
The antiviral activities of compounds 44–49 and 51 (Table 2) confirmed that the installation of a bromine (44) or a chlorine (45) at the 5-position on the benzimidazole core resulted in a 10-fold gain in potency relative to compound 2 (EC50 = 0.5 and 0.7 nM, respectively) with no change in toxicity (CC50 > 20 μM). These compounds were ~70-fold more potent than BMS-433771 and approximately equipotent with JNJ-53718678. On the other hand, the installation of a fluorine (46) resulted in a significant loss in potency (EC50 > 250 nM). These compounds were also analyzed for their stability in HLMs. The introduction of a halogen reduced the t1/2 of compounds 44 and 45 about 5-fold relative to compound 2, likely due to an increase in cLogD.
Table 2.
Antiviral Activities and Stability Data of Compounds Featuring a Halogen on the Benzimidazole
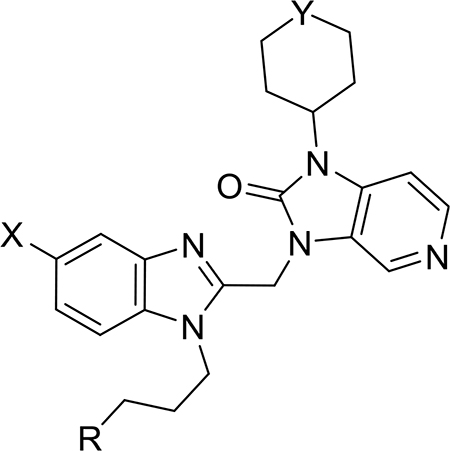 | ||||||||
|---|---|---|---|---|---|---|---|---|
| compd | R | X | Y | EC50 (nM) | EC90 (nM) | CC50 (nM) | therapeutic index | HLM (t1/2) (min) |
| 2 | CN | H | O | 5 | 470 | >300 000 | >60 000 | 84.4 |
| 44 | CN | Br | O | 0.5 | 2.0 | >20 000 | >40 000 | 15 |
| 45 | CN | Cl | O | 0.7 | 1.3 | >20 000 | >28 571 | 16.8 |
| 46 | CN | F | O | >250 | >250 | 15 710 | >62 | 37.9 |
| 47 | CN | Br | SO2 | 3.1 | 10.4 | >20 000 | >6451 | 150.7 |
| 48 | SO2Me | Br | O | 8.3 | 18.2 | >20 000 | >2409 | 22.4 |
| 49 | CF3 | Br | O | 2.5 | 15.3 | >20 000 | >8000 | 3.6 |
| 51 | CH2OH | Br | O | 1.3 | 52.2 | >20 000 | >15 385 | 19.2 |
| BMS-433771 | 34.5 | >2000 | >300 000 | >8695 | 64.2 | |||
| JNJ-53718678 | 0.9 | 1.1 | 16 056 | 17 840 | 52.9 | |||
Of the 5 compounds described in Table 1, compounds 1 and 3, featuring a thietane-1,1-dioxide in place of the tetrahydropyran of compound 2, exhibited greater stability in HLMs. With this in mind, we synthesized compound 47 featuring a tetrahydro-2H-thiopyran-1,1-dioxide. While this did not result in the same subnanomolar potencies seen with compounds 44 and 45, compound 47 exhibited a significantly improved t1/2 of 151 min.
Finally, compounds 48, 49, and 51 were synthesized to assess the importance of the nitrile group for antiviral activity. Compound 48 features the methyl sulfone thought to be responsible for the protein–ligand water-mediated hydrogen bonding interactions, as described for JNJ-5371876.34 The introduction of the methyl sulfone on our series of compounds, however, resulted in a slight loss in potency relative to compounds 44 and 45. The introduction of the trifluoromethyl group (49) and the hydroxyl group (51) had no significant impact on potency, with EC50 values of 3 and 1 nM, respectively. Compounds 48, 49, and 51 all exhibited t1/2 values of <30 min.
In this assay, our compounds, while extremely potent against wild-type RSV, lose significant potency against the viral escape mutant, D489E.33 Although its clinical impact remains unclear, this mutation, as well as other resistance hot spots that are observed clinically,33,34 reminds us of the important lesson learned in treating viral diseases over the past three decades; i.e., success is rarely achieved using only monotherapy. Moreover, while the clinical utility of RSV fusion inhibitors remains unclear (e.g., a recently completed phase 2 clinical trial with presatovir in adult hematopoietic cell transplant recipients has revealed no therapeutic benefit),35 several other fusion inhibitors are currently undergoing clinical evaluations, and the results of these studies will likely tell the tale of the potential of this class of drugs.
By employing a machine learning algorithm, we were able to rapidly identify a series of novel compounds as highly potent inhibitors of the RSV F protein. In this series, compound 2 was found to be 7-fold more potent than BMS-433771. Subsequently, we could further improve the potency by introducing a halogen onto the benzimidazole portion of our compounds to exploit a halogen bonding interaction with Thr397. This approach proved to be successful, and potency improved by an additional 10-fold. Our work provides important proof-of-concept for the use of a machine learning algorithm-based strategy to achieve chemotype diversification and lead optimization.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to express their gratitude to Dr. Sameshnee Pelly for her assistance in reviewing this manuscript. This work was supported, in part, by Public Health Service grants AI071002 (to R.K.P.) and HD079327 (to R.K.P.) from the NIH/NIAID and NIH/NICHD, respectively. The funders had no role in the manuscript preparation or the decision to submit the work for publication.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.9b00524.
Complete description of the experimental conditions and characterization data of all of the compounds described in the manuscript; detailed description of the in vitro biological assessments (PDF)
Detailed data of the compounds (TXT)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.9b00524
Contributor Information
Nicole Pribut, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Thomas M. Kaiser, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Robert J. Wilson, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Edgars Jecs, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Zackery W. Dentmon, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Stephen C. Pelly, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States
Savita Sharma, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Perry W. Bartsch, III, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Pieter B. Burger, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States
Soyon S. Hwang, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States
Thalia Le, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
Julien Sourimant, Institute for Biomedical Sciences, Georgia State University, Atlanta, Georgia 30303, United States.
Jeong-Joong Yoon, Institute for Biomedical Sciences, Georgia State University, Atlanta, Georgia 30303, United States.
Richard K. Plemper, Institute for Biomedical Sciences, Georgia State University, Atlanta, Georgia 30303, United States
Dennis C. Liotta, Department of Chemistry, Emory University, Atlanta, Georgia 30322, United States.
REFERENCES
- (1).Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, and Fukuda K (2003) Mortality associated with influenza and respiratory syncytial virus in the united states. J. Am. Med. Assoc 289, 179–186. [DOI] [PubMed] [Google Scholar]
- (2).Heylen E, Neyts J, and Jochmans D (2017) Drug candidates and model systems in respiratory syncytial virus antiviral drug discovery. Biochem. Pharmacol 127, 1–12. [DOI] [PubMed] [Google Scholar]
- (3).Borchers AT, Chang C, Gershwin ME, and Gershwin LJ (2013) Respiratory syncytial virus—a comprehensive review. Clin. Rev. Allergy Immunol 45, 331–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Paes BA, Mitchell I, Banerji A, Lanctôt KL, and Langley JM (2011) A decade of respiratory syncytial virus epidemiology and prophylaxis: Translating evidence into everyday clinical practice. Can. Respir. J 18, e10–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, and Campbell H (2010) Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet 375, 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shi T, Mcallister DA, O’brien KL, Simoes E. a. F., Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, De Cuellar E, Dang D-A, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, De Freitas Lázaro Emediato CC, Fasce RA, Feikin DR, Feng L, Gentile A, Gordon A, Goswami D, Goyet S, Groome M, Halasa N, Hirve S, Homaira N, Howie SRC, Jara J, Jroundi I, Kartasasmita CB, Khuri-Bulos N, Kotloff KL, Krishnan A, Libster R, Lopez O, Lucero MG, Lucion F, Lupisan SP, Marcone DN, Mccracken JP, Mejia M, Moisi JC, Montgomery JM, Moore DP, Moraleda C, Moyes J, Munywoki P, Mutyara K, Nicol MP, Nokes DJ, Nymadawa P, Da Costa Oliveira MT, Oshitani H, Pandey N, Paranhos-Baccala G, Phillips LN, Picot VS, Rahman M, Rakoto-Andrianarivelo M, Rasmussen ZA, Rath BA, Robinson A, Romero C, Russomando G, Salimi V, Sawatwong P, Scheltema N, Schweiger B, Scott J. a. G., Seidenberg P, Shen K, Singleton R, Sotomayor V, Strand TA, Sutanto A, Sylla M, Tapia MD, Thamthitiwat S, Thomas ED, Tokarz R, Turner C, Venter M, Waicharoen S, Wang J, Watthanaworawit W, Yoshida L-M, Yu H, Zar HJ, Campbell H, and Nair H (2017) Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet 390, 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jha A, Jarvis H, Fraser C, and Openshaw PJM (2016) Respiratory syncytial virus In SARS, MERS and other viral lung infections (Hui DS, Rossi GA, and Johnston SL, Eds.), European Respiratory Society. [PubMed] [Google Scholar]
- (8).Pilie P, Werbel WA, Riddell J IV, Shu X, Schaubel D, and Gregg KS (2015) Adult patients with respiratory syncytial virus infection: Impact of solid organ and hematopoietic stem cell transplantation on outcomes. Transpl Infect. Dis 17, 551–557. [DOI] [PubMed] [Google Scholar]
- (9).Scheltema NM, Gentile A, Lucion F, Nokes DJ, Munywoki PK, Madhi SA, Groome MJ, Cohen C, Moyes J, Thorburn K, Thamthitiwat S, Oshitani H, Lupisan SP, Gordon A, Sánchez JF, O’brien KL, Group PS, Gessner BD, Sutanto A, Mejias A, Ramilo O, Khuri-Bulos N, Halasa N, De-Paris F, Pires MR, Spaeder MC, Paes BA, Simões E. a. F., Leung TF, Da Costa Oliveira MT, De Freitas Lázaro Emediato CC, Bassat Q, Butt W, Chi H, Aamir UB, Ali A, Lucero MG, Fasce RA, Lopez O, Rath BA, Polack FP, Papenburg J, Roglić S, Ito H, Goka EA, Grobbee DE, Nair H, and Bont LJ (2017) Global respiratory syncytial virus-associated mortality in young children (rsv gold): A retrospective case series. Lancet. Global Health 5, e984–e991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Falsey AR, Hennessey PA, Formica MA, Cox C, and Walsh EE (2005) Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med 352, 1749–1759. [DOI] [PubMed] [Google Scholar]
- (11).Falsey AR, and Walsh EE (2000) Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev 13, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhou H, Thompson WW, Viboud CG, Ringholz CM, Cheng P-Y, Steiner C, Abedi GR, Anderson LJ, Brammer L, and Shay DK (2012) Hospitalizations associated with influenza and respiratory syncytial virus in the united states, 1993–2008. Clin. Infect. Dis 54, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Turner TL, Kopp BT, Paul G, Landgrave LC, Hayes D Jr., and Thompson R (2014) Respiratory syncytial virus: Current and emerging treatment options. Clinicoecon. Outcomes. Res 6, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brady MT (2014) Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 134, 415–420. [DOI] [PubMed] [Google Scholar]
- (15).Pollack P, Groothuis JR, and Barbarotto GM (2002) Development and use of palivizumab (synagis): A passive immunoprophylactic agent for rsv. J. Infect. Chemother 8, 201–206. [DOI] [PubMed] [Google Scholar]
- (16).Simões E. a. F., Devincenzo JP, Boeckh M, Bont L, Crowe JE, Griffiths P, Hayden FG, Hodinka RL, Smyth RL, Spencer K, Thirstrup S, Walsh EE, and Whitley RJ (2015) Challenges and opportunities in developing respiratory syncytial virus therapeutics. J. Infect. Dis 211, S1–S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Batonick M, and Wertz GW (2011) Requirements for human respiratory syncytial virus glycoproteins in assembly and egress from infected cells. Adv. Virol 2011, 343408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Meanwell NA, Cianci CW, and Krystal MR (2011) Discovery and development of orally active rsv fursion inhibitors In Antiviral drugs: From basic discovery through clinical trials (Kazmierski WM, Ed.), John Wiley & Sons, Inc. [Google Scholar]
- (19).Battles MB, Langedijk JP, Furmanova-Hollenstein P, Chaiwatpongsakorn S, Costello HM, Kwanten L, Vranckx L, Vink P, Jaensch S, Jonckers THM, Koul A, Arnoult E, Peeples ME, Roymans D, and Mclellan JS (2016) Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat. Chem. Biol 12, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Perron M, Stray K, Kinkade A, Theodore D, Lee G, Eisenberg E, Sangi M, Gilbert BE, Jordan R, Piedra PA, Toms GL, Mackman R, and Cihlar T (2016) Gs-5806 inhibits a broad range of respiratory syncytial virus clinical isolates by blocking the virus-cell fusion process. Antimicrob. Agents Chemother 60, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Polack FP (2015) The changing landscape of respiratory syncytial virus. Vaccine 33, 6473–6478. [DOI] [PubMed] [Google Scholar]
- (22).Stevens M, Rusch S, Devincenzo J, Kim Y-I, Harrison L, Meals EA, Boyers A, Fok-Seang J, Huntjens D, Lounis N, Marien K, Remmerie B, Roymans D, Koul A, and Verloës R (2018) Antiviral activity of oral jnj-53718678 in healthy adult volunteers challenged with respiratory syncytial virus: A placebocontrolled study. J. Infect. Dis 218, 748–756. [DOI] [PubMed] [Google Scholar]
- (23).Shi Q, Kaiser TM, Dentmon ZW, Ceruso M, Vullo D, Supuran CT, and Snyder JP (2015) Design and validation of fresh, a drug discovery paradigm resting on robust chemical synthesis. ACS Med. Chem. Lett 6, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kaiser TM, Burger PB, Butch CJ, Pelly SC, and Liotta DC (2018) A machine learning approach for predicting hiv reverse transcriptase mutation susceptibility of biologically active compounds. J. Chem. Inf. Model 58, 1544–1552. [DOI] [PubMed] [Google Scholar]
- (25).Kaiser TM, Dentmon ZW, Dalloul CE, Sharma SK, and Liotta DC (2020) Accelerated discovery of novel ponatinib analogs with improved properties for the treatment of parkinson’s disease. ACS Med. Chem. Lett 11, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yu K-L, Sin N, Civiello RL, Wang XA, Combrink KD, Gulgeze HB, Venables BL, Wright JJK, Dalterio RA, Zadjura L, Marino A, Dando S, D’arienzo C, Kadow KF, Cianci CW, Li Z, Clarke J, Genovesi EV, Medina I, Lamb L, Colonno RJ, Yang Z, Krystal M, and Meanwell NA (2007) Respiratory syncytial virus fusion inhibitors. Part 4: Optimization for oral bioavailability. Bioorg. Med. Chem. Lett 17, 895–901. [DOI] [PubMed] [Google Scholar]
- (27).Papadatos G, Gaulton A, Hersey A, and Overington JP (2015) Activity, assay and target data curation and quality in the chembl database. J. Comput.-Aided Mol. Des 29, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gao L, Guo L, Liang C, Wang B, Wang L, Yun H, Zhang W, and Zheng X (2015) Novel aza-oxo-indoles for the treatment and prophylaxis of respiratory syncytial virus infection. WO 2015022263 A1.
- (29).Kaiser TM, and Burger PB (2019) Error tolerance of machine learning algorithms across contemporary biological targets. Molecules 24, E2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sterling T, and Irwin JJ (2015) Zinc 15 - ligand discovery for everyone. J. Chem. Inf. Model 55, 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Combrink KD, Gulgeze HB, Thuring JW, Yu K-L, Civiello RL, Zhang Y, Pearce BC, Yin Z, Langley DR, Kadow KF, Cianci CW, Li Z, Clarke J, Genovesi EV, Medina I, Lamb L, Yang Z, Zadjura L, Krystal M, and Meanwell NA (2007) Respiratory syncytial virus fusion inhibitors. Part 6: An examination of the effect of structural variation of the benzimidazol-2-one heterocycle moiety. Bioorg. Med. Chem. Lett 17, 4784–4790. [DOI] [PubMed] [Google Scholar]
- (32).Sin N, Venables BL, Combrink KD, Gulgeze HB, Yu K-L, Civiello RL, Thuring J, Wang XA, Yang Z, Zadjura L, Marino A, Kadow KF, Cianci CW, Clarke J, Genovesi EV, Medina I, Lamb L, Krystal M, and Meanwell NA (2009) Respiratory syncytial virus fusion inhibitors. Part 7: Structure-activity relationships associated with a series of isatin oximes that demonstrate antiviral activity in vivo. Bioorg. Med. Chem. Lett 19, 4857–4862. [DOI] [PubMed] [Google Scholar]
- (33).Yan D, Lee S, Thakkar VD, Luo M, Moore ML, and Plemper RK (2014) Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc. Natl. Acad. Sci. U. S. A 111, E3441–E3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Roymans D, Alnajjar SS, Battles MB, Sitthicharoenchai P, Furmanova-Hollenstein P, Rigaux P, Berg JVD, Kwanten L, Ginderen MV, Verheyen N, Vranckx L, Jaensch S, Arnoult E, Voorzaat R, Gallup JM, Larios-Mora A, Crabbe M, Huntjens D, Raboisson P, Langedijk JP, Ackermann MR, Mclellan JS, Vendeville S, and Koul A (2017) Therapeutic efficacy of a respiratory syncytial virus fusion inhibitor. Nat. Commun 8, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chemaly RF, Dadwal SS, Bergeron A, Ljungman P, Kim YJ, Cheng GS, Pipavath SN, Limaye AP, Blanchard E, Winston DJ, Stiff PJ, Zuckerman T, Lachance S, Rahav G, Small CB, Mullane KM, Patron RL, Lee DG, Hirsch HH, Waghmare A, Mckevitt M, Jordan R, Guo Y, German P, Porter DP, Gossage DL, Watkins TR, Marty FM, Chien JW, and Boeckh M (2019) A phase 2, randomized, double-blind, placebocontrolled trial of presatovir for the treatment of respiratory syncytial virus upper respiratory tract infection in hematopoietic-cell transplant recipients. Clin. Infect. Dis, ciz1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.