Abstract
The prevalence and outcomes of patients who had re-activation of coronavirus disease 2019 (COVID-19) after discharge remain poorly understood. We included 126 consecutively confirmed cases of COVID-19 with 2-month follow-up data after discharge in this retrospective study. The upper respiratory specimen using a reverse-transcription polymerase chain reaction test of three patients (71 years [60–76]) were positive within 11–20 days after their discharge, with an event rate of 19.8 (95%CI 2.60–42.1) per 1,000,000 patient-days. Moreover, all re-positive patients were asymptomatic. Our findings suggest that few recovered patients may still be virus carriers even after reaching the discharge criteria.
Electronic supplementary material
The online version of this article (10.1007/s10096-020-04024-1) contains supplementary material, which is available to authorized users.
Keywords: Coronavirus disease 2019, Re-positive, Infectivity, Follow-up, Outcome
Introduction
In December 2019, patients presenting with viral pneumonia due to 2019 novel coronavirus (2019-nCoV) were firstly reported in Wuhan, China [1]. Although the coronavirus disease 2019 (COVID-19) pandemic has been under considerable control in China, a few discharged patients were reported to have re-positivation of 2019-nCoV during their follow-up visits [2, 3]. However, studies on the prevalence, clinical features, timing of viral nucleic acid re-emergence, and infectivity in these patients are very rare. Monitoring of the coronavirus disease 2019 (COVID-19) prognosis and effective control of the “second wave of an outbreak” of the epidemic remain a huge challenge to the public health.
We aimed to describe the prevalence, demographics, clinical features, and laboratory data of viral nucleic acid re-emergence by investigating the follow-up data of discharged COVID-19 patients. Our findings may help to better understand the follow-up management of discharged COVID-19 patients.
Methods
Participants and data collection
We collected the demographic and clinical data of patients with confirmed COVID-19 who had been admitted to the Tumor Center of Union Hospital (Wuhan, China) between February 15 and March 14, 2020. We obtained and clarified data by direct communication with attending doctors and other healthcare providers when data were missing or uncertain from the medical records. Two physicians (J.C and X.C) extracted the epidemiological, demographic, clinical, laboratory data on admission, and treatment data using a standardized data collection form. The 2019-nCoV was detected by a real-time reverse-transcription polymerase chain reaction (RT-PCR) kit (Bio-Germ, Shanghai, China) targeting the open reading frame 1ab (ORF1ab) gene and nucleocapsid protein (N) gene, recommended by the Chinese Center for Disease Control and Prevention [4]. Viral RNA was extracted from the nasopharynx and oropharynx swab using a viral RNA extraction kit (Tianlong Scientific Company, Xi’an, China). It was defined positive when the cycle threshold (Ct) value less than 37 of both the ORF1ab gene and the N gene fragment. A Ct value of more than 40 was defined as a negative test. For a Ct value ranging from 37 to 40, a second test was required and weakly positive was reported as a recurrence of Ct value of 37–40. The re-positive results were confirmed on the sample after re-extraction and on a subsequent sample. Chest computed tomography findings were reviewed by a physician (L.G.) and a radiologist (H.L). Two authors (X.P and S.F) followed up all patients through telephone interviews until May 31, 2020. Patients could be discharged based on the China National Health Commission discharge criteria [5]: two consecutively negative 2019-nCoV molecular tests, normal body temperature, resolution of respiratory symptoms, with the improvement of lung CT imaging; and patients were required to quarantine for 14 days in a designated allocation. Their upper respiratory specimens were usually collected on the 7th and 14th days during the quarantine after discharge.
Outcomes
Our primary outcome was a re-positive result of 2019-nCoV nucleic acid test during the follow-up after discharge.
Statistics
Continuous variables were summarized as means and standard deviations or medians with interquartile ranges (IQR) as appropriate. Categorical variables were expressed as counts with percentages. We calculated the Kaplan-Meier probability of a re-emergence of 2019-nCoV detected by RT-PCR during follow-up. All statistical analyses were performed with SPSS statistical software version 22.0 (IBM Inc).
Results
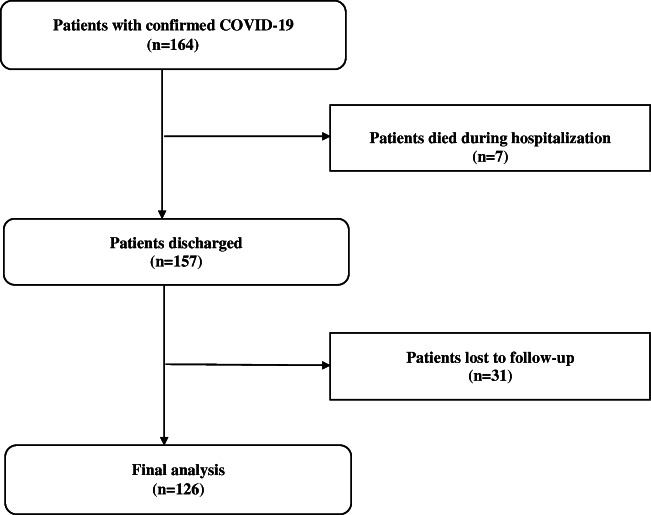
A total of 126 patients with a median age of 66 (54–69) were included in the final analysis (Fig. 1), providing 15,200 patient-days follow-up of data. Compared with those with follow-up data, patients lost to follow-up were similar in age (67 [55–70] vs 63 [53–70], p = 0.726), to be male (16 [51.6%] vs 61 [48.4%], p = 0.749), exposure to wet seafood market (1 [3.2%] vs 1 [0.8%], p = 0.851), and time interval between symptom onset and hospital admission (17 [9–25] vs 13 [7–20], p = 0.140). Patients lost to follow-up had shorter hospital stay (21 [16–27] vs 26 [18–33], p = 0.032) and were more likely to have severe COVID-19 (8 [25.8%] vs 14 [11.1%], p = 0.035) illness.
Fig. 1.
Flow chart of patient selection
One male and two female patients with a median age of 71 (60–76) were re-detectable positive for 2019-nCoV, with an event rate of 19.8 (95%CI 2.60–42.1) per 1,000,000 patient-days. All re-positive patients were asymptomatic. The demographics and clinical and radiological characteristics between patients with and without re-positive findings are shown in Table 1. Two re-positive patients had increased serum lactate dehydrogenase (LDH) and C-reaction protein (CRP) levels. All three re-positive patients were treated with antiviral drugs and Chinese traditional medicine. The time interval between anti-2019-nCoV treatment discontinuation and RT-PCR re-positivation was 10–18 days. The re-positive patients did not report contact with any person who had a fever and respiratory symptoms after discharge. No family member infection was reported. The dynamic results of RT-PCR in the re-positive patients after discharge are shown in Supplemental Table 1.
Table 1.
Characteristics at baseline in COVID-19 patients with and without re-positive to 2019-nCoV nucleic test
| Total (n = 126) | Re-positive (n = 3) | Non-re-positive (n = 123) | |
|---|---|---|---|
| Age (years), median (IQR) | 66 (54–69) | 71 (60–76) | 62 (53–69) |
| Male, n (%) | 61 (48.4) | 1 (33.3) | 60 (48.8) |
| Current smoker, n (%) | 13 (10.3) | 0 (0) | 13 (10.6) |
| Often drinker, n (%) | 3 (2.4) | 0 (0) | 3 (2.4) |
| Hypertension, n (%) | 39 (31.0) | 1 (33.3) | 38 (30.9) |
| Diabetes, n (%) | 26 (20.6) | 1 (33.3) | 25 (20.3) |
| COPD, n (%) | 5 (4.0) | 1 (33.3) | 4 (3.3) |
| CHD, n (%) | 16 (12.7) | 0 (0) | 16 (13.0) |
| Digestive disease, n (%) | 13 (10.3) | 1 (33.3) | 12 (9.8) |
| Previous tumor, n (%) | 8 (6.3) | 0 (0) | 8 (6.5) |
| Immunosuppressive drugs, n (%) | 2 (1.6) | 0 (0) | 2 (1.6) |
| Renal impairment, n (%) | 16 (12.7) | 1 (33.3) | 15 (12.2) |
| Wet market exposure, n (%) | 1 (0.8) | 0 (0) | 1 (0.8) |
| Clinical manifestation | |||
| Fever, n (%) | 87 (69.0) | 2 (66.7) | 85 (69.1) |
| Dry cough, n (%) | 75 (59.5) | 2 (66.7) | 73 (59.3) |
| Productive cough, n (%) | 18 (14.3) | 0 (0) | 18 (14.6) |
| Fatigue, n (%) | 49 (38.9) | 0 (0) | 49 (39.8) |
| Muscle or joint ache, n (%) | 16 (12.7) | 0 (0) | 16 (13.0) |
| Thoracalgia, n (%) | 26 (20.6) | 1 (33.3) | 25 (20.3) |
| Sore throat, n (%) | 17 (13.5) | 0 (0) | 17 (13.8) |
| Diarrhea, n (%) | 11 (8.7) | 1 (33.3) | 10 (8.1) |
| Catarrh, n (%) | 4 (3.2) | 0 (0) | 4 (3.3) |
| Anorexia, n (%) | 41 (32.5) | 0 (0) | 41 (33.3) |
| Shortness of breath, n (%) | 49 (38.9) | 1 (33.3) | 48 (39.0) |
| Headache, n (%) | 15 (11.9) | 1 (33.3) | 14 (11.4) |
| Total symptoms (IQR) | 3 (2–4) | 3 (2–4) | 3 (2–4) |
| Routine blood examinations | |||
| Decreased leukocytes, n (%) | 8 (6.3) | 1 (33.3) | 7 (5.7) |
| Decreased lymphocytes, n (%) | 39 (31.0) | 1 (33.3) | 38 (30.9) |
| Decreased hemoglobin, n (%) | 31 (24.6) | 2 (66.7) | 29 (23.6) |
| Decreased platelets, n (%) | 9 (7.1) | 1 (33.3) | 8 (6.5) |
| ALT > 40 U/L | 37 (29.4) | 0 | 37 (30.1) |
| AST > 40 U/L | 25 (19.8) | 0 | 25 (20.3) |
| Albumin <30 g/L | 7 (5.7) | 0 | 7 (5.7) |
| LDH > 245 g/L | 39 (31.0) | 2 (66.7) | 37 (30.1) |
| CRP > 4 mg/L (data available in 125 patients) | 65 (52.0) | 2 (100) | 63 (51.2) |
| CT findings, n (%) | |||
| Unilateral pneumonia, n (%) | 18 (14.3) | 1 (33.3) | 17 (13.8) |
| Bilateral pneumonia, n (%) | 73 (57.9) | 1 (33.3) | 72 (58.5) |
| Multiple mottling and ground-glass opacity, n (%) | 35 (27.8) | 1 (33.3) | 34 (27.6) |
| Treated with steroid, n (%) | 12 (9.5) | 0 | 12 (9.8) |
| Antiviral, n (%) | 123 (97.6) | 3 (100) | 120 (97.6) |
| Treated with CTM, n (%) | 121 (96.0) | 3 (100) | 118 (95.9) |
| Antibacterial, n (%) | 97 (72.4) | 2 (66.7) | 95 (72.5) |
| Severe COVID-19, n (%) | 13 (10.6) | 1(33.3) | 14 (11.1) |
| Onset to admission (day), median (IQR) | 13 (7–20) | 10 (9–12) | 13 (8–20) |
| Hospital stay (day), median (IQR) | 26 (18–33) | 26 (19–31) | 26 (18–33) |
Decreased means below the lower limit of the normal range. Leukocytes (× 109/L; normal range 3.5–9.5); lymphocytes (× 109/L; normal range 1.1–3.2); platelets (× 109 /L; normal range 125–350); hemoglobin (g/L; normal range 130–175); ALT and AST (U/L; normal range 0–40); LDH (U/L; normal range 109–245); CRP (mg/L; normal range < 4.0)
IQR, interquartile range; COVID-19, coronavirus disease 2019; COPD, chronic obstructive pulmonary disease; CHD, coronary heart disease; ALT, alanine transaminase; AST, alanine aminotransferase; LDH, lactate dehydrogenase; CRP, C-reactive protein; CT, computed tomography; CTM, Chines traditional medicine
Discussion
Our main finding was that patients with COVID-19 after hospital discharge had a low chance to be tested re-positive for 2019-nCoV. Moreover, all re-positive patients were asymptomatic.
The time interval between discharge and re-positive RT-PCR results in our cohort was 11–20 days, which was longer than previously reported [2, 6, 7]. It was reported that the viral shedding duration lasted for 65 days in a recovered COVID-19 patient [8]. Together these findings raise concerns about the shedding window of COVID-19 and the current criteria for discontinuation of quarantine.
The 2019-nCoV nucleic acid detection in patients may fluctuate due to the possible occurrence of false-negative nucleic test findings and the operator’s experience in collecting the sample [9]. However, the re-positive cases in our cohort were likely to have a real re-activation of the infection after three consecutively negative molecular tests of samples collected by trained doctors in addition to symptom resolution. Few studies reported the changes of exact viral load (Ct value or copies/mL) in discharged COVID-19 patients. A case study showed that two discharged COVID-19 patients had decreased Ct values (compared with baseline Ct values) when they became symptomatic with COVID-19 again [10]. Another case study reported the recurrent presence of 2019-nCoV RNA with fluctuating Ct values in a 33-year-old patient who were symptomatic after discharge [11]. Interestingly, 2019-nCoV viral load may be similar in asymptomatic as symptomatic patients [12]. The relationship of baseline 2019-nCoV viral load with re-activation needs to be addressed in future studies.
Re-positive cases pose a major public health concern since little is known about the infectivity of this population. A positive RT-PCR result of 2019-nCoV nucleic acid does not necessarily mean that the virus is infectious. All re-positive patients in our cohort were asymptomatic, with no evidence of infectivity. The recent Wuhan mass COVID-19 screening reported only 300 asymptomatic cases of 9,899,828 participants. None of the samples has cultivated a live virus in the sputum samples and throat swabs from 106 asymptomatic cases [13]. This promising finding might add key information for the improved management of patients recovered from COVID-19.
The limitations of this retrospective observational study include the small number of patients from a single center. Additionally, near 20% of patients were lost to follow-up, which brings selection bias. Due to the tiny number of re-positive samples, we cannot statistically compare the difference between patients with and without re-positivity. A previous study showed that cough accompanying with expectoration and chest congestion accompanying with dyspnea were associated with an increased risk of nucleic acid re-positivity [7]. The clinical risk factors for the re-activation of 2019-nCoV need to be investigated by further large sample–sized studies.
In conclusion, our study indicates that few discharged patients with COVID-19 may have re-positive results of 2019-nCoV detection. The infectivity of this population needs to be studied urgently.
Electronic supplementary material
(PDF 56 kb)
Acknowledgments
We thank all the patients with their data for this analysis. We thank the medical workers who are on the front line of caring for patients. We thank Dr. Jin Wei (College of Health Science, University of Tasmania, Australia, Email: jwei10@utas.edu.cn) for language improvement.
Code availability
SPSS statistical software version 22.0 (IBM Inc)
Author contributions
Drs. Du and Liu had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Drs. Du and Liu.
Acquisition, analysis, or interpretation of data: Drs. H Du J, Chen, X Pan, X Chen, L Gao, C Li, and N Liu.
Drafting of the manuscript: Drs. H Du, J Chen, X Pan, X Chen, and N Liu.
Critical revision of the manuscript for important intellectual content: Drs. X Li, P Xia, and L Chen.
Statistical analysis: Dr. X Li and Dr. P Xia.
Funding
This work was supported by the Fujian Natural Science Foundation (2018J01309), the Joint Funds for the Innovation of Science and Technology, Fujian Province (2019Y9099), and the Fujian Provincial Special Foundation for Natural Science Innovation Project (2016B014).
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author at xieheliunan1984@fjmu.edu.cn on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was approved by the Ethics Committee of Fujian Medical University Union Hospital, which is a member of the National Medical Team Support Wuhan for COVID-19. Written informed consent was waived to collect the data from a medical database due to the nature of our retrospective study of routine clinical data. Oral informed consent was obtained in view of follow-up data.
Consent to participate
Not applicable
Consent for publication
Not applicable
Disclaimer
The funders had no role in the study design and the collection, analysis, and interpretation of data or drafting of the article and the decision to submit it for publication. The researchers confirm their independence from the funder.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hou-wei Du and Jun-nian Chen contributed equally to this work.
References
- 1.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lan L, Xu D, Ye G et al (2020) Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 10.1001/jama.2020.2783 Accessed 27 Feb 2020 [DOI] [PMC free article] [PubMed]
- 3.Hu R, Jiang Z, Gao H, et al. Recurrent positive reverse transcriptase-polymerase chain reaction results for coronavirus disease 2019 in patients discharged from a hospital in China. JAMA Netw Open. 2020;3:e2010475. doi: 10.1001/jamanetworkopen.2020.10475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xing Y, Mo P, Xiao Y, et al. Post-discharge surveillance and positive virus detection in two medical staff recovered from coronavirus disease 2019 (COVID-19), China, January to February 2020. Euro Surveill. 2020;25:2000191. doi: 10.2807/1560-7917.ES.2020.25.10.2000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.China National Health Commission (2020) Diagnosis and treatment of 2019-nCoV pneumonia in China (version7) [in Chinese]. http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml. Accessed 5 March 2020
- 6.Zhu H, Fu L, Jin Y, et al. Clinical features of COVID-19 convalescent patients with re-positive nucleic acid detection. J Clin Lab Anal. 2020;34:e23392. doi: 10.1002/jcla.23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan N, Wang W, Gao Y, et al. Medium term follow-up of 337 patients with coronavirus disease 2019 (COVID-19) in a Fangcang shelter hospital in Wuhan, China. Front Med. 2020;7:373. doi: 10.3389/fmed.2020.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Cai ZB, Huang JS, et al. Positive SARS-CoV-2 RNA recurs repeatedly in a case recovered from COVID-19: dynamic results from 108 days of follow-up. Pathog Dis. 2020;78:ftaa031. doi: 10.1093/femspd/ftaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen D, Xu W, Lei Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis. 2020;93:297–299. doi: 10.1016/j.ijid.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lafaie L, Celarier T, Goethals L et al (2020) Recurrence or relapse of COVID-19 in older patients: a description of three cases. J Am Geriatr Soc. 10.1111/jgs.16728 Accessed 7 July 2020 [DOI] [PMC free article] [PubMed]
- 11.Wang P (2020) Recurrent presence of SARS-CoV-2 RNA in a 33-year-old man. Med Virol. 10.1002/jmv.26334 Accessed 22 July 2020 [DOI] [PMC free article] [PubMed]
- 12.Lee S, Kim T, Lee E et al (2020) Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med. 10.1001/jamainternmed.2020.3862 Accessed 6 August 2020 [DOI] [PMC free article] [PubMed]
- 13.Wuhan National Health Commission (2020) http://www.wuhan.gov.cn/sy/whyw/202006/t20200603_1359620.shtml. Accessed 3 June 2020
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 56 kb)
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author at xieheliunan1984@fjmu.edu.cn on reasonable request.