Abstract
Objective
Gastrointestinal microbiota may be involved in Helicobacter pylori-associated gastric cancer development. The aim of this study was to explore the possible microbial mechanisms in gastric carcinogenesis and potential dysbiosis arising from H. pylori infection.
Design
Deep sequencing of the microbial 16S ribosomal RNA gene was used to investigate alterations in paired gastric biopsies and stool samples in 58 subjects with successful and 57 subjects with failed anti-H. pylori treatment, relative to 49 H. pylori negative subjects.
Results
In H. pylori positive subjects, richness and Shannon indexes increased significantly (both p<0.001) after successful eradication and showed no difference to those of negative subjects (p=0.493 for richness and p=0.420 for Shannon index). Differential taxa analysis identified 18 significantly altered gastric genera after eradication. The combination of these genera into a Microbial Dysbiosis Index revealed that the dysbiotic microbiota in H. pylori positive mucosa was associated with advanced gastric lesions (chronic atrophic gastritis and intestinal metaplasia/dysplasia) and could be reversed by eradication. Strong coexcluding interactions between Helicobacter and Fusobacterium, Neisseria, Prevotella, Veillonella, Rothia were found only in advanced gastric lesion patients, and were absent in normal/superficial gastritis group. Changes in faecal microbiota included increased Bifidobacterium after successful H. pylori eradication and more upregulated drug-resistant functional orthologs after failed treatment.
Conclusion
H. pylori infection contributes significantly to gastric microbial dysbiosis that may be involved in carcinogenesis. Successful H. pylori eradication potentially restores gastric microbiota to a similar status as found in uninfected individuals, and shows beneficial effects on gut microbiota.
Keywords: helicobacter pylori - treatment, gastric diseases, gastric pre-cancer, bacterial interactions
Significance of this study.
What is already known on this subject?
Helicobacter pylori eradication was recommended by WHO-International Agency for Research on Cancer as an effective gastric cancer prevention strategy.
The previous retrospective studies suggest that H. pylori and some other interactive gastric commensals may play roles in gastric carcinogenesis, while the effects of H. pylori and antibiotic treatment on the entire gastrointestinal microecosystem need further well-designed prospective studies.
What are the new findings?
H. pylori is one of the main factors in gastric microbial dysbiosis and successful eradication can lead to the restoration of gastric microbiota to a similar status of negative subjects.
Strong coexcluding interactions in gastric microbiota between Helicobacter and Fusobacterium, Neisseria, Prevotella, Veillonella, Rothia were found only in patients with advanced gastric lesions, and were absent in normal/superficial gastritis group.
Successful H. pylori eradication exerts more beneficial effects on gut microbiota than failed treatment, including increased abundance of probiotic bacteria and putative downregulation of drug-resistance mechanisms.
How might it impact on clinical practice in the foreseeable future?
Our findings provide new insights into the microbial mechanism of H. pylori-associated gastric carcinogenesis and new evidences for beneficial effects of prevention strategy.
Introduction
Helicobacter pylori infection is a major risk factor for gastric cancer (GC).1 The effectiveness of H. pylori eradication for GC and precancerous lesion prevention has been proven in different populations.2–4 Therefore, eradication was recommended by WHO-International Agency for Research on Cancer as a prevention strategy.5 However, uncertainties remain concerning possible effects on non-H. pylori bacterial species and potentially adverse consequences of anti-H. pylori treatment, such as gut microbiota dysbiosis, metabolic effects and increased prevalence of antibiotic-resistant pathogens.
The previous retrospective studies reported dysbiotic microbial communities, altered bacterial interactions and the over-representation of intestinal commensals in GC and precancerous lesions.6 7 GC-specific microbiota characterised by decreased abundance of Helicobacter and enrichment of other bacterial taxa suggested non-H. pylori gastric bacteria may also play an important role in gastric carcinogenesis, although the exact mechanism is yet to be established.
Microbial network analysis showed that the presence of H. pylori influences the strength of gastric microbial interactions, which may be associated with GC progression.6 However, whether H. pylori can function as a bacterial driver and interact with other gastric bacteria, which are subsequently involved in the process of carcinogenesis, remains unknown. Investigation of gastrointestinal microbiota based on a prospective study design following anti-H. pylori treatment may be helpful in exploring the role of H. pylori and its interaction with other bacteria in gastric carcinogenesis. Until now, studies outlining changes in the entire gastric and gut microecosystem on H. pylori eradication are limited.8–11
We designed a prospective population-based study, where gastric biopsies and stool samples were taken from subjects at baseline and follow-up time points after successful or failed anti-H. pylori therapy. Gastric and faecal microbiota were compared before and after treatment using deep sequencing of the 16S ribosomal RNA (16S rRNA) gene, with negative subjects as reference. We were also interested in analysing differential taxa for their interactions with H. pylori and associations with precancerous gastric lesions. This study provided us with a unique opportunity to unravel the relationship between H. pylori and other gastrointestinal bacteria in the process of GC development, and evaluate the potential health effects of eradication therapy on gastrointestinal microbiota.
Methods
Patient and public involvement
Linqu, a rural county in Shandong province, China, possesses one of the highest GC mortality rates worldwide (age-adjusted rates per 100 000 were 55 for men and 19 for women in 1980–1982.12 The present study was conducted within the framework of the National Upper Gastrointestinal Cancer Early Detection Project in Linqu county.
In December 2016, 332 project volunteers were screened using 13C-urea breath test (13C-UBT) with 186 subjects identified as H. pylori positive and 146 as negative. The positive subjects were invited for a 10 day quadruple anti-H. pylori treatment including omeprazole (20 mg two times per day), tetracycline (750 mg three times per day), metronidazole (400 mg three times per day) and bismuth citrate (300 mg two times per day). Six months after the treatment, participants were followed up by repeated 13C-UBT, endoscopic examination and drug intake/adverse effect interview. The participants with completed drug intake records and positive follow-up results of 13C-UBT were defined as failed treatment. A total of 145 participants completed baseline and follow-up endoscopic examinations and agreed to provide gastric biopsies, stool samples and general health information using a structured questionnaire on age, sex, cigarette and alcohol consumption habits, and antibiotics use history (using of any kind of antibiotic at least 1 day within 6 months before the baseline interview).
For the present study, 115 anti-H. pylori treatment participants were enrolled (58 subjects with successful eradication and 57 subjects with failed treatment) for possessing eligible paired baseline and follow-up gastric biopsy and stool samples. For comparison, 49 H. pylori negative subjects were enrolled from the 146 negative subjects according to the initial 13C-UBT screening. Using the same procedure, general information, gastric biopsies and stool samples were collected from these negative subjects.
Upper endoscopic examination and histopathology
Upper endoscopic examinations were conducted by two experienced gastroenterologists using video endoscopes (Olympus). Gastric mucosa was examined and at least two biopsies were obtained from less curve of antrum, one for DNA extraction and one for pathological diagnosis. The gastric mucosa specimens were reviewed blindly by two pathologists according to the criteria proposed by the Chinese Association of Gastric Cancer13 and Updated Sydney System.14 Each biopsy was diagnosed as normal, superficial gastritis (SG), chronic atrophic gastritis (CAG), intestinal metaplasia (IM) or dysplasia (DYS) based on the most severe histology.
DNA extraction and 16S rRNA gene sequencing
All biopsies and stool samples were frozen immediately after collection at −80℃ until DNA extraction. DNA was extracted from biopsies using the QIAamp DNA Mini Kit and from stool samples using the QIAamp Fast DNA Stool Mini Kit (Qiagen, California, USA) according to the manufacturer’s instructions. The hypervariable region V3-V4 of microbial 16S rRNA gene was amplified using universal primers (341F, 5'-CCTACGGGNBGCASCAG-3'; 805R, 5'-GACTACNVGGGTATCTAAT CC-3'). The PCR products were purified using QIAquick Gel Extraction Kit (Qiagen). The resulting amplicon library was sequenced on Illumina Hiseq 2500 PE250 platform.
Sequencing data analysis
Raw sequence reads were processed using IMNGS (www.imngs.org),15 a UPARSE-based pipeline.16 USEARCH V.8.0 was used for pairing, quality filtering and operational taxonomic units (OTUs) clustering.17 OTU with a relative abundance ≥0.1% in at least one sample were clustered at 97% sequence similarity. Taxonomic classification was assigned by RDP classifier V.2.11 training set 15.18
Microbial diversity analysis
Diversity analysis was performed using Rhea19 based on R software and illustrated by richness and Shannon indexes. Wilcoxon signed-rank test was performed to compare diversity indexes before and after anti-H. pylori treatment. Multivariate logistic regression was performed for the comparison of diversity indexes between post-treatment and H. pylori negative groups, adjusted by age, sex and gastric lesions. The generalised UniFrac distance was used for microbial community structure comparison and non-metric multidimensional scaling plots were generated. P values were calculated by the permutational multivariate analysis of variance (PERMANOVA) test for dissimilarities between groups and adjusted for multiple comparisons by the false discovery rate (FDR). The corresponding q values <0.05 were considered statistically significant.20
Detection of differential taxa
To identify differential microbial taxa after treatment, paired t-tests were performed after arcsine transformation21 of relative abundance data and q values were used for multiple testing adjustment with a significance threshold of 0.05. The fold change of relative abundances was calculated as the ratio of the mean value after treatment to the mean value before treatment.
Prediction of metagenomic functions
Tax4Fun22 was used to predict the metabolic pathways and functional orthologs of microbial communities based on the SILVA SSU rRNA database23 and the Kyoto Encyclopedia of Genes and Genomes.24 Differential functional orthologs and pathways after treatment were selected by paired t-test after arcsine transformation of relative frequency with q values <0.05 as statistical significance.
Microbial interaction network analysis
Spearman correlation analysis was performed to calculate the correlation coefficients (r values) between the specific disease-related genera in the gastric mucosa. Cytoscape V.3.6.1 was used for visualisation of networks with significant correlations between genera based on r values showing p<0.05. Two genera with correlations of r>0 and p<0.05 were defined as co-occurring genera, while those with r<0 and p<0.05 were defined as coexcluding genera. Kruskal-Wallis test was conducted for comparison of interaction strengths between different gastric lesion groups. A p<0.05 was considered statistically significant.
Results
General characteristics of study participants
The baseline characteristics of successful eradication, failed treatment and H. pylori negative control groups are presented in online supplementary table 1. No significant differences in baseline age, body mass index (BMI), smoking habits, alcohol consumption, antibiotics use or gastric lesions were found between successful eradication and failed treatment groups (all p>0.05). There was a higher frequency of male subjects in the failed treatment group than in the successful eradication group (70.2% vs 51.7%, p=0.043). Compared with negative controls, higher BMI, increased incidences of smoking and advanced gastric lesions (CAG and IM/DYS) were observed in each of the two anti-H. pylori treatment groups (all p<0.05).
gutjnl-2019-319696supp001.pdf (416.7KB, pdf)
Alterations in gastrointestinal microbial diversity after anti-H. pylori treatment
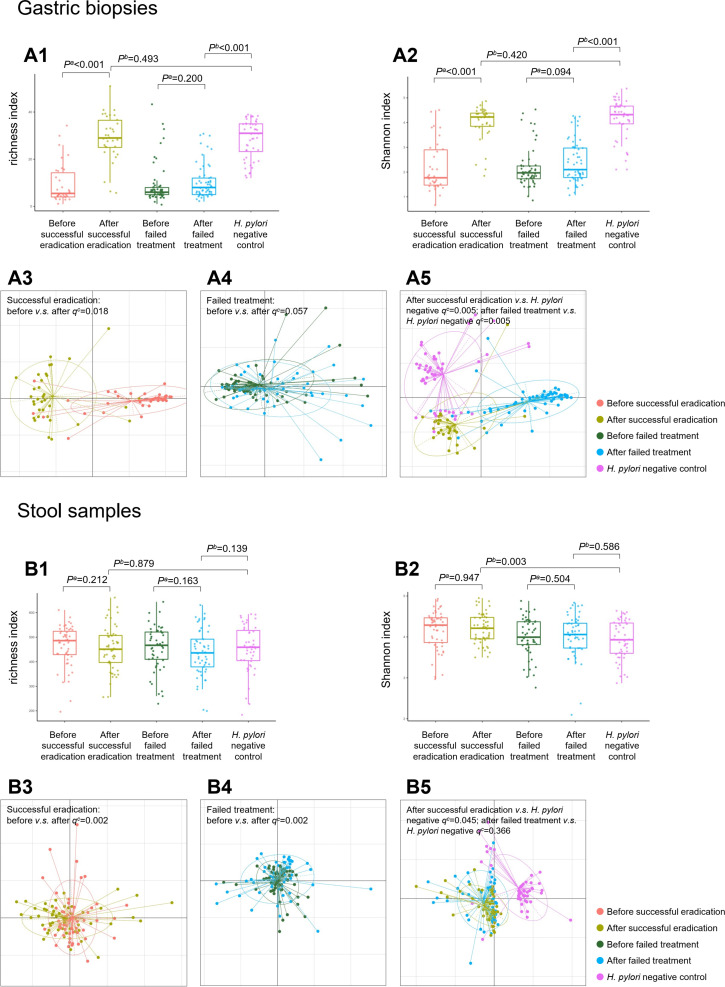
16S rRNA sequencing was finally completed for 230 gastric biopsies: 34 pairs before and after successful eradication, 57 pairs before and after failed treatment and 48 negative controls. Microbial diversity analysis found that richness and Shannon indexes were increased significantly after successful eradication (both p<0.001). These indexes after successful eradication showed no differences to negative controls (p=0.493 for richness and p=0.420 for Shannon index). In participants who failed to clear H. pylori, gastric microbial richness and Shannon indexes were not significantly changed by therapy (p=0.200 for richness and p=0.094 for Shannon index), which were still lower than those of negative controls (both p<0.001, figures 1 A1–2). The trends in microbial richness and Shannon indexes before and after treatment are shown in online supplementary figure 1 A1–4.
Figure 1.
Microbial diversity and community structure in gastric biopsy and stool samples. Box plots showing (A1) increased gastric richness index following successful eradication compared with that of baseline and showing no difference to that of Helicobacter pylori negative subjects; (A2) increased gastric Shannon index following successful eradication compared with that of baseline and showing no difference to that of H. pylori negative subjects. Dissimilarities of microbial community structure in gastric biopsies showing (A3) a significant difference before and after successful eradication; (A4) no change before and after failed treatment; (A5) significant differences when the biopsies after successful or failed treatment were compared with H. pylori negative subjects. Box plots showing (B1) no significant change in faecal richness index before and after anti-H. pylori treatment; (B2) no significant change in faecal Shannon index before and after anti-H. pylori treatment. Dissimilarities of microbial community structure in stool samples showing (B3) a significant difference before and after successful eradication; (B4) a significant difference before and after failed treatment; (B5) marginal difference between after successful eradication and H. pylori negative subjects. aWilcoxon signed-rank test. bLogistic regression adjusted for age, sex and gastric lesion. cPERMANOVA test.
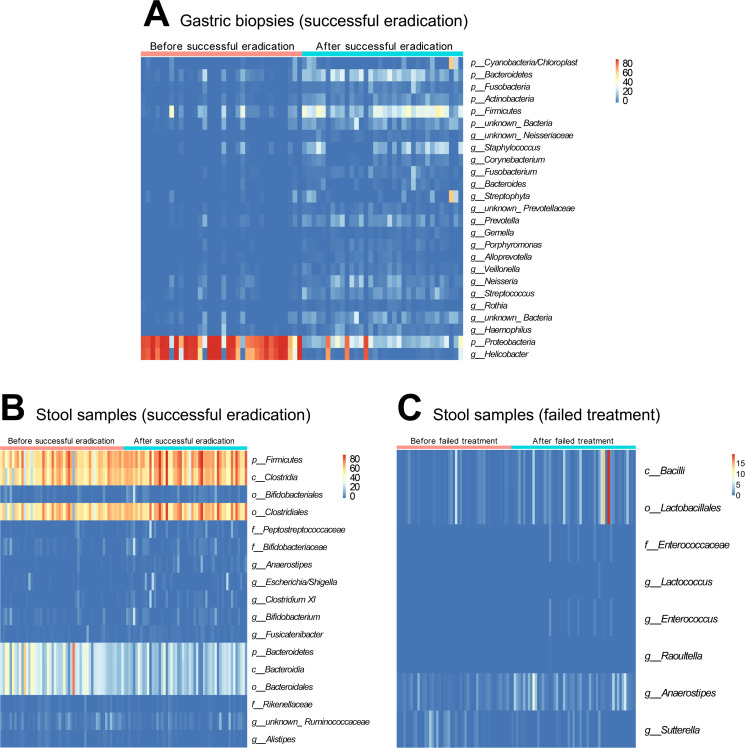
Figure 2.
Changes in gastrointestinal taxa after anti-Helicobacter pylori treatment. To identify differential microbial taxa after treatment, paired t-tests were performed after arcsine transformation of relative abundance data and Q values were used for multiple testing adjustment with a significance threshold of 0.05. Significantly changed taxa after successful eradication were shown (A) in gastric biopsies at phylum and genus levels with mean relative abundance >1%; (B) in stool samples at all levels with mean relative abundance >1%. Only eight taxa with low baseline abundances (<1%) were changed significantly in stool samples after failed treatment (C).
Differences in microbial community structure were further evaluated using generalised UniFrac distance. A significant difference in gastric microbial community before and after successful eradication was observed (q=0.018 adjusted for multiple comparison by the FDR method), with no corresponding change before and after failed treatment (q=0.057). When we compared the biopsies after successful or failed treatment with negative controls, significant differences with both q=0.005 were observed (figure 1 A3–5).
Faecal microbial diversity analysis was completed for 259 stool samples: 53 pairs before and after successful eradication, 53 pairs before and after failed treatment and 47 negative controls. Compared with baseline stool samples, no significant changes were observed in microbial richness and Shannon indexes after successful (p=0.212 and 0.947) and failed treatment (p=0.163 and 0.504). No differences in microbial richness were detected between stools after treatment and negative controls, although Shannon indexes were different in stools after successful eradication compared with negative controls (p=0.003, figure 1 B1–2). The trends in faecal microbial richness and Shannon indexes before and after treatment are shown in online supplementary figure 1 B1–4.
The main differences in faecal microbial community structure were found before and after anti-H. pylori treatment (both successful eradication and failed treatment groups q=0.002). Marginal differences were observed between successful eradication and negative control groups (q=0.045), while no difference was found between failed treatment and negative control groups (q=0.366, figure 1 B3–5).
Changes in gastrointestinal taxa after anti-H. pylori treatment
The sequencing of gastric biopsies revealed 194 bacterial taxa with changes in abundance after successful H. pylori eradication. A total of 5 taxa were significantly decreased by eradication, which were all H. pylori related including Proteobacteria (phylum), Epsilonproteobacteria (class), Campylobacterales (order), Helicobacteraceae (family) and Helicobacter (genus). In the 189 significantly increased taxa, 60 with relative abundances>1% after eradication were selected (online supplementary table 2). At phylum level, significant increases were observed in Cyanobacteria/Chloroplast, Bacteroidetes, Fusobacteria, Actinobacteria and Firmicutes (figure 2A). In addition, 18 differential genera after successful eradication were selected for further analysis including 17 increased genera and one decreased genus, that is, Helicobacter (figure 2A). In failed treatment subjects, no differentially distributed taxa were found in gastric biopsies.
In paired stool samples, 21 taxa showed changes in abundance following successful eradication: 13 increased and 8 decreased. Among them, 17 with relative abundances >1% were selected (figure 2B). The increased taxa were mainly Clostridiales related, including Firmicutes (phylum), Clostridia (class), Clostridiales (order) and Bifidobacterium related, including Bifidobacteriales (order), Bifidobacteriaceae (family), Bifidobacterium (genus). The decreased taxa were mainly Bacteroidales related: Bacteroidetes (phylum), Bacteroidia (class), Bacteroidales (order) (online supplementary table 2). In the failed treatment group, only eight taxa with low baseline abundances (<1%) changed significantly following treatment, seven of which increased and one decreased (figure 2C). The abundances of Bacilli, Lactobacillales and Anaerostipes were increased to >1% (online supplementary table 3).
Changes in predicted microbiota functional capacity after anti-H. pylori treatment
In addition to identifying changes in specific taxa, we predicted the functional capacity changes of microbial communities using Tax4Fun.22 After successful H. pylori eradication, 253 metabolic pathways were significantly changed (q<0.05) in gastric mucosa. Reduced pathways included flagellar assembly, bacterial chemotaxis and bacterial secretion system. Upregulated pathways included protein digestion and absorption, glycosaminoglycan and other glycan degradation, gastric acid secretion, and carbohydrate digestion and absorption (online supplementary table 4). No significant changes were detected in predicted gastric microbiota functional capacity after failed treatment.
In stool samples, 71 predicted functional pathways were changed significantly after successful eradication and 48 were changed after failed treatment (q<0.05, online supplementary table 5). Exploration of drug-resistance-related functional orthologs revealed over-representation of multidrug-resistance protein, methicillin-resistance regulatory protein, vancomycin-resistance sensor histidine kinase, tetracycline-resistance protein and chloramphenicol-resistance protein after failed treatment compared with baseline. Although an over-representation was also found after successful eradication in chloramphenicol-resistance protein (major facilitator superfamily (MFS) transporter drug:H+ antiporter (DHA)1 family) and multidrug-resistance protein (MFS transporter aromatic compound/drug exporter family), others were downregulated including multidrug-resistance protein (multidrug and toxin extrusion family, MFS transporter DHA2 family and protein A) and fosmidomycin-resistance protein (MFS transporter fosmidomycin resistance family, table 1).
Table 1.
Significant changes in predicted drug-resistance-related orthologs using KEGG after anti-Helicobacter pylori treatment in stool samples
| Drug-resistance-related orthologs | Alteration trend after treatment | Mean relative frequency before treatment | Mean relative frequency after treatment | Fold change* | Q value† |
| Successful eradication group | |||||
| K08164 MFS transporter, DHA1 family, chloramphenicol-resistance protein | ↑ | 4.85E-07 | 1.11E-06 | 2.28 | 0.024 |
| K08221 MFS transporter, aromatic compound/drug exporter family, multidrug-resistance protein | ↑ | 4.79E-07 | 1.08E-06 | 2.26 | 0.026 |
| K03327 multidrug-resistance protein, multidrug and toxin extrusion family | ↓ | 9.64E-04 | 8.26E-04 | 0.86 | 0.011 |
| K03543 multidrug-resistance protein A | ↓ | 2.06E-04 | 1.57E-04 | 0.76 | 0.008 |
| K08169 MFS transporter, DHA2 family, multidrug-resistance protein | ↓ | 1.76E-04 | 1.30E-04 | 0.74 | 0.024 |
| K08223 MFS transporter, fosmidomycin resistance family, fosmidomycin-resistance protein | ↓ | 2.53E-04 | 1.84E-04 | 0.73 | 0.007 |
| Failed treatment group | |||||
| K08164 MFS transporter, DHA1 family, chloramphenicol-resistance protein | ↑ | 4.27E-07 | 1.21E-06 | 2.82 | 0.005 |
| K08221 MFS transporter, aromatic compound/drug exporter family, multidrug-resistance protein | ↑ | 4.01E-07 | 1.17E-06 | 2.92 | 0.005 |
| K02546 methicillin-resistance regulatory protein | ↑ | 1.12E-08 | 4.10E-08 | 3.64 | 0.007 |
| K07681 two-component system, NarL family, vancomycin-resistance sensor histidine kinase VraS(EC:2.7.13.3) | ↑ | 7.10E-07 | 2.56E-06 | 3.61 | 0.007 |
| K08151 MFS transporter, DHA1 family, tetracycline-resistance protein | ↑ | 2.00E-06 | 6.58E-06 | 3.29 | 0.009 |
| K08152 MFS transporter, DHA1 family, multidrug-resistance protein B | ↑ | 9.71E-06 | 1.44E-05 | 1.48 | 0.025 |
| K08153 MFS transporter, DHA1 family, multidrug-resistance protein | ↑ | 3.52E-06 | 7.81E-06 | 2.22 | 0.009 |
| K08161 MFS transporter, DHA1 family, multidrug-resistance protein | ↑ | 3.01E-05 | 5.75E-05 | 1.91 | 0.017 |
| K08170 MFS transporter, DHA2 family, multidrug-resistance protein | ↑ | 3.97E-06 | 1.29E-05 | 3.25 | 0.007 |
| K11814 multidrug-resistance protein EbrA | ↑ | 1.70E-09 | 1.90E-08 | 10.61 | 0.005 |
| K11815 multidrug-resistance protein EbrB | ↑ | 6.52E-09 | 2.77E-08 | 4.21 | 0.020 |
*The fold change was calculated as the ratio of the mean relative frequency after treatment to the mean relative frequency before treatment.
†Paired t-test after arcsine transformation of relative frequency and q values were used after adjustment for multiple comparison by the FDR.
DHA, drug:H+ antiporter; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; MFS, major facilitator superfamily.
Associations of gastric microbial dysbiosis with anti-H. pylori treatment and gastric lesions
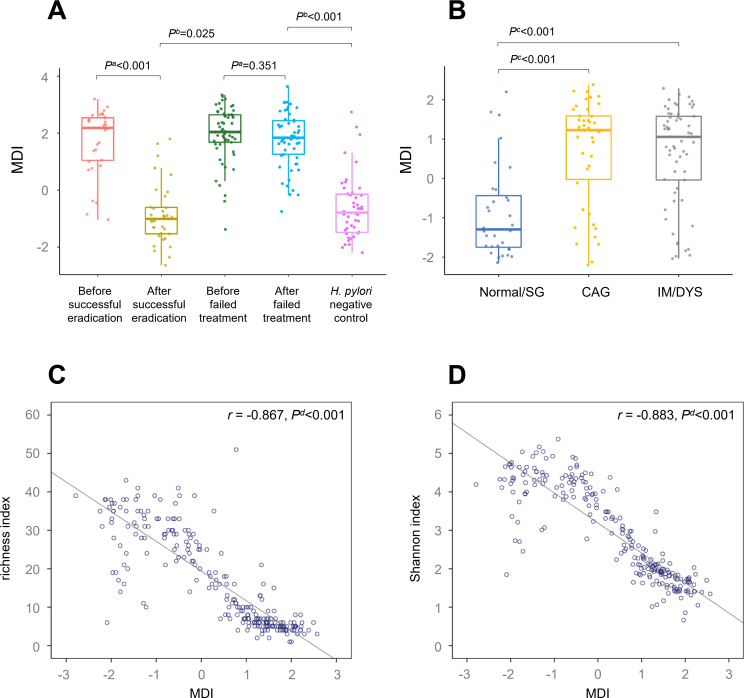
In order to investigate if microbial dysbiosis changed on anti-H. pylori treatment, the 18 genera with changed abundance in gastric biopsies after successful eradication (figure 2A) were combined into Microbial Dysbiosis Index (MDI) for each biopsy using follow formula: MDI=log(total abundance of genera decreased after successful eradication/total abundance of genera increased after successful eradication).7
In comparison with H. pylori positive baseline gastric mucosa, MDI was decreased significantly after successful eradication (p<0.001), and remained higher than that of the negative group (p=0.025). Similarly, high MDIs were found both before and after failed treatment, with no statistical difference between them (p=0.351, figure 3A).
Figure 3.
Gastric microbial dysbiosis is associated with Helicobacter pylori infection and gastric lesions. Box plot showing (A) decreased gastric MDI following successful eradication compared with that of baseline; (B) increased gastric MDIs in CAG and IM/DYS groups compared with that of normal/SG group. Scatter plot showing inverse correlations between (C) gastric MDI and richness index; (D) gastric MDI and Shannon index. aWilcoxon signed-rank test. bLogistic regression adjusted for age, sex and gastric lesion. cKruskal-Wallis test. dSpearman correlation analysis. CAG, chronic atrophic gastritis; DYS, dysplasia; IM, intestinal metaplasia; MDI, Microbial Dysbiosis Index; SG, superficial gastritis.
To further investigate the relationship between gastric microbial dysbiosis and precancerous lesions, 87 H. pylori positive baseline biopsies and 48 negative controls with pathological diagnosis were categorised into normal/SG, CAG and IM/DYS groups. Compared with normal/SG group, MDI was increased significantly in CAG and IM/DYS groups (both p<0.001, figure 3B). When we further analysed associations between MDI and anti-H. pylori treatment by baseline gastric lesions, similar higher MDIs were found before treatment and can be decreased by successful eradication in all the three baseline gastric lesion subgroups (online supplementary figure 2). Gastric MDIs showed an inverse correlation with microbial diversity (r for richness index=−0.867, p<0.001; r for Shannon index=−0.883, p<0.001, figure 3C–D).
Associations of specific genera and their interactions with gastric lesions
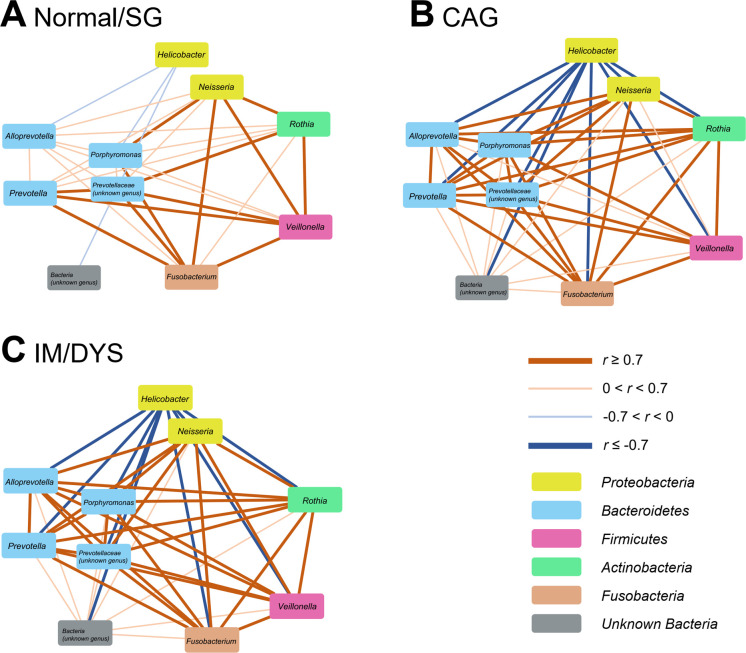
The relative abundances of the same 18 genera (see section "Changes in gastrointestinal taxa after anti-H. pylori treatment") were compared among different baseline gastric lesion groups. Using multiple logistic regression adjusted for age and gender, we found that Helicobacter was significantly increased in CAG and IM/DYS groups compared with normal/SG group (both p<0.001). Nine genera were decreased both in CAG and IM/DYS groups (all p<0.05), including Alloprevotella, Fusobacterium, Neisseria, Porphyromonas, Prevotella, Rothia, Veillonella, unknown_Bacteria and unknown_Prevotellaceae (online supplementary table 6).
The interactions among these 10 genera (Helicobacter and 9 genera above) were further explored by correlation network construction. Significantly stronger correlations were observed in CAG and IM/DYS groups than in normal/SG group (CAG vs normal/SG, p=0.040; IM/DYS vs normal/SG, p=0.003). Coexcluding interactions occurred between Helicobacter and the other nine genera, while co-occurring interactions existed among the nine non-Helicobacter genera (figure 4A–C). Particularly strong coexcluding interactions (all p<0.001) between Helicobacter and Fusobacterium (r=−0.84 in CAG; r=−0.80 in IM/DYS), Neisseria (r=−0.82 in CAG; r=−0.82 in IM/DYS), Prevotella (r=−0.88 in CAG; r=−0.83 in IM/DYS), Veillonella (r=−0.71 in CAG; r=−0.72 in IM/DYS) and Rothia (r=−0.78 in CAG; r=−0.75 in IM/DYS) were noticed only in individuals with advanced gastric lesions. However, such significant interactions were absent in normal/SG group.
Figure 4.
Correlation networks of specific gastric genera in different gastric lesion groups. Spearman correlation analysis was performed to calculate the correlation coefficients (R values) between specific gastric genera. Visualised networks of significant correlations (p<0.05) by Cytoscape V.3.6.1 showed stronger correlation strengths of specific gastric genera in (B) CAG and (C) IM/DYS groups than in (A) normal/SG group. CAG, chronic atrophic gastritis; DYS, dysplasia; IM, intestinal metaplasia; SG, superficial gastritis.
Discussion
Our prospective population-based study observed that H. pylori induces gastric microbial dysbiosis, and that specific H. pylori interactive taxa may be associated with precancerous lesions. Successful eradication reverses the gastric dysbiosis to a similar level as negative controls, and exerts more beneficial effects on gut microbiota than failed treatment, including increased probiotic and putative downregulation of drug-resistance mechanisms.
H. pylori is a well-studied pathogen dominant in stomach. Consistent with decreased microbial diversity in H. pylori positive subjects,25–28 our study indicates that successful eradication can significantly elevate the low gastric microbial richness and evenness in H. pylori positive subjects to similar levels as those in negative controls. While, the different microbial community structure between subjects after successful eradication and negative controls still implies some effects of antibiotics. In contrast, no changes were found in microbial diversity and community structure after failed treatment. Our design including the subjects who failed to clear H. pylori after the same treatment regimen can help to identify important alterations after successful eradication as H. pylori related. These results suggest that H. pylori changes gastric microbial diversity and community structure, which can be reversed by successful eradication.
Besides gastric microbiota composition, successful H. pylori eradication significantly changed the abundances of 194 taxa, while failed treatment did not lead to such alterations. Similar to a small sample size intervention trial,29 our study demonstrates that only H. pylori-related taxa were reduced dramatically after eradication. Increased taxa after eradication were the commonly dominant commensals, including Firmicutes, Bacteroidetes, Actinobacteria, Cyanobacteria and Fusobacteria. Our study tentatively suggests H. pylori as the main disruptor for homoeostasis of gastric commensals. Short-term (10 days) quadruple treatment alone may not disturb gastric microbiota 6 months later.
Functional capacity prediction of gastric microbiota helped us to better understand the possible bacterial function shift by H. pylori eradication. The downregulated functions included flagellar assembly, bacterial chemotaxis and bacterial secretion system, which is reasonable and consistent with the decrease of H. pylori infection after eradication. The upregulated pathways after eradication covered a wide range. Among them, increased gastric acid secretion pathway was confirmed by the significant decrease of gastric pH 6 months after eradication (data not shown). Combination of these evidences supports that H. pylori infection is an important factor for inducing high gastric pH through chronic inflammation, parietal cell loss and microbial metabolic products.30 Other increased bacterial functions following successful eradication included glycan degradation, carbohydrate and protein digestion and absorption, which showed complicated associations with protective production of short chain fatty acids, butyrate and modulation of inflammation.31 While further validation is needed for the predicted functional capacity changes.
Since H. pylori was suggested as the main inducer for gastric dysbiosis, we integrated the 18 differential genera after successful eradication to calculate MDI. The MDI was previously used to monitor the microbial dysbiosis in different diseases.7 In our study, MDIs show strong inverse correlations with microbial diversity and illustrate high degree of dysbiosis in H. pylori positive mucosa and advanced lesions (CAG and IM/DYS). Reversal of dysbiosis and gastritis activity (data not shown) by successful eradication suggested that gastric dysbiosis induced by H. pylori may be involved in gastric lesion progression.
In addition to the dysbiosis by H. pylori, we also revealed that an increase of Helicobacter with concomitant decrease of nine specific genera may be associated with advanced gastric lesions. Increased strength of correlations among these differential genera was shown in advanced lesion groups. Particularly, strong coexcluding interactions (significant negative correlations, r<0 and p<0.05) were found between Helicobacter and Fusobacterium, Neisseria, Prevotella, Veillonella, Rothia only in CAG and IM/DYS rather than in normal/SG group. Our study identified the interactions of Helicobacter and other gastric disease-related bacteria in carcinogenesis process, although the mechanisms remain unclear. The five H. pylori interactive taxa were among the common genera characterising healthy conditions of gastric microbiota. The coexcluding interactions of Neisseria and Prevotella with Helicobacter in advanced lesions, together with their previously reported depletion in GC,7 suggest possible roles in gastric carcinogenesis. The mutual modulation between gastric ecology and host immune status may serve as one of the mechanisms, for example, the link between immunosuppression and Prevotella depletion.32
Stool samples collected before and after treatment and from H. pylori negative subjects provided us an opportunity to assess gut as well as gastric microbiota. Previous multiple time point follow-up studies10 33 reported a short significant decrease of faecal microbial diversity one or 2 weeks after H. pylori eradication, and slow restoration in long term. These findings can help to explain our observations of no significant microbial diversity alterations as a consequence of slow restoration of faecal microbial community 6 months after treatment.
Despite the relatively stable faecal microbial community, we identified some differential taxa after successful eradication. The decreased Bacteroidetes-to-Firmicutes ratio 6 months after eradication may provide one possible reason for weight gain as a potential adverse effect, since Firmicutes and Bacteroidetes were found related to lipid and bile acid metabolism and energy homoeostasis in host.34 Reversal of the ratio 18 months after eradication10 suggested the temporary nature and restorability of the imbalance. We also noticed that a well-known probiotic for inhibiting pathogens, Bifidobacterium-related taxa,35 was enriched only after successful eradication. This result may preliminarily suggest a beneficial effect of H. pylori eradication, since the influences on gut microbiota are complicated and large-scale confirmation is needed.
One of the main concerns about anti-H. pylori treatment is antibiotic resistance. We identified more upregulated drug-resistance-related functional orthologs in failed tetracycline containing treatment group, including multidrug-resistance protein, methicillin-resistance regulatory protein, vancomycin-resistance sensor histidine kinase, chloramphenicol-resistance protein as well as tetracycline-resistance protein. This phenomenon may serve as one of the numerous reasons for failed consequence and suggested complicated effects of antibiotics on gut microbiota. On the contrary, most of the significantly altered drug-resistance functional orthologs were downregulated after successful eradication, suggesting one-time successful eradication may have beneficial effects on faecal microbiota, including probiotic enrichment and downregulation of drug-resistant mechanism.
Our study has several strengths. Different from previous retrospective studies, our prospective design with self-comparison before and after successful and failed treatment can effectively control the individual differences and complex confounding factors, and investigate the effects of H. pylori infection and medication therapy on microbiota in combination or separately. The simultaneous collection of gastric biopsies and stool samples helped us to comprehensively understand the alterations of gastric and gut microbiota by anti-H. pylori treatment in the whole organism. However, our study’s limitations include a modest sample size and a lack of validation at various gastric lesions and GC. Furthermore, mechanism investigation is needed for a deeper understanding of the relationship between specific bacteria and H. pylori-associated gastric carcinogenesis.
In conclusion, our study identified H. pylori as one of the main factors for gastric microbial dysbiosis. Coexcluding interactions between Helicobacter and Fusobacterium, Neisseria, Prevotella, Veillonella, Rothia may be involved in the progression of gastric lesions or carcinogenesis, although validation is needed. Successful H. pylori eradication can lead to the restoration of gastric microbiota to a similar status of negative subjects, and may exert more beneficial effects on gut microbiota than failed treatment, such as probiotic enrichment and downregulation of drug-resistant mechanism. Our findings provide new insights into the microbial mechanism of H. pylori-associated gastric carcinogenesis and prevention strategy.
Acknowledgments
We thank all the individuals who participated in this study and donated samples.
Footnotes
YG and YZ contributed equally.
Contributors: K-FP and W-CY contributed design of the study. MB and SS conducted upper endoscopy examinations. LZ, J-LM, W-DL and Z-XL contributed to subject recruitment and sample collection. MV, KU, MQ, RS and MC completed histological diagnoses. YZ, J-JG, YG and TZ carried out experiments. YG analysed experimental results. YZ and YG wrote the draft of the manuscript. MG, RML, W-QL and K-FP revised the manuscript. All authors read and approved the submitted version.
Funding: National Key R&D Program of China (2018YFC1313100), National Natural Science Foundation of China (81572811), International (regional) Cooperation and Exchange Project (NSFC-DFG, 81861138041), German Federal Ministry of Education and Research (BMBF) (German Research Presence in Asia: 01DO17022) to MG, German Research Foundation (DFG) (SFB 1371) to MG, Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20181102).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the Institutional Review Boards of Peking University Cancer Hospital and Institute.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as online supplementary information. The extradata about bacterial taxa are available from the responsible author for reuse under the permission of the Chinese Human Genetic Resources Administration Office.
References
- 1. Schistosomes, liver flukes and Helicobacter pylori. IARC Working group on the evaluation of carcinogenic risks to humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 2. Ma J-L, Zhang L, Brown LM, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst 2012;104:488–92. 10.1093/jnci/djs003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li W-Q, Ma J-L, Zhang L, et al. Effects of Helicobacter pylori treatment on gastric cancer incidence and mortality in subgroups. J Natl Cancer Inst 2014;106:dju116 10.1093/jnci/dju116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong BCY, Zhang L, Ma J-ling, et al. Effects of selective COX-2 inhibitor and Helicobacter pylori eradication on precancerous gastric lesions. Gut 2012;61:812–8. 10.1136/gutjnl-2011-300154 [DOI] [PubMed] [Google Scholar]
- 5. International Agency for Research on Cancer Helicobacter pylori eradication as a strategy for preventing gastric cancer. IARC working group reports Volume 8. Available: http://www.iarc.fr/en/publications/pdfs-online/wrk/wrk8/index.php
- 6. Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67:1024–32. 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67:226–36. 10.1136/gutjnl-2017-314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gotoda T, Takano C, Kusano C, et al. Gut microbiome can be restored without adverse events after Helicobacter pylori eradication therapy in teenagers. Helicobacter 2018;23:e12541 10.1111/hel.12541 [DOI] [PubMed] [Google Scholar]
- 9. Hsu P-I, Pan C-Y, Kao JY, et al. Helicobacter pylori eradication with bismuth quadruple therapy leads to dysbiosis of gut microbiota with an increased relative abundance of Proteobacteria and decreased relative abundances of Bacteroidetes and Actinobacteria. Helicobacter 2018;23:e12498 10.1111/hel.12498 [DOI] [PubMed] [Google Scholar]
- 10. Yap TW-C, Gan H-M, Lee Y-P, et al. Helicobacter pylori eradication causes perturbation of the human gut microbiome in young adults. PLoS One 2016;11:e0151893 10.1371/journal.pone.0151893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jakobsson HE, Jernberg C, Andersson AF, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 2010;5:e9836 10.1371/journal.pone.0009836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. You WC, Blot WJ, Chang YS, et al. Diet and high risk of stomach cancer in Shandong, China. Cancer Res 1988;48:3518–23. [PubMed] [Google Scholar]
- 13. You WC, Blot WJ, Li JY, et al. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res 1993;53:1317–21. [PubMed] [Google Scholar]
- 14. Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney system. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol 1996;20:1161–81. 10.1097/00000478-199610000-00001 [DOI] [PubMed] [Google Scholar]
- 15. Lagkouvardos I, Joseph D, Kapfhammer M, et al. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep 2016;6:33721 10.1038/srep33721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edgar RC. UPARSE: highly accurate otu sequences from microbial amplicon reads. Nat Methods 2013;10:996–8. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 17. Edgar RC. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010;26:2460–1. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 18. Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lagkouvardos I, Fischer S, Kumar N, et al. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017;5:e2836 10.7717/peerj.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 2001;29:1165–88. [Google Scholar]
- 21. Sokal RR, Rohlf JF. Biometry: the principles and practice of statistics in biological research. New York: Freeman, 1995. [Google Scholar]
- 22. Aßhauer KP, Wemheuer B, Daniel R, et al. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data: fig. 1. Bioinformatics 2015;31:2882–4. 10.1093/bioinformatics/btv287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quast C, Pruesse E, Yilmaz P, et al. The Silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42:D199–205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schulz C, Schütte K, Koch N, et al. The active bacterial assemblages of the upper GI tract in individuals with and without Helicobacter infection. Gut 2018;67:216–25. 10.1136/gutjnl-2016-312904 [DOI] [PubMed] [Google Scholar]
- 26. Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103:732–7. 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parsons BN, Ijaz UZ, D'Amore R, et al. Comparison of the human gastric microbiota in hypochlorhydric states arising as a result of Helicobacter pylori-induced atrophic gastritis, autoimmune atrophic gastritis and proton pump inhibitor use. PLoS Pathog 2017;13:e1006653 10.1371/journal.ppat.1006653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klymiuk I, Bilgilier C, Stadlmann A, et al. The Human Gastric Microbiome Is Predicated upon Infection with Helicobacter pylori. Front Microbiol 2017;8:2508 10.3389/fmicb.2017.02508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li TH, Qin Y, Sham PC, et al. Alterations in gastric microbiota after H. pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep 2017;7:44935 10.1038/srep44935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waldum HL, Kleveland PM, Sørdal Øystein F. Helicobacter pylori and gastric acid: an intimate and reciprocal relationship. Therap Adv Gastroenterol 2016;9:836–44. 10.1177/1756283X16663395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong JMW, de Souza R, Kendall CWC, et al. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 2006;40:235–43. 10.1097/00004836-200603000-00015 [DOI] [PubMed] [Google Scholar]
- 32. Espinoza JL, Matsumoto A, Tanaka H, et al. Gastric microbiota: an emerging player in Helicobacter pylori-induced gastric malignancies. Cancer Lett 2018;414:147–52. 10.1016/j.canlet.2017.11.009 [DOI] [PubMed] [Google Scholar]
- 33. Liou J-M, Chen C-C, Chang C-M, et al. Long-Term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect Dis 2019;19:1109–20. 10.1016/S1473-3099(19)30272-5 [DOI] [PubMed] [Google Scholar]
- 34. Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–3. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 35. Hidalgo-Cantabrana C, Delgado S, Ruiz L, et al. Bifidobacteria and their health-promoting effects. Microbiol Spectr 2017;5 10.1128/microbiolspec.BAD-0010-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319696supp001.pdf (416.7KB, pdf)