Abstract
Increased concern has recently emerged pertaining to the occurrence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in aquatic environment during the current coronavirus disease 2019 (COVID-19) pandemic. While infectious SARS-CoV-2 has yet to be identified in the aquatic environment, the virus potentially enters the wastewater stream from patient excretions and a precautionary approach dictates evaluating transmission pathways to ensure public health and safety. Although enveloped viruses have presumed low persistence in water and are generally susceptible to inactivation by environmental stressors, previously identified enveloped viruses persist in the aqueous environment from days to several weeks. Our analysis suggests that not only the surface water, but also groundwater, represent SARS-CoV-2 control points through possible leaching and infiltrations of effluents from health care facilities, sewage, and drainage water. Most fecally transmitted viruses are highly persistent in the aquatic environment, and therefore, the persistence of SARS-CoV-2 in water is essential to inform its fate in water, wastewater and groundwater and subsequent human exposure.
Keywords: Coronavirus, COVID-19, Pathways, Water, Groundwater
1. Background
The World Health Organization (WHO) declared coronavirus disease 2019 (COVID-19) as a public health emergency of international concern and the causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected over 13 million people by 16th July 2020 in a total of 195 countries. SARS-CoV-2 belongs to the family Coronaviridae, a type of lipid bi-enveloped coronavirus. This virus family causes diverse disease symptoms, including common cold, with SARS-CoV-2 causing acute pneumonia (Chen et al., 2020). With an incubation period ranging from 3–24 days, fever and cough were reported to be the most common symptoms (Guan et al., 2020). SARS-CoV-2 is highly contagious with an effective reproduction number (Ro) ranging from 2 to 4 and the rate of infections becomes double in every 5 days (Inglesby, 2020). The major route of SARS-CoV-2 transmission is reported to be through aerosols, fomites (Guan et al., 2020), and respiratory droplets (Kampf et al., 2020). Generally, there is a lack of knowledge on the occurrence, viability, and transmission of SARS-CoV-2 in the aquatic environment. A previous study reported the detection of viral RNA of severe acute respiratory syndrome coronavirus (SARS-CoV), which is closely related to SARS-CoV-2, in sewage and hospital wastewater (Leung et al., 2003). Recently, SARS-CoV-2 genetic material was detected in the rectal swab, blood, and oropharyngeal samples in many patients who were tested positive (Gu et al., 2020, Peng et al., 2020). Recent studies have also identified SARS-CoV-2 RNA in wastewater, primarily for epidemiological monitoring purposes (Ahmed et al., 2020, Haramoto et al., 2020, Sherchan et al., 2020; Foladori et al., 2020).
The emergence of high consequence viruses in the last two decades has raised subsequent concerns pertaining to the source, fate, and transport pathways of these viruses in the environment (Bibby et al., 2017). With the experience of the COVID-19 pandemic, it becomes more important than ever to explore each aspect of the potential pathway of virus transmission in the biotic/abiotic environment (Brainard et al., 2017). For example, traces of the Ebola Virus in latrines restricted the people from using it to allow for viral inactivation, ranging from 7 to 28 days (Bibby et al., 2015; Kelly et al., 2018). Therefore, a better understanding of the fate of SARS-CoV-2 in water and wastewater is critical. Under these considerations, we evaluated the propensity and repercussions of SARS-CoV-2 transmission in the aquatic environment, with a particular emphasis on the fate and transport of SARS-CoV-2 in groundwater. The perspective of SARS-CoV-2 entering the aquatic ecosystem from inadequately treated wastewater have been more critically reviewed since RNA presence of SARS-CoV-2 in wastewater have reached public domain (Ahmed et al., 2020, Haramoto et al., 2020, Sherchan et al., 2020, Annalaura et al.; 2020). Although occurrence of fecal-oral route transmission and infectivity of SARS-CoV-2 in wastewater are still uncertain, there are growing concerns on exposure risk of SARS-CoV-2 in natural water bodies. We have particularly tried to study the transport perspectives of the SARS-CoV-2 along the surface and sub-surface water in a qualitative manner. This study aims to spread awareness on multiple aspects of SARS-CoV-2 in the environment and assist concerned authorities and policymakers to formulate appropriate guidelines to prevent waterborne viral spread in an epidemic.
2. Similarities of SARS-CoV-2 with other pathogens detected in the aquatic environment
In order to assess the similarity and to identify a potential surrogate to SARS-CoV-2, information on various enveloped and non-enveloped viruses in comparison with SARS-CoV-2, were collected and summarized in Table 1 . The persistence of enveloped viruses like influenza viruses and herpes simplex viruses in the water was found to be around 200 days at 4 °C (Dublineau et al., 2011) and 24 h in distilled water (Nerurkar et al., 1983). While the T90 (time required for the initial viral titer to decrease by 90%) values for several enveloped viruses in wastewater range from 20 to 40 days in wastewater (Ye, 2018), the T90 value for human coronavirus (hCoV-229E) was reported to vary between 200 to 400 days at 4 °C in various water matrices including buffer, surface water, groundwater, and tap water (Table 1). In Bibby et al. (2011) 9 HCoV 229E and 1 HCoV HKU1 sequences were detected in biosolids from a wastewater treatment facility. Another study by Bibby and Peccia (2013) confirmed the presence coronavirus in 83% of the influent and effluent sewage samples. The HCoVs were rarely detected in the aquatic environment due to very low recovery efficiency in the present detection methods (Annalaura et al., 2020). Wang et al., 2020 studied the presence of SARS-CoV-2 at different steps of wastewater treatment, around a hospital setting by RT-qPCR and infectivity assay. Several studies regarding the presence of SARS-CoV-2 RNA in wastewater samples with RT-qPCR have also recently been carried out in last months (Medema et al., 2020, Ahmed et al., 2020, Haramoto et al., 2020, Sherchan et al., 2020, Kumar et al., 2020b). This apparent increased persistence has motivated investigations evaluating SARS-CoV-2 transmission and occurrence in the aquatic environment.
Table 1.
Similarity Index of SARS-CoV-2 with other viruses (non-enveloped and enveloped) which are detected in water bodies, stool and urine samples.
| Genome/Species | Virus | Similar Characteristics with SARS-CoV-2 (Davg = 120 nm) (Genome Size 26.4 – 31.7 kilobases) |
Isoeletric points | Presence in different water bodies |
Wastewater/stool samples | Major Diseases/Outbreaks | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Diameter (Average) | Genome Type | Genome Size | River/Coastal | Lake/Urban Catchment | Groundwater | |||||
| Enteroviruses | Polioviruses | 30 nm | +ve sense, Single Strand (SS) RNA | 7440 bases | 6.9 (PV-1)22 | 62%1(California, 2000) 0%6(Florida Beach, 2009) 50%9(Germany, 2010) 36%11 (Chicago, 2009) 50%16(Wuhan, 2010–11) |
29–76%3(Germany, 2002–03)3 –7%7(Michigan, USA) 40%12(Singapore 2006–09) |
23%10Wisconsin (2003–04) 50%13(England, 2009) 8.8%15(Korea, 2007–08) |
– | Polio Outbreaks |
| Coxsackievirus | 30 nm | SS RNA | 7400 bases | 4.8 & 6.122 | HFMD Outbreak | |||||
| Echovirus | 24–30 nm | SS RNA | 7500 bases | 4.0–6.422 | Liver Failure, Mycocarditis | |||||
| Enetrovirus | 25–30 nm | +ve sense, SS RNA | 7200–8500 bases | 4.0–5.522 | Herpangina, Pleurodynia, Aseptic meningitis | |||||
| Hepevirus | HEV/HAV | 27–34 nm | +ve sense, single strand RNA | 7200 bases | 2.822 | 76%1(California, 2000), 57%2(Georgia, 2002) 0%6(Florida, 2009) |
5–20%3(Germany, 2002–03) 8.9%12(Singapore 2006–09) |
– | – | Hepatitis E, Jaundice |
| Norovirus | Norovirus | 23–40 nm | +ve sense, single strand RNA | 7500 bases | 5.922 | 0%6(Florida, 2009) 25.7%9(Germany, 2010) 31%17(Finland, 2005) |
15–53%3(Germany, 2002–03) 54.8%(GI) and 69%(GII)12(Singapore 2006–09) |
50%13(England, 2009) 15.38%14(Italy, 2009) 8.1%15(Korea, 2007–08) |
– | Vomiting Diarrhea |
| Astrovirus | Astrovirus | 28–35 nm | +ve sense, single strand RNA | 6800–7900 bases | 24–42%3(Germany, 2002–03 50%12(Singapore 2006–09) |
Diarrhea, Malaise, Nausea, Vomiting, fever | ||||
| Sapovirus | Sapovirus | 27–40 nm | +ve sense, single strand RNA | 7700 bases | – | – | – | – | Gastroenteritis | |
| Orthoreovirus | Orthoreovirus | 70–85 nm | Double strand RNA | 23,500 bases | 3.8 & 3.922 | – | – | Respiratory Tract Disease, Gastroenteritis | ||
| Rotavirus | Rotavirus | 76.5 nm | Double strand RNA | 37,100 bases | 8.022 | 50%9(Germany, 2010) 100%16(Wuhan, 2010–11) |
3–24%3(Germany, 2002–03)0 –9%7(Michigan, 2007) |
4.8%15(Korea, 2007–08) 7.2%20(Stool)(Brazil 2016–17) |
– | Diarrhoeaand Gastroenteritis |
| Adenovirus | Adenovirus | 90–100 nm | Double strand DNA | 26000–48000 bases | 4.522 | 52%1(California, 2000) 37%2(Georgia, 2002) 90%5(Barcelona, 2007–08) 50%9(Germany, 2010) 54%8(Newzealand Sea, 2002–07) 20%11(Chicago, 2009) 100%16(Wuhan, 2010–11) |
20%3(Germany, 2002–03) 24%4(Michigan, 2004) 39–49%7(Michigan, 2007) 35.7%12(Singapore 2006–09) |
50%13(England, 2009 3.2%15(Korea, 2007–08) |
Respiratory Tract and Intestinal Tract | |
| Influenza A Virus | H1N1 | 80–120 nm | Single RNA | 13,500 bases | 6.5–7.022 | 40%18(Netherland, 2009–10) | 10%18(Influent)(Netherland, 200,910) | |||
| Coronavirus/Coronaviridae | SARS | Davg = 78 nm | +ve sense single strand RNA | 30,000 | 6.2423 | – | – | Present21(CNK/tap) | 50%19(stool) | Respiratory Problems, Severe pneumonia, Gastroenteritis |
| MERS | – | +ve sense single strand RNA | – | – | 103gc/c19(stool) | |||||
| Human | – | +ve sense single strand RNA | 27,500–30,700 | – | – | Present21(CNK/tap) | 2.3%19(stool) | Bronchitis, Acute Pneumonia, Respiratory Issues | ||
| Feline | – | – | – | – | – | Present21(CNK/Tap) | 7.2%20(BCoV, Calves stool)(Brazil, 2016–17) | Intestinal effects | ||
| TGEV(Transmissible Gastroenteritis Virus) | 100–150 nm | +ve sense single strand RNA | 28,600 | – | – | – | – | Epidemic Murine Illness | ||
| MHV (Murine Hepatitis Virus) | – | – | 31,357 | – | – | – | – | Diarrhea | ||
| Flavirvirus | Zika | 50 nm | +ve sense single strand RNA | 11,000 | – | – | – | – | 49%19(septic Tanks) | Zika |
| Dengue | 50 nm | +ve sense single strand RNA | 11,000 | – | – | – | – | 50%19(urine) | Dengue | |
| West Nile | 40–50 nm | +ve sense single strand RNA | 11,000 | – | – | – | – | 44%19(urine) | West nile Disease | |
Resource: Gibson, 2014, ‘–’ means ‘Not Available’,CNK-Concentration Not Known1Jiang and Chu, 2004, 2Fong and Lipp, 2005, 3Pusch et al., 2005, 4Xagoraraki et al., 2007, 5Albinana-Gimenez et al., 2009, 6Abdelzaher et al., 2010, 7Wong et al., 2009, 8Dong et al., 2010, 9Jurzik et al., 2010, 10Borchardt et al., 2007, 11Dorevitch et al., 2011, 12Aw and Gin, 2011, 13Charles et al., 2009, 14Gabrieli et al., 2009, 15Park et al., 2010, 16Ye et al., 2012, 17Maunula et al., 2012, 18Heijnen and Medema, 2011, 19Ye, 2018, 20Cruvinel et al., 2020, 21Kitajima et al., 2020. Non Enveloped (Enterovirus, Hepevirus, Norovirus, Rotavirus, Orthovirus, Sapovirus, Astrovirus, Adenovirus) and Enveloped (Influenza A Virus, Coronavirus, Flavivirus), 22Michen and Graule, 2010, 23Kumar, 2020.
Structural similarities between SARS-CoV and SARS-CoV-2 suggest the potential to use previously developed environmental data for SARS-CoV and its surrogates to inform the environmental fate of SARS-CoV-2 while environmental persistence data for SARS-CoV-2 is developed. Casanova et al. (2009) analyzed the survivability of surrogate coronaviruses in water and sewage. The SARS-CoV persistence in sewage ranges between 2 to 14 days depending upon the temperature, and the RNA of these viruses may survive for a longer period of time (Wang et al., 2005). Both SARS-CoV and SARS-CoV-2 RNA have been identified in feces (106 gc (genome copies)/swab) and rectal swabs (105 gc/swab) (Hung et al., 2004, Woelfel et al., 2020), suggesting the potential of environmental fecal contamination from these viruses. . The study by (Wu et al., 2020a) states that the 5% of total fecal samples of infected people in treatment facility catchment tested positive for SARS-CoV-2. The viral genome which has been detected in the fecal material ranges from 600,000 viral genomes per mL (Zhang et al., 2020) to 30,000,000 per mL (Wölfel et al., 2020) in fecal sample from infected individuals. Animal coronaviruses remain infectious for up to a year in water and wastewater, depending on temperature conditions (Mullis et al., 2012). Some studies reported that at around −60 °C some CoVs (coronaviruses) can survive for years, keeping intact its infectious nature (Andries et al., 1978) (McIntosh et al., 1974). Enteric viruses have a size ranging between 25 and 100 nm, potentially allowing them to infiltrate into aquifers more easily than larger bacteria or protozoa (Borchardt et al., 2003). SARS-CoV-2 is elliptical and pleomorphic in shape, with diameters between 60 and 140 nm (Cascella et al., 2020), can thus affect the mobility of the virus. Enteric viruses that have been detected to date in groundwater include poliovirus, echovirus, coxsackievirus, norovirus, rotavirus, reovirus, adenovirus, and hepatitis A virus (Table 1). The viruses mentioned above do have a strong potential to contaminate a wide range of environmental components, including the water cycle, as they are tolerant to hostile environments and may survive a range of water treatments (Grassi et al., 2010). However, the research on the fate and survival of the enveloped virus in groundwater have not yet been reported.
3. Propensity of Migration – I: Sources (of SARS-CoV-2) to the aquatic environment
The propensity of any contaminant migration depends upon source and pathways. The primary source of enteric viruses into the aquatic environment is the release of the partially treated effluents to the surface waters. A recent study identified RNAs in treated wastewater with no detectable bacterial indicators (Wu et al., 2020b), and 5 out of 274 samples of reclaimed water from wastewater in Arizona, tested by ICC/PCR, were found positive for poliovirus (Reynolds, 2000). Several researchers have confirmed the occurrence of SARS-CoV-2 RNA in wastewater samples from different parts of the world (Ahmed et al., 2020, Medema et al., 2020, Lodder and de Roda Husman, 2020, Haramoto et al., 2020, Sherchan et al., 2020; Nemudryi et al., 2020; Wurtzer et al., 2020). The study focused on collecting the wastewater effluent from a health facility WWTPs (wastewater treatment plants) (02). There was a pre-treatment of the sewage samples before the analysis. The samples were pasteurized at 60 °C for 90 min in order to inactivate the virus. The samples were also filtered through 0.2 μm membrane filter in order to remove bacterial cells and debris. The quantification was done with RT-qPCR followed by direct DNA sequencing method. The samples were collected from an urban WWTP in Massachusetts suggesting the population residing in an urban setting. Most of these studies are focused on detection techniques, and wastewater-based epidemiology; however, these detections raise questions regarding potential infectious viral presence and transmission via the water environment. Another important sources of viral contamination to the water environment is the leakage from sewage pipes. Other sources of contamination include the recreational bathing activities, leakages from septic tanks, and cesspits and inadequately treated municipal wastes (Reynolds, 2000, Kumar et al., 2019, Kumar et al., 2020a). The main virus detection sites noted in this study were heavily disinfected tertiary effluents; 10-m deep monitoring wells and other reclaimed water facilities.
4. Propensity of Migration – II: Integration of probable pathways
We have integrated modern water and wastewater management technologies and traditional hydrology to prepare a probable holistic pathway for SARS-CoV-2 transmission in the subsurface system (Fig. 1 ). The fate of SARS-CoV-2 in urban water cycle and its effect on human health has yet to be clearly defined (Naddeo and Liu, 2020); however, viral transmission via sewage is at least plausible. For example, during the SARS outbreak in 2003 in Hong Kong, infections were documented when contaminated water from a leaking sewage pipe was transformed into aerosols (Hung, 2003). In many places of the globe, groundwater storage in urban water settings has become increasingly vulnerable to influence by sanitary sewers, sewage treatment plants, and leakages from pipelines, latrines, dumpsites, crematories, and landfills (Marsalek et al., 2008). Despite removing suspended solids, organic carbon and nutrients from wastewaters, a significant fraction of virus may survive the treatment and may infiltrate through the soil and reach the aquifers, due to high infiltration rates, which may be between 30 and 110 m/yr (Marsalek et al., 2008). For irrigation and potable purpose, a 6-log reduction and 12-log reduction is suggested normally (Gerba et al., 2017).
Fig. 1.
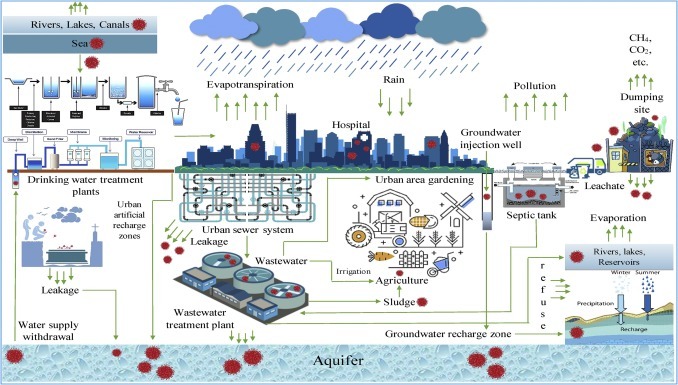
Components of urban water cycle and probable pathway of the novel coronavirus in water environment.
Some of the paramount processes that govern the transport of virions in the aquifer include (i) adsorption to soil particles, which affects the virus survival and its transport; (ii) aggregation among pathogens making them more resistant to external stressors; (iii) temperature, which governs the inactivation rate of the viruses; (iv) microbial activity, which contributes to viral inactivation due to extracellular enzymatic activity; (v) moisture content: more active in saturated soil; (vi) pH: affects viral adhesion to different surfaces and inactivation rates; (vii) dissolved salts, which affects the activity and mobility of viruses; (viii) organic content; (ix) hydrogeological properties: gradient affects transport; (x) virus structure, including viral diameter, genome content, and whether the virus is enveloped (Berg, 1972, Bixby and O’Brien, 1979, Drewry and Eliassen, 1968, Gerba and Bitton, 1984, Nestor and Costin, 1971, Yates et al., 1985).
Enveloped viruses, including SARS-CoV-2, differ from non-enveloped viruses due to the presence of an additional lipid membrane surrounding its capsid protein (Kumar et al., 2020c; Kumar et al., 2020d). The presence of several functional groups and spike protein most likely impact viral partitioning and, subsequently, viral fate and transport in the soil. Even though lipid layers are generally more susceptible to detergents and other organic solvents, two modeled enveloped viruses (MHV and ϕ6) have shown greater adsorption potential to negatively charged solid fractions compared to two non-enveloped bacteriophages (MS2 and T3 with isoelectric point (IP) < 6) (Ye et al., 2016). In the case of non-enveloped viruses, electrostatic force of interactions and hydrophobic effects between the viral capsid proteins and the sorbent surfaces affects the rate of adsorption. However, van der Waals force of attraction and steric interactions plays an insignificant role (Armanious et al., 2016). Such observation highlights the possibility of a strong association of SARS-CoV-2 due to its amphoteric nature with negatively charged solid fraction when sufficient equilibrium time is provided. Thus, the migration of this virus is expected to be slow compared to non-enveloped viruses. However, with an increase in the flow rate, an increase in the viral transport was observed in saturated soils compared to unsaturated ones under anoxic condition (Funderburg et al., 1981, Jin et al., 2000, Williamson et al., 2005, Betancourt et al., 2019). Under saturated soil conditions, the interstitial spaces filled with water, which allows faster transport of viruses without coming in contact with the soil particle. Under the unsaturated condition, the virus possibly gets adsorbed onto soil restricting its movement through the soil. However, increased rainfall and reduction in soil ionic strength can cause the virus to desorb and aid in its migration in the subsurface system (Yates et al., 1987).
Instead of filtration, it is believed that adsorption to the soil is the major removal mechanism for viruses in aquifers. However, smaller size of the virus compared to the soil pores prevent them from being trapped in the pore space. Several studies have highlighted the importance of the texture of the soil, which can significantly attenuate the virus. For example, fine-grained soil has showed greater potential in virus retention compared to coarse-grained soil. Similarly, soil with a higher fraction of clay content showed higher adsorption capacity, which may prevent the transport of viruses to groundwater. Sandy soil coated with a higher concentration of aluminum oxide with a high electrostatic force of attraction showed greater potential for φX174, MS2, and Aichi virus adsorption compared to sandy soil with a higher concentration of goethite or sandy soil alone (Attinti et al., 2010). Presence of preferential flow in the structured clay loam soil enhanced the movement of water and viruses (Sobsey et al., 1995, Morales et al., 2014). Enveloped coronavirus with nominal diameter 50–200 nm (Chen et al., 2020) is relatively larger than most human enteric viruses, and its isoelectric point-pI (∼6.67) is relatively low compared to IP of the enteric viruses (4.5–7.5) (Betancourt et al., 2019). Hence, the transport of SARS-CoV-2 through the soil may be possible though interstitial column velocity. Alignment of the virion long axis with the effective vertical flow field may result in preferential flow paths, which may facilitate the transport of virus in the soil, as reported in other studies (Betancourt et al., 2019). Additionally, hydrophobic interaction leading to an aggregation of enveloped viruses with small-sized negatively charged soil or sediment particle (small hetero-aggregates) may enhance the transport of virus in the porous medium as has been observed for bacteriophage φ6, (a model enveloped virus) in sediments of the colloidal size range in the presence of goethite, montmorillonite, illite, and kaolinite (Katz et al., 2018). Even with the fragile nature of an enveloped virus, a large number of virions have sustained their viability, indicating the risk of infection through the groundwater.
The most common transport pathways for pathogenic viruses in groundwater are through the fractures in the aquifers. Well-bores and imperfection in well casing are other issues facilitating the movement of viruses into the aquifers. Other sources could be abandoned tube wells and bore wells, which are not functioning anymore but are directly linked to the aquifers. Recharge zones, where surface waters are introduced to restore aquifer volume, also play critical roles in the probable introduction of viruses to the aquifers. Due to the smaller size of viruses and their capability to survive in extreme conditions, viral contamination often is transported further and longer than bacteria and protozoans in groundwater. Dowd et al. (1998) conducted the batch column test for bacteriophage, where virus transport in soil (95% sand, 3% silt and 2% clay) columns were studied with the help of mathematical modeling equations. The factors which were involved in the governing equations were dispersion coefficient, porosity, specific discharge, decay rate, mast concentration, time, and rate coefficients/retardation factor of the pathogen release in soil. For smaller viruses, isoelectric point was the major controlling factor, while for larger ones, size/lipid content was the controlling factor, as may be in the case of CoV.
5. Propensity of Migration – III: Similar instances of CoV presence in other compartments
There have been instances where CoVs have been detected in the various compartments of the urban water cycle. The first instance being reported by Annalaura et al. (2020) is the study by Derbyshire and Brown (1978) where CoVs were detected in the cattle slurry and groundwater with primary cell culture method. The second major study was by Wang et al. (2005), where the detection of SARS-CoV was carried out different type of water samples including hospital wastewater, by RT-PCR. Here, the hospital wastewater if remain inadequately treated may end up contaminating the natural water bodies, if the environmental condition favors.
6. Repercussions: Secondary outbreak and treatment prospects
SARS-CoV-2 has already infected millions of people globally and with a high mortality rate, a rise in burials of the deceased have been taken place. The amount of sewage of infected people from hospitals, nursing homes, and quarantine canters since the beginning of 2020 is significant. The enteric involvement of the virus is yet to be determined as the fecal-oral mode of transmission is yet to be definitively demonstrated (Yeo et al., 2020; Gu et al., 2020). Even though the concentration of viral RNA of SARS-CoV-2 is lower in fecal samples compared with that of enteric viruses, the documented transport of enteric viruses to the aquatic environment, including aquifers, suggests that the potential transport to water environments should be fully evaluated in terms of risk management strategies of contaminated water. This threat is especially pronounced in developing countries in the tropics, with high rainfall, sewage leakage and overflow, and limited wastewater treatment facilities. While in more developed water management systems, the presence of multiple water protection barriers will be protective of SARS-CoV-2 release, and the situation may be more challenging in locations lacking adequate sewage treatment systems.
Historically, wastewater disinfection has been used as the standard for pathogen removal. Other advanced methods, for example, the use of membrane bioreactors (MBRs) may provide enhanced viral removal compared to conventional systems (Hai et al., 2014). Ozonation is also effective for viral removal (Wang et al., 2018). Decentralized wastewater treatment plants with advancements such as UV-based advanced oxidation processes, might play an important role to limit high-consequence virus release in outbreak response. Despite the uncertainty surrounding the potential for fecal-oral transmission of SARS-CoV-2, it is important to manage adequately the wastes containing feces and the wastewater out of infected patients, as well as other potential viral sources to the water system (e.g., the dead bodies of infected casualties), to protect against further transmission.
7. Conclusions
Two major aspects, such as survival and migration, control the virus fate in the aquatic environment. In general, a virus with longer persistence has a high capability of causing infection once it reaches the aquatic environment.
-
•
Occurrence of viruses in the urban water cycle has been documented in the past few decades and both enveloped and non-enveloped viruses are reported in the ambient water i.e. wastewater, lake, rivers and even in groundwater.
-
•
Enveloped viruses like SARS-CoV-2 may have significant mobility in subsurface systems; however, limited data is available to assess this and additional research is necessary to evaluate its persistence and transport in such a system.
-
•
Main transport mechanism and pathways affecting the drinking water source will be surface-groundwater interaction in hyporheic zones and leaching infiltration, and several factors like pH, ionic strength, and soil properties will be affecting the adsorption of viruses onto the subsurface aquifer systems.
-
•
Beyond the COVID-19 pandemic, there may be a possibility of the emergence of other novel viruses in the future; thus, our water management strategies should include further considerations to protect against the introduction of viruses, including emerging viruses, into the water system.
Conflicts of interest
The authors declare no competing financial interest.
References
- Abdelzaher A.M., Wright M.E., Ortega C., Solo-Gabriele H.M., Miller G., Elmir S., Newman X., Shih P., Bonilla J.A., Bonilla T.D., Palmer C.J. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 2010;76(3):724–732. doi: 10.1128/AEM.02127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020:138764. doi: 10.1016/J.SCITOTENV.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albinana-Gimenez N., Miagostovich M.P., Calgua B., Huguet J.M., Matia L., Girones R. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009;43(7):2011–2019. doi: 10.1016/j.watres.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Andries K., Pensaert M., Callebaut P. Pathogenicity of hemagglutinating encephalomyelitis (vomiting and wasting disease) virus of pigs, using different routes of inoculation. Zentralbl. Veterinarmed. B. 1978;25(6):461–468. doi: 10.1111/j.1439-0450.1978.tb00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annalaura C., Ileana F., Dasheng L., Marco V. Making waves: Coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020:115907. doi: 10.1016/j.watres.2020.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanious A., Aeppli M., Jacak R., Refardt D., Sigstam T., Kohn T., Sander M. Viruses at solid–water interfaces: a systematic assessment of interactions driving adsorption. Environ. Sci. Technol. 2016;50(2):732–743. doi: 10.1021/acs.est.5b04644. [DOI] [PubMed] [Google Scholar]
- Attinti R., Wei J., Kniel K., Sims J.T., Jin Y. Virus’(MS2, ϕX174, and Aichi) attachment on sand measured by atomic force microscopy and their transport through sand columns. Environ. Sci. Technol. 2010;44(7):2426–2432. doi: 10.1021/es903221p. [DOI] [PubMed] [Google Scholar]
- Aw T.G., Gin K.H. Prevalence and genetic diversity of waterborne pathogenic viruses in surface waters of tropical urban catchments. J. Appl. Microbiol. 2011;110(4):903–914. doi: 10.1111/j.1365-2672.2011.04947.x. [DOI] [PubMed] [Google Scholar]
- Berg G. 1972. Reassessment of the Virus Problem in Sewage and in Surface and Renovated Waters. [Google Scholar]
- Betancourt W.Q., Schijven J., Regnery J., Wing A., Morrison C.M., Drewes J.E., Gerba C.P. Variable non-linear removal of viruses during transport through a saturated soil column. J. Contam. Hydrol. 2019;223:p.103479. doi: 10.1016/j.jconhyd.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Bibby K., Viau E., Peccia J. Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Letters Appl. Microbiol. 2011;52(4):386–392. doi: 10.1111/j.1472-765X.2011.03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47(4):1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby K., Aquino De Carvalho N., Wigginton K. Research needs for wastewater handling in virus outbreak response. Environ. Sci. Technol. 2017 doi: 10.1021/acs.est.6b06492. [DOI] [PubMed] [Google Scholar]
- Bibby K., Fischer R.J., Casson L.W., Stachler E., Haas C.N., Munster V.J. Persistence of Ebola virus in sterilized wastewater. Environ. Sci. Technol. Lett. 2015;2(9):245–249. doi: 10.1021/acs.estlett.5b00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby R.L., O’Brien D.J. Influence of fulvic acid on bacteriophage adsorption and complexation in soil. Appl. Environ. Microbiol. 1979;38(5):840–845. doi: 10.1128/aem.38.5.840-845.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M.A., Bertz P.D., Spencer S.K., Battigelli D.A. Incidence of enteric viruses in groundwater from household wells in Wisconsin. Appl. Environ. Microbiol. 2003;69(2):1172–1180. doi: 10.1128/AEM.69.2.1172-1180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt M.A., Bradbury K.R., Gotkowitz M.B., Cherry J.A., Parker B.L. Human enteric viruses in groundwater from a confined bedrock aquifer. Environ. Sci. Technol. 2007;41(18):6606–6612. doi: 10.1021/es071110+. [DOI] [PubMed] [Google Scholar]
- Brainard J., Pond K., Hunter P.R. Censored regression modeling to predict virus inactivation in wastewaters. Environ. Sci. Technol. 2017;51(3):1795–1801. doi: 10.1021/acs.est.6b05190. [DOI] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls [Internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- Charles K.J., Shore J., Sellwood J., Laverick M., Hart A., Pedley S. Assessment of the stability of human viruses and coliphage in groundwater by PCR and infectivity methods. J. Appl. Microbiol. 2009;106(6):1827–1837. doi: 10.1111/j.1365-2672.2009.04150.x. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruvinel L.B., Ayres H., Zapa D.M.B., Nicaretta J.E., Couto L.F.M., Heller L.M., Bastos T.S.A., Cruz B.C., Soares V.E., Teixeira W.F., de Oliveira J.S. Prevalence and risk factors for agents causing diarrhea (Coronavirus, Rotavirus, Cryptosporidium spp., Eimeria spp., and nematodes helminthes) according to age in dairy calves from Brazil. Trop. Anim. Health Prod. 2020;52(2):777–791. doi: 10.1007/s11250-019-02069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire J.B., Brown E.G. Isolation of animal viruses from farm livestock waste, soil and water. Epidemiol. Infect. 1978;81(2):295–302. doi: 10.1017/s0022172400025134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Kim J., Lewis G.D. Evaluation of methodology for detection of human adenoviruses in wastewater, drinking water, stream water and recreational waters. J. Appl. Microbiol. 2010;108(3):800–809. doi: 10.1111/j.1365-2672.2009.04477.x. [DOI] [PubMed] [Google Scholar]
- Dorevitch S., Panthi S., Huang Y., Li H., Michalek A.M., Pratap P., Wroblewski M., Liu L., Scheff P.A., Li A. Water ingestion during water recreation. Water Res. 2011;45:2020–2028. doi: 10.1016/j.watres.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Drewry W.A., Eliassen R. Virus movement in groundwater. J. (Water Pollut. Control Federation) 1968:R257–R271. [PubMed] [Google Scholar]
- Dublineau A., Batejat C., Pinon A., Burguiere A.M., Manuguerra J.C. Persistence of the 2009 pandemic influenza A (H1N1) virus in water and on non-porous surface. PLoS ONE. 2011;6(11) doi: 10.1371/journal.pone.0028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S.E., Pillai S.D., Wang S., Corapcioglu M.Y. Delineating the specific influence of virus isoelectric point and size on virus adsorption and transport through sandy soils. Appl. Environ. Microbiol. 1998;64(2):405–410. doi: 10.1128/aem.64.2.405-410.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69(2):357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburg S.W., Moore B.E., Sagik B.P., Sorber C.A. Viral transport through soil columns under conditions of saturated flow. Water Res. 1981;15(6):703–711. [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli R., Maccari F., Ruta A., Pana A., Divizia M. Norovirus detection in groundwater. Food Environ. Virol. 2009;1(2):92–96. [Google Scholar]
- Gerba C.P., Bitton G. John Wiley and Sons; New York, New York 1984: 1984. Microbial Pollutants: Their Survival and Transport Pattern to Groundwater. Groundwater Pollution Microbiology; pp. 65–88. 3 fig, 4 tab, 123 ref.Gerba, C.P., 1984. Applied and theoretical aspects of virus adsorption to surfaces. In Advances in applied microbiology (Vol. 30, pp. 133–168). Academic Press. [Google Scholar]
- Gerba C.P., Betancourt W.Q., Kitajima M. How much reduction of virus is needed for recycled water: a continuous changing need for assessment? Water Res. 2017;108:25–31. doi: 10.1016/j.watres.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.E. Viral pathogens in water: occurrence, public health impact, and available control strategies. Curr. Opin. Virol. 2014;4:50–57. doi: 10.1016/j.coviro.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi T., Bagordo F., Idolo A., Lugoli F., Gabutti G., De Donno A. Rotavirus detection in environmental water samples by tangential flow ultrafiltration and RT-nested PCR. Environ. Monit. Assess. 2010;164(1–4):199–205. doi: 10.1007/s10661-009-0885-x. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S., Du B. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv. 2020 [Google Scholar]
- Gu J., Han B., Wang J. COVID-19: gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai F.I., Riley T., Shawkat S., Magram S.F., Yamamoto K. Removal of pathogens by membrane bioreactors: a review of the mechanisms, influencing factors and reduction in chemical disinfectant dosing. Water. 2014;6(12):3603–3630. [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health. 2011;9(3):434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Hung L.S. The SARS epidemic in Hong Kong: what lessons have we learned? J. R. Soc. Med. 2003;96(8):374–378. doi: 10.1258/jrsm.96.8.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F.N., Cheng V.C.C., Wu A.K.L., Tang B.S.F., Chan K.H., Chu C.M., Wong M.M.L., Hui W.T., Poon L.L.M., Tse D.M.W., Chan K.S. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 2004;10(9):1550. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby T.V. Public health measures and the reproduction number of SARS-CoV-2. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.7878. [DOI] [PubMed] [Google Scholar]
- Jiang S.C., Chu W. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 2004;97(1):17–28. doi: 10.1111/j.1365-2672.2004.02269.x. [DOI] [PubMed] [Google Scholar]
- Jin Y., Chu Y., Li Y. Virus removal and transport in saturated and unsaturated sand columns. J. Contam. Hydrol. 2000;43(2):111–128. [Google Scholar]
- Jurzik L., Hamza I.A., Puchert W., Überla K., Wilhelm M. Chemical and microbiological parameters as possible indicators for human enteric viruses in surface water. Int. J. Hyg. Environ. Health. 2010;213(3):210–216. doi: 10.1016/j.ijheh.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J. Hosp. Infect. 2020 doi: 10.1016/j.jhin.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Peña S., Alimova A., Gottlieb P., Xu M., Block K.A. Heteroaggregation of an enveloped bacteriophage with colloidal sediments and effect on virus viability. Sci. Total Environ. 2018;637:104–111. doi: 10.1016/j.scitotenv.2018.04.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.D., Barrie M.B., Mesman A.W., Karku S., Quiwa K., Drasher M., Schlough G.W., Dierberg K., Koedoyoma S., Lindan C.P., Jones J.H. Anatomy of a hotspot: chain and seroepidemiology of Ebola virus transmission, Sukudu, Sierra Leone, 2015–16. J. Infect. Dis. 2018;217(8):1214–1221. doi: 10.1093/infdis/jiy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020:139076. doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Ram B., Honda R., Poopipattana C., Canh V.D., Chaminda T., Furumai H. Concurrence of antibiotic resistant bacteria (ARB), viruses, pharmaceuticals and personal care products (PPCPs) in ambient waters of Guwahati, India: urban vulnerability and resilience perspective. Sci. Total Environ. 2019;693:133640. doi: 10.1016/j.scitotenv.2019.133640. [DOI] [PubMed] [Google Scholar]
- Kumar M., Taki K., Gahlot R., Sharma A., Dhangar K. A chronicle of SARS-CoV-2: Part-I-Epidemiology, diagnosis, prognosis, transmission and treatment. Sci. Total Environ. 2020:139278. doi: 10.1016/j.scitotenv.2020.139278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. The first proof of the capability of wastewater surveillance for COVID-19 in India through the detection of the genetic material of SARS-CoV-2. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Dhangar K., Mazumder P., Sonne C., Rinklebe J., Kitajima M. Potential emergence of antiviral-resistant pandemic viruses via environmental drug exposure of animal reservoirs. Environ. Sci. Technol. 2020;54(14):8503–8505. doi: 10.1021/acs.est.0c03105. [DOI] [PubMed] [Google Scholar]
- Kumar M., Mohapatra S., Mazumder P., Singh A., Honda R., Kumari R., Goswami R., Jha P.K., Vithanage M., Kuroda K., Chuxia Making Waves Perspectives of Modeling and Monitoring of SARS-CoV-2 in Aquatic Environment for COVID -19 Pandemic. Curr. Pollut. Reports. 2020:2. doi: 10.1007/s40726-020-00161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. 2020. Drug and Vaccine Design Against Novel Coronavirus (2019-nCoV) Spike Protein Through Computational Approach. Preprints (www.preprints.org)[Internet] [Google Scholar]
- Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N., Yuen K.Y., Sung J.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020 doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunula L., Söderberg K., Vahtera H., Vuorilehto V.P., von Bonsdorff C.H., Valtari M., Laakso T., Lahti K. Presence of human noro-and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J. Water Health. 2012;10(1):87–99. doi: 10.2166/wh.2011.095. [DOI] [PubMed] [Google Scholar]
- Marsalek J., Cisneros B.J., Karamouz M., Malmquist P.A., Goldenfum J.A., Chocat B. vol. 2. CRC Press; 2008. (Urban Water Cycle Processes and Interactions: Urban Water Series-UNESCO-IHP). [Google Scholar]
- McIntosh K., Chao R.K., Krause H.E., Wasil R., Mocega H.E., Mufson M.A. Coronavirus infection in acute lower respiratory tract disease of infants. J. Infect. Dis. 1974;130(5):502–507. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109(2):388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Morales I., Atoyan J.A., Amador J.A., Boving T. Transport of pathogen surrogates in soil treatment units: numerical modeling. Water. 2014;6(4):818–838. [Google Scholar]
- Mullis L., Saif L.J., Zhang Y., Zhang X., Azevedo M.S. Stability of bovine coronavirus on lettuce surfaces under household refrigeration conditions. Food Microbiol. 2012;30(1):180–186. doi: 10.1016/j.fm.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci.: Water Res. Technol. 2020 [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. 2020. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. medRxiv 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerurkar L.S., West F., May M., Madden D.L., Sever J.L. Survival of herpes simplex virus in water specimens collected from hot tubs in spa facilities and on plastic surfaces. J. Am. Med. Assoc. 1983;250(22):3081–3083. [PubMed] [Google Scholar]
- Nestor I., Costin L. The removal of Cox-sackie virus from water by sand obtained from the rapid sand filters of water-plants. J. Hyg. Epidemiol. Microbiol. Immunol. 1971;15(2):129–136. [PubMed] [Google Scholar]
- Park S.H., Kim E.J., Yun T.H., Lee J.H., Kim C.K., Seo Y.H., Oh S.A., Choi S.S., Cho S.J., Kim M.S., Han G.Y. Human enteric viruses in groundwater. Food Environ. Virol. 2010;2(2):69–73. [Google Scholar]
- Peng L., Liu J., Xu W., Luo Q., Chen D., Lei Z., Huang Z., Li X., Deng K., Lin B., Gao Z. SARS-CoV-2 can be detected in urine, blood, anal swabs and oropharyngeal swabs specimens. J. Med. Virol. 2020 doi: 10.1002/jmv.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch D., Oh D.Y., Wolf S., Dumke R., Schröter-Bobsin U., Höhne M., Röske I., Schreier E. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 2005;150(5):929–947. doi: 10.1007/s00705-004-0467-8. [DOI] [PubMed] [Google Scholar]
- Reynolds K.A. Human viruses found in groundwater recharge sites. Water Cond. Purif. 2000;42(2):148–150. [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020:140621. doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobsey M.D., Hall R.M., Hazard R.L. Comparative reductions of hepatitis A virus, enteroviruses and coliphage MS2 in miniature soil columns. Water Sci. Technol. 1995;31(5–6):203. [Google Scholar]
- Wang X.W., Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Xiao W.J., Zhu X.M. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital. J. Virol. Methods. 2005;128(1–2):156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sikora P., Rutgersson C., Lindh M., Brodin T., Björlenius B., Larsson D.G.J., Norder H. Differential removal of human pathogenic viruses from sewage by conventional and ozone treatments. Int. J. Hyg. Environ. Health. 2018;221(3):479–488. doi: 10.1016/j.ijheh.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv. 2020 [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K.E., Radosevich M., Wommack K.E. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 2005;71(6):3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wong M., Kumar L., Jenkins T.M., Xagoraraki I., Phanikumar M.S., Rose J.B. Evaluation of public health risks at recreational beaches in Lake Michigan via detection of enteric viruses and a human-specific bacteriological marker. Water Res. 2009;43(4):1137–1149. doi: 10.1016/j.watres.2008.11.051. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., GU X., Lee W., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Pr C., Thompson J., Alm E. SARSCoV-2 titers in wastewater are higher than expected from clinically confirmed cases. medRxiv. 2020 doi: 10.1128/mSystems.00614-20. 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Greaves J., Arp L., Stone D., Bibby K. Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environ. Int. 2020;136:105452. doi: 10.1016/j.envint.2019.105452. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 2020.04.12.20062679. [Google Scholar]
- Xagoraraki I., Kuo D.H.W., Wong K., Wong M., Rose J.B. Occurrence of human adenoviruses at two recreational beaches of the Great Lakes. Appl. Environ. Microbiol. 2007;73(24):7874–7881. doi: 10.1128/AEM.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates M.V., Gerba C.P., Kelley L.M. Virus persistence in groundwater. Appl. Environ. Microbiol. 1985;49(4):778–781. doi: 10.1128/aem.49.4.778-781.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates M.V., Yates S.R., Wagner J., Gerba C.P. Modeling virus survival and transport in the subsurface. J. Contam. Hydrol. 1987;1(3):329–345. [Google Scholar]
- Ye X.Y., Ming X., Zhang Y.L., Xiao W.Q., Huang X.N., Cao Y.G., Gu K.D. Real-time PCR detection of enteric viruses in source water and treated drinking water in Wuhan, China. Curr. Microbiol. 2012;65(3):244–253. doi: 10.1007/s00284-012-0152-1. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Ye Y. University of Michigan; 2018. The Detection and Fate of Enveloped Viruses in Water Environment. PhD Thesis. [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5(4):335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Gong Y., Meng F., Bi Y., Yang P., Wang F. Virus shedding patterns in nasopharyngeal and fecal specimens of COVID-19 patients. medRxiv. 2020 doi: 10.1007/s11427-020-1783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


