Abstract
New plant breeding techniques (NPBTs) aim to overcome traditional breeding limits for fruit tree species, in order to obtain new varieties with improved organoleptic traits and resistance to biotic and abiotic stress, and to maintain fruit quality achieved over centuries by (clonal) selection. Knowledge on the gene(s) controlling a specific trait is essential for the use of NPBTs, such as genome editing and cisgenesis. In the framework of the international scientific community working on fruit tree species, including citrus, NPBTs have mainly been applied to address pathogen threats. Citrus could take advantage of NPBTs because of its complex species biology (seedlessness, apomixis, high heterozygosity, and long juvenility phase) and aptitude for in vitro manipulation. To our knowledge, genome editing in citrus via transgenesis has successful for induced resistance to Citrus bacterial canker in sweet orange and grapefruit using the resistance gene CsLOB1. In the future, NPBTs will also be used to improve fruit traits, making them healthier. The regeneration of plants following the application of NPBTs is a bottleneck, making it necessary to optimize the efficiency of current protocols. The strengths and weaknesses of using explants from young in vitro plantlets, and from mature plants, will be discussed. Other major issues addressed in this review are related to the requirement for marker-free systems and shortening the long juvenility phase. This review aims to summarize methods and approaches available in the literature that are suitable to citrus, focusing on the principles observed before the use of NPBTs.
Keywords: Citrus, editing, cisgenesis, fruit quality, transformation, regeneration, marker-free vectors, early flowering
Introduction
Citrus belongs to Rutaceae family and is among the most important fruit crops in the world. Citrus fruits represent a source of macro- and micronutrients (Ting, 1980) and of dietary fiber (Marín et al., 2007). They are also rich in antioxidants compounds (Liu et al., 2012), reveal anticancer, and anti-inflammatory properties (Reviewed in Ma et al., 2020), and are effective at reducing the risk of cardiovascular disease, osteoporosis, and type-2 diabetes (Yuan et al., 2003; Tripoli et al., 2007; Benavente-García and Castillo, 2008; Sugiura et al., 2012; Cirmi et al., 2016; Sugiura et al., 2016).
Citrus, like other woody plants, has a long juvenile phase, extensive hybridization and outcrossing, and reduced population structure. Most cultivated citrus species were domesticated from their wild ancestors or underwent crossbreeding. Recently, an increasing number of studies have focused on the domestication of Citrus, helping us to elucidate the origin of cultivated species. This provides a comprehensive resource for how wild resources can contribute to improve the existing varieties (Wu et al., 2014; Wang et al., 2017; Wu et al., 2018; Ahmed et al., 2019).
Traditional breeding is one of the main strategies used to improve agronomic traits. In many Citrus species, several varieties have developed through conventional methods, such as mutagenesis, inter- and intra-specific crosses, and clonal selection (Caruso et al., 2020). The aim is to produce high-quality citrus fruits (in terms of size, sugar and acidity balance, juice yield, and seedlessness), that are healthy and rich in antioxidant compounds, tolerant or resistant to different abiotic and biotic threats, and with high productivity. Conventional citrus breeding is a long-term and expensive process; long time and resources to obtain progenies and to evaluate their traits are needed. In addition, sexual breeding is not always feasible because some cultivars to be used in crosses are incompatible, sterile, or polyembrionic (Talon and Gmitter, 2008). Moreover, in many cases, after breeding, backcrosses are required to recover elite features of the improved cultivar, lengthening even more breeding programs. This process can be also longer in the case of rootstock breeding (25 years and more). Despite their relatively low efficiency, traditional breeding methods so far enabled the release of most of the new varieties and rootstocks in citriculture.
Since the 1990s, new biotechnology techniques including the use of molecular markers, genome mapping, sequencing, and in vitro culture have been applied to breeding, providing efficient alternatives to traditional methods for the improvement of novel varieties. In addition, transgenesis had enabled the release of many commercial varieties and new rootstocks in citriculture (Gentile et al., 2007; La Malfa et al., 2011), in particular, with improved resistance to biotic and abiotic stresses. This has been possible through the development of transformation protocols involving Agrobacterium tumefaciens infection or polyethylene glycol (PEG) mediated DNA uptake process, starting from many source of explants, such as epicotyls and internodes or embryogenic cell suspensions and protoplasts.
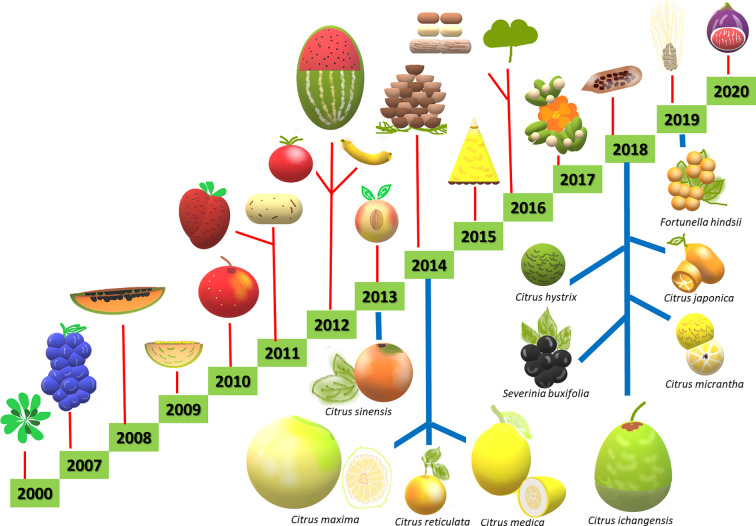
More recently, a number of new techniques has been developed and classified as new plant breeding techniques (NPBTs). These include (I) zinc finger nuclease technology, (II) oligonucleotide directed mutagenesis, (III) cisgenesis, (IV) intragenesis, (V) RNA-dependent DNA methylation, (VI) grafting on genetically modified rootstock, (VII) reverse breeding, (VIII) agro-infiltration, and (IX) synthetic genomics (Lusser et al., 2011). However, the efficiency of NPBTs requires knowledge on the genetic control of horticulturally important traits, which remains limited in citrus compared to other major crops. In the last 20 years, the development of different technologies and sequencing platforms has resulted in the publication of genomes from several horticultural species ( Figure 1 ). Regarding woody plants, to date, one of the main applications of NPBTs is the improvement of agronomic traits related to biotic and abiotic stress resistance ( Figure 2 ). So far, NPBTs are ruled as genetically modified organisms (GMO) according to the GMO 2001/18 legislation, because they are included among the recombinant DNA technologies. In Europe several debates are ongoing in order to establish if the NPBTs methodologies themselves, or their products, could undergo to different protocols for their acceptance, autonomous from GMO regulation.
Figure 1.
Some of the main plant genomes sequenced from years 2000 to 2020, including Citrus and related genera (bottom, indicated with blue lines) and other crops (up, indicated with red lines).
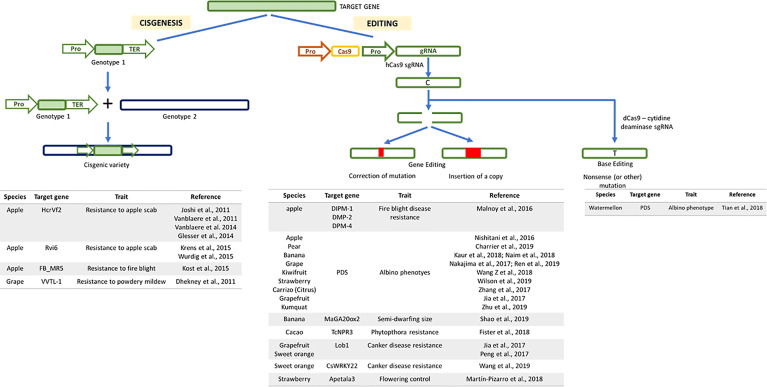
Figure 2.
Genome modifications obtained by the application of new plant breeding techniques approaches illustrated through schematic workflow. In “cisgenesis” the new trait is derived from a sexually compatible species and it is transferred as it is, with its promoter and terminator, to the recipient. The “gene editing” requires the use of a Cas9 to modify the guide RNA (gRNA), specifically designed on the target gene. “Base editing” consists in the action of a chimeric protein composed of a dead or nickase Cas9 with a conserved specific DNA binding capacity and a base modifying enzyme such as adenosine and cytidine deaminase. Below each approach, a list of the main target genes controlling specific traits of several fruit tree crops are reported with corresponding references.
The objective of the present review was to describe the current state of the art and recent advances on the use of NPBTs, in particular regarding genome editing and cisgenesis, on citrus, and other fruit tree species. We highlighted technical, applicative, and legislative aspects of these technologies and the potential advantages and disadvantages of applying NPBTs to improve qualitative traits of fruit, making them healthier due to a higher content of antioxidants.
Current Status of New Plant Breeding Techniques Worldwide
NPBTs provide alternative methods for advancing biotic and abiotic resistance, nutritional quality, and crop performance (Cao et al., 2016; Lassoued et al., 2018); among the others, genome editing and cisgenesis represent two of the most promising strategies to develop genetically improved tree crops. Genome editing involves the production of specific, stable, and inheritable mutations in a precise position of the genome, through DNA repair systems in the cell, with a low probability of inducing undesired errors (off-target effects) without leaving exogenous DNA. Cisgenesis involves the transfer of genes resulting from cross-compatible species. NPBTs represent innovative alternative methods to conventional breeding, with a shortened process (although this is true exclusively for tree fruit and woody plants), and for their precise mechanisms of action. NPBTs produce targeted and minimal modifications to selected genotypes, such as elite cultivars, which are highly valued by consumers for their quality and productivity but that can be even further improved. Unlike traditional breeding, and similarly to transgenesis approaches, NPBTs do not alter the genetic background and this is particularly important for elite cultivars. Advances in in vitro culture, in genome sequencing, and in functional studies have helped to improve the application of molecular breeding techniques in many crops. Many of the methodologies and protocols that make the use of NPBTs in plants feasible, including the availability of engineered plasmids, efficient Agrobacterium-mediated transformation and regeneration protocols from mature plants have been developed for the transgenic approach.
Recently, European and non-European countries, independently or coordinately, have supported the use of NPBTs to improve traits of several crops. GENIUS (2012–2019, https://www6.inra.genius-project_eng/) and BIOTECH (2018–2021, https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/9613) are projects in France and Italy, respectively, which showcase current NPBTs applied to traditional local crop plants. GENIUS addresses the use of genome editing for traits involved in the reduced use of pesticides and in the mitigation of climate change for more sustainable agriculture. The traits actually under evaluation used as proof of concept consist of resistance to biotic and abiotic stresses, flowering time and plant architecture control, plant reproductive mechanisms in several herbaceous, horticultural, ornamental, and fruit species, such as rice, wheat, maize, tomato, potato, oilseed rape, rose, poplar, and apple). BIOTECH is focused on the application of genome editing and cisgenesis to improve fruit qualitative traits, resistance to biotic and abiotic stress, and the architecture of main Mediterranean crops, such as rice, wheat, barley, tomato, eggplant, basil, grape, citrus, peach, apple, and poplar. GENIUS is founded by the French National Research Agency, while BIOTECH is funded by the Italian Agricultural Ministry. Moreover, Russia has also announced a proximal federal program aiming to create 10 new varieties of gene-edited among crops and animals within 2020, and another 20 crops by 2027 (Dobrovidova, 2019). Conversely, a Dutch project (2006–2016, https://www.wur.nl/en/Research-Results/Research-Institutes/plant-research/DuRPh.htm), DuRPh (durable resistance against Phytophthora infestans), aims to create cisgenic potatoes carrying multiple late blight-resistance genes from crossable wild species. In the framework of a European panel, PlantED (2019–2023, https://plantgenomeediting.eu/ CA18111) is the cost action aiming to assess the full innovation potential and impact of plant genome editing, setting the future direction of research priorities, promoting the link between research and innovation in a socially responsible manner, and examining the synergistic interactions between closely related fields. Moreover, also the International Community is working for developing short, medium, and long-term strategies in order to prevent and, eventually, to face the risk related to Huanglongbing (HLB), representing the most devastating citrus disease in the world. The identification of resistance and susceptible genes and the application of NPBTs to produce, in the near proximal future, resistance citrus plants represent only some of the objectives of the Horizon 2020 Project “preHLB”—preventing HLB epidemics for ensuring citrus survival in Europe (2019–2023). Finally, the United States Department of Agriculture (USDA) has financed projects aiming to engineer nanomaterials, or to identify methods to deliver CRISPR-Cas9 vectors and/or ribonucleoproteins (RNPs) to the plant nucleus, aiming to speed up the development of new crop varieties (https://nifa.usda.gov/program/plant-breeding-genetics-genomics-programs).
The regulation of NPBTs has been a topic of worldwide discussion. A critical point of discussion involves two aspects, considering either the used method or the characteristics of the end product. The resolution of critical issues on both aspects enables to process of NPBT or product assimilated to be considered, and, as consequence, ruled as a GMO or not. Eckerstorfer et al. (2019) analyzed regulatory frameworks for GMOs in different countries, and evaluated whether process- or product-addressed NPBT is more helpful for the regulation of NPBT applications. This led to the conclusion that neither system can be regarded as superior. In addition, NPBTs are differentially regulated throughout the world (Eckerstorfer et al., 2019; Friedrichs et al., 2019). In the USA, NPBT crops are exempt from the strict rules and regulations of the USDA (Waltz, 2016), considering genome editing as a method of conventional breeding, only faster. However, the USDA declared that products resulting from genome editing approaches should be considered on a case by case basis (Lusser and Davies, 2013; Voytas and Gao, 2014; Jones, 2015). Conversely, in Europe, the Court of Justice of the European Union (CJEU) (case C-528/16, ruling issued 25th July 2018) declared that NPBTs should be considered GMOs. In this way, NPBTs are subjected to the obligations of Directive 2001/18/EC, whereby GMOs are organisms whose genetic material has been altered and does not derive by natural reproduction and/or recombination.
Examples of the Use of Cisgenesis in Fruit Tree Species
The term cisgenesis was first introduced by Schouten et al., in 2006, who defined it as “the genetic modification of plants using genes that originate only from the species itself or from a species that can be crossed conventionally with this species.” According to the above definition, cisgenesis involves the transfer of a gene (introns included) along with its controlling sequences (promoter and terminator, in the sense direction) from one genotype to another of the same or of a sexually compatible species (Schouten et al., 2006; Lusser and Davies, 2013). Cisgenesis can overcome the major bottleneck of traditional breeding, termed the ‘linkage drag’ (unwanted gene transfer along with the gene of interest), allowing the transfer of the gene of interest without other genetic regions controlling undesirable traits (Jacobsen and Schouten, 2007). Therefore, the gene pool considered by cisgenesis can also be theoretically transferred through classical breeding approaches (Holme et al., 2013). However, cisgenesis has several drawbacks, which limit its wider application. In particular, the casual insertion of the cisgene in the host genome could induce a negative effect (Vanblaere et al., 2014) and potentially interrupt or modify genic or intergenic relevant sequences. The many deposited genomes give information on genes and related annotations that can be used for the cisgenic approach; however, in many cases, the lack of efficient promoters and selectable markers remain the main bottleneck in the application of this technology (Limera et al., 2017). Moreover, the number of gene copies that will be integrated into the host genome may represent an additional drawback, even if, as for transgenesis and intragenesis, any clear correlation has been reported (Jones et al., 1987; Zanek et al., 2007; Zeng et al., 2009; Joshi et al., 2011). To date, few examples of cisgenic plants have been reported, and these are found almost exclusively in apple and grape, and aim to induce resistance to scab (Joshi et al., 2011; Vanblaere et al., 2011; Vanblaere et al., 2014; Gessler et al., 2014) and fire blight in apple (Krens et al., 2015; Würdig et al., 2015), as well as powdery mildew in grape (Dhekney et al., 2011) ( Figure 2 ).
Techniques and Methodologies for Genome Editing
Genome editing technologies represent effective tools for the accurate handling of targeted sequences for crop improvement (Arora and Narula, 2017; Limera et al., 2017). These technologies allow to edit, delete, and replace a specific sequence within a target site of a genome site without introducing extra DNA. These technologies induce double-strand breaks (DSB), then “repaired” through non-homologous end-joining (NHEJ) and homology-directed repair (HDR) processes (Gaj et al., 2013; Voytas, 2013; Fauser et al., 2014; Rinaldo and Ayliffe, 2015). Zinc finger (ZF) and transcription activator-like effector (TALE) represent first engineered nucleases adopted for targeted mutagenesis. These nucleases recognize specific target DNA sequences of one (TALE) or three (ZF) nucleotides, providing a nuclease that disrupts DNA adjacent to the recognition zones (Gaj et al., 2013). CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat-associated protein 9) represents a revolutionary molecular tool that was originally discovered for defense against viral infection (Mojica et al., 2000). This technology works through guide RNAs (gRNAs), characterized by specific sequences designed on the basis of their targets in the genome. The Cas nuclease (commonly indicated as Cas9), when guided by gRNA, produces a DSB adjacent to the gRNA annealing location, permitting target-specific mutagenesis. CRISPR/Cpf1 (another CRISPR/Cas system) is more efficient (Ledford, 2015; Zetsche et al., 2015; Fonfara et al., 2016; Jia et al., 2019), overcoming several Cas9 limitations.
In plants, naturally occurring DNA double-strand breaks occur, and they are rejoined predominantly by NHEJ in absence of foreign donor sequences. This process is prone to errors, determining small mutations (such as frameshift caused by insertions and deletions) in the original sequence and inducing, as a consequence, the loss of function of the target gene and eventually a mutated phenotype. This is similar to the process of induced mutagenesis (by chemical or physical mutagens); however, in the genome editing approach, the induced mutations are not random (as in classical mutagenesis) but limited to specific genes of known function. Moreover, in conventional random mutagenesis, in addition to unwanted mutations, a large-scale screen of mutagenized populations is also needed to detect plants carrying mutations of interest. Conversely, CRISPR/Cas9 can also be associated with specific DNA fragments homologous to the target sequences.
One of the key advantages of CRISPR/Cas9 technology consists in its (relatively) ease of engineering, with modification of only 17–20 bp of the CRISPR RNA (crRNA), part of the guide RNA (gRNA), specificity, and complementary to the target DNA of the gene of interest. Several tools are available to assist with gRNA design, and for the in silico assembly of the construct to be used for plant transformation. Some of the most commonly used tools are Benchling (www.benchling.com), GoldenBraid 4.0 (www.gbcloning.upv.es), CRISPR-P 2.0 (www.crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR), CRISPRdirect (www.crispr.dbcls.jp), Chop-Chop (www.chopchop.cbu.uib.no), E-CRISPR (www.e-crisp.org), CRISPR-GE (www.crdd.osdd.net/servers/crisprge), CRISPR RGEN Tools (http://www.rgenome.net), and CRISPOR (http://www.crispor.tefor.net).
However, the use of CRISPR/Cas9 is limited by the occurrence of off-target mutations, which can generate unintended genetic changes in other regions of the genome. This phenomenon is likely due to the non-specific design of the gRNA and represents a critical limiting factor considering that Cas9 tolerates mismatches between gRNA and target DNA at different positions in a sequence-dependent manner (Hsu et al., 2013). Several tools, such as Cas-OFFinder (Bae et al., 2014) and VARSCOT (Wilson et al., 2019b), assist researchers regarding the location and number of potential off-targets that may hinder the genome editing approach. However, this approach can only be used for species with available genome information. Recently, Hajiahmadi et al. (2019) reported and discussed various methods addressed to reduce such off-targets without the use of external elicitant factors (such as temperature-independent methods), or expensive equipment.
To improve CRISPR/Cas technology and reduce limitations several different Cas, which differ in the position and sequence of the protospacer adjacent motif (PAM) recognition site, have been identified. Among these, Cas9 orthologs from Streptococcus thermophilus (St1Cas9) and Staphylococcus aureus (SaCas9) (Steinert et al., 2015), and Cas12a (formerly named Cpf1), which was identified in Francisella and Pretovella bacteria (Zetsche et al., 2015), are the most common.
To date, the main traits modified through gene editing are related to the albino phenotype, which is used as a proof-of-concept for established protocols, since it allows early phenotype screening. The phytoene desaturase (PDS) gene has been modified in many species, such as apple (Nishitani et al., 2016), pear (Charrier et al., 2019), banana (Kaur et al., 2018; Naim et al., 2018), grape (Nakajima et al., 2017; Ren et al., 2019), kiwifruit (Wang et al., 2018), strawberry (Wilson et al., 2019a), and citrus (Jia et al., 2017a; Zhang et al., 2017; Zhu et al., 2019). In addition to the albino trait, gene editing has been used to induce resistance to important pathogens for some fruit tree species ( Figure 2 ). Erwinia amylovora is the causative agent of fire blight, an invasive disease that threatens apple and several commercial and ornamental Rosaceae. Editing the DPM-1, DPM-2-, and DPM-4 genes, which are disease-specific genes that provide susceptibility to E. amylovora, allowed the release of apple resistant genotypes to the disease (Malnoy et al., 2016). In the framework of fungal diseases, editing of cacao non-expressor of pathogenesis-related 3 (NPR3) gene, a negative regulator which represses the immune system of the plant, increased the resistance in planta to Phytophthora tropicalis (Fister et al., 2018). Furthermore, the Cas9 protein can be engineered (Fauser et al., 2014), and with this aim, two different enzymes with modified nuclease activity have been produced: nickase Cas9 (nCas9) and dead Cas9 (dCas9). nCas9 lacks nuclease activity in one between RuvC or HNH domains, retaining the ability to cleave only one strand, producing site-specific nicks (Fauser et al., 2014). dCas9 has lost its nuclease activity due to a point mutation in either the RuvC and HNH domain resulting in a loss of ability to break DNA. In this way, dCas9 exclusively recognizes target DNA without inducing double (or single) strand breaks (Qi et al., 2013). In 2016, a cytidine deaminase was fused with dCas9 and nCas9 to produce a cytosine base editor (CBE) that changes C:G to T:A (Hua et al., 2019). Similarly, Gaudelli et al., 2018 reported adenine base editors (ABEs) converting A.T to G.C. In this way, the mutated Cas-base editors produce single mutations in the target DNA specifically and precisely (base editing). Base editing has been adopted in plant species ( Figure 2 ), such as watermelon (Li et al., 2017; Lu and Zhu, 2017; Ren et al., 2018; Tian et al., 2018), to produce transgene-free homozygous als (acetolactate synthase) mutant plants, resistant to the herbicide tribenuron (Tian et al., 2018). In addition to base editing, other applications of CRISPR/dCas9 in plants have been recently reviewed, including gene induction and suppression, epigenetic variations, and DNA-free modifications (Moradpour and Abdulah, 2020). More recently a highly versatile and accurate method has been developed, determining the presence of new genetic information in a specific DNA site. This approach is based on the use of nCas9 merged with a reverse transcriptase, engineered with a prime editing guide RNA (pegRNA). Both nCas9 and pegRNA specify the target site and encode the desired edit, inducing lower levels of off-target editing compared with Cas9 nuclease (Anzalone et al., 2019). To our knowledge, prime editing has been applied on annual crops, resulting in 21.8% prime-edited regenerated plants (Lin et al., 2020).
Contribution of Horizontal Gene Transfer
Mutation mapping approaches have shown how similar mutations in orthologue genes among species that are not genetically related, exert similar effects on the phenotype. For example: i) the PETALOSA gene, when mutated in the microRNA172 target site, induces a dominant double flower phenotype in peach, rose, and carnation, and the phenotype has been already reproduced in tobacco by genome editing (Gattolin et al., 2020); ii) the ALS gene is the target gene of one of the most widely used herbicide classes, which inhibits the ALS enzyme. Many species developed independent resistance induced by a point mutation in the ALS sequence, with minimal effect on the catalytic function of the enzyme (Tranel and Wright, 2002). This knowledge allowed to replicate a point mutation in the ALS gene, by base editing, to produce herbicide-resistance crops (Shimatani et al., 2017); iii) finally, attention has been paid to the Terminal Flower gene family (FT/FTL), whose function is to guide the transition from vegetative to floral meristem and is preserved in many species (Pillitteri et al., 2004). Horizontal gene know-how transfer in fruit tree species has resulted in the production of marker-free vectors and in the reduction of juvenility based on ALS resistance and FT/FTL manipulation, respectively, as discussed in the next paragraphs.
Marker-Free Vectors and the Development of New Reporter Genes
To be scientifically useful and relevant, modifications induced through NPBTs should be permanent in the plant. The main limitation in the use of stable transformants is the presence of selectable marker genes (SMGs), which can confer resistance to a toxin (in the case of antibiotics) or can metabolize specific products required by cells for growth. The use of marker genes for resistance to antibiotics (i.e., nptII) or herbicides is not accepted by the public, and, consequently, since December 31 2004, the European Food Safety Authority has forbidden the use of resistance markers antibiotics in Europe (Directive 2001/18/EC), in contrast to the USA and other countries. In annual crops “host” genes, deriving from a genome editing approach, can be discarded using traditional crosses; on the contrary this method is rather impossible in perennial species, due to the long time period needed, and the cross itself will not maintain the identity of the clonally propagated variety (Song et al., 2019a).
To overcome the limits associated with GMOs, several strategies have been tried to eliminate marker genes, consisting in the use of marker-free constructs and recombination systems to excise SMGs (Krens et al., 2015). In the first case, a gene editing can be performed using transient transformation without stable integration of the CRISPR/Cas9 components (Chen et al., 2018), although identification of targeted mutant cells is challenging due to a lack of SMGs for transformed plant cells (Song et al., 2019a). In these cases, it is essential to produce a large number of regenerants so to make it possible a large-scale selection; however, in most of fruit tree species the lack of efficient protocols represents a limiting factor (Rugini et al., 2020). In the second approach the marker gene, bordered by two recognition sequences for a recombinase, can be chemically (using dexamethasone) or physically (by heat-shock) activated, allowing the excision of SMGs. Several recombination loci bordering the unwanted sequences are available: cre-lox (Dale and Ow, 1991), R/Rs (Schaart et al., 2004), and FLP/FRT (Lyznik et al., 1993). Recombination/excision systems have been shown to effectively produce SMG-free apple (Kost et al., 2015; Krens et al., 2015), apricot (Petri et al., 2012), and citrus (Zou et al., 2013). Recently, Pompili et al. (2020) reported the use, in apple, of a CRISPR/Cas9-FLP/FRT gene editing system aimed at the production of regenerated plants with low content of residues of exogenous DNA.
Another attempt to produce marker-free plants was made in citrus (Ballester et al., 2008) and apricot (López-Noguera et al., 2009) consisting in the use of the multi-auto-transformation (MAT) vector associated with the isopentenyl transferase (ipt) gene for positive selection due to site-specific recombination. This method involves the removal of undesired sequences, resulting in the integrated of useful genes only. In citrus, 65% efficiency was demonstrated for ‘Pineapple’ sweet orange plant; however, the results were not appropriate for the ‘Carrizo’ citrange, due to the genotype specificity of this approach (Ballester et al., 2008).
Rather than removing integrated exogenous DNA, an approach to avoid the stable integration of foreign DNA was recently proposed. Thus, in several annual crops, transgene-free plants were produced directly by inducing point mutations through base editing in the gene of interest and in the ALS gene. The latter confers stable resistance to the herbicide, such that selection made by the selective agent, versus no-edited plants, does not impose any integration of foreign DNA. This approach is limited to traits reproducible by a point mutation (Danilo et al., 2019; Veillet et al., 2019; Zhang et al., 2019).
There has also been progress in the development of reporter genes belonging to the MYB transcription factor (TF) family, specifically involved in the activation of anthocyanin pigmentation in plants (Elomaa et al., 2003). In particular, Kandel et al. (2016) compared VvMybA1 of grapevine with conventional reporter genes GFP and GUS, showing that the MybA1 (used as reporter gene) is appropriate to evaluate the gene expression at cellular level. Similarly, in citrus Dutt et al. (2018b) used the embryo-specific Dc3 gene promoter to drive the VvMybA1, inducing a anthocyanin-color based selection.
State of The Art On Citrus
Applications of New Plant Breeding Techniques in Citrus
Most cultivated citrus species originated from a complex admixture between four basic or primary species: citron (Citrus medica), pummelo (Citrus maxima), mandarin (Citrus reticulata), and Citrus micrantha (Xu et al., 2013; Velasco and Licciardello, 2014; Wu et al., 2014; Curk et al., 2016; Wu et al., 2018). Within the primary and derived species (including sweet orange and lemon), most of the worldwide cultivated citrus varieties are derived from somatic mutations of a single ancestor, which accumulated over time in the different growing areas (Wu et al., 2014; Caruso et al., 2016; Curk et al., 2016). Traditional breeding methods used for genetic improvement of citrus cultivars and create new varieties, or to improve desired traits require a long time and is hampered by many difficulties and restrictions related with plant size and long juvenile phase of citrus, as well as female and/or male sterility, polyembryony, heterozygosity, and parthenocarpy.
The partial availability and knowledge of the genes that control the traits of interest, has slowed recently, and the possibility to explore and make use of NPBTs in citrus is feasible. The genes and markers reported herein refer to well-known and characterized traits, and information on these genes and markers were developed after the availability of the Citrus genomes (Xu et al., 2013; Wu et al., 2014; Curk et al., 2016; Wang et al., 2017; Wu et al., 2018). To date, Ruby (a MYB-like TF) and its promoter 3′LTR, represents one of the main genes responsible for the control of purple pigmentation due to the presence of anthocyanins in citrus fruits (Butelli et al., 2012); CsLOB1 (lateral organ boundaries gene family) and CsWRKY22 are genes associated with susceptibility to citrus bacterial canker (CBC) in sweet orange (Hu et al., 2014; Wang et al., 2019); CitRWP controls polyembryony in sweet orange, grapefruit, lemon, and mandarin (Wang et al., 2017); Noemi represents Myc-like, which along with Ruby, controls anthocyanins pigmentation in Citrus and is also responsible for the acidless trait in citrus fruits (Butelli et al., 2019).
As for other fruit tree species, the application of NPBTs in Citrus has mainly focused on the generation of plants resistant to biotic stresses (varieties and rootstocks). Thus, the successful application of genome editing in citrus was first based on the use of CsLOB1, the gene responsible for susceptibility to CBC (Hu et al., 2014; Duan et al., 2018). CBC is a severe quarantine disease caused by the bacterium Xanthomonas citri pathovar citri (Xcc) and aurantifolii (Xca), which are found globally, except for the Mediterranean basin (Gottwald et al., 2001). Genetic engineering is the best approach to induce resistance to CBC (Zhang et al., 2010). Considering the difficulty to introgress resistance through traditional breeding, a recent method involved the use of NPBTs to intervene in the mechanism of action involved in the host-pathogen interaction. Considering the bacterium, X. citri strains are characterized by specific pathotypes (PthA4, PthA*, PthAw, PthB, and PthC), which are distinguished based on the conservation of repeated variable diresidues (RVDs), encoding transcription activator-like (TAL) effectors. These recognize the corresponding effector binding element (EBE) in the promoter of susceptibility plant genes, such as LOB and Sugar Transport TFs (Hutin et al., 2015). Genome editing of a single EBE allele (type 1) in the promoter of CsLOB1 in ‘Duncan’ grapefruit (Jia et al., 2016) allowed the generation of transgenic lines resistant to a mutated Xcc strain, but susceptible to wild type-Xcc. Mutations in both EBEs alleles (type I and type II) of CsLOB1 in “Duncan” grapefruit and “Wanjincheng” orange, result in reduced symptoms in transformed plants caused by wild-type Xcc infection (Jia et al., 2016; Jia et al., 2017b; Peng et al., 2017). Furthermore, to improve resistance through CRISPR/Cas9 approach on the CsLOB1 promoter (Zhou et al., 2017), homozygous mutants have been generated directly from citrus explants decreasing the susceptibility to CBC knocking out the CsWRKY22 a marker gene for pathogen-triggered immunity in ‘Wanjincheng’ orange (Wang et al., 2019). Moreover, the availability of Cas12a has been successfully used in Citrus. The efficiency of CRISPR/Cas12a has been examined for editing CsPDS in ‘Duncan’ grapefruit via Xcc-facilitated agroinfiltration to modify two alleles of EBEPthA4-CsLOBPs. One of seven transformed ‘Duncan’ plants has been found to contain the highest mutation rate, demonstrating reduced canker susceptibility (Jia et al., 2019). Recently, Zhu et al. (2019) used Fortunella hindsii, a wild Citrus species characterized by a juvenile phase of about 8 months and a dwarf habit, to observe the effects of a successful CRISPR/Cas9 experiment using Agrobacterium-mediated transformation. Two gRNAs were synthesized to edit the PDS gene and five transgenic lines exhibited targeted mutagenesis sites, resulting in a global and mosaic albino phenotype (Zhu et al., 2019). These results suggest that editing or cisgenesis to induce early flowering could be a successful strategy to speed up gene characterization in functional genomic studies, especially for characters related to reproductive biology and to fruits.
Compared to genome editing approaches, there were fewer applications of cisgenesis in citrus. This was due to technical difficulties and limited knowledge on the function of specific genes and on their promoters. The only example in citrus involved the use of the Ruby gene, which is intronless, flanked by the CaMV 35S promoter and terminator (a classical intragenesis experiment), with the aim to produce ‘Mexican’ lime (Citrus aurantifolia) fruits highly enriched in anthocyanins content (Dutt et al., 2016).
It is noteworthy to remark that the application of NPBTs in citrus strictly relies on the availability of a transformation process that needs the use of selectable marker gene, in most of cases nptII gene. So far, no marker-free vectors have been used for cisgenesis and genome editing in citrus and this limits the possibility to apply NPBTs for these species.
Transformation and Regeneration: The Main Bottlenecks of New Plant Breeding Techniques for Citrus Genetic Improvement
Regeneration and transformation represent the main bottlenecks for the use of molecular breeding and NPBTs, also in citrus. Stable or transient Agrobacterium-mediated transformation of juvenile tissues is the preferred approach to introduce foreign DNA, and has been performed on many citrus genotypes. To date, transformation and regeneration protocols are available for: ‘Carrizo’ citrange and ‘Swingle’ citrumelo (Cervera et al., 1998b), two rootstocks that can be considered model plants; ‘Pineapple’ (Peña et al., 1995), ‘Hamlin,’ and ‘Valencia’ sweet oranges (Dutt and Grosser, 2010); ‘Femminello Siracusano’ lemon (Gentile et al., 2007); ‘Duncan’ grapefruit (Jia et al., 2017b); clementine (Cervera et al., 2008), Mexican lime (Dutt et al., 2016); F. hindsii (Zhu et al., 2019). However, owing to the large number of important varieties belonging to different species and hybrids, the lack of specific protocols especially for commercial cultivars remains an issue.
Many factors affect the stable transformation process mediated by A. tumefaciens that usually involves explants pre-incubation, co-cultivation with the engineered bacteria, callus induction, and selection of the regenerated shoots. A balanced composition of plant growth regulators in the regeneration medium is important for inducing the proliferation of transformed cells. In particular, the presence of cytokinin in the callus-induction medium promotes callus formation (Guo et al., 2002; Khan et al., 2009). The regeneration rate is tightly correlated with photoperiod (Moreira-Dias et al., 2000; Marutani-Hert et al., 2012) and growth of non-transformed buds need to be avoided using the appropriate antibiotics (Ballester et al., 2008; Rodríguez et al., 2008). Furthermore, the rate of A. tumefaciens transformation is affected by explant source and type, and conditions by the duration of cocultivation (Dutt and Grosser, 2009). In ‘Tarocco’ orange tissues, a limited explant pre-incubation period with hormone-rich liquid medium was found to increase the transformation efficiency (Peng et al., 2019). Transient gene expression assays, performed via Agrobacterium infiltration in grapefruit leaves, have demonstrated that transient transformation efficiency can be influenced by different factors, such as infiltration medium, Agrobacterium concentration, and leaf development stage (Figueiredo et al., 2011). Li et al. (2017) showed that sweet orange leaves approaching full maturity were at an optimal, in terms of transient protein expression using Xcc-facilitated agroinfiltration (Jia and Wang, 2014).
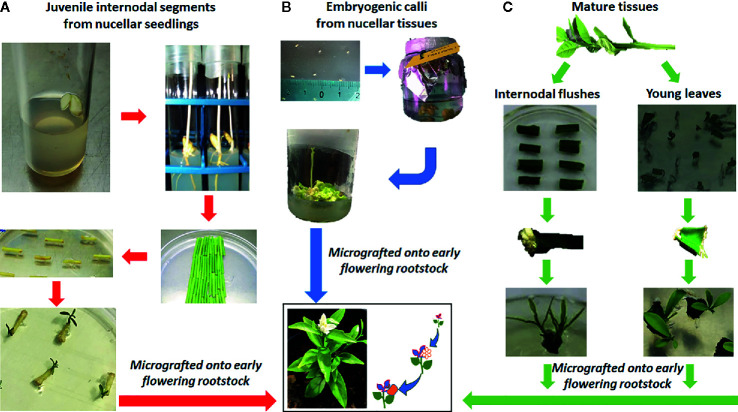
The choice between in-vitro juvenile and mature plants should also be considered, and it has to be approached differently if dealing with seedy or seedless fruits ( Figure 3 ). Moreover, the polyembryonic nature of many citrus varieties enables the transformation of nucellar seedlings, leading to the production of transgenic plants that maintain the genetic background of the mother plant. Young tissues such as epicotyl explants (Peña et al., 1995; Dutt and Grosser, 2009) and leaf segments (Khan et al., 2009; Khan et al., 2012) have been widely used in the transformation mediated by A. tumefaciens, and high percentage of transformation efficiencies were achieved; however, plants regenerated from these tissues usually exhibit the long juvenility and require many years before bearing fruits, delaying the evaluation of horticultural characteristics of the transformed plants.
Figure 3.
Different approaches for Agrobacterium-mediated transformation of citrus varieties starting from different tissue sources. (A) Juvenile internodal segments from nucellar seedlings. (B) Embryogenic callus obtained from unfertilized ovules. (C) Mature tissues, such as internodal segments from flushes and leaves. All the shoots or embryos, obtained from regeneration of transformed explants, are grafted onto vigorous rootstocks in order to induce early flowering, with the final aim to shorten the evaluation of the modified traits.
The use of explants from adult mature plants could overcome these limits through the use of ex vitro invigoration of source plant material, that consists in grafting mature buds onto juvenile rootstocks (Cervera et al., 1998a; Cervera et al., 2008; Orbović et al., 2015); however, the transformation efficiency of adult material, in citrus as well as in other fruit tree species, is several-fold lower than that obtained from juvenile citrus tissues (Almeida et al., 2003; He et al., 2011) genotype-dependent ( Table 1 ) and experiment requires a high number of explants, together with high contamination rate that can complicate the process. Despite different efforts being made, the transformation efficiency of mature internode explants remains very low and does not exceed 6.1% ( Table 1 ), except for ‘Hamlin’ sweet orange that shows 12.8% of transformed shoots (Orbović et al., 2015); the transformation of nodes with removed buds led to a transformation efficiency of 11.7% (Peng et al., 2019) and when thin section are used as explants, reaches 35% in ‘Pera’ sweet orange (Kobayashi et al., 2017). In addition, the different response of each cultivar to different regeneration conditions (Rodríguez et al., 2008) highlights the need to increase the number of citrus regeneration protocols; also in order to successfully apply transgenesis and NPBTs to citrus there is a need to optimize organogenic response of commercial varieties.
Table 1.
List of genotypes, number of explants (internodes) from mature tissues, and transformation efficiency (TE%).
| Genotype (species) | N° internodes cultured | TE% | Reference |
|---|---|---|---|
| Pineapple (Citrus sinensis) | 294 | 6.1 | Cervera et al., 1998a |
| Clementine (C. clementina) | 600 | 3 | Cervera et al., 2008 |
| US-942 | 449 | 3.96 | Marutani-Hert et al., 2012 |
| (C. reticulata Sunki x Poncirus trifoliata Flying Dragon) | |||
| Valencia (C. sinensis) | 1,368 | 3.7 | Fávero et al., 2012 |
| Pera (C. sinensis) | 1,104 | 1.4 | Fávero et al., 2012 |
| Hamlin (C. sinensis) | 942 | 2.5 | Fávero et al., 2012 |
| Etrog (C. medica) | 431 | 1.49 | Marutani-Hert et al., 2012 |
| Ruby Red (C. paradisi) | 484 | 1.05 | Marutani-Hert et al., 2012 |
| Hamlin (C. sinensis) | 530 | 12.8 | Orbović et al., 2015 |
| Pera (C. sinensis) | Not reported | 35 | Kobayashi et al., 2017 |
| Tarocco (C. sinensis) | Not reported | 11.7 | Peng et al., 2019 |
An alternative to Agrobacterium-mediated transformation is the direct transformation of cells from suspension cultures obtained from citrus embryogenic calli derived from unfertilized ovules (Li et al., 2002; Dutt and Grosser, 2010; Omar et al., 2016) and of protoplasts (Guo et al., 2005; Dutt et al., 2018a; Omar et al., 2018).
These alternative strategies can be useful to transform recalcitrant genotypes and difficult-to-transform varieties such as some mandarin and lemon cultivars (Dutt and Grosser, 2010).
For both protoplasts and cells, the regenerated somatic embryos derive from a single cell, so that chimeras are rarely observed; on the other hand, this material requires longer time for the recovery of the regenerated plants and these will display juvenile nature (Dutt et al., 2018a). In citrus, the use of embryogenic cell culture and of protoplast technology has been exploited since 1990; many somatic hybrids have been generated through protoplast fusion (Grosser and Gmitter, 1990; Grosser and Gmitter, 2011; Grosser et al., 1992; Grosser et al., 2000) and transgenic plants have been recovered from transformed protoplasts (Guo et al., 2005; Omar et al., 2007; Dutt et al., 2018b; Omar et al., 2018) and calli (Dutt et al., 2018b). As for the application of NPBTs, citrus protoplasts could be used for a direct delivery of purified CRISPR/Cas9 ribonucleoproteins (RNPs) for efficient targeted mutagenesis, as already performed in grapevine and apple for transient cell transformation (Malnoy et al., 2016).
Inducing Early Flowering as a Strategy to Shorten the Long Juvenility Phase in Citrus
The use of NPBTs in citriculture is hampered by limits in both the transformation and regeneration phases, as well as the long juvenility phase, which characterize citrus as well other fruit tree species (Hanke et al., 2007). Juvenility is adopted to reach faster a critical size to be able to compete in the environment. The photosynthesis products are directed toward the growth of the crown until the plant is not able to support the reproductive efforts (Hackett, 2011). Several approaches have been performed to active the precocious flowering in Citrus. The availability of a large germplasm collection and cross populations represent a source of precious material, which can be useful for individual natural early flowering plants. F. hindsii is a species that is phylogenetically close to the Citrus genus, which is particularly interesting because it produces flowers much earlier (around 8 months) than other common Citrus species (Zhu et al., 2019). In particular, F. hindsii needs around 5 months from seeds to T0 plantlets and about another 10 months to obtain the following T1 generation after the editing in PDS gene, supporting the evidence to be really promising using F. hindsii for functional studies (Zhu et al., 2019).
In the framework of biotechnological uses, it may be possible to reduce the duration of the breeding cycle acting on the endogenous genetic flowering pathways by using inducible promoters that activate transgene expression, in addition to several approaches used to transmit the transgenic stimulus through grafting (van Nocker and Gardiner, 2014). Evidence indicates that the genetic mechanisms of flowering are widely conserved among flowering plants (Albani and Coupland, 2010; Amasino, 2010), although the most promising is the FT/FTL1 family, encoding proteins that act as promoter or repressors of flowering. These proteins are highly similar to phosphatidyl ethanolamine-binding ones. These have been used widely in many woody plants, such as apple (Kotoda et al., 2010), plum (Srinivasan et al., 2012), and olive (Cerezo et al., 2014). In citrus for the first time Peña et al. (2001) used flowering involved genes (APETALA and LEAFY) to induce early flowering and fruiting. Later, Pons et al. (2014) highlighted the usefulness of the overexpression of early flowering genes as a tool to accelerate the evaluation of traits related to fruit quality. Moreover, the effect of grafting on the precious floral induction has been reported, up to now, in apple, where the M9 rootstock (a spontaneous mutant overexpressing FT genes) induces early flowering in juvenile apple scion (Foster et al., 2014). Similar results have been reported in Arabidopsis, tomato, and cassava (Lifschitz et al., 2006; Corbesier et al., 2007; Notaguchi et al., 2008; Silva Souza et al., 2018). In blueberry, the phenomenon was artificially reproduced using an FT transgenic rootstock and a non-transgenic scion (Song et al., 2019b).
Very recently, several examples have shown how the use of genome editing can also address the induction of early flowering, such as in kiwifruit (Varkonyi-Gasic et al., 2019). In tobacco the same approach was used to induce early flowering and precocious anthocyanin pigmentation (Liu et al., 2019). No published data are still available in citrus.
In plants, viral vectors have been used for the suppression of endogenous genes by virus-induced gene silencing (VIGS) and the expression of foreign genes (Senthil-Kumar and Mysore, 2011). An example is a viral vector based on the citrus leaf blotch virus (CLBV) developed by Velázquez et al. (2016). This vector carries the FT gene from Arabidopsis thaliana and Citrus. CLBV is a valid tool, because it does not produce symptoms in most commercial cultivars (Galipienso et al., 2000), it infects only the phloem (Agüero et al., 2013), and it is not transmitted by pests. Moreover, inoculated plants show normal reproductive biology producing flowers, viable pollen, and fruits (Velázquez et al., 2016). The viral vector cannot be integrated into the genome of the inoculated plant so is not transmitted by pollen (Vives et al., 2008). The CLBV vector carrying the FT gene anticipates flowering in young citrus plants at 4 months post-infection and for a period of at least 5 years, depending on the genotype and season. This vector is a biotechnological instrument with tremendous practical applications to speed-up traditional breeding programs and genetic studies.
Shortening the juvenile phase is useful for accelerating the selection process in citrus plants, in which resistance to pathogens is induced through the use of NPBTs. Recently, new sources of resistance to HLB have been identified in Eremocitrus and Microcitrus spp. (Ramadugu et al., 2016). The international citrus community is focused on the identification of specific genes that can be transferred using a cisgenic approach, and on the identification of susceptible genes to be edited. There is a risk of losing the world’s citrus crop if a solution is not promptly identified.
Use of the viral vector in combination with the NPBTs could substantially reduce the time needed to evaluate citrus plants edited for qualitative traits. Very few papers have reported the use of NPBTs to improve fruits or vegetables from a qualitative point of view. The major limitation of this approach is that the antioxidant, nutritional and healthy properties are under the control of several genes, generally multilocus, working cooperatively, producing complex, often under epigenetic control, not easy to be managed. From a genetic point of view, MYBs TFs have been widely reported to control polyphenol and carotenoid biosynthesis, in addition to other quality traits, such as flavor and texture, as reviewed in Allan and Espley (2018). In tomato, the concomitant overexpression of Rosea1 (MYB) together with Delila (bHLH) produced very high levels of anthocyanin and purple fruit (Butelli et al., 2008). These engineered tomatoes were used in a mouse feeding trial and demonstrated a protective effect against cancer progression in Trp53–/– knockout mice (Butelli et al., 2008). Similarly, in apple, MYB10 overexpression induced the large accumulation of anthocyanin in the flesh, and resulted in reduced inflammatory markers in mice (Espley et al., 2014). In Citrus, the slimming effect of pigmented Moro juice in obese mice (Titta et al., 2010) supported similar observations for other fruit, in addition to the fact that the Ruby gene is the MYB TF, which controls anthocyanins pigmentation in citrus fruits (Butelli et al., 2012; Butelli et al., 2017). The use of MYB TFs to enhance the levels of phytochemical levels in fruit, making them healthier, offers the potential to produce new crops that are not genetically modified. In this case, there is a need to observe the final phenotype in term of the quality and quantity of sugar, micro and macronutrients, vitamins, and seeds.
Concluding Remarks
NPBTs are powerful tools used to improve quality traits without modifying the characteristics of elites in citrus, and to protect the world citriculture from the most aggressive and devastating disease, like HLB. Citrus species and relatives do not represent model species for the application of NPBTs due to the complexity of their biology; this is supported by a lack of confirmed data. To date, scientific research has placed scientists in a position to reduce the historical gap compared to other woody plants. Despite this, technical advances made in other woody species have involved the induction of plant regeneration from protoplast cells using the gRNA-Cas9/Cpf1 RNP complex in grapevine (Malnoy et al., 2016) as well as the release of CRISPR/Cas9 RNPs to induce DNA-free targeted mutations in apple and grapevine protoplasts (Osakabe et al., 2018); these data would be also be applied to citrus in order to establish more rapid transformation approaches. In particular, genome editing approaches could display their effectiveness especially for those characters for which the down regulation or the reduction of the expression levels of a single gene could determine a modification of a given trait.
Thus, overcoming genotype specificity and source explant problems, the availability of transformation platforms, and the adoption of new vectors and editing strategies, may provide citrus breeders reliable tools to quickly introgress genes related to fruit quality and other important agronomic traits.
Author Contributions
FS prepared the main parts of the manuscript being the application of new plant breeding techniques in citrus the scope of his PhD project. AC focused on the paragraphs regarding methodologies for base editing, the contribution of horizontal gene transfer and on the induction of early flowering. LP and HP contributed to manuscript elaboration for the transformation and regeneration paragraph. SM and CL coordinated and wrote the manuscript.
Funding
This work has been granted by Italian Ministry of Agriculture Food and Forestry through the project «BIOTECH Biotecnologie sostenibili per l’agricoltura italiana» (DM 15930/7305/2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marco Caruso (CREA - Research Centre for Olive, Fruit and Citrus Crops) for his comments aimed at improving the manuscript. We also would like to thank Editage (www.editage.com) for English language editing.
References
- Agüero J., Vives M. C., Velázquez K., Ruiz-Ruiz S., Juárez J., Navarro L., et al. (2013). Citrus leaf blotch virus invades meristematic regions in Nicotiana benthamiana and citrus . Mol. Plant Pathol. 14, 610–616. 10.1111/mpp.12031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed D., Comte A., Curk F., Costantino G., Luro F., Dereeper A., et al. (2019). Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: A case study in diploid and polyploid citrus. Ann. Bot. 123, 1231–1251. 10.1093/aob/mcz029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani M. C., Coupland G. (2010). “Comparative Analysis of Flowering in Annual and Perennial Plants,” in Current Topics in Developmental Biology. Ed. Timmermans M. C. P. (Cambridge, MA: Elsevier; ), 323–348. 10.1016/S0070-2153(10)91011-9 [DOI] [PubMed] [Google Scholar]
- Allan A. C., Espley R. V. (2018). MYBs Drive Novel Consumer Traits in Fruits and Vegetables. Trends Plant Sci. 23, 693–705. 10.1016/j.tplants.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Almeida W. A. B., Mourão Filho F. A. A., Pino L. E., Boscariol R. L., Rodriguez A. P. M., Mendes B. M. J. (2003). Genetic transformation and plant recovery from mature tissues of Citrus sinensis L. Osbeck. Plant Sci. 164, 203–211. 10.1016/S0168-9452(02)00401-6 [DOI] [Google Scholar]
- Amasino R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. 10.1111/j.1365-313X.2010.04148.x [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora L., Narula A. (2017). Gene editing and crop improvement using CRISPR-cas9 system. Front. Plant Sci. 8:1932. 10.3389/fpls.2017.01932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S., Park J., Kim J.-S. (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. 10.1093/bioinformatics/btu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester A., Cervera M., Peña L. (2008). Evaluation of selection strategies alternative to nptII in genetic transformation of citrus. Plant Cell Rep. 27, 1005–1015. 10.1007/s00299-008-0523-z [DOI] [PubMed] [Google Scholar]
- Benavente-García O., Castillo J. (2008). Update on uses and properties of citrus flavonoids: new findings in anticancer, cardiovascular, and anti-inflammatory activity. J. Agric. Food Chem. 56, 6185–6205. 10.1021/jf8006568 [DOI] [PubMed] [Google Scholar]
- Butelli E., Titta L., Giorgio M., Mock H.-P., Matros A., Peterek S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. 10.1038/nbt.1506 [DOI] [PubMed] [Google Scholar]
- Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. 10.1105/tpc.111.095232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Garcia-Lor A., Licciardello C., Las Casas G., Hill L., Reforgiato Recupero G., et al. (2017). Changes in anthocyanin production during domestication of citrus. Plant Physiol. 173, 2225–2242. 10.1104/pp.16.01701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelli E., Licciardello C., Ramadugu C., Durand-Hulak M., Celant A., Reforgiato Recupero G., et al. (2019). Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr. Biol. 29, 158–164. 10.1016/j.cub.2018.11.040 [DOI] [PubMed] [Google Scholar]
- Cao H. X., Wenqin W., Hien T. T. L., Giang T. H. V. (2016). The power of CRISPR-CAS9-induced genome editing to speed up plant breeding. Int. J. Genomics 2016, 1–10. 10.1155/2016/5078796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso M., Ferlito F., Licciardello C., Allegra M., Strano M. C., Di Silvestro S., et al. (2016). Pomological diversity of the Italian blood orange germplasm. Sci. Hortic. 213, 331–339. 10.1016/j.scienta.2016.10.044 [DOI] [Google Scholar]
- Caruso M., Smith M. W., Froelicher Y., Russo G., Gmitter F. G., Jr (2020). “Traditional breeding,” in The Genus Citrus, 1st edition Eds. Talon M., Caruso M., Gmitter F. G. (Cambridge, UK: Elsevier; ), 129–148. [Google Scholar]
- Cerezo S., Barceló A., Samach A., Mercado J. A., Pliego-Alfaro F. (2014). Over-expression of an FT-homologous gene of Medicago truncatula induces early flowering and a modified branching habit in olive plants Proceedings of the 1st International Rapid Cycle Crop Breeding Conference, 7–9 January 2014, Leesburg, VA, USA [Google Scholar]
- Cervera M., Juárez J., Navarro A., Pina J. A., Durán-Vila N., Navarro L., et al. (1998. a). Genetic transformation and regeneration of mature tissues of woody fruit plants bypassing the juvenile stage. Transgenic Res. 7, 51–59. 10.1023/A:1008855922283 [DOI] [Google Scholar]
- Cervera M., Pina J. A., Juárez J., Navarro L., Peña L. (1998. b). Agrobacterium-mediated transformation of citrange: factors affecting transformation and regeneration. Plant Cell Rep. 18, 271–278. 10.1007/s002990050570 [DOI] [PubMed] [Google Scholar]
- Cervera M., Navarro A., Navarro L., Peña L. (2008). Production of transgenic adult plants from clementine mandarin by enhancing cell competence for transformation and regeneration. Tree Physiol. 28, 55–66. 10.1093/treephys/28.1.55 [DOI] [PubMed] [Google Scholar]
- Charrier A., Vergne E., Dousset N., Richer A., Petiteau A., Chevreau E. (2019). Efficient targeted mutagenesis in apple and first-time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10, 40. 10.3389/fpls.2019.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li W., Katin-Grazzini L., Ding J., Gu X., Li Y., et al. (2018). A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 5, 13. 10.1038/s41438-018-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirmi S., Ferlazzo N., Lombardo G., Maugeri A., Calapai G., Gangemi S., et al. (2016). Chemopreventive agents and inhibitors of cancer hallmarks: may citrus offer new perspectives? Nutrients 8, 698. 10.3390/nu8110698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Curk F., Ollitrault F., Garcia-Lor A., Luro F., Navarro L., Ollitrault P. (2016). Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 117, 565–583. 10.1093/aob/mcw005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale E. C., Ow D. W. (1991). Gene transfer with subsequent removal of the selection gene from the host genome. Proc. Natl. Acad. Sci. 88, 10558–10562. 10.1073/pnas.88.23.10558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilo B., Perrot L., Mara K., Botton E., Nogué F., Mazier M. (2019). Efficient and transgene-free gene targeting using Agrobacterium-mediated delivery of the CRISPR/Cas9 system in tomato. Plant Cell Rep. 38, 459–462. 10.1007/s00299-019-02373-6 [DOI] [PubMed] [Google Scholar]
- Dhekney S. A., Li Z. T., Gray D. J. (2011). Grapevines engineered to express cisgenic Vitis vinifera thaumatin-like protein exhibit fungal disease resistance. Vitr. Cell. Dev. Biol. Plant 47, 458–466. 10.1007/s11627-011-9358-3 [DOI] [Google Scholar]
- Dobrovidova O. (2019). Russia joins in global gene-editing bonanza. Nature 569, 319–320. 10.1038/d41586-019-01519-6 [DOI] [PubMed] [Google Scholar]
- Duan S., Jia H., Pang Z., Teper D., White F., Jones J., et al. (2018). Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 19 (8), 1908–1916. 10.1111/mpp.12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt M., Grosser J. W. (2009). Evaluation of parameters affecting Agrobacterium-mediated transformation of citrus. Plant Cell Tissue Organ. Cult. 98, 331–340. 10.1007/s11240-009-9567-1 [DOI] [Google Scholar]
- Dutt M., Grosser J. W. (2010). An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep. 29, 1251–1260. 10.1007/s00299-010-0910-0 [DOI] [PubMed] [Google Scholar]
- Dutt M., Stanton D., Grosser J. W. (2016). Ornacitrus: Development of genetically modified anthocyanin-expressing citrus with both ornamental and fresh fruit potential. J. Am. Soc Hortic. Sci. 14, 54–61. 10.21273/JASHS.141.1.54 [DOI] [Google Scholar]
- Dutt M., Erpen L., Grosser J. W. (2018. a). Genetic transformation of the ‘W Murcott’ tangor: comparison between different techniques. Sci. Hortic. 242, 90–94. 10.1016/j.scienta.2018.07.026 [DOI] [Google Scholar]
- Dutt M., Zambon F. T., Erpen L., Soriano L., Grosser J. (2018. b). Embryo-specific expression of a visual reporter gene as a selection system for citrus transformation. PLoS One 13, e0190413. 10.1371/journal.pone.0190413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerstorfer M. F., Dolezel M., Heissenberger A., Miklau M., Reichenbecher W., Steinbrecher R. A., et al. (2019). An EU perspective on biosafety considerations for plants developed by genome editing and other new genetic modification techniques (nGMs). Front. Bioeng. Biotechnol. 7, 31. 10.3389/fbioe.2019.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa P., Uimari A., Mehto M., Albert V. A., Laitinen R. A. E., Teeri T. H. (2003). Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 133, 1831–1842. 10.1104/pp.103.026039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley R. V., Butts C. A., Laing W. A., Martell S., Smith H., McGhie T. K., et al. (2014). Dietary flavonoids from modified apple reduce inflammation markers and modulate gut microbiota in mice. J. Nutr. 144, 146–154. 10.3945/jn.113.182659 [DOI] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. 10.1111/tpj.12554 [DOI] [PubMed] [Google Scholar]
- Fávero P., de Alves Mourão Filho F. A., Stipp L. C. L., Mendes B. M. J. (2012). Genetic transformation of three sweet orange cultivars from explants of adult plants. Acta Physiol. Plant 34, 471–477. 10.1007/s11738-011-0843-4 [DOI] [Google Scholar]
- Figueiredo J. F. L., Römer P., Lahaye T., Graham J. H., White F. F., Jones J. B. (2011). Agrobacterium-mediated transient expression in citrus leaves: A rapid tool for gene expression and functional gene assay. Plant Cell Rep. 30, 1339–1345. 10.1007/s00299-011-1045-7 [DOI] [PubMed] [Google Scholar]
- Fister A. S., Landherr L., Maximova S. N., Guiltinan M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao . Front. Plant Sci. 9, 268. 10.3389/fpls.2018.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I., Richter H., Bratovič M., Le Rhun A., Charpentier E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. 10.1038/nature17945 [DOI] [PubMed] [Google Scholar]
- Foster T. M., Watson A. E., van Hooijdonk B. M., Schaffer R. J. (2014). Key flowering genes including FT-like genes are upregulated in the vasculature of apple dwarfing rootstocks. Tree Genet. Genomes 10, 189–202. 10.1007/s11295-013-0675-z [DOI] [Google Scholar]
- Friedrichs S., Takasu Y., Kearns P., Dagallier B., Oshima R., Schofield J., et al. (2019). An overview of regulatory approaches to genome editing in agriculture. Biotechnol. Res. Innov. 3, 208–220. 10.1016/j.biori.2019.07.001 [DOI] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipienso L., Navarro L., Ballester-Olmos J. F., Pina J. A., Moreno P., Guerri J. (2000). Host range and symptomatology of a graft-transmissible pathogen causing bud union crease of citrus on trifoliate rootstocks. Plant Pathol. 49, 308–314. 10.1046/j.1365-3059.2000.00449.x [DOI] [Google Scholar]
- Gattolin S., Cirilli M., Chessa S., Stella A., Bassi D., Rossini L. (2020). Mutations in orthologous PETALOSA TOE-type genes cause dominant double-flower phenotype in phylogenetically distant eudicots. J. Exp. Bot. 71 (9), 2585–2595. 10.1093/jxb/eraa032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N. M., Komor A. C., Rees H. A., Packer M. S., Badran A. H., Bryson D., II, et al. (2018). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644.Programmable [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A., Deng Z., La Malfa S., Distefano G., Domina F., Vitale A., et al. (2007). Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 126, 146–151. 10.1111/j.1439-0523.2007.01297.x [DOI] [Google Scholar]
- Gessler C., Vanblaere T., Parravicini G., Broggini G. A. L. (2014). Cisgenic ‘Gala’ containing the scab resistance gene from Malus floribunda 821 and the fire blight resistance genes from m. ‘Evereste’. Acta Hortic. 1048, 43–49. 10.17660/ActaHortic.2014.1048.4 [DOI] [Google Scholar]
- Gottwald T. R., Hughes G., Graham J. H., Sun X., Riley T. (2001). The Citrus canker epidemic in Florida: the scientific basis of regulatory eradication policy for an invasive species. Phytopathology 91, 30–34. 10.1094/PHYTO.2001.91.1.30 [DOI] [PubMed] [Google Scholar]
- Grosser J. W., Gmitter F. G., Jr. (2011). Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell Tiss. Org. Cult. 104 (3), 343–357. 10.1007/s11240-010-9823-4 [DOI] [Google Scholar]
- Grosser J. W., Gmitter F. G., Jr. (1990). Protoplast fusion and citrus improvement. Plant Breed. Rev. 8, 339–374. 10.1002/9781118061053.ch10 [DOI] [Google Scholar]
- Grosser I. W., Gmitter F. G., Jr., Sesto F., Deng X. X., Chandler J. L. (1992). Six new somatic citrus hybrids and their potential for cultivar improvement. J. Amer. Soc. Hort. Sci. 117, 169–173. 10.21273/JASHS.117.1.169 [DOI] [Google Scholar]
- Grosser J. W., Ollitrault P., Olivares-Fuster O. (2000). Somatic hybridization in citrus: an effective tool to facilitate variety improvement. In Vitro Cell Dev. Biol. Plant 36, 434–449. 10.1007/s11627-000-0080-9 [DOI] [Google Scholar]
- Guo W. W., Cheng Y. J., Deng X. X. (2002). Regeneration and molecular characterization of intergeneric somatic hybrids between Citrus reticulata and Poncirus trifoliata . Plant Cell Rep. 20, 829–834. 10.1007/s00299-001-0399-7 [DOI] [Google Scholar]
- Guo W., Duan Y., Fuster O. O., Wu Z., Arias C. R., Burns J. K., et al. (2005). Protoplast transformation and regeneration of transgenic Valencia sweet orange plants containing a juice quality-related pectin methylesterase gene. Plant Cell Rep. 24, 482–486. 10.1007/s00299-005-0952-x [DOI] [PubMed] [Google Scholar]
- Hackett W. P. (2011). Juvenility, maturation, and rejuvenation in woody plants,” in Horticultural Reviews. Ed. Janick J. (Hoboken, NJ, USA: John Wiley & Sons, Inc; ), 109–155. 10.1002/9781118060735.ch3 [DOI] [Google Scholar]
- Hajiahmadi Z., Movahedi A., Wei H., Li D., Orooji Y., Ruan H., et al. (2019). Strategies to increase on-target and reduce off-target effects of the CRISPR/Cas9 system in plants. Int. J. Mol. Sci. 20, 3719. 10.3390/ijms20153719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M. V., Flachowsky H., Peil A., Hättasch C. (2007). No flower no fruit—genetic potentials to trigger flowering in fruit trees. Genes Genomes Genomics 1 (1), 1–20. [Google Scholar]
- He Y., Chen S., Peng A., Zou X., Xu L., Lei T., et al. (2011). Production and evaluation of transgenic sweet orange (Citrus sinensis Osbeck) containing bivalent antibacterial peptide genes (Shiva A and Cecropin B) via a novel Agrobacterium-mediated transformation of mature axillary buds. Sci. Hortic. 128, 99–107. 10.1016/j.scienta.2011.01.002 [DOI] [Google Scholar]
- Holme I. B., Wendt T., Holm P. B. (2013). Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 11, 395–407. 10.1111/pbi.12055 [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., Agarwala V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 (9), 827–832. 10.1038/NPBT.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. 111, 521–529. 10.1073/pnas.1313271111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Tao X., Zhu J. K. (2019). Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 17, 499–504. 10.1111/pbi.12993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin M., Pérez-Quintero A. L., Lopez C., Szurek B. (2015). MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front. Plant Sci. 6, 535. 10.3389/fpls.2015.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen E., Schouten H. J. (2007). Cisgenesis strongly improves introgression breeding and induced translocation breeding of plants. Trends Biotechnol. 25, 219–223. 10.1016/j.tibtech.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Jia H., Wang N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9, e93806. 10.1371/journal.pone.0093806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Orbović V., Jones J. B., Wang N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating Xcc pthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. 10.1111/pbi.12495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Xu J., Orbović V., Zhang Y., Wang N. (2017. a). Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 8, 2135. 10.3389/fpls.2017.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Zhang Y., Orbovic V., Xu J., White F. F., Jones J. B., et al. (2017. b). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. 10.1111/pbi.12677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Orbović V., Wang N. (2019). CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 17, 1928–1937. 10.1111/pbi.13109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. G., Gilbert D. E., Grady K. L., Jorgensen R. A. (1987). T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol. Gen. Genet. 207, 478–485. 10.1007/BF00331618 [DOI] [Google Scholar]
- Jones H. D. (2015). Regulatory uncertainty over genome editing. Nat. Plants. 1, 14011. 10.1038/nplants.2014.11 [DOI] [PubMed] [Google Scholar]
- Joshi S. G., Schaart J. G., Groenwold R., Jacobsen E., Schouten H. J., Krens F. A. (2011). Functional analysis and expression profiling of HcrVf1 and HcrVf2 for development of scab resistant cisgenic and intragenic apples. Plant Mol. Biol. 75, 579–591. 10.1007/s11103-011-9749-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel R., Bergey D. R., Dutt M., Sitther V., Li Z. T., Gray D. J., et al. (2016). Evaluation of a grapevine-derived reporter gene system for precision breeding of Vitis . Plant Cell Tissue Organ Cult. 124, 599–609. 10.1007/s11240-015-0918-9 [DOI] [Google Scholar]
- Kaur N., Alok A., Shivani, Kaur N., Pandey P., Awasthi P., et al. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics 18, 89–99. 10.1007/s10142-017-0577-5 [DOI] [PubMed] [Google Scholar]
- Khan E. U., Fu X. Z., Wang J., Fan Q. J., Huang X. S., Zhang G. N., et al. (2009). Regeneration and characterization of plants derived from leaf in vitro culture of two sweet orange (Citrus sinensis (L.) Osbeck) cultivars. Sci. Hortic. 120, 70–76. 10.1016/j.scienta.2008.10.004 [DOI] [Google Scholar]
- Khan E. U., Fu X.-Z., Liu J.-H. (2012). Agrobacterium-mediated genetic transformation and regeneration of transgenic plants using leaf segments as explants in Valencia sweet orange. Plant Cell Tissue Organ Cult. 109, 383–390. 10.1007/s11240-011-0092-7 [DOI] [Google Scholar]
- Kobayashi A. K., Vieira L. G. E., Bespalhok Filho J. C., Leite R. P. Jr., Pereira L. F. P., Molinari H. B. C. (2017). Enhanced resistance to citrus canker in transgenic sweet orange expressing the sarcotoxin IA gene. Eur. J. Plant Pathol. 149, 865–873. 10.1007/s10658-017-1234-5 [DOI] [Google Scholar]
- Kost T. D., Gessler C., Jänsch M., Flachowsky H., Patocchi A., Broggini G. A. L. (2015). Development of the first cisgenic apple with increased resistance to fire blight. PLoS One 10, e0143980. 10.1371/journal.pone.0143980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoda N., Hayashi H., Suzuki M., Igarashi M., Hatsuyama Y., Kidou S., et al. (2010). Molecular characterization of FLOWERING LOCUS T-Like genes of apple (Malus × domestica Borkh.). Plant Cell Physiol. 51, 561–575. 10.1093/pcp/pcq021 [DOI] [PubMed] [Google Scholar]
- Krens F. A., Schaart J. G., van der Burgh A. M., Tinnenbroek-Capel I. E. M., Groenwold R., Kodde L. P., et al. (2015). Cisgenic apple trees; development, characterization, and performance. Front. Plant Sci. 6, 286. 10.3389/fpls.2015.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Malfa S., Distefano G., Domina F., Nicolosi E., Toscano V., Gentile A. (2011). Evaluation of citrus rootstock transgenic for rolabc genes. Acta Hortic. 892, 131–140. 10.17660/ActaHortic.2011.892.16 [DOI] [Google Scholar]
- Lassoued R., Hesseln H., Phillips P. W. B., Smyth S. J. (2018). Top plant breeding techniques for improving food security: an expert Delphi survey of the opportunities and challenges. Int. J. Agric. Resour. Gov. Ecol. 14, 321–337. 10.1504/IJARGE.2018.097986 [DOI] [Google Scholar]
- Ledford H. (2015). Alternative CRISPR system could improve genome editing. Nature 526, 17–17. 10.1038/nature.2015.18432 [DOI] [PubMed] [Google Scholar]
- Li D. D., Shi W., Deng X. X. (2002). Agrobacterium-mediated transformation of embryogenic calluses of Ponkan mandarin and the regeneration of plants containing the chimeric ribonuclease gene. Plant Cell Rep. 21, 153–156. 10.1007/s00299-002-0492-6 [DOI] [Google Scholar]
- Li F., Dai S., Deng Z., Li D., Long G., Li N., et al. (2017). Evaluation of parameters affecting Agrobacterium-mediated transient expression in citrus. J. Integr. Agric. 15, 60345–60347. 10.1016/S2095-3119(16)61460-0 [DOI] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., et al. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. U.S.A. 103, 6398–6403. 10.1073/pnas.0601620103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limera C., Sabbadini S., Sweet J. B., Mezzetti B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8, 1418. 10.3389/fpls.2017.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. 10.1038/s41587-020-0455-x [DOI] [PubMed] [Google Scholar]
- Liu Y., Heying E., Tanumihardjo S. A. (2012). History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 11, 530–545. 10.1111/j.1541-4337.2012.00201.x [DOI] [Google Scholar]
- Liu Y., Zeng J., Yuan C., Guo Y., Yu H., Li Y., et al. (2019). Cas9-PF, an early flowering and visual selection marker system, enhances the frequency of editing event occurrence and expedites the isolation of genome-edited and transgene-free plants. Plant Biotechnol. J. 17, 1191–1193. 10.1111/pbi.13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Noguera S., Petri C., Burgos L. (2009). Combining a regeneration-promoting ipt gene and site-specific recombination allows a more efficient apricot transformation and the elimination of marker genes. Plant Cell Rep. 28, 1781–1790. 10.1007/s00299-009-0778-z [DOI] [PubMed] [Google Scholar]
- Lu Y., Zhu J.-K. (2017). Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 10, 523–525. 10.1016/j.molp.2016.11.013 [DOI] [PubMed] [Google Scholar]
- Lusser M., Davies H. V. (2013). Comparative regulatory approaches for groups of new plant breeding techniques. New Biotechnol. 30, 437–446. 10.1016/j.nbt.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Lusser M., Parisi C., Plan D., Cerezo E. R. (2011). “New plant breeding techniques. State-of-the-art and prospects for commercial development,” in JRC Scientific and Technical Reports. (Luxembourg: Publications Office of the European Union; ), pp. 1–220. 10.2791/54761 [DOI] [Google Scholar]
- Lyznik L. A., Mitchell J. C., Hirayama L., Hodges T. K. (1993). Activity of yeast FLP recombinase in maize and rice protoplasts. Nucleic Acids Res. 21, 969–975. 10.1093/nar/21.4.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G., Zhang L., Sugiura M., Kato M. (2020). “Citrus and health,” in The Genus Citrus, 1st edition Eds. Talon M.,., Caruso M., Gmitter F. G. (Cambridge, UK: Elsevier; ), 495–511. [Google Scholar]
- Malnoy M., Viola R., Jung M. H., Koo O. J., Kim S., Kim J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. 10.3389/fpls.2016.01904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín F. R., Soler-Rivas C., Benavente-García O., Castillo J., Pérez-Alvarez J. A. (2007). By-products from different citrus processes as a source of customized functional fibres. Food Chem. 100, 736–741. 10.1016/j.foodchem.2005.04.040 [DOI] [Google Scholar]
- Marutani-Hert M., Bowman K. D., McCollum G. T., Mirkov T. E., Evens T. J., Niedz R. P. (2012). A dark incubation period is important for Agrobacterium-mediated transformation of mature internode explants of sweet orange, grapefruit, citron, and a citrange rootstock. PLoS One 7, e47426. 10.1371/journal.pone.0047426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J., Díez-Villaseñor C., Soria E., Juez G. (2000). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36, 244–246. 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- Moradpour M., Abdulah S. N. A. (2020). CRISPR/dCas9 platforms in plants: strategies and applications beyond genome editing. Plant Biotechnol. J. 18, 32–44. 10.1111/pbi.13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Dias J. M., Molina R. V., Bordón Y., Guardiola J. L., García-Luis A. (2000). Direct and indirect shoot organogenic pathways in epicotyl cuttings of Troyer citrange differ in hormone requirements and in their response to light. Ann. Bot. 85, 103–110. 10.1006/anbo.2000.1001 [DOI] [Google Scholar]
- Naim F., Dugdale B., Kleidon J., Brinin A., Shand K., Waterhouse P., et al. (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. 10.1007/s11248-018-0083-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima I., Ban Y., Azuma A., Onoue N., Moriguchi T., Yamamoto T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS One 12, e0177966. 10.1371/journal.pone.0177966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani C., Hirai N., Komori S., Wada M., Okada K., Osakabe K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481. 10.1038/srep31481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaguchi M., Abe M., Kimura T., Daimon Y., Kobayashi T., Yamaguchi A., et al. (2008). Long-distance, graft-transmissible action of Arabidopsis FLOWERING LOCUS T protein to promote flowering. Plant Cell Physiol. 49, 1645–1658. 10.1093/pcp/pcn154 [DOI] [PubMed] [Google Scholar]
- Omar A. A., Song W. Y., Grosser J. W. (2007). Introduction of the Xa21, Xanthomonas resistance gene from rice, into ‘Hamlin’ sweet orange [Citrus Sinensis (L.) Osbeck] using protoplast/GFP co-transformation and single plasmid transformation. J. Hortic. Sci. Biotechnol. 82 (6), 914–923. 10.1080/14620316.2007.11512326 [DOI] [Google Scholar]
- Omar A. A., Dutt M., Gmitter F. G., Grosser J. W. (2016). “Somatic Embryogenesis: Still a Relevant Technique in Citrus Improvement,” in In Vitro Embryogenesis in Higher Plants, 1st edition Eds. Germanà M. A., Lambardi M. (New York, NY: Springer Science + Business Media; ), 289–327. 10.1007/978-1-4939-3061-6_13 [DOI] [PubMed] [Google Scholar]
- Omar A. A., Murata M. M., El-Shamy H. A., Graham J. H., Grosser J. W. (2018). Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 27, 179–191. 10.1007/s11248-018-0065-2 [DOI] [PubMed] [Google Scholar]
- Orbović V., Shankar A., Peeples M., Hubbard C., Zale J. (2015). “Citrus transformation using mature tissue explants,” in Agrobacterium protocols Methods in molecular biology, vol. 2 (Clifton, N.J: ), pp. 7–8. [DOI] [PubMed] [Google Scholar]
- Osakabe Y., Liang Z., Ren C., Nishitani C., Osakabe K., Wada M., et al. (2018). CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 13, 2844–2863. 10.1038/s41596-018-0067-9 [DOI] [PubMed] [Google Scholar]
- Peña L., Cervera M., Juárez J., Navarro A., Pina J. A., Durán-Vila N., et al. (1995). Agrobacterium-mediated transformation of sweet orange and regeneration of transgenic plants. Plant Cell Rep. 14, 616–619. 10.1007/BF00232724 [DOI] [PubMed] [Google Scholar]
- Peña L., Martín-Trillo M., Juárez J., Pina J. A., Navarro L., Martínez-Zapater J. M. (2001). Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 19, 263–267. 10.1038/85719 [DOI] [PubMed] [Google Scholar]
- Peng A., Chen S., Lei T., Xu L., He Y., Wu L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. 10.1111/pbi.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., Zou X., Xu L., He Y., Lei T., Yao L., et al. (2019). Improved protocol for the transformation of adult Citrus sinensis Osbeck ‘Tarocco’ blood orange tissues. Vitr. Cell. Dev. Biol. - Plant 55, 659–667. 10.1007/s11627-019-10011-9 [DOI] [Google Scholar]
- Petri C., López-Noguera S., Wang H., García-Almodóvar C., Alburquerque N., Burgos L. (2012). A chemical-inducible Cre-LoxP system allows for elimination of selection marker genes in transgenic apricot. Plant Cell Tissue Organ Cult. 110, 337–346. 10.1007/s11240-012-0155-4 [DOI] [Google Scholar]
- Pillitteri L. J., Lovatt C. J., Walling L. L. (2004). Isolation and characterization of a terminal flower homolog and its correlation with juvenility in citrus. Plant Physiol. 135, 1540–1551. 10.1104/pp.103.036178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompili V., Dalla Costa L., Piazza S., Pindo M., Malnoy M. (2020). Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 18, 845–858. 10.1111/pbi.13253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons E., Alquézar B., Rodriguez A., Martorell P., Genovés S., Ramon D., et al. (2014). Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 12, 17–27. 10.1111/pbi.12112 [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadugu C., Keremane M. L., Halbert S. E., Duan Y. P., Roose M. L., Stover E., et al. (2016). Long-term field evaluation reveals Huanglongbing resistance in citrus relatives. Plant Dis. 100, 1858–1869. 10.1094/PDIS-03-16-0271-RE [DOI] [PubMed] [Google Scholar]
- Ren B., Yan F., Kuang Y., Li N., Zhang D., Zhou X., et al. (2018). Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant 11, 623–626. 10.1016/j.molp.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Ren F., Ren C., Zhang Z., Duan W., Lecourieux D., Li S., et al. (2019). Efficiency optimization of CRISPR/CAS9-mediated targeted mutagenesis in grape. Front. Plant Sci. 10, 612. 10.3389/fpls.2019.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo A. R., Ayliffe M. (2015). Gene targeting and editing in crop plants: a new era of precision opportunities. Mol. Breed. 35, 40. 10.1007/s11032-015-0210-z [DOI] [Google Scholar]
- Rodríguez A., Cervera M., Peris J. E., Peña L. (2008). The same treatment for transgenic shoot regeneration elicits the opposite effect in mature explants from two closely related sweet orange (Citrus sinensis (L.) Osb.) genotypes. Plant Cell. Tissue Organ Cult 93, 97–106. 10.1007/s11240-008-9347-3 [DOI] [Google Scholar]
- Rugini E., Bashir M. A., Cristofori V., Ruggiero B., Silvestri C. (2020). A review of genetic improvement of main fruit trees through modern biotechnological tools and considerations of the cultivation and research of the engineered plant restrictions. Pakistan J. Agric. Sci. 57 (1), 17–42. 10.21162/PAKJAS/20.8361 [DOI] [Google Scholar]
- Schaart J. G., Krens F. A., Pelgrom K. T. B., Mendes O., Rouwendal G. J. A. (2004). Effective production of marker-free transgenic strawberry plants using inducible site-specific recombination and a bifunctional selectable marker gene. Plant Biotechnol. J. 2, 233–240. 10.1111/j.1467-7652.2004.00067.x [DOI] [PubMed] [Google Scholar]
- Schouten H. J., Krens F. A., Jacobsen E. (2006). Cisgenic plants are similar to traditionally bred plants: International regulations for genetically modified organisms should be altered to exempt cisgenesis. EMBO Rep. 7, 750–753. 10.1038/sj.embor.7400769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M., Mysore K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 16, 656–665. 10.1016/j.tplants.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., et al. (2017). Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443. 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- Silva Souza L., Diniz R. P., Neves R., de J., Alves A. A. C., de Oliveira E. J. (2018). Grafting as a strategy to increase flowering of cassava. Sci. Hortic. 240, 544–551. 10.1016/j.scienta.2018.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., Prieto H., Orbović V. (2019. a). Agrobacterium-mediated transformation of tree fruit crops: methods, progress, and challenges. Front. Plant Sci. 10, 226. 10.3389/fpls.2019.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.-q., Walworth A., Lin T., Chen Q., Han X., Zaharia L., II, et al. (2019. b). VcFT-induced mobile florigenic signals in transgenic and transgrafted blueberries. Hortic. Res. 6 (105), 1–15. 10.1038/s41438-019-0188-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan C., Dardick C., Callahan A., Scorza R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7, e40715. 10.1371/journal.pone.0040715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert J., Schiml S., Fauser F., Puchta H. (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus . Plant J. 84, 1295–1305. 10.1111/tpj.13078 [DOI] [PubMed] [Google Scholar]
- Sugiura M., Nakamura M., Ogawa K., Ikoma Y., Yano M. (2012). High serum carotenoids associated with lower risk for bone loss and osteoporosis in post-menopausal Japanese female subjects: prospective cohort study. PLoS One 7, e52643. 10.1371/journal.pone.0052643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Nakamura M., Ogawa K., Ikoma Y., Yano M. (2016). High vitamin C intake with high serum β-cryptoxanthin associated with lower risk for osteoporosis in post-menopausal Japanese female subjects: Mikkabi cohort study. J. Nutr. Sci. Vitaminol. 62, 185–191. 10.3177/jnsv.62.185 [DOI] [PubMed] [Google Scholar]
- Talon M., Gmitter F. G. (2008). Citrus genomics. Int. J. Plant Genomics, 1–17. 10.1155/2008/528361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Jiang L., Cui X., Zhang J., Guo S., Li M., et al. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37, 1353–1356. 10.1007/s00299-018-2299-0 [DOI] [PubMed] [Google Scholar]
- Ting S. V. (1980). “Nutrients and Nutrition of Citrus Fruits,” in Citrus nutrition and quality. Eds. NagyJohn S., Attaway A. (Washington, DC. USA: American chemical society; ), 3–24. 10.1021/bk-1980-0143.ch001 [DOI] [Google Scholar]
- Titta L., Trinei M., Stendardo M., Berniakovich I., Petroni K., Tonelli C., et al. (2010). Blood orange juice inhibits fat accumulation in mice. Int. J. Obes. 34, 578–588. 10.1038/ijo.2009.266 [DOI] [PubMed] [Google Scholar]
- Tranel P. J., Wright T. R. (2002). Resistance of weeds to ALS-inhibiting herbicides: what have we learned? Weed Sci. 50, 700–712. 10.1614/0043-1745(2002)050[0700:RROWTA]2.0.CO;2 [DOI] [Google Scholar]
- Tripoli E., La Guardia M., Giammanco S., Di Majo D., Giammanco M. (2007). Citrus flavonoids: molecular structure, biological activity and nutritional properties: A review. Food Chem. 104, 466–479. 10.1016/j.foodchem.2006.11.054 [DOI] [Google Scholar]
- van Nocker S., Gardiner S. E. (2014). Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic. Res. 1, 14022. 10.1038/hortres.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanblaere T., Szankowski I., Schaart J., Schouten H., Flachowsky H., Broggini G. A. L., et al. (2011). The development of a cisgenic apple plant. J. Biotechnol. 154, 304–311. 10.1016/j.jbiotec.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Vanblaere T., Flachowsky H., Gessler C., Broggini G. A. L. (2014). Molecular characterization of cisgenic lines of apple ‘Gala’ carrying the Rvi6 scab resistance gene. Plant Biotechnol. J. 12, 2–9. 10.1111/pbi.12110 [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wang T., Voogd C., Jeon S., Drummond R. S. M., Gleave A. P., et al. (2019). Mutagenesis of kiwifruit CENTRORADIALIS -like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 17, 869–880. 10.1111/pbi.13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet F., Perrot L., Chauvin L., Kermarrec M.-P., Guyon-Debast A., Chauvin J.-E., et al. (2019). Transgene-Free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a crispr/cas9 cytidine base editor. Int. J. Mol. Sci. 20, 402. 10.3390/ijms20020402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., Licciardello C. (2014). A genealogy of the citrus family. Nat. Biotechnol. 32, 640–642. 10.1038/nbt.2954 [DOI] [PubMed] [Google Scholar]
- Velázquez K., Agüero J., Vives M. C., Aleza P., Pina J. A., Moreno P., et al. (2016). Precocious flowering of juvenile citrus induced by a viral vector based on Citrus leaf blotch virus: a new tool for genetics and breeding. Plant Biotechnol. J. 14, 1976–1985. 10.1111/pbi.12555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives M. C., Martín S., Ambrós S., Renovell Á., Navarro L., Pina J. A., et al. (2008). Development of a full-genome cDNA clone of Citrus leaf blotch virus and infection of citrus plants. Mol. Plant Pathol. 9, 787–797. 10.1111/j.1364-3703.2008.00501.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F., Gao C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12, e1001877. 10.1371/journal.pbio.1001877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. 10.1146/annurev-arplant-042811-105552 [DOI] [PubMed] [Google Scholar]
- Waltz E. (2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532, 293. 10.1038/nature.2016.19754 [DOI] [PubMed] [Google Scholar]
- Wang X., Xu Y., Zhang S., Cao L., Huang Y., Cheng J., et al. (2017). Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 49, 765–772. 10.1038/ng.3839 [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang S., Li D., Zhang Q., Li L., Zhong C., et al. (2018). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. 10.1111/pbi.12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen S., Peng A., Xie Z., He Y., Zou X. (2019). CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 13, 501–510. 10.1007/s11816-019-00556-x [DOI] [Google Scholar]
- Wilson F. M., Harrison K., Armitage A. D., Simkin A. J., Harrison R. J. (2019. a). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45. 10.1186/s13007-019-0428-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L. O. W., Hetzel S., Pockrandt C., Reinert K., Bauer D. C. (2019. b). VARSCOT: variant-aware detection and scoring enables sensitive and personalized off-target detection for CRISPR-Cas9. BMC Biotechnol. 19, 40. 10.1186/s12896-019-0535-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. A., Prochnik S., Jenkins J., Salse J., Hellsten U., Murat F., et al. (2014). Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662. 10.1038/nbt.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. A., Terol J., Ibanez V., López-García A., Pérez-Román E., Borredá C., et al. (2018). Genomics of the origin and evolution of Citrus. Nature 554, 311–316. 10.1038/nature25447 [DOI] [PubMed] [Google Scholar]
- Würdig J., Flachowsky H., Saß A., Peil A., Hanke M.-V. (2015). Improving resistance of different apple cultivars using the Rvi6 scab resistance gene in a cisgenic approach based on the Flp/FRT recombinase system. Mol. Breed. 35, 95. 10.1007/s11032-015-0291-8 [DOI] [Google Scholar]
- Xu Q., Chen L.-L., Ruan X., Chen D., Zhu A., Chen C., et al. (2013). The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66. 10.1038/ng.2472 [DOI] [PubMed] [Google Scholar]
- Yuan J. M., Stram D. O., Arakawa K., Lee H. P., Yu M. C. (2003). Dietary cryptoxanthin and reduced risk of lung cancer: The Singapore Chinese health study. Cancer Epidemiol. Biomarkers Prev. 12, 890–898. [PubMed] [Google Scholar]
- Zanek M. C., Reyes C. A., Cervera M., Peña E. J., Velázquez K., Costa N., et al. (2007). Genetic transformation of sweet orange with the coat protein gene of Citrus psorosis virus and evaluation of resistance against the virus. Plant Cell Rep. 27, 57–66. 10.1007/s00299-007-0422-8 [DOI] [PubMed] [Google Scholar]
- Zeng F., Zhan Y., Nan N., Xin Y., Qi F., Yang C. (2009). Expression of bgt gene in transgenic birch (Betula platyphylla Suk.). Afr. J. Biotechnol. 8, 3392–3398. 10.5897/AJB2009.000-9330 [DOI] [Google Scholar]
- Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., Essletzbichler P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Francis M., II, Dawson W. O., Graham J. H., Orbović V., Triplett E. W., et al. (2010). Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 128, 91–100. 10.1007/s10658-010-9633-x [DOI] [Google Scholar]
- Zhang F., LeBlanc C., Irish V. F., Jacob Y. (2017). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36, 1883–1887. 10.1007/s00299-017-2202-4 [DOI] [PubMed] [Google Scholar]
- Zhang R., Liu J., Chai Z., Chen S., Bai Y., Zong Y., et al. (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5, 480–485. 10.1038/s41477-019-0405-0 [DOI] [PubMed] [Google Scholar]
- Zhou P., Jia R., Chen S., Xu L., Peng A., Lei T., et al. (2017). Cloning and expression analysis of four citrus WRKY genes responding to Xanthomon asaxonopodis pv. citri . Acta Hortic. Sin. 44, 452–462. 10.16420/j.issn.0513-353x.2016-0577 [DOI] [Google Scholar]
- Zhu C., Zheng X., Huang Y., Ye J., Chen P., Zhang C., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17, 2199–2210. 10.1111/pbi.13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Peng A., Xu L., Liu X., Lei T., Yao L., et al. (2013). Efficient auto-excision of a selectable marker gene from transgenic citrus by combining the Cre/loxP system and ipt selection. Plant Cell Rep. 32, 1601–1613. 10.1007/s00299-013-1470-x [DOI] [PubMed] [Google Scholar]